Summary
The purpose of the current study was to alter the broad native tropism of human adenovirus for virus targeting to c-erbB2-positive cancer cells. First, we engineered a single-chain antibody (scFv) against the c-erbB2 oncoprotein into minor capsid protein IX (pIX) of adenovirus serotype 5 (Ad5) in a manner commensurate with virion integrity and binding to the soluble extracellular c-erbB2 domain. To ablate native viral tropism and facilitate binding of the pIX-incorporated scFv to cellular c-erbB2 we replaced the Ad5 fiber with the Ad41 short (41s) fiber devoid of all known cell-binding determinants. The resultant Ad5F41sIX6.5 vector demonstrated increased cell binding and gene transfer as compared to the Ad5F41s control, however, this augmentation of virus infectivity was not c-erbB2-specific. Incorporation of a six histidine (His6) peptide into the C-terminus of the 41s fiber protein resulted in markedly increased Ad5F41s6H infectivity in 293AR cells, which express a membrane-anchored scFv against the C-terminal oligo-histidine tag, as compared to the Ad5F41s vector and the parental 293 cells. These data suggested that a 41s fiber-incorporated His6 tag could serve for attachment of an adapter protein designed to guide Ad5F41s6H infection in a c-erbB2-specific manner. We therefore engineered a bispecific scFv diabody (scDb) combining affinities for both c-erbB2 and the His6 tag and showed its ability to provide up to 25-fold increase of Ad5F41s6H infectivity in c-erbB2-positive cells. Thus, Ad5 fiber replacement by a His6–tagged 41s fiber coupled with virus targeting mediated by an scDb adapter represents a promising strategy to confer Ad5 vector tropism for c-erbB2-positive cancer cells.
Keywords: Adenovirus, c-erbB2, serotype 41 short fiber, capsid protein IX, single-chain diabody
Introduction
The human adenovirus (Ad) family includes 51 serotypes divided into six species (A-F) that represent a large family of nonenveloped viruses containing a linear double-stranded DNA genome of approximately 36 kb 1 packaged into an icosahedral capsid 2; 3. The major capsid components are the trimeric hexon protein and the penton that is a noncovalent complex between the pentameric penton base and the trimeric fiber protein. The interactions between Ad capsid proteins and host cell have been extensively characterized in vitro revealing two distinct sequential steps critical for initiation of Ad infection. Ad attachment to the cell is mediated by fiber binding with its C-terminal knob domain to a primary cellular receptor 4. Subsequent interaction of the αvβ3/5-integrins, the secondary cellular receptor, with an Arg–Gly–Asp (RGD) sequence within a protein loop extended from the penton base is required to trigger endocytosis resulting in virus internalization 5; 6. Most Ad of species B have been shown to utilize human membrane cofactor CD46 as the predominant binding site 7; 8 for infection of dendritic cells, hematopoietic stem cells, and some malignant cell types. The primary high-affinity cellular attachment receptor for many representatives of species A, C, D, E, and F has been identified 9 as the coxsackievirus group B and Ad receptor 10, called CAR. The presence of CAR on human airway epithelia determines the susceptibility of epithelial cells to Ad5 infection both in vitro and in vivo 11; 12. Other receptors including heparan sulfate proteoglycans (HSPG) 13; 14 and class I histocompatibility complex 15 have also been described for Ad serotype 2 (Ad2) and 5 (Ad5) from species C that are most extensively used for vector development.
The delineation of key aspects of the Ad cellular entry pathway in vitro, wherein cell attachment is distinct from subsequent virus internalization, suggested that binding of Ad to the primary receptor determines the host cell range and native viral tropism 16; 17; 18. The modification of residues critical for Ad binding to CAR 19; 20; 21 and/or integrins 22; 23; 24 have resulted in a dramatic decrease of Ad5 infectivity in vitro. However, these mutations were shown to have only a marginal effect on the biodistribution pattern of the virus in vivo, including its predominant accumulation in hepatic tissue after intravascular delivery 24; 25; 26; 27; 28; 29, an important route of administration for many clinical applications. On the other hand, a significant impact on vector-mediated liver transduction has been demonstrated in several animal species by mutating the KKTK motif, a putative HSPG-binding site found in the Ad5 fiber shaft 30; 31, or by pseudotyping the Ad5 capsid with short-shafted fibers from Ad serotype 35 30; 32; 33; 34, 40 35, or 41 36; 37 that lack both CAR-binding and the KKTK motif. Furthermore, a number of plasma proteins including coagulation factors VII, IX, X, protein C, and complement component C4-binding protein have been shown to facilitate CAR-independent infection of hepatic cells by virtue of binding to either trimeric fiber knob 38 or hexon 39; 40 and bridging Ad particles to the cell surface in vitro and in vivo. Therefore, improving the therapeutic potential of Ad vectors requires the elimination of the natural viral tropism and introduction of a novel mechanism of selective cell recognition that would allow directed virus localization to the target tissue.
Several strategies including the use of bispecific adapter molecules and the genetic incorporation of targeting ligands into capsid proteins have been developed to redirect Ad5 infection via non-native pathways (reviewed in 41). Originally, Ad targeting was demonstrated using a bispecific protein complex containing chemically conjugated virus- and receptor-binding moieties to achieve indirect virus linkage with the cells of interest via the folate receptor 42. This approach was further translated to confer Ad5 vector targeting capabilities toward receptors upregulated in the cells of interest using a variety of targeting ligands that include Fab fragments of monoclonal antibodies 43; 44, engineered single-chain antibody (scFv) molecules 45; 46, and growth factors 47; 48; 49. Evaluation of adapter-mediated Ad targeting in vivo provided compelling evidence of a substantial reduction of virus uptake in the liver and a significant increase in Ad vector delivery to the target organ 44; 50; 51 or tumors 52; 53 in a receptor-specific manner.
Since single-component vector systems are favored for application in clinical settings, genetic ligand incorporation into the viral capsid has been extensively explored to achieve Ad targeting. While engineering of the Ad5 fiber knob domain to incorporate peptide ligands derived from phage display libraries has been shown to be feasible 54; 55; 56, subsequent attempts to employ various natural ligands with augmented affinity have been challenged by the limited tolerance of the fiber for genetic modifications 57. In contrast, the carboxy terminus of the minor capsid protein IX (pIX) has been identified 58; 59 as a locale amenable to incorporation of various heterologous polypeptides including fluorescent proteins 60; 61; 62, enzymes 63; 64; 65, an scFv 66, and single-chain T-cell receptor 67 with minimal effect on viral function.
Herein, we explored the utility of the pIX ectodomain to display ligands with augmented size/complexity for retargeting of Ad infection via cellular markers associated with malignant cell transformation. This study was designed to restrict the broad native tropism of Ad5 to HER-2-positive cancer cells by means of genetic incorporation of anti-c-erbB2 scFv into pIX and replacing the Ad5 fiber with the Ad41 short (41s) fiber, which is devoid of all known cell-binding determinants. We showed that the pIX-incorporated scFv failed to provide the receptor-specific virus infection while the 41s fiber modification to contain a six histidine (His6) peptide allowed efficient c-erbB2-targeting using a bispecific scDb adapter, an scFv-derived diabody molecule that was engineered with affinities for both c-erbB2 and the His6 tag.
Results
Genetic scFv incorporation into Ad5 capsid
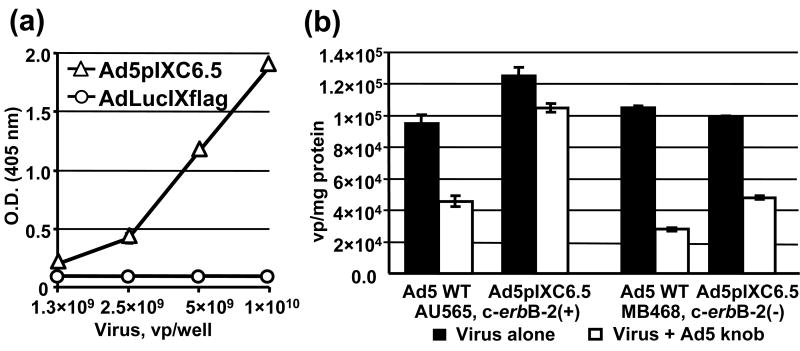
To incorporate the scFv ligand into the viral capsid we engineered the pIX gene in the Ad5 genome to encode a C-terminal Flag peptide followed by anti-c-erbB-2 scFv C6.5 68 fused to the pIX ectodomain. The replication-competent Ad5pIXC6.5 vector was rescued and the presence of protein with molecular mass of approximately 45 kD, as predicted for the pIXflagC6.5 polypeptide, was confirmed by western blotting (Fig. 2a) suggesting that the full-length scFv fused to the pIX ectodomain had been incorporated into the capsid. Electron microscopy of negatively stained viral particles (Fig. 2b) did not reveal any significant changes in Ad5pIXC6.5 virion morphology as compared to control AdLucIXflag and Ad5 WT viruses, thus indicating that scFv incorporation into pIX did not interfere with the structural integrity of the Ad5 capsid. To assess the ability of the pIX-fused C6.5 scFv to recognize the c-erbB2 oncoprotein, we carried out an ELISA using recombinant ErbB2/Fc protein containing the extracellular c-erbB2 domain. The ErbB2/Fc protein demonstrated dose-dependent binding to Ad5pIXC6.5 particles but not to the control AdLucIXflag virus (Fig. 3a), thereby validating that the capsid-incorporated scFv C6.5 retained its antigen-binding specificity. Then, we carried out a binding assay using radiolabeled virus in the presence of the Ad5 fiber knob to block CAR-dependent Ad attachment to c-erbB2-positive and -negative cells and assess Ad5pIXC6.5 binding to the cellular c-erbB2. While the efficiency of Ad5pIXC6.5 binding to c-erbB2-negative cells and Ad5 WT binding to both cell types was inhibited at least 50%, binding of Ad5pIXC6.5 vector to c-erbB2-positive cells was not significantly affected by Ad5 knob (Fig. 3b). These data suggested that the genetic modification of Ad5 fiber eliminating CAR-dependent viral tropism could allow Ad5pIXC6.5 vector to achieve c-erbB2-specific cell recognition.
Fig 2.
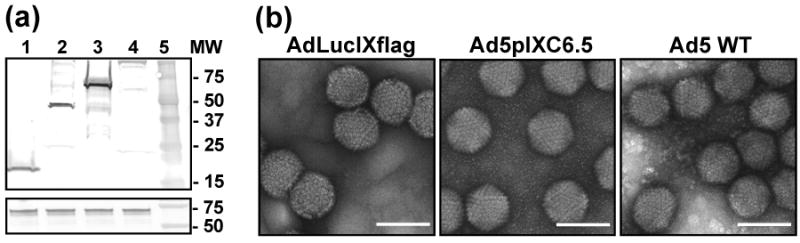
Structural analysis of Ad5pIXC6.5 vector. (a) Validation of pIXflagC6.5 fusion protein incorporation into Ad capsid. Samples of AdLucIXflag (lane 1), Ad5pIXC6.5 (lane 2), Ad5FHIFlag (lane 3) and Ad5 WT (lane 4) were analyzed by western blot using anti-Flag mAb M2 (upper panel) and penton-base specific rabbit sera (lower panel). Molecular masses of protein standards (lane 5) are indicated in kDa on the right. (b) Evaluation of Ad virion integrity by electron microscopy. Samples of AdLucIXflag, Ad5pIXC6.5 and Ad5 WT were negatively stained and examined with transmission EM. Representative images of the viral particles are shown at a magnification of 100,000×. The scale bar represents 100 nm.
Fig. 3.
Binding of Ad-incorporated C6.5 scFv to c-erbB-2 oncoprotein. (a) Ad5pIXC6.5 and AdLucIXflag viral particles adsorbed on an ELISA plate at the indicated concentrations were incubated with ErbB2/Fc chimeric protein at a concentration of 50 ng/ml. Bound ErbB2/Fc protein was detected with an AP-conjugated antibody against human IgG1 Fc fragment using the chromogenic PNPP substrate and a plate reader set at 405 nm. Each point represents the cumulative mean of triplicate determinations (error bars depicting standard deviations are smaller than the symbols). (b) Samples of Tc-99m-labeled Ad5pIXC6.5 or Ad5 WT containing 6 × 108 vp were added to AU-565 or MDA-MB-468 cell monolayers that were preincubated with Ad5 knob protein (Virus + Ad5 knob) or PBS (Virus alone). After washing and harvesting the cells, bound radioactivity was detected with a gamma-counter and used to calculate the total number of bound Ad5pIXC6.5 or Ad5 WT particles, which was normalized for the total protein concentration determined in each sample. Each bar represents the cumulative mean of triplicate vp/mg protein determinations ±SD.
Pseudotyping of Ad5pIXC6.5 capsid with 41s fiber
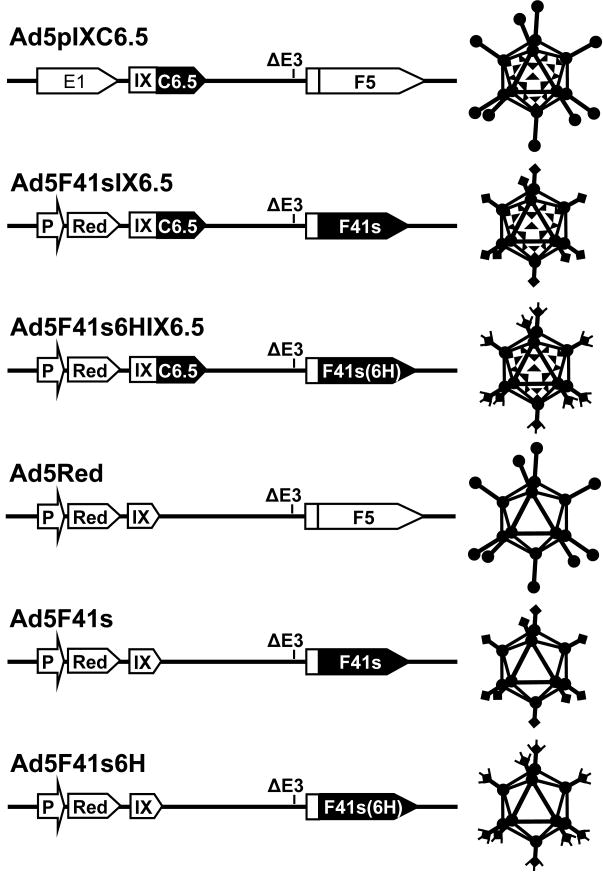
To improve the capacity of the pIX-incorporated C6.5 scFv to interact with the cellular c-erbB2, we choose to eliminate the fiber-mediated virus-cell recognition by genetically replacing the Ad5 fiber in the Ad5pIXC6.5 capsid with the short Ad41 (41s) fiber, which lacks both CAR and HSPG binding 36; 37; 69. To promote incorporation of the 41s fiber into the Ad5 capsid, we engineered a recombinant gene encoding the Ad5 fiber tail region fused with the shaft and knob domains of the 41s fiber. The 3′-end of the recombinant 41s fiber gene was modified to contain a coding sequence for a His6 tag to facilitate virus infection and propagation using previously described 293AR cells, which express an artificial anti-His6 viral attachment receptor 70. We generated Ad vectors encoding pIX-C6.5 fusion and the recombinant 41s fiber with or without the His6 tag, designated Ad5F41sIX6.5 and Ad5F41s6HIX6.5 respectively, as well as control Ad5F41s6H and Ad5F41s vectors containing the wild-type pIX gene. These replication-deficient Ad vectors were constructed to contain the CMV-driven DsRed2 gene encoding red fluorescent protein in place of the early viral E1 region.
The presence of the 41s fiber in the capsid of Ad5F41s6HIX6.5, Ad5F41sIX6.5, Ad5F41s6H, and Ad5F41s vector was confirmed by western blotting of CsCl-purified viral particles using mAb 4D2 that recognizes the tail portion of fiber protein 71. Major protein bands with molecular masses of 40 kD, corresponding to the short fiber of Ad41, were detected in the Ad5F41s6HIX6.5, Ad5F41sIX6.5, Ad5F41s6H, and Ad5F41s samples (Fig. 4, upper panel), thus validating the efficient incorporation of recombinant 41s fibers in these Ad vector capsids. As can be seen in Fig. 4 (bottom panel), western blot analysis using anti-Flag M2 mAb revealed a markedly improved efficiency of incorporation of the pIX-C6.5 fusion protein into the Ad5F41s6HIX6.5 and Ad5F41sIX6.5 capsids as compared to Ad5pIXC6.5 vector. The presence of the His6 tag in the fibers of Ad5F41s6HIX6.5 and Ad5F41s6H vectors was confirmed by ELISA (data not shown).
Fig. 4.
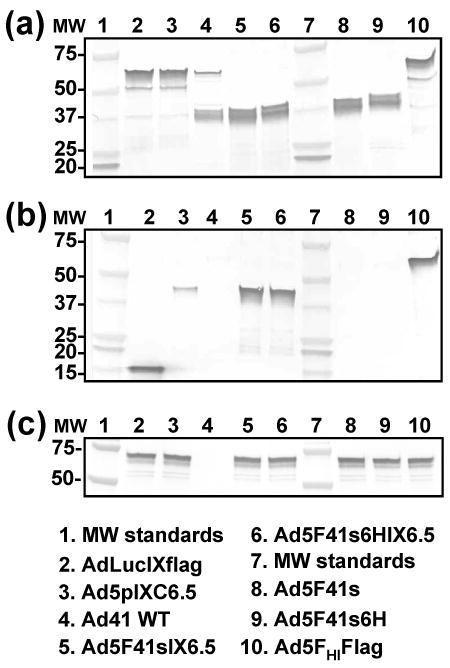
Western blot analysis of fiber and pIX modifications introduced into generated Ad vectors. Electrophoretically resolved viral proteins were transferred to a PVDF membrane and probed with mAb 4D2, mAb M2, or penton-base specific rabbit sera to detect the presence of fiber (a), pIX (b), or penton base (c), respectively, in the capsids of Ad5F41sIX6.5, Ad5F41s6HIX6.5, Ad5F41s, and Ad5F41s6H. AdLucIXflag, Ad5pIXC6.5, Ad41, and Ad5FHIFlag vectors served as controls. The numbers on the left indicate molecular masses of protein standards (lanes 1 and 7) in kDa.
Assessment of ability of Ad5F41sIX6.5 to recognize c-erbB2
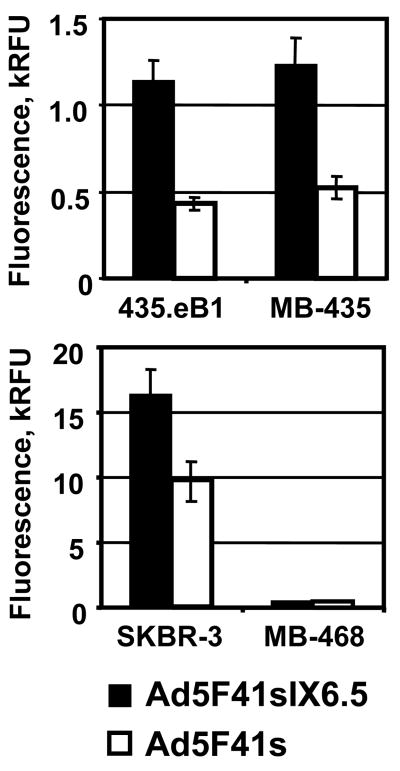
To test whether replacement of the Ad5 fiber with the 41s fiber allowed Ad5F41sIX6.5 vector to achieve c-erbB2-dependent cell recognition, we compared its gene transfer efficiencies with Ad5F41s vector in c-erbB2-negative MDA-MB-435 cells 72 and their derivative 435.eB1 cells overexpressing the c-erbB2-oncoprotein 73; 74. As shown in Fig. 5, Ad5F41sIX6.5 exhibited 2.5-fold higher transgene expression level then Ad5F41s vector in both cell lines. Ad5F41sIX6.5 also showed 1.7-fold increased gene transfer in c-erbB2-overexpressing SKBR-3 cells and the same transgene expression in c-erbB2-negative MDA-MB-468 cells as compared to the Ad5F41s control (Fig. 5). Since these results did not meet our expectations regarding the magnitude of infectivity enhancement achievable via the capsid-incorporated scFv, we assessed the specificity of Ad5F41sIX6.5 and Ad5F41s binding to SKBR-3 cells in the presence of the scFv C6.5 as a competitor. The use of a 1000-fold molar excess of scFv C6.5 with respect to amount of virus particles did not reveal any significant inhibition of Ad5F41sIX6.5 vector binding to SKBR-3 cells (data not shown). The ability of the Ad5F41sIX6.5-incorporated C6.5 scFv to recognize the cognate receptor was validated by ELISA with lyzate of c-erbB2-overexpressing SKBR-3 cells (Fig 6a) and by immunoprecipitation of Ad5F41sIX6.5 particles using ErbB2/Fc protein immobilized on Protein A gel (Fig 6b). These experiments demonstrated that pIX-incorporated scFv C6.5 is able of maintaining functional folding and the antigen-binding capacity that, however, was not translated to scFv-mediated Ad recognition of c-erbB2-positive cells.
Fig. 5.
Evaluation of Ad5F41sIX6.5 infection via pIX-incorporated scFv. Monolayers of 435.eB1 and MDA-MB-435 cells (upper panel) or SKBR-3 and MDA-MB-468 cells (lower panel) were infected with Ad5F41sIX6.5 or control Ad5F41s vector at a dose of 100 vp/cell and incubated for two days to allow expression of the DsRed2 reporter gene. The fluorescent light intensity was measured in plate reader, using 560 nm emission and 620 nm excitation filters, and relative fluorescent units (RFU) detected in infected cells are presented as kRFU after subtracting the background light signal detected in uninfected cells. Each bar represents the cumulative mean of triplicate determinations ±SD.
Fig. 6.
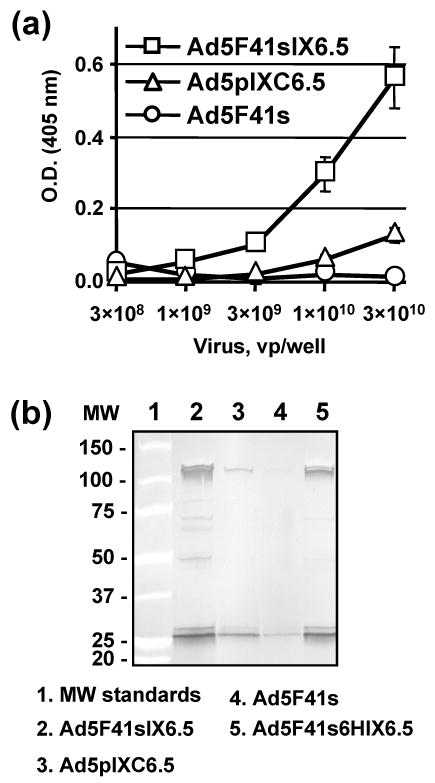
Validation of binding of Ad5F41sIX6.5-incorporated scFv to c-erbB2. (a) Ad5F41sIX6.5, Ad5pIXC6.5, and Ad5F41s viral particles were adsorbed on an ELISA plate at the indicated concentrations and incubated with 50-μl aliquots of SKBR-3 cell lysate. Bound c-erbB2 oncoprotein was detected with mAb cocktail against the cytoplasmic c-erbB2 domain followed by AP-conjugated goat anti-mouse secondary antibody, and the plate was read at 405 nm. Each data point represents the cumulative mean ±SD of triplicate determinations (some error bars are smaller than the symbols). (b) The indicated Ad vectors were incubated with ErbB2/Fc protein that was immobilized on protein A gel. The viral particles bound to ErbB2 were immunoprecipitated, eluted from the gel and analyzed by western blot using rabbit anti-Ad5 serum. The numbers on the left indicate molecular masses of protein standards in kDa.
Analysis of efficiency of infection of Ad5F41s6H via CAR and artificial receptor
To evaluate whether the Ad5 fiber replacement maneuver results in ablation of viral tropism in vitro, we determined the infection efficiencies of Ad5F41s, Ad5F41s6H, and control Ad5Red vectors. The gene transfer assay showed that the infectivity of the Ad5F41s and Ad5F41s6H vectors was reduced by 50- and 100-fold relative to the Ad5Red control in CAR-positive 293 cells (Fig. 7a). This decrease in gene transfer efficacy correlated with substantially reduced cytopathic effects (CPE) observed in cell monolayers infected with Ad5F41s and Ad5F41s6H vectors as compared to the Ad5Red control (Fig. 7b, upper panel). Thus, both assays confirmed that the Ad5 capsid pseudotyping with the 41s fiber ablated host cell recognition via CAR-dependent mechanism while resulting in a significant decrease of virus infection efficiency.
Fig. 7.
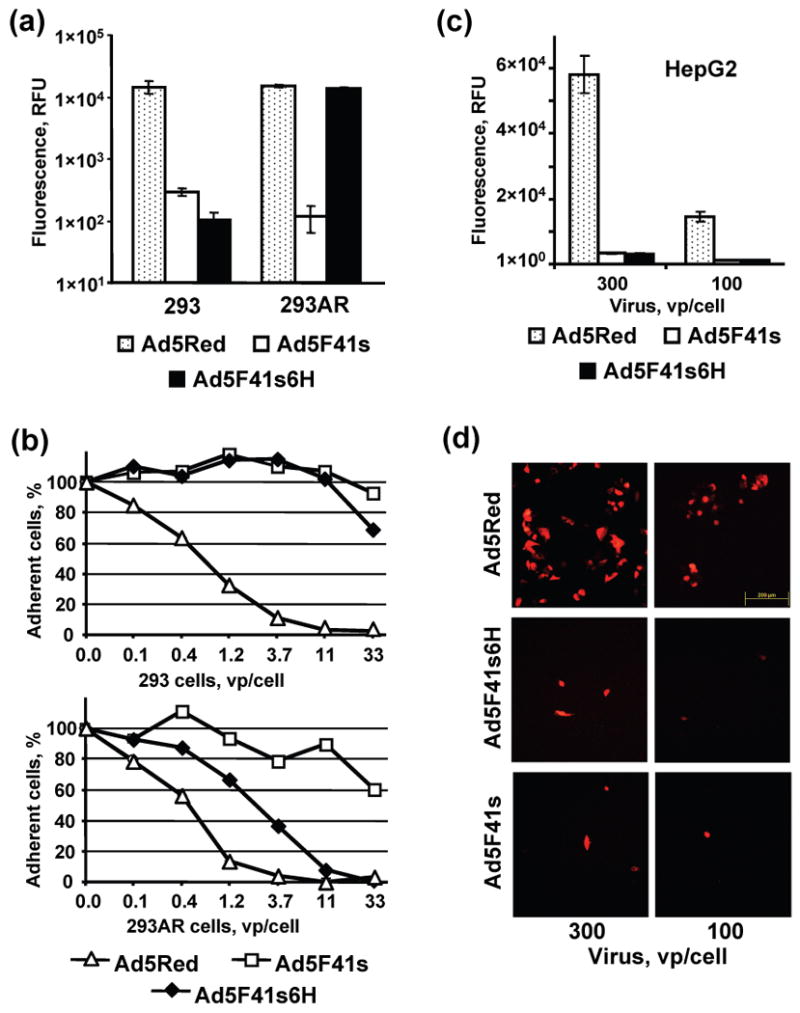
Analysis of efficiency of infection of Ad5F41s6H, Ad5F41s, and Ad5Red vectors in 293, 293AR and HepG2 cells. (a) Monolayers of 293 and 293AR cells were infected with Ad5F41s6H, Ad5F41s, or Ad5Red vector. The fluorescent light intensity was measured 20 hours postinfection in plate reader, using 560 nm emission and 620 nm excitation filters, and relative fluorescent units (RFU) detected in infected cells are presented after subtracting the background light signal detected in uninfected cells. Each bar represents the cumulative mean ±SD of triplicate determinations. (b) Monolayers of 293 and 293AR cells were infected with Ad5F41s6H, Ad5F41s, or Ad5Red vector at the indicated MOIs. The virus-induced CPE was monitored 4 days postinfection by staining adherent cells with crystal violet and then scanning wells using a plate reader set at 565 nm. The data are presented as the percentages of cells in monolayers infected with each viral dose that were determined with respect to the uninfected control ±SD (some error bars are smaller than the symbols). (c) HepG2 cells were incubated with 100 or 300 vp/cell of the indicated Ad vectors and the RFU values detected in the cell monolayers 40 hours postinfection are presented after subtracting the background light signal detected in uninfected cells. Each bar represents the cumulative mean ±SD of triplicate determinations. (d) Monolayers of HepG2 cells incubated with 100 or 300 vp/cell of the indicated Ad vectors were imaged 40 h postinfection using fluorescence microscopy. The representative images of fluorescent cells are shown at a magnification of 100×. The scale bar represents 200 μm.
Further, we evaluated if elimination of Ad5 fiber-mediated virus-cell interaction, which was allowed by fiber pseudotyping, could also attenuate Ad5 propensity for hepatic cell types. To this end, we used HepG2 human hepatocellular carcinoma cells that are highly permissive for Ad5 infection to assess the levels of gene transfer and transgene expression mediated by Ad5F41s and Ad5F41s6H vectors with respect to Ad5Red control. As shown in Fig. 7, both 41s fiber-pseudotyped Ad5 vectors revealed at least 20-fold lower transgene expression (Fig. 7c), which correlated with marked decrease in numbers of reporter-positive cells infected with Ad5F41s and Ad5F41s6H vector (Fig. 7d) as compared to Ad5Red. These data demonstrated a substantial level of un-targeting of Ad vectors pseudotyped with of 41s fibers from hepatic cells in vitro.
To test if Ad5 tropism ablated via fiber replacement could be substituted by an alternative mechanism permitting receptor-specific cell recognition, we used 293AR cells. We previously demonstrated that a membrane-anchored scFv against a C-terminal oligo-histidine sequence 75 expressed on the surface of 293AR cells can serve as an artificial receptor for virions incorporating His6-tagged Ad5 fibers 70. As shown in Fig. 7a, the Ad5F41s6H vector achieved a 120-fold increased gene transfer in 293AR cells as compared to the Ad5F41s vector, demonstrating virtually the same infection efficiency as the Ad5Red control. This enhancement of Ad5F41s6H vector infectivity in 293AR cells resulted in markedly improved CPE (Fig. 7b, bottom panel) compared to the Ad5F41s vector and 293 cells.
Taken together, these data validated abrogation of CAR-dependent host cell recognition by Ad5F41s and Ad5F41s6H vectors while suggesting the means to achieve retargeting of Ad5F41s6H vector via alternative receptors by employing an adapter-based approach.
Generation of Ad5F41s6H-targeting bispecific adapter protein
We hypothesized that incorporation of the His6 tag into the 41s fiber could be exploited to achieve indirect linkage of Ad5F41s6H vector with c-erbB2-positive cells using bispecific adapter protein. To test our hypothesis, we employed the so-called bispecific single-chain diabody (scDb) format 76; 77 to design an adapter protein consisting of anti-c-erbB2 scFv C6.5 68 and scFv 3D5 against the oligo-histidine sequence 75. To this end, the VH and VL chains from scFvs of different specificity were joined by linker sequences into a single-chain (VL1-VH2)-(VL2-VH1) polypeptide (Fig. 8a), which folded head-to-tail into a diabody-like structure containing two antigen–binding sites as illustrated in Fig. 8b. We constructed an expression cassette encoding the murine Ig kappa-chain leader followed by the influenza hemagglutin (HA) epitope, scDb 3D5×C6.5, and Strep-tag II 78 peptide under the transcriptional control of the CMV promoter (Fig. 8a). The resultant 3D5×C6.5 gene was expressed in stably transfected Hela cells and the secreted scDb protein was purified by affinity chromatography as described in the Materials and Methods. Western blot analysis detected the presence of both HA and Strep-tag epitopes in the purified protein validating that the full-length scDb polypeptide with a molecular mass of 57 kDa, as predicted for the secretory 3D5×C6.5 molecule, was produced (Fig. 8c).
Fig. 8.
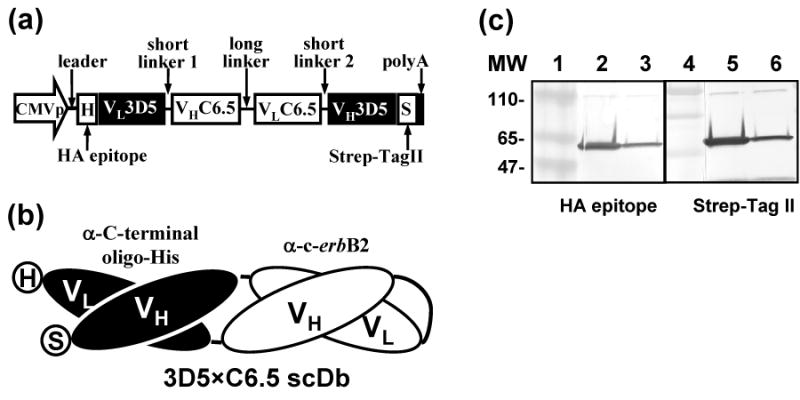
Generation of bispecific 3D5×C6.5 scDb adapter. (a) The recombinant 3D5×C6.5 gene was assembled to encode the following elements: leader signal from murine Ig kappa-chain (Leader), HA epitope (H), variable light chain of 3D5 scFv (VL3D5), 5-aa linker (short linker 1), variable heavy chain of C6.5 scFv (VHC6.5), 15-aa (G4S)3 linker (long linker), variable light chain of C6.5 scFv (VLC6.5), 5-aa linker (short linker 2), variable heavy chain of 3D5 scFv (VH3D5), and Strep-tag II peptide (S). The constructed gene was placed under the transcriptional control of the CMV promoter to produce the scDb in mammalian cells. (b) Schematic representation of 3D5×C6.5 polypeptide folded head-to-tail into a diabody-like structure to form antigen–binding sites for the C-terminal oligo-His tag and the c-erbB2 oncoprotein. (c) The secreted 3D5×C6.5 scDb was purified on Strep-Tactin affinity column and the protein characterized by western blot using an anti-HA mAb (lanes 2 and 3) and Strep-Tactin (lanes 5 and 6), both conjugated with AP. The samples in lanes 2 and 5 were boiled to denature the scDb protein to monomers, while proteins in lanes 3 and 6 were not denatured. The numbers on the left indicate molecular masses of protein standards (lanes 1 and 4) in kDa.
Analysis of scDb binding specificities
We tested whether the VH and VL domains of 3D5 and C6.5 scFv incorporated into the scDb molecules were able to fold correctly and form antigen–binding sites of corresponding specificities. The specificity of binding of the 3D5×C6.5 scDb to the His6 tag introduced into the C-terminus of the fiber protein was validated by western blotting of Ad5F41s6H and Ad5F6H vectors using purified scDb as a primary antibody. Fig. 9a shows that single major bands of approximately 66 and 41 kDa, which correspond to the predicted molecular masses of the His6-tagged Ad5 and 41s fibers, respectively, were detected with 3D5×C6.5 scDb in the Ad5F6H and Ad5F41s6H samples but not in the Ad5 and Ad5F41s controls. The 3D5×C6.5 scDb molecule also demonstrated dose-dependent binding to Ad5F41s6H viral particles immobilized on an ELISA plate, while no binding was detected with the control Ad5F41s virus (Fig. 9b). The c-erbB2-binding specificity of 3D5×C6.5 scDb was confirmed by an ELISA using ErbB2/Fc chimeric protein, as illustrated in Fig. 9c. The affinities of 3D5×C6.5 scDb binding to c-erbB2 and C-terminal His6 tag determined by an inhibition ELISA method 79 were very close to those obtained for the parental scFvs 68, 75, as indicated by Kd values of 23 and 390 nM, respectively. These data validated the dual binding capacity of the generated 3D5×C6.5 scDb, thus suggesting that it could serve as an adapter molecule bridging Ad5F41s6H viral particles with the cellular c-erbB2 oncoprotein.
Fig. 9.
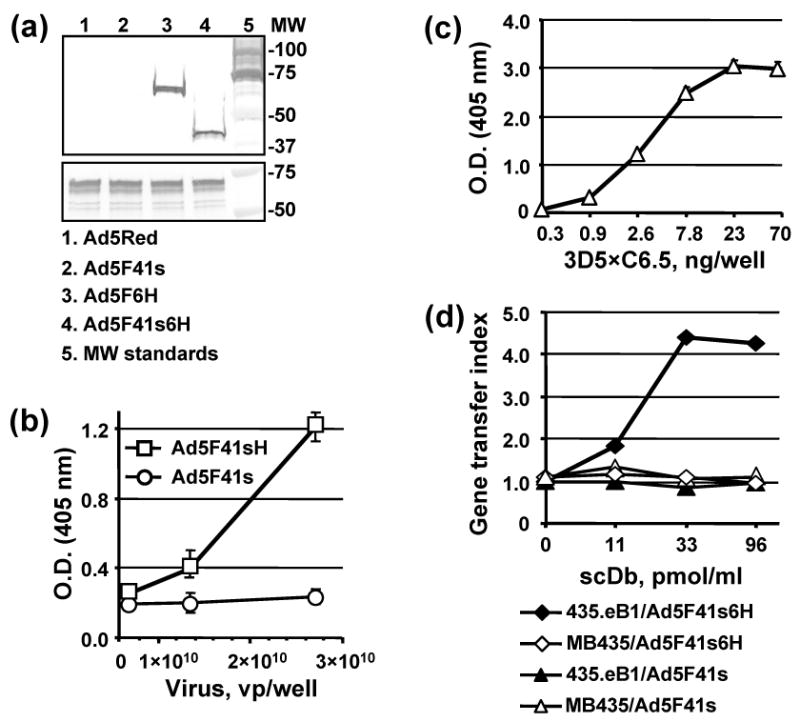
Characterization of 3D5×C6.5 scDb binding specificity. (a) Electrophoretically resolved viral proteins were transferred to a PVDF membrane and probed with 3D5×C6.5 scDb, diluted to a concentration of 1 μg/ml, or penton-base specific rabbit sera to detect the presence of His6-tagged fiber proteins (upper panel) or penton base (lower panel) respectively. The numbers on the right indicate molecular masses of protein standards (lane 5) in kDa. (b) Ad5F41s6H or Ad5F41s virions immobilized on an ELISA plate at the indicated concentrations were incubated with 100 ng/ml of 3D5×C6.5 scDb. Bound scDb protein was detected with an anti-HA mAb conjugated with AP. (c) 3D5×C6.5 scDb was diluted to the indicated concentrations, adsorbed on an ELISA plate, and incubated with 50 μl aliquots of SKBR-3 cell lysate. Bound c-erbB2 oncoprotein was detected with mAb cocktail against the cytoplasmic c-erbB2 domain followed by AP-conjugated goat anti-mouse secondary antibody, and the plate was read at 405 nm. (d) Samples of A5F41s6H and A5F41s vectors were preincubated with the indicated dilutions of 3D5×C6.5 scDb for 30 min and then used to infect 435.eB1 or MDA-MB-435 cells at a dose of 100 vp/cell. The level of DsRed2 reporter gene expression was assessed two days postinfection by determining the RFU values using 560 nm emission and 620 nm excitation filters. The ratios of RFU detected in the cells infected in the presence of scDb adapter to RFU detected in cells infected with virus alone were calculated and are presented as gene transfer indices.
To test whether the use of 3D5×C6.5 scDb could promote Ad5F41s6H infection in a receptor-specific manner, by virtue of binding both the His6-tagged 41s fiber and the ectodomain of c-erbB2, we used 435.eB1 cells overexpressing c-erbB2/HER-2/neu gene 74 and c-erbB2-negative MDA-MB-435 cells. When infection of these cell lines with either Ad5F41s6H or Ad5F41s vector was carried out in the presence of varying concentrations of 3D5×C6.5 scDb, only Ad5F41s6H-infected 435.eB1 cells demonstrated scDb dose-dependent enhancement of infection (Fig. 9d). As measured by improvements of virus-mediated gene transfer, the use of the 3D5×C6.5 adapter provided more then 4-fold increased Ad5F41s6H infection efficiency as compared to virus alone, thereby demonstrating the feasibility of Ad5F41s6H vector targeting to c-erbB2-positive cells.
Analysis of efficiency and specificity of 3D5×C6.5-mediated A5F41s6H vector targeting
To further assess the potential of the 3D5×C6.5 scDb for Ad5F41s6H vector targeting via a c-erbB2-dependent route, we used cell lines established from breast and ovarian carcinomas. The magnitude of gene transfer augmentation that was achieved by complexing of the Ad5F41s6H vector with increasing amounts of 3D5×C6.5 protein is illustrated in Fig. 10, along with gene transfer levels of the control Ad5F41s and Ad5Red vectors alone or in the presence of the same scDb concentrations. The infectivities of Ad5F41s and Ad5F41s6H vectors alone were somewhat similar and at least 15-fold lower than the Ad5Red vector in the selected cell lines. While preincubation with 3D5×C6.5 scDb had no effect on the infectivity of Ad5F41s and Ad5Red, the Ad5F41s6H vector showed an scDb dose-dependent gene transfer increase in all c-erbB2-positive cells tested (Fig. 10a). The use of the 3D5×C6.5 protein at a concentration of 22 μg/ml provided 12-, 16-, and 25-fold enhancement of Ad5F41s6H gene transfer efficiency in AU-565, SKBR-3, and SKOV3 cells, respectively. The scDb-mediated enhancement of Ad5F41s6H infectivity in c-erbB2-positive cancer cells achieved an efficiency of infection similar to that of the control Ad5Red vector via a CAR-independent pathway.
Fig. 10.
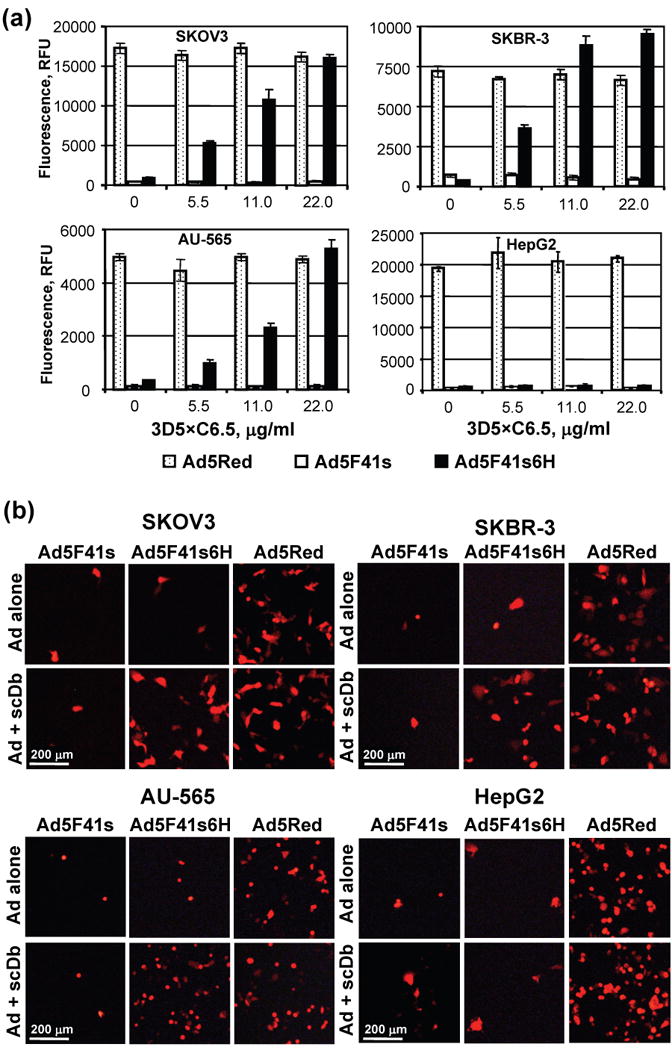
Analysis of efficiency and specificity of 3D5×C6.5-mediated A5F41s6H targeting via c-erbB2. (a) Aliquots of A5F41s6H, Ad5F41s, and Ad5Red were incubated with the 3D5×C6.5 scDb at the indicated concentrations and added to monolayers of SKBR-3, AU-565, SKOV3, MDA-MB-468 cells at a dose of 100 vp/cell. The efficiency of Ad-mediated DsRed2 reporter gene transfer was determined 40 hours postinfection based on the RFU values that were detected in infected cells (background signal detected in uninfected cells was subtracted) using plate reader equipped with a 560/620 nm emission/excitation filter set. The data are presented as RFU detected in cells infected with the indicated Ad vectors preincubated with the same concentration of 3D5×C6.5 protein. Each bar represents the cumulative mean ±SD of triplicate determinations. (b) Cell monolayers infected with indicated viruses either alone or in the presence of scDb adapter protein at a concentration of 22 μg/ml were imaged 40 h postinfection using fluorescence microscopy. The representative images of fluorescent cells are shown at a magnification of 40×. The scale bar represents 200 μm.
To validate that the transgene expression enhancement achieved by Ad5F41s6H vector in the presence targeting scDb adapter was associated with increased number of infected cells, we used fluorescence microscopy to image Ad-mediated expression of red fluorescent protein. Consistent with gene transfer assay, Ad5Red vector demonstrated relatively high levels of reporter-positive cells, while transduction with Ad5F41s resulted in few fluorescent cells regardless of presence or absence of scDb adapter in all cell lines tested (Fig. 10b). In contrast, markedly increased numbers of fluorescent cells were detected in monolayers of SKOV3, SKBR-3, and AU-565 cells, but not in c-erbB2-negative HepG2 cells, infected with Ad5F41s6H vector that was complexed with scDb adapter as compared to virus alone (Fig. 10b).
These data therefore clearly demonstrate that pseudotyping the Ad5 capsid with a His6-tagged 41s fiber coupled with 3D5×C6.5-guided receptor recognition resulted in c-erbB2-targeted modification of Ad vector tropism in vitro.
Discussion
Targeted Ad vectors hold the promise to expand the types of diseases that can be treated by gene therapy and to make the therapeutic applications of Ad vectors more effective. The increased specificity achieved by targeting virus infection to cells of interest will ultimately allow lower and safer doses of Ad vectors to be provided when regional or systemic delivery is contemplated in the future. The purpose of this study was to design an Ad vector that embodies genetic modifications to allow targeting to cancer cells.
It is widely acknowledged that genetic modifications of the Ad capsid to ablate endogenous mechanisms of host cell recognition while introducing novel infection specificity via alternative cellular receptors would represent the optimal means to achieve the goal of cell-specific Ad retargeting. To this end, genetic engineering of the Ad fiber protein appears the most straightforward way to eliminate virus binding to the primary cellular receptor and generate Ad vectors with novel tropism. The feasibility of this concept has been demonstrated using peptides derived from phage display libraries 54; 55, however, this revealed the limited capacity of the trimeric fiber for accommodating heterologous polypeptides. A key consideration in this context is the requirement to preserve both Ad virion integrity and ligand functionality while surmounting biosynthetic incompatibilities between viral capsid assembly in the nucleus and posttranslational processing of secretory ligand molecules.
The shortcomings of genetic Ad targeting have been addressed by the use of a bispecific adapter approach, the replacement of the fiber or its knob domain by exogenous trimerization motifs to improve the capacity to incorporate ligands, and the design of artificial ligands with superior biosynthetic compatibility. In this regard, the development of the so-called Affibody ligand 80, which was derived from the 58 amino acid residue Z domain of staphylococcal protein A, has facilitated the development of c-erbB2-targeted Ad5 vectors. The Affibody has been successfully incorporated into the HI loop of an Ad5 fiber knob ablated for CAR binding 81 or into a T4 fibritin-trimerized chimeric knobless fiber 82. The use of a knobless fiber to display a single-chain T-cell receptor (scTCR), generated against melanoma-associated antigen (MAGE)-A1 in complex with the HLA class I molecule, has enabled Ad retargeting to melanoma cells expressing HLA-A1/MAGE-A1 83. Despite these major advancements illustrating the potential of genetic Ad targeting in vitro, efforts to employ high-affinity ligands including growth factors and scFvs have mostly been unsuccessful, thus frustrating targeting of Ad vectors to many attractive cellular markers.
In order to overcome the low tolerance of the Ad5 fiber toward genetic modification, pIX has been extensively explored to achieve the display of heterologous polypeptides of augmented complexity on the viral capsid (reviewed in 84; 85). We have previously demonstrated the feasibility of genetic modification of the C-terminus of pIX 58. The capacity of pIX for protein incorporation has been assessed by integrating fluorescent proteins 60; 61; 62; 86, herpes simplex virus type 1 thymidine kinase (HSV-1 TK) 64, HSV-1 TK fused with firefly luciferase 65, and a hyper-stable scFv against E. coli β-galactosidase 66 into the capsid with minimal effect on viral function. Moreover, pIX has been successfully employed for incorporation of a functional scTCR allowing efficient Ad5 vector targeting to HLA-A1/MAGE-A1-positive melanoma cells 67. These data strongly suggested that the ectodomain of pIX could offer an improved capacity for the incorporation of targeting ligands as compared to sites in the fiber and hexon 87; 88; 89.
Herein we studied whether modification of pIX to incorporate scFv C6.5 68 against c-erbB2 could be tolerated by the Ad5 capsid and allow the folding of functional a scFv, which is essential for antigen recognition. We successfully rescued a vector, Ad5pIXC6.5, incorporating a pIX-fused C6.5 scFv in a manner commensurate with virion structural integrity. In order to eliminate native viral tropism and facilitate binding of the capsid-incorporated scFv to cellular c-erbB2, we chose to replace the Ad5 fiber in this Ad5pIXC6.5 vector with the short fiber Ad41. It had previously been demonstrated that the short fibers of Ad from subgroup F lack all cell-binding determinants described to date for other Ad serotypes 35; 37 and substantially reduce ectopic sequestration of fiber-pseudotyped Ad5 vectors in vivo 69. The Ad5F41sIX6.5 and Ad5F41s6HIX6.5 vectors that we generated showed levels of incorporation of recombinant 41s fiber similar to the wild-type Ad5 fiber, while revealing a significantly increased capacity to encapsidate the pIX-scFv fusion protein as compared to the Ad5pIXC6.5 vector. The ability of the capsid-incorporated C6.5 scFv to recognize the c-erbB2 ectodomain was validated by an ELISA and an immunoprecipitation assay using lysates of c-erbB2-overexpressing SKBR-3 cells and recombinant ErbB2/Fc chimeric protein, respectively. These results corroborated that pIX is a capsid locale with structural and biosynthetic compatibility with a diverse repertoire of heterologous proteins, including conventional scFvs.
Despite the ability of the capsid-incorporated scFv to bind the soluble extracellular c-erbB2 domain, Ad5pIXC6.5 and Ad5F41sIX6.5 vectors failed to achieve c-erbB2-specific enhancement of either cell binding or vector-mediated gene transfer as compared to control Ad vectors. The lack of ligand-mediated virus attachment to target cells suggested that the interaction between the pIX-incorporated scFv and cellular c-erbB2 was affected by factors other than ligand folding. In this regard, several studies have reported that insertion of an alpha-helical spacer between the ligand moiety and the pIX ectodomain to position the ligand sequence distal to the capsid surface might be critical to provide sufficient ligand flexibility to allow proper recognition of target receptors 59; 66; 67. Accordingly, genetic incorporation of a biotin acceptor peptide into pIX to allow metabolic biotinylation and virus retargeting using avidin in combination with various high affinity ligands has been proven ineffective in contrast to a biotinylation of the fiber in an Ad5 vector 63; 89. Campos and Barry 89 speculated that pIX may be preferentially used for the attachment of low-affinity ligands, which are able of dissociating quickly from their receptors subsequent to virus internalization, to avoid entrapment of viral complexes in the endosomes and aberrant intracellular trafficking. In this regard, the recent study by Corjon et al. 90 showed that chemical coupling of large high-affinity ligands to the C-terminal cysteines introduced to pIX can impair intracellular virus trafficking while demonstrating that dissociation of pIX-conjugated ligand moiety from its receptor after endocytosis is an important prerequisite for achieving successful targeting. In the aggregate, these studies highlighted that, despite its superior capacity for heterologous protein accommodation, pIX has a limited ability to display targeting ligand in a manner required for efficient interaction with a cellular receptor, cell entry, and subsequent virion trafficking to the nucleus.
We previously showed that incorporation of six-histidine residues at the C-terminal of the fiber protein enables Ad infection of nonpermissive cell types displaying an artificial receptor consisting of a membrane-anchored anti-His sFv 70; 91. Herein, we demonstrated that the introduction of the His6 tag the 41s fiber, which lacks any known cell-binding determinants, markedly improved Ad5F41s6H vector infectivity via this artificial receptor expressed in 293AR cells 70 as compared to the Ad5F41s control vector. Modifications of the Ad41 short fiber knob to incorporate the RGD-4C peptide motif within surface-exposed protein loops and at the C-terminus have been employed to retarget a fiber-pseudotyped Ad5 vector to primary and established melanoma cells via cellular integrins 92. In this report, we assessed the utility of the 41s fiber displaying a C-terminal His6 tag for vector retargeting using an adapter-based approach. To confer targeting of the Ad5F41s6H vector toward c-erbB2-positive cancer cells, we generated the 3D5×C6.5 scDb with affinities for both c-erbB2 and the His6 tag and showed its ability to increase Ad5F41s6H infectivity up to 25-fold in a c-erbB2-specific manner. The use of ascDb-dased MelAd adapter binding both the Ad5 fiber knob and the high molecular weight melanoma-associated antigen has been previously reported to target a CAR binding-ablated Ad5 vector to melanoma cells with a high level of efficiency and specificity 93. The utility of adapter molecules constructed using an anti-Ad5 knob scFv or the extracellular CAR domain is obviously limited to Ad5 and other CAR-binding Ad serotypes. Our data provide a rationale for the development of a new class of protein adapters capable of Ad vector targeting by virtue of binding to a fiber-incorporated His6 peptide. The use of such a serotype-independent targeting modality could provide the technical means for testing the ability of vectors derived from representatives of various Ad species to localize to the tissue of interest while overcoming native Ad5 hepatotropism 94.
The nature of the virus–host interactions that dictate the fate of systemically administered Ad5 vectors has come under considerable scrutiny in recent years. Initially, the fiber protein was implicated as the major structural determinant responsible for Ad5 liver tropism in vivo (reviewed by Nicklin et al. 29). More recently, Shayakhmetov et al. 38 uncovered a novel role for coagulation factor IX (FIX) and complement component C4-binding protein (C4BP) in hepatocyte and Kupffer cell uptake of intravenously injected Ad. This finding refocused research into the biology of interactions between Ad5 capsid components and host blood factors and their influence on systemic virus biodistribution 95. Further studies revealed the ability of the vitamin K-dependent coagulation factors VII, IX, X, and protein C to bind trimeric hexon in the viral capsid and facilitate CAR-independent infection of hepatic cells following intravascular Ad5 vector administration 39; 40. These efforts serve to highlight the complexity of virus/host interplay in the artificial bloodborne environment and have identified modifications of the fiber 38 and hexon 89; 96 proteins that significantly decrease infection and virus-induced toxicity in the liver. Thus, it is recognized that the infection pathway of systemically administered Ad5 is mediated via multiple mechanisms involving blood factors rather then direct virus interaction with cellular receptors. On this basis, it becomes increasingly apparent that engineering of capsid proteins to overcome ectopic Ad5 sequestration in the liver coupled with virus retargeting via non-native infection pathway, which is mediated by bispecific adapter proteins represents a rational strategy to direct Ad vector localization to the tissue of interest following systemic administration.
While single-component vector systems have been favored for employment in human trials, rigorous analysis of the pharmaco-dynamics and systemic stability of vector-adapter complexes could provide the rationale for clinical translation. In this respect, previous in vivo studies using various Ad5 fiber knob-binding adapters have provided compelling evidence of reduced ectopic liver transduction and receptor-specific vector delivery to target organs 44; 50; 51 or tumors 52; 53. Thus, our novel Ad tropism modification maneuver that embodies concepts of ‘de-targeting’ and ‘re-targeting’ by combining elements of genetic capsid modification and adapter-based approaches has encouraging implications for further development of advanced delivery vehicles.
Materials and Methods
Cells
The 293 human kidney cell line transformed with Ad5 DNA was purchased from Microbix (Toronto, Ontario, Canada). 293AR cells were generated previously 70 to express an artificial receptor derived from 3D5 scFv 75 specific for a C-terminal oligo-histidine tag. The SKOV3 human ovarian cancer, MDA-MB-468, AU-565, and SKBR-3 human breast cancer, and HepG2 human hepatocellular carcinoma cell lines were obtained from the American Type Culture Collection (Manassas, VA). MDA-MB-435 breast adenocarcinoma cells and their derivative stably transfected with the HER-2/neu gene, namely 435.eB1 cells 73; 74, were kindly provided by Dr. Dihua Yu (The University of Texas M. D. Anderson Cancer Center, Houston, TX). All cell lines were maintained in recommended growth media supplied by Mediatech (Herndon, VA.) containing 10% FBS (HyClone, Logan, UT) and 2 mM glutamine at 37°C in a humidified atmosphere of 5% CO2.
Ad vector engineering
The engineering of the Ad5FHIFlag vector containing the Flag octapeptide (DYKDDDDK) within the Ad5 fiber knob HI loop has been described elsewhere 97. The replication-deficient AdLucIXflag vector encoding the Flag peptide fused to the C-terminus of pIX was generated previously 58. To construct the genome of a replication-competent Ad vector encoding I/C6.5 scFv fused to the C-terminus of pIX we used the previously described pShlpIXNhe vector 58 as follows. The BamHI-MfeI DNA fragment (3942 bp) containing the Ad5 early E1 region was isolated from plasmid pTG3602 98 and ligated with the BamHI-MfeI fragment of pShlpIXNhe generating plasmid pShE1pIXNhe. The sequence encoding I/C6.5 scFv was PCR-amplified from plasmid pFBsCARfC6.5 46 using primers (5′-AGG AAA CCG GTG GTC TAG ATC AGG TGC AGC T-3′ and 5′-AGT ATC TAG AGG GAA CTA GTA CGG TCA GCT TGG TCC CTC-3′) designed to contain the XbaI recognition sites (underlined) and was cloned into NheI-digested pShE1pIXNhe. The resultant plasmid pShE1pIXc6.5 was used for homologous recombination with the pAdEasy-1 plasmid in E.coli strain BJ5183 (Stratagene, La Jolla, CA) as described elsewhere 99, to generate plasmid pAd5pIXC6.5 containing the genome of the Ad5pIXC6.5 vector.
To construct the recombinant fiber gene, the EcoRI-Acc65I restriction fragment (3544 bp) from pTG3602 was cloned between the EcoRI and Acc65I sites in the pNEB193 plasmid (New England BioLabs, Beverly, MA). The resultant pNebAd5F plasmid was used for PCR primer-directed mutagenesis to introduce the DraIII and ClaI restriction sites into the DNA sequences coding for the tail-shaft juncture and the C-terminus of Ad5 fiber, respectively (details are available upon request). The DNA fragment encoding shaft and knob regions of the short Ad41 fiber was PCR-amplified from the Ad41 genome of strain TAK (ATCC) using primers (5′-GGA AGA ACC ACC TGG TGT TTT AGC AC-3′ and 5′-CCA TCG ATT GTT CAG TTA TGT AGC AAA-3′) containing the DraIII and ClaI recognition sites (underlined), respectively, and was cloned into pNebAd5F generating plasmid pNF5t41s. To incorporate a His6 tag into the C-terminus of the recombinant Ad41 short fiber oligonucleotides 5′-CGG GCC ATC ACC ATC ACC ATC ACT AG-3′ and 5′-CGC TAG TGA TGG TGA TGG TGA TGG CC-3′ were annealed to form a DNA duplex containing ClaI-compatible 5′-cohesive ends (underlined) and then ligated with ClaI-digested pNF5t41s, resulting in plasmid pNF5t41s6H. Plasmids pNF5t41s and pNF5t41s6H were used for homologous recombination with plasmid pVK50 as described previously 97 to generate pAd5F41s or pAd5F41s6H genomes carrying genes encoding the recombinant 41s or 41sHis6 fiber protein, respectively. The DsRed2 gene encoding red fluorescent protein (Clontech Laboratories, Mountain View, CA) was cloned into pShuttle-CMV 99 and then, resultant plasmid was recombined with pAd5F41s, pAd5F41s6H, or pTG3602 98 to construct the corresponding plasmid derivatives containing the DsRed2 reporter gene under transcriptional control of the human cytomegalovirus (CMV) immediate-early promoter in place of the early viral E1 region for the further generation of Ad5F41s, Ad5F41s6H, and Ad5Red vectors respectively. The pShE1pIXc6.5 plasmid was also modified to introduce the CMV-driven DsRed2 gene in E1 and then was recombined with the genomes in pAd5F41s and pAd5F41s6H to generate pAd5F41sIX6.5 and pAd5F41s6HIX6.5 plasmids, respectively.
To generate recombinant Ad vectors, the plasmid constructs were digested with PacI to release the inverted terminal repeats of the viral genomic DNA and were then used to transfect 293 cells to rescue the viral genomes. All newly rescued Ad vectors as well as Ad5 and Ad41 wild type (WT) viruses were propagated on 293 cells, purified by centrifugation on CsCl gradients according to standard protocol and dialyzed against PBS (8 mM Na2HPO4, 2 mM KH2PO4 [pH 7.4], 137 mM NaCl, and 2.7 mM KCl) containing 10% glycerol. The titers of physical viral particles (vp) were determined by the methods of Maizel et al. 100
The titers of infectious viral particles were determined by plaque assay using 293 cells as descried by Mittereder et al. 101. The ratios of vp to plaque forming units (pfu) determined for Ad5Red, Ad5pIXC6.5, Ad5F41s, Ad5F41s6H, Ad5F41sIX6.5, and Ad5F41s6HIX6.5 were 14, 957, 200, 220, 2640, and 2555 vp/pfu respectively. All viruses were propagated at yields comparable to the control Ad5Red vector, with the exception of Ad5pIXC6.5. However, the pIX-modified Ad5pIXC6.5, Ad5F41sIX6.5, and Ad5F41s6HIX6.5 vectors were deficient in their plaque forming ability as reflected by high vp/pfu values. Ad5 capsid pseudotyping with the 41s fiber resulted in a substantial attenuation of Ad5F41s and Ad5F41s6H infectivity in 293 cells and a corresponding increase of vp/pfu ratios as compared to the control Ad5Red vector The diagrams of generated Ad constructs denoting the incorporation of pIX-scFv, Ad5 fiber, and 41s fiber with or without His6 sequence are schematically presented in Fig. 1.
Fig 1.
Graphical representation of generated Ad vectors. All generated Ad-based vectors have the early viral E3 genes deleted (ΔE3) and are replication incompetent, with the exception of Ad5pIXC6.5, and contain the DsRed2 reporter gene (Red) under control of the CMV promoter (P) in place of the early viral E1 region (E1). The protein IX gene (IX) is modified in Ad5pIXC6.5, Ad5F41sIXC6.5, and Ad5F41s6HIXC6.5 vectors to encode the C-terminal scFv C6.5 (C6.5). Ad5F41sIXC6.5, Ad5F41s6HIXC6.5, Ad5F41s, and Ad5F41s6H contain the Ad5 fiber (F5) shaft and knob domain-coding sequences replaced with those of Ad41 short fiber gene (F41s). The C-terminal six-histidine-coding sequence (6H) is incorporated into the recombinant fiber gene encoded by Ad5F41s6HIXC6.5 and Ad5F41s6H vectors.
Electron microscopy
Samples of CsCl-purified Ad5pIXC6.5, Ad5 WT, or the previously described AdLucIXflag vector 58 were adsorbed for 5 min onto Formvar/carbon coated copper grids (300 mesh). Grids were rinsed in TBS (20 mM Tris-HCl [pH 7.8] and 150 mM NaCl) for 2 min, stained with 2% uranyl acetate and examined with transmission EM. Micrographs were taken at 100,000× magnification from areas selected at low magnification.
Generation of 3D5×C6.5 scDb
To construct the gene encoding the secretory 3D5×C6.5 scDb we used the previously described pHissFv.rec plasmid 70, which codes for membrane-anchored 3D5 scFv against the C-terminal His tag, in order to amplify the 3D5 scFv VL and VH fragments. The DNA sequence encoding the signal leader peptide and 3D5 VL fragment was PCR-amplified with the primer pair 3D5Vl.F (5′-AAT TAA GCT TGT CGA CAT CCA CCA TGG AGA CAG AC-3′) and 3D5Vl.R (5′-CCC ACT TCC GGA GCC ACT ACC ACG TTT TAT TTC-3′), while the VH-coding DNA sequence was amplified with the primer pair 3D5Vh.F (5′-TAG TGG CTC CGG AAG TGG GTC CCA GGT TCA G-3′) and 3D5Vh.R (5′-GAC ACT CGA GGC CGC AGA GAC AG-3′). These two DNA fragments were joined by a second PCR using the 3D5Vl.F and 3D5Vh.R primers which contain HindIII and XhoI recognition sites (underlined), respectively, and the resultant 866-bp DNA fragment was cloned between the HindIII and XhoI sites of plasmid pDisplay (Invitrogen, Carlsbad, CA), resulting in plasmid pD3D5. The I/C6.5 scFv-coding sequence was PCR-amplified from plasmid pFBsCARfC6.5 46 using primers 5′-TCA ATC CGG ACA GGT TCA GCT GGT GCA GTC-3′ and 5′-GCG GTC CGG ATA GTA CAG TCA GCT TGG TC-3′ containing the BspEI recognition sites (underlined) and cloned into BspEI-digested pD3D5. The resultant pD3D5-C6.5 plasmid was digested with XhoI and ligated with a DNA duplex encoding the Strep-Tag II epitope, which was made of annealed 5′-TCG ACT GAT TAT AAG GAT GAC GAT GAC AAA CCG GTG GGG ATC CCC TGG AGC CAC CCG CAG TTC GAG AAA TAA C-3′ and 5′-TCG AGT TAT TTC TCG AAC TGC GGG TGG CTC CAG GGG ATC CCC ACC GGT TTG TCA TCG TCA TCC TTA TA TCA G-3′ oligonucleotides, containing XhoI-compatible 5′-cohesive ends (underlined).
The resultant plasmid designated as pD3D5-C6.5s was transfected into Hela cells and single-cell clones stably expressing the scDb gene were then isolated by selection in the presence of G418 as we described previously 70. The clones were expanded to produce the secretory 3D5×C6.5 protein that was purified from culture media by affinity chromatography on Strep-Tactin column (IBA US, St. Louis, MO) as recommended by the manufacturer.
Western blot analysis
Samples of CsCl-purified Ad vectors were boiled in Laemmli loading buffer and 1.0 × 109 vp of each virus were loaded on a 4-20% gradient SDS-PAGE gel (Pierce, Rockford, IL). Electrophoretically resolved viral proteins were transferred to a PVDF membrane and analyzed for the presence of modified pIX or fiber proteins as described previously 58; 97. Briefly, the pIXflagC6.5 fusion protein was detected with anti-Flag monoclonal antibody (mAb) M2 (Sigma, St. Louis, MO), while mAb 4D2 71 against the tail region was used to detect the recombinant 41s fiber protein. The Penta-His mAb (Qiagen, Valencia, CA) was used to detect the His6 tag at the C-terminus of the 41s fiber. All primary mAbs were used at a dilution of 1:1000 for overnight incubation at 4°C. Bound mAbs were detected with a secondary goat anti-mouse antibody conjugated with alkaline phosphatase (AP) (Sigma) and developed with an AP substrate kit (Bio-Rad Laboratories, Hercules, CA).
To assess the binding of the Ad capsid-incorporated C6.5 scFv with the c-erbB2 oncoprotein we used recombinant ErbB2/Fc protein (R&D Systems Inc. Minneapolis, MN) consisting of the extracellular c-erbB2 domain fused to the Fc fragment of human IgG. To this end, the ErbB2/Fc protein was non-covalently immobilized on Protein A gel (Pierce, Rockford, IL) as recommended by the manufacturer and incubated with 5 × 1010 vp of Ad5F41sIX6.5, Ad5pIXC6.5, Ad5F41s, or Ad5F41s6HIX6.5 vector. Viral particles bound to the immobilized ErbB2/Fc were immunoprecipitated, eluted from the gel according to the manufacturer's recommendations, and subjected to SDS-PAGE. Electrophoretically resolved proteins transferred to a PVDF membrane were incubated with rabbit serum against Ad5, and then detected with secondary AP-conjugated goat anti-rabbit antibodies (Pierce) and an AP substrate kit (Bio-Rad Laboratories).
To characterize the purified 3D5×C6.5 protein, it was run on a 4-20% gradient SDS-PAGE gel, transferred to a PVDF membrane, and probed with AP-conjugated anti-HA epitope HA7 mAb (Sigma) or AP-conjugated Strep-Tactin (Bio-Rad Laboratories). To validate 3D5×C6.5 scDb binding to the His6 tag incorporated into the C-terminus of the fiber protein, 1.0 × 109 vp of Ad vectors were subjected to SDS-PAGE and transfer to a PVDF membrane as described above. The membrane was incubated with the purified scDb at a concentration of 1 μg/ml for 1 hour at room temperature. Bound scDbs were detected using AP-conjugated Strep-Tactin.
To validate that equal number of viral particles of each Ad vector was loaded per lane we used penton-base specific sera at a dilution of 1:1000 and a secondary AP-conjugated goat anti-rabbit antibody (Sigma).
ELISA
To assess the ability of the Ad capsid-incorporated C6.5 scFv to recognize the extracellular c-erbB2 domain, we carried out a solid-phase binding enzyme-linked immunosorbent assay (ELISA) as follows. CsCl-purified Ad5pIXC6.5 and control AdLucIXflag virions were diluted in 50 mM NaHCO3 [pH 9.6] and adsorbed on Nunc-Maxisorp ELISA plate wells at concentrations ranging from 1.3 × 109 to 1.0 × 1010 vp/well in triplicates for 2 hours at 4°C. The wells were blocked with PBS containing 0.05% Tween 20 and 2% BSA, and then washed three times with PBS containing 0.05% Tween 20. Recombinant ErbB2/Fc protein diluted in blocking buffer was added to the wells at a concentration of 5 ng/well and the plate was incubated overnight at 4°C. Bound ErbB2/Fc protein was detected by incubation with rabbit anti-human IgG(Fc) antibody conjugated with biotin (Pierce) for 1 hour at room temperature, followed by 1-hour incubation with AP-conjugated streptavidin (Bio-Rad Laboratories).
To test binding of the Ad5F41sIX6.5-incorporated C6.5 scFv to the c-erbB2 oncoprotein, Ad5F41sIX6.5, Ad5pIXC6.5 and Ad5F41s virions were adsorbed on an ELISA plate at increasing concentrations (3.0 × 108 – 3.0 × 1010 vp/well) essentially as described above. Immobilized viral particles were incubated overnight with 50 μl/well of lyzate of SKBR-3 cells, a line that expresses high levels of c-erbB2 46, which was prepared as follows. The cells were solubilized at 1.0 × 107 cells/ml in lysis buffer (1% NP-40, 20 mM Tris [pH 8.0], 137 mM NaCl, 10% glycerol, 2 mM EDTA, 10 μg/ml Aprotinin, and 10 μg/ml Leupeptin) by gentle pipetting at 4°C for 30 min and then centrifuged at 14,000 × g for 5 min to remove insoluble cellular debris. To detect bound c-erbB2 oncoprotein, the plates were sequentially incubated for 1 hour with Ab-17 mAb cocktail (Thermoscientific, Fremont, CA) against the cytoplasmic c-erbB2 domain and secondary AP-conjugated goat anti-mouse antibody (Sigma, St. Louis, MO).
To validate 3D5×C6.5 scDb binding to the His6 tag incorporated into the 41s fiber, Ad5F41s6H or Ad5F41s virions were adsorbed on an ELISA plate at varying concentrations (6.7 × 109 – 2.7 × 1010 vp/well) and were incubated overnight with 10 ng/well of 3D5×C6.5 scDb diluted in blocking buffer. Bound scDb protein was detected with AP-conjugated Strep-Tactin. To validate binding of 3D5×C6.5 scDb to the extracellular c-erbB2 domain, purified scDb protein was adsorbed on ELISA plate at concentrations ranging from 0.3 to 70 ng/well. The immobilized scDb was incubated overnight with 50 μl/well of SKBR-3 cell lysate, and bound c-erbB2 oncoprotein was detected essentially as described above.
The ELISA plates were washed three times with PBS containing 0.05% Tween 20 following each incubation and developed with p-nitrophenyl phosphate (PNPP) for 1 hour. The absorbance at 405 nm was measured using a Synergy HT plate reader (Bio-Tek Instruments, Winooski, VT).
Ad binding assay
Radiolabeling of pAd5IXc6.5 and Ad5 WT with technetium-99m (99mTc) was carried out as described previously 102 yielding 99mTc-labeled viral preparations with specific activities of 7.5 × 10-3 and 3.3 × 10-3 CPM/vp, respectively. To assess binding of 99mTc-labeled Ad5pIXC6.5 and Ad5 WT to c-erbB2-positive AU-565 and c-erbB2-negative MDA-MB-468 cells, cell monolayers grown in a 24-well plate (5 × 105 cells/well) were preincubated for 30 min at room temperature with 200 μl of 2% FBS-containing growth media with or without Ad5 knob protein (50 μg/ml). Then, 200-μl aliquots containing 6 × 108 vp of 99mTc-labeled Ad5pIXC6.5 or Ad5 WT (4.5 × 106 and 2 × 106 CPM, respectively) were added to triplicate cell monolayers in the presence or absence of knob protein and incubated for 1 h. Bound radioactivity was determined using a gamma-counter after washing the cell monolayers 3 times with 1 ml of PBS and harvesting the cells. The total number of bound Ad5pIXC6.5 or Ad5 WT particles was calculated for each cell monolayer and was normalized for the total protein content that was determined using the Bio-Rad Dc protein assay kit (Bio-Rad Laboratories).
Analysis of Ad5F41sIX6.5 vector binding to c-erbB2-positive cells via the capsid-incorporated C6.5 scFv was carried out as previously described 58. Briefly, 100-μl aliquots of SKBR-3 cells resuspended to a final concentration of 5 × 106 cells/ml in binding media (DMEM/F12, 20 mM HEPES, 0.5% BSA) were mixed with 100-μl aliquots of C6.5 scFv (2 μg/ml) diluted in binding media or media alone in 5-ml test tubes and incubated for 30 min at 4°C. Then, 100-μl aliquots containing 5×107 vp of either Ad5F41sIX6.5 or Ad5F41s vector were added to the cells in the presence or absence of C6.5 scFv and incubation was continued for 1 h to allow binding. Cells were diluted with 4 ml of binding media, centrifuged, and supernatants containing unbound virus were aspirated. Total DNA was isolated from the cell pellets with QIAamp DNA Mini Kit (Qiagen, Valencia, CA) and was used to determine the viral and cellular genomic DNA contents by quantitative PCR assay in the LightCycler System (Roche Diagnostics, Indianapolis, IN). The Ad genome copy number was determined with primers and probe specific for the Ad5 hexon gene and normalized by the amount of β-actin DNA detected in each sample.
Ad-mediated gene transfer assay
The efficiency of Ad vector infection was assessed by gene transfer assay using the DsRed2 reporter gene encoding red fluorescent protein. To address a technical issue associated with different vp/pfu values of Ad5F41s6H and Ad5Red vectors, we validated that the use of equal amounts of Ad5F41s6H and Ad5Red viral particles could provide a similar gene transfer effects in 293AR cells. On this basis, we choose to use viral particle titers instead of plaque forming units in order to normalize the concentrations of Ad5Red, Ad5F41s6H, and Ad5F41s vectors in all gene transfer experiments.
Cell monolayers grown in a 24-well plate (3 – 5 × 105 cells/well) were incubated for 30 min at room temperature with 100 vp/cell of Ad vector diluted in 200-μl of culture media containing 2% FBS. The infection media was aspirated, cells were washed with PBS, and then incubated in phenol red-free media containing 5% FBS for at least 40 hours at 37°C to allow reporter gene expression. The fluorescent light intensities in cell monolayers were measured in the multifunctional Synergy HT plate reader (Bio-Tek Instruments) using 560 nm emission and 620 nm excitation filters. The data are presented as relative fluorescent units (RFU) detected in triplicate infected cells after the background light signal detected in uninfected cells was subtracted.
To visualize the levels of Ad-mediated gene transfer of the DsRed2 reporter gene cell monolayers were imaged using fluorescence microscopy with an inverted IX-70 microscope (Olympus, Melville, NY) equipped with a MagnaFire digital CCD camera (Optronics, Goleta, CA). Images were acquired with a ×10 objective using 2 sec exposure and processed with Optronics MagnaFire imaging software v. 1.0.
Ad-induced CPE assay
To assess the cytopathic effect (CPE) induced by virus propagation, monolayers of 293 and 293AR cells grown in a 96-well plate (3 - 5 × 103 cells/well) were infected in triplicates with Ad5F41s6H, Ad5F41s, or Ad5Red vector at MOIs ranging from 0.1 to 33 vp/cell. Plates were incubated for 4 days at 37°C and the virus-induced CPEs were assessed by staining attached cells with crystal violet and then scanning wells using a Synergy HT plate reader (Bio-Tek Instruments) set at 565 nm. The data are presented as the percentage of optical density detected in infected cell monolayers with respect to the uninfected control.
Analysis of scDb-mediated Ad targeting
Analyses of 3D5×C6.5 scDb-mediated Ad vector targeting to c-erbB2-positive cells were performed as follows. Fifty microliter aliquots of A5F41s6H vector were mixed with 50 μl-aliquots of 3D5×C6.5 scDb diluted in PBS to concentrations ranging from 5 to 22 μg/ml or with PBS alone and incubated for 15 min at room temperature. The A5F41s6H/scDb mixtures were diluted 10-fold with growth media containing 2% FBS, and 200-μl aliquots were then added to cell monolayers grown in a 24-well plate (3 – 5 × 105 cells/well) at a dose of 100 vp/cell. Ad5Red and Ad5F41s vectors preincubated with scDb protein as above were used as controls. After a 30-min exposure at room temperature, the Ad/scDb mixtures were aspirated, the cells were washed with PBS, and then incubated in a phenol red-free media containing 5% FBS at 37°C for at least 40 hours to allow reporter gene expression. The efficiency of c-erbB2-specific A5F41s6H vector targeting mediated by 3D5×C6.5 scDb adapter was assessed by measuring the gene transfer level (essentially as described above) in cells infected with A5F41s6H, Ad5Red, or Ad5F41s in the presence of the scDb as compared to Ad alone. Infected cell monolayers were imaged using fluorescence microscopy as described above. Images were acquired with a ×4 objective using 1 sec exposure and processed with Optronics MagnaFire imaging software v. 1.0.
Acknowledgments
This work was supported by grants R21CA116525 (to Dr. Dmitriev, I.P.), R01CA083821 (to Dr. Curiel, D.T.), and R01CA108585 (to Dr. Douglas, J.T.) from the National Cancer Institute and DAMD17-03-1-0629 (to Dr. Dmitriev, I.P.) from the Department of Defense Breast Cancer Research Program.
We are grateful to Leigh Millican from the High Resolution Imaging Facility at University of Alabama Birmingham for her help with electron microscopy. We would like to disclose that David T. Curiel and Joanne T. Douglas hold equity in VectorLogics, Inc. and Igor P. Dmitriev has Intellectual Property related to this publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shenk T. Adenoviridae: The Viruses and Their Replication. In: Fields BN, Knipe DM, H PM, et al., editors. Fields Virology. Third. Vol. 2. Lippincott - Raven Publishers; Philadelphia: 1996. pp. 2111–2148. [Google Scholar]
- 2.Fabry CM, Rosa-Calatrava M, Conway JF, Zubieta C, Cusack S, Ruigrok RW, Schoehn G. A quasi-atomic model of human adenovirus type 5 capsid. Embo J. 2005 doi: 10.1038/sj.emboj.7600653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart PL. Adenovirus Structure. In: Curiel DT, Douglas JT, editors. Adenoviral Vectors for Gene Therapy. Academic Press; San Diego, Ca: 2002. pp. 1–18. [Google Scholar]
- 4.Henry LJ, Xia D, Wilke ME, Deisenhofer J, Gerard RD. Characterization of the knob domain of the adenovirus type 5 fiber protein expressed in Escherichia coli. J Virol. 1994;68:5239–46. doi: 10.1128/jvi.68.8.5239-5246.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greber UF, Willetts M, Webster P, Helenius A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell. 1993;75:477–86. doi: 10.1016/0092-8674(93)90382-z. [DOI] [PubMed] [Google Scholar]
- 6.Wickham TJ, Mathias P, Cheresh DA, Nemerow GR. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–19. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 7.Gaggar A, Shayakhmetov DM, Lieber A. CD46 is a cellular receptor for group B adenoviruses. Nat Med. 2003;9:1408–12. doi: 10.1038/nm952. [DOI] [PubMed] [Google Scholar]
- 8.Sirena D, Lilienfeld B, Eisenhut M, Kalin S, Boucke K, Beerli RR, Vogt L, Ruedl C, Bachmann MF, Greber UF, Hemmi S. The human membrane cofactor CD46 is a receptor for species B adenovirus serotype 3. J Virol. 2004;78:4454–62. doi: 10.1128/JVI.78.9.4454-4462.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roelvink PW, Lizonova A, Lee JG, Li Y, Bergelson JM, Finberg RW, Brough DE, Kovesdi I, Wickham TJ. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J Virol. 1998;72:7909–15. doi: 10.1128/jvi.72.10.7909-7915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS, Horwitz MS, Crowell RL, Finberg RW. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–3. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 11.Walters RW, Freimuth P, Moninger TO, Ganske I, Zabner J, Welsh MJ. Adenovirus fiber disrupts CAR-mediated intercellular adhesion allowing virus escape. Cell. 2002;110:789–99. doi: 10.1016/s0092-8674(02)00912-1. [DOI] [PubMed] [Google Scholar]
- 12.Walters RW, van't Hof W, Yi SM, Schroth MK, Zabner J, Crystal RG, Welsh MJ. Apical localization of the coxsackie-adenovirus receptor by glycosyl-phosphatidylinositol modification is sufficient for adenovirus-mediated gene transfer through the apical surface of human airway epithelia. J Virol. 2001;75:7703–11. doi: 10.1128/JVI.75.16.7703-7711.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dechecchi MC, Melotti P, Bonizzato A, Santacatterina M, Chilosi M, Cabrini G. Heparan sulfate glycosaminoglycans are receptors sufficient to mediate the initial binding of adenovirus types 2 and 5. J Virol. 2001;75:8772–80. doi: 10.1128/JVI.75.18.8772-8780.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dechecchi MC, Tamanini A, Bonizzato A, Cabrini G. Heparan sulfate glycosaminoglycans are involved in adenovirus type 5 and 2-host cell interactions. Virology. 2000;268:382–90. doi: 10.1006/viro.1999.0171. [DOI] [PubMed] [Google Scholar]
- 15.Hong SS, Karayan L, Tournier J, Curiel DT, Boulanger PA. Adenovirus type 5 fiber knob binds to MHC class I alpha2 domain at the surface of human epithelial and B lymphoblastoid cells. Embo J. 1997;16:2294–306. doi: 10.1093/emboj/16.9.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.You Z, Fischer DC, Tong X, Hasenburg A, Aguilar-Cordova E, Kieback DG. Coxsackievirus-adenovirus receptor expression in ovarian cancer cell lines is associated with increased adenovirus transduction efficiency and transgene expression. Cancer Gene Ther. 2001;8:168–75. doi: 10.1038/sj.cgt.7700284. [DOI] [PubMed] [Google Scholar]
- 17.Hidaka C, Milano E, Leopold PL, Bergelson JM, Hackett NR, Finberg RW, Wickham TJ, Kovesdi I, Roelvink P, Crystal RG. CAR-dependent and CAR-independent pathways of adenovirus vector-mediated gene transfer and expression in human fibroblasts. J Clin Invest. 1999;103:579–87. doi: 10.1172/JCI5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fechner H, Wang X, Wang H, Jansen A, Pauschinger M, Scherubl H, Bergelson JM, Schultheiss HP, Poller W. Trans-complementation of vector replication versus Coxsackie-adenovirus-receptor overexpression to improve transgene expression in poorly permissive cancer cells. Gene Ther. 2000;7:1954–68. doi: 10.1038/sj.gt.3301321. [DOI] [PubMed] [Google Scholar]
- 19.Kirby I, Davison E, Beavil AJ, Soh CP, Wickham TJ, Roelvink PW, Kovesdi I, Sutton BJ, Santis G. Mutations in the DG loop of adenovirus type 5 fiber knob protein abolish high-affinity binding to its cellular receptor CAR. J Virol. 1999;73:9508–14. doi: 10.1128/jvi.73.11.9508-9514.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirby I, Davison E, Beavil AJ, Soh CP, Wickham TJ, Roelvink PW, Kovesdi I, Sutton BJ, Santis G. Identification of contact residues and definition of the CAR-binding site of adenovirus type 5 fiber protein. J Virol. 2000;74:2804–13. doi: 10.1128/jvi.74.6.2804-2813.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jakubczak JL, Rollence ML, Stewart DA, Jafari JD, Von Seggern DJ, Nemerow GR, Stevenson SC, Hallenbeck PL. Adenovirus type 5 viral particles pseudotyped with mutagenized fiber proteins show diminished infectivity of coxsackie B-adenovirus receptor- bearing cells. J Virol. 2001;75:2972–81. doi: 10.1128/JVI.75.6.2972-2981.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Einfeld DA, Schroeder R, Roelvink PW, Lizonova A, King CR, Kovesdi I, Wickham TJ. Reducing the Native Tropism of Adenovirus Vectors Requires Removal of both CAR and Integrin Interactions. J Virol. 2001;75:11284–91. doi: 10.1128/JVI.75.23.11284-11291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akiyama M, Thorne S, Kirn D, Roelvink PW, Einfeld DA, King CR, Wickham TJ. Ablating CAR and integrin binding in adenovirus vectors reduces nontarget organ transduction and permits sustained bloodstream persistence following intraperitoneal administration. Mol Ther. 2004;9:218–30. doi: 10.1016/j.ymthe.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Martin K, Brie A, Saulnier P, Perricaudet M, Yeh P, Vigne E. Simultaneous CAR- and alpha V integrin-binding ablation fails to reduce Ad5 liver tropism. Mol Ther. 2003;8:485–94. doi: 10.1016/s1525-0016(03)00182-5. [DOI] [PubMed] [Google Scholar]
- 25.Alemany R, Curiel DT. CAR-binding ablation does not change biodistribution and toxicity of adenoviral vectors. Gene Ther. 2001;8:1347–53. doi: 10.1038/sj.gt.3301515. [DOI] [PubMed] [Google Scholar]
- 26.Smith T, Idamakanti N, Kylefjord H, Rollence M, King L, Kaloss M, Kaleko M, Stevenson SC. In vivo hepatic adenoviral gene delivery occurs independently of the coxsackievirus-adenovirus receptor. Mol Ther. 2002;5:770–9. doi: 10.1006/mthe.2002.0613. [DOI] [PubMed] [Google Scholar]
- 27.Mizuguchi H, Koizumi N, Hosono T, Ishii-Watabe A, Uchida E, Utoguchi N, Watanabe Y, Hayakawa T. CAR- or alphav integrin-binding ablated adenovirus vectors, but not fiber-modified vectors containing RGD peptide, do not change the systemic gene transfer properties in mice. Gene Ther. 2002;9:769–76. doi: 10.1038/sj.gt.3301701. [DOI] [PubMed] [Google Scholar]
- 28.Leissner P, Legrand V, Schlesinger Y, Hadji DA, van Raaij M, Cusack S, Pavirani A, Mehtali M. Influence of adenoviral fiber mutations on viral encapsidation, infectivity and in vivo tropism. Gene Ther. 2001;8:49–57. doi: 10.1038/sj.gt.3301343. [DOI] [PubMed] [Google Scholar]
- 29.Nicklin SA, Wu E, Nemerow GR, Baker AH. The influence of adenovirus fiber structure and function on vector development for gene therapy. Mol Ther. 2005;12:384–93. doi: 10.1016/j.ymthe.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Smith TA, Idamakanti N, Rollence ML, Marshall-Neff J, Kim J, Mulgrew K, Nemerow GR, Kaleko M, Stevenson SC. Adenovirus serotype 5 fiber shaft influences in vivo gene transfer in mice. Hum Gene Ther. 2003;14:777–87. doi: 10.1089/104303403765255165. [DOI] [PubMed] [Google Scholar]
- 31.Bayo-Puxan N, Cascallo M, Gros A, Huch M, Fillat C, Alemany R. Role of the putative heparan sulfate glycosaminoglycan-binding site of the adenovirus type 5 fiber shaft on liver detargeting and knob-mediated retargeting. J Gen Virol. 2006;87:2487–95. doi: 10.1099/vir.0.81889-0. [DOI] [PubMed] [Google Scholar]
- 32.Shayakhmetov DM, Li ZY, Ni S, Lieber A. Analysis of adenovirus sequestration in the liver, transduction of hepatic cells, and innate toxicity after injection of fiber-modified vectors. J Virol. 2004;78:5368–81. doi: 10.1128/JVI.78.10.5368-5381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ni S, Bernt K, Gaggar A, Li ZY, Kiem HP, Lieber A. Evaluation of biodistribution and safety of adenovirus vectors containing group B fibers after intravenous injection into baboons. Hum Gene Ther. 2005;16:664–77. doi: 10.1089/hum.2005.16.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koizumi N, Mizuguchi H, Sakurai F, Yamaguchi T, Watanabe Y, Hayakawa T. Reduction of natural adenovirus tropism to mouse liver by fiber-shaft exchange in combination with both CAR- and alphav integrin-binding ablation. J Virol. 2003;77:13062–72. doi: 10.1128/JVI.77.24.13062-13072.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamura T, Sato K, Hamada H. Reduction of natural adenovirus tropism to the liver by both ablation of fiber-coxsackievirus and adenovirus receptor interaction and use of replaceable short fiber. J Virol. 2003;77:2512–21. doi: 10.1128/JVI.77.4.2512-2521.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicol CG, Graham D, Miller WH, White SJ, Smith TA, Nicklin SA, Stevenson SC, Baker AH. Effect of adenovirus serotype 5 fiber and penton modifications on in vivo tropism in rats. Mol Ther. 2004;10:344–54. doi: 10.1016/j.ymthe.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 37.Schoggins JW, Gall JG, Falck-Pedersen E. Subgroup B and f fiber chimeras eliminate normal adenovirus type 5 vector transduction in vitro and in vivo. J Virol. 2003;77:1039–48. doi: 10.1128/JVI.77.2.1039-1048.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shayakhmetov DM, Gaggar A, Ni S, Li ZY, Lieber A. Adenovirus binding to blood factors results in liver cell infection and hepatotoxicity. J Virol. 2005;79:7478–91. doi: 10.1128/JVI.79.12.7478-7491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalyuzhniy O, Di Paolo NC, Silvestry M, Hofherr SE, Barry MA, Stewart PL, Shayakhmetov DM. Adenovirus serotype 5 hexon is critical for virus infection of hepatocytes in vivo. Proc Natl Acad Sci U S A. 2008;105:5483–8. doi: 10.1073/pnas.0711757105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waddington SN, McVey JH, Bhella D, Parker AL, Barker K, Atoda H, Pink R, Buckley SM, Greig JA, Denby L, Custers J, Morita T, Francischetti IM, Monteiro RQ, Barouch DH, van Rooijen N, Napoli C, Havenga MJ, Nicklin SA, Baker AH. Adenovirus serotype 5 hexon mediates liver gene transfer. Cell. 2008;132:397–409. doi: 10.1016/j.cell.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 41.Waehler R, Russell SJ, Curiel DT. Engineering targeted viral vectors for gene therapy. Nat Rev Genet. 2007;8:573–87. doi: 10.1038/nrg2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Douglas JT, Rogers BE, Rosenfeld ME, Michael SI, Feng M, Curiel DT. Targeted gene delivery by tropism-modified adenoviral vectors. Nat Biotechnol. 1996;14:1574–8. doi: 10.1038/nbt1196-1574. [DOI] [PubMed] [Google Scholar]
- 43.Miller CR, Buchsbaum DJ, Reynolds PN, Douglas JT, Gillespie GY, Mayo MS, Raben D, Curiel DT. Differential susceptibility of primary and established human glioma cells to adenovirus infection: targeting via the epidermal growth factor receptor achieves fiber receptor-independent gene transfer. Cancer Res. 1998;58:5738–48. [PubMed] [Google Scholar]
- 44.Reynolds PN, Zinn KR, Gavrilyuk VD, Balyasnikova IV, Rogers BE, Buchsbaum DJ, Wang MH, Miletich DJ, Grizzle WE, Douglas JT, Danilov SM, Curiel DT. A targetable, injectable adenoviral vector for selective gene delivery to pulmonary endothelium in vivo. Mol Ther. 2000;2:562–78. doi: 10.1006/mthe.2000.0205. [DOI] [PubMed] [Google Scholar]
- 45.Haisma HJ, Grill J, Curiel DT, Hoogeland S, Van Beusechem VW, Pinedo HM, Gerritsen WR. Targeting of adenoviral vectors through a bispecific single-chain antibody. Cancer Gene Therapy. 2000;7:901–4. doi: 10.1038/sj.cgt.7700198. [DOI] [PubMed] [Google Scholar]
- 46.Kashentseva EA, Seki T, Curiel DT, Dmitriev IP. Adenovirus targeting to c-erbB-2 oncoprotein by single-chain antibody fused to trimeric form of adenovirus receptor ectodomain. Cancer Res. 2002;62:609–16. [PubMed] [Google Scholar]
- 47.Goldman CK, Rogers BE, Douglas JT, Sosnowski BA, Ying W, Siegal GP, Baird A, Campain JA, Curiel DT. Targeted gene delivery to Kaposi's sarcoma cells via the fibroblast growth factor receptor. Cancer Res. 1997;57:1447–51. [PubMed] [Google Scholar]
- 48.Watkins SJ, Mesyanzhinov VV, Kurochkina LP, Hawkins RE. The ‘adenobody’ approach to viral targeting: specific and enhanced adenoviral gene delivery. Gene Ther. 1997;4:1004–12. doi: 10.1038/sj.gt.3300511. [DOI] [PubMed] [Google Scholar]
- 49.Dmitriev I, Kashentseva E, Rogers BE, Krasnykh V, Curiel DT. Ectodomain of coxsackievirus and adenovirus receptor genetically fused to epidermal growth factor mediates adenovirus targeting to epidermal growth factor receptor-positive cells. J Virol. 2000;74:6875–84. doi: 10.1128/jvi.74.15.6875-6884.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reynolds PN, Nicklin SA, Kaliberova L, Boatman BG, Grizzle WE, Balyasnikova IV, Baker AH, Danilov SM, Curiel DT. Combined transductional and transcriptional targeting improves the specificity of transgene expression in vivo. Nat Biotechnol. 2001;19:838–42. doi: 10.1038/nbt0901-838. [DOI] [PubMed] [Google Scholar]
- 51.Everts M, Kim-Park SA, Preuss MA, Passineau MJ, Glasgow JN, Pereboev AV, Mahasreshti PJ, Grizzle WE, Reynolds PN, Curiel DT. Selective induction of tumor-associated antigens in murine pulmonary vasculature using double-targeted adenoviral vectors. Gene Ther. 2005;12:1042–8. doi: 10.1038/sj.gt.3302491. [DOI] [PubMed] [Google Scholar]
- 52.Liang Q, Dmitriev I, Kashentseva E, Curiel DT, Herschman HR. Noninvasive of adenovirus tumor retargeting in living subjects by a soluble adenovirus receptor-epidermal growth factor (sCAR-EGF) fusion protein. Mol Imaging Biol. 2004;6:385–94. doi: 10.1016/j.mibio.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 53.Li HJ, Everts M, Pereboeva L, Komarova S, Idan A, Curiel DT, Herschman HR. Adenovirus Tumor Targeting and Hepatic Untargeting by a Coxsackie/Adenovirus Receptor Ectodomain Anti-Carcinoembryonic Antigen Bispecific Adapter. Cancer Res. 2007;67:5354–5361. doi: 10.1158/0008-5472.CAN-06-4679. [DOI] [PubMed] [Google Scholar]
- 54.Krasnykh VN, Douglas JT, van Beusechem VW. Genetic targeting of adenoviral vectors. Mol Ther. 2000;1:391–405. doi: 10.1006/mthe.2000.0062. [DOI] [PubMed] [Google Scholar]
- 55.Wickham TJ. Targeting adenovirus. Gene Ther. 2000;7:110–4. doi: 10.1038/sj.gt.3301115. [DOI] [PubMed] [Google Scholar]
- 56.Nicklin SA, Von Seggern DJ, Work LM, Pek DC, Dominiczak AF, Nemerow GR, Baker AH. Ablating adenovirus type 5 fiber-CAR binding and HI loop insertion of the SIGYPLP peptide generate an endothelial cell-selective adenovirus. Mol Ther. 2001;4:534–42. doi: 10.1006/mthe.2001.0489. [DOI] [PubMed] [Google Scholar]
- 57.Magnusson MK, Hong SS, Henning P, Boulanger P, Lindholm L. Genetic retargeting of adenovirus vectors: functionality of targeting ligands and their influence on virus viability. J Gene Med. 2002;4:356–70. doi: 10.1002/jgm.285. [DOI] [PubMed] [Google Scholar]
- 58.Dmitriev IP, Kashentseva EA, Curiel DT. Engineering of adenovirus vectors containing heterologous peptide sequences in the C terminus of capsid protein IX. J Virol. 2002;76:6893–9. doi: 10.1128/JVI.76.14.6893-6899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vellinga J, Rabelink MJ, Cramer SJ, van den Wollenberg DJ, Van der Meulen H, Leppard KN, Fallaux FJ, Hoeben RC. Spacers increase the accessibility of peptide ligands linked to the carboxyl terminus of adenovirus minor capsid protein IX. J Virol. 2004;78:3470–9. doi: 10.1128/JVI.78.7.3470-3479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Le LP, Everts M, Dmitriev IP, Davydova JG, Yamamoto M, Curiel DT. Fluorescently labeled adenovirus with pIX-EGFP for vector detection. Mol Imaging. 2004;3:105–16. doi: 10.1162/15353500200404100. [DOI] [PubMed] [Google Scholar]
- 61.Le LP, Le HN, Dmitriev IP, Davydova JG, Gavrikova T, Yamamoto S, Curiel DT, Yamamoto M. Dynamic monitoring of oncolytic adenovirus in vivo by genetic capsid labeling. J Natl Cancer Inst. 2006;98:203–14. doi: 10.1093/jnci/djj022. [DOI] [PubMed] [Google Scholar]
- 62.Meulenbroek RA, Sargent KL, Lunde J, Jasmin BJ, Parks RJ. Use of adenovirus protein IX (pIX) to display large polypeptides on the virion--generation of fluorescent virus through the incorporation of pIX-GFP. Mol Ther. 2004;9:617–24. doi: 10.1016/j.ymthe.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 63.Campos SK, Parrott MB, Barry MA. Avidin-based targeting and purification of a protein IX-modified, metabolically biotinylated adenoviral vector. Mol Ther. 2004;9:942–54. doi: 10.1016/j.ymthe.2004.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li J, Le L, Sibley DA, Mathis JM, Curiel DT. Genetic incorporation of HSV-1 thymidine kinase into the adenovirus protein IX for functional display on the virion. Virology. 2005;338:247–58. doi: 10.1016/j.virol.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 65.Matthews QL, Sibley DA, Wu H, Li J, Stoff-Khalili MA, Waehler R, Mathis JM, Curiel DT. Genetic incorporation of a herpes simplex virus type 1 thymidine kinase and firefly luciferase fusion into the adenovirus protein IX for functional display on the virion. Mol Imaging. 2006;5:510–9. [PMC free article] [PubMed] [Google Scholar]
- 66.Vellinga J, de Vrij J, Myhre S, Uil T, Martineau P, Lindholm L, Hoeben RC. Efficient incorporation of a functional hyper-stable single-chain antibody fragment protein-IX fusion in the adenovirus capsid. Gene Ther. 2007;14:664–70. doi: 10.1038/sj.gt.3302908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Vrij J, Uil TG, van den Hengel SK, Cramer SJ, Koppers-Lalic D, Verweij MC, Wiertz EJ, Vellinga J, Willemsen RA, Hoeben RC. Adenovirus targeting to HLA-A1/MAGE-A1-positive tumor cells by fusing a single-chain T-cell receptor with minor capsid protein IX. Gene Ther. 2008 doi: 10.1038/gt.2008.26. [DOI] [PubMed] [Google Scholar]
- 68.Schier R, McCall A, Adams GP, Marshall KW, Merritt H, Yim M, Crawford RS, Weiner LM, Marks C, Marks JD. Isolation of picomolar affinity anti-c-erbB-2 single-chain Fv by molecular evolution of the complementarity determining regions in the center of the antibody binding site. J Mol Biol. 1996;263:551–67. doi: 10.1006/jmbi.1996.0598. [DOI] [PubMed] [Google Scholar]
- 69.Schoggins JW, Nociari M, Philpott N, Falck-Pedersen E. Influence of fiber detargeting on adenovirus-mediated innate and adaptive immune activation. J Virol. 2005;79:11627–37. doi: 10.1128/JVI.79.18.11627-11637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Douglas JT, Miller CR, Kim M, Dmitriev I, Mikheeva G, Krasnykh V, Curiel DT. A system for the propagation of adenoviral vectors with genetically modified receptor specificities. Nat Biotechnol. 1999;17:470–5. doi: 10.1038/8647. [DOI] [PubMed] [Google Scholar]
- 71.Michael SI, Hong JS, Curiel DT, Engler JA. Addition of a short peptide ligand to the adenovirus fiber protein. Gene Ther. 1995;2:660–8. [PubMed] [Google Scholar]
- 72.Siciliano MJ, Barker PE, Cailleau R. Mutually exclusive genetic signatures of human breast tumor cell lines with a common chromosomal marker. Cancer Res. 1979;39:919–22. [PubMed] [Google Scholar]
- 73.Yu D, Liu B, Jing T, Sun D, Price JE, Singletary SE, Ibrahim N, Hortobagyi GN, Hung MC. Overexpression of both p185c-erbB2 and p170mdr-1 renders breast cancer cells highly resistant to taxol. Oncogene. 1998;16:2087–94. doi: 10.1038/sj.onc.1201729. [DOI] [PubMed] [Google Scholar]
- 74.Tan M, Yao J, Yu D. Overexpression of the c-erbB-2 gene enhanced intrinsic metastasis potential in human breast cancer cells without increasing their transformation abilities. Cancer Res. 1997;57:1199–205. [PubMed] [Google Scholar]
- 75.Lindner P, Bauer K, Krebber A, Nieba L, Kremmer E, Krebber C, Honegger A, Klinger B, Mocikat R, Pluckthun A. Specific detection of his-tagged proteins with recombinant anti-His tag scFv-phosphatase or scFv-phage fusions. Biotechniques. 1997;22:140–9. doi: 10.2144/97221rr01. [DOI] [PubMed] [Google Scholar]
- 76.Kipriyanov SM, Moldenhauer G, Schuhmacher J, Cochlovius B, Von der Lieth CW, Matys ER, Little M. Bispecific tandem diabody for tumor therapy with improved antigen binding and pharmacokinetics. J Mol Biol. 1999;293:41–56. doi: 10.1006/jmbi.1999.3156. [DOI] [PubMed] [Google Scholar]
- 77.Kontermann RE, Muller R. Intracellular and cell surface displayed single-chain diabodies. J Immunol Methods. 1999;226:179–88. doi: 10.1016/s0022-1759(99)00062-9. [DOI] [PubMed] [Google Scholar]
- 78.Voss S, Skerra A. Mutagenesis of a flexible loop in streptavidin leads to higher affinity for the Strep-tag II peptide and improved performance in recombinant protein purification. Protein Eng. 1997;10:975–82. doi: 10.1093/protein/10.8.975. [DOI] [PubMed] [Google Scholar]
- 79.Friguet B, Chaffotte AF, Djavadi-Ohaniance L, Goldberg ME. Measurements of the true affinity constant in solution of antigen-antibody complexes by enzyme-linked immunosorbent assay. J Immunol Methods. 1985;77:305–19. doi: 10.1016/0022-1759(85)90044-4. [DOI] [PubMed] [Google Scholar]
- 80.Wikman M, Steffen AC, Gunneriusson E, Tolmachev V, Adams GP, Carlsson J, Stahl S. Selection and characterization of HER2/neu-binding affibody ligands. Protein Eng Des Sel. 2004;17:455–62. doi: 10.1093/protein/gzh053. [DOI] [PubMed] [Google Scholar]
- 81.Magnusson MK, Henning P, Myhre S, Wikman M, Uil TG, Friedman M, Andersson KM, Hong SS, Hoeben RC, Habib NA, Stahl S, Boulanger P, Lindholm L. Adenovirus 5 vector genetically re-targeted by an Affibody molecule with specificity for tumor antigen HER2/neu. Cancer Gene Ther. 2007;14:468–79. doi: 10.1038/sj.cgt.7701027. [DOI] [PubMed] [Google Scholar]
- 82.Belousova N, Mikheeva G, Gelovani J, Krasnykh V. Modification of adenovirus capsid with a designed protein ligand yields a gene vector targeted to a major molecular marker of cancer. J Virol. 2008;82:630–7. doi: 10.1128/JVI.01896-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sebestyen Z, de Vrij J, Magnusson M, Debets R, Willemsen R. An oncolytic adenovirus redirected with a tumor-specific T-cell receptor. Cancer Res. 2007;67:11309–16. doi: 10.1158/0008-5472.CAN-07-0739. [DOI] [PubMed] [Google Scholar]
- 84.Parks RJ. Adenovirus protein IX: a new look at an old protein. Mol Ther. 2005;11:19–25. doi: 10.1016/j.ymthe.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 85.Hedley SJ, Chen J, Mountz JD, Li J, Curiel DT, Korokhov N, Kovesdi I. Targeted and shielded adenovectors for cancer therapy. Cancer Immunol Immunother. 2006;55:1412–9. doi: 10.1007/s00262-006-0158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marsh MP, Campos SK, Baker ML, Chen CY, Chiu W, Barry MA. Cryoelectron microscopy of protein IX-modified adenoviruses suggests a new position for the C terminus of protein IX. J Virol. 2006;80:11881–6. doi: 10.1128/JVI.01471-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vigne E, Mahfouz I, Dedieu JF, Brie A, Perricaudet M, Yeh P. RGD inclusion in the hexon monomer provides adenovirus type 5-based vectors with a fiber knob-independent pathway for infection. J Virol. 1999;73:5156–61. doi: 10.1128/jvi.73.6.5156-5161.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu H, Han T, Belousova N, Krasnykh V, Kashentseva E, Dmitriev I, Kataram M, Mahasreshti PJ, Curiel DT. Identification of sites in adenovirus hexon for foreign peptide incorporation. J Virol. 2005;79:3382–90. doi: 10.1128/JVI.79.6.3382-3390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Campos SK, Barry MA. Comparison of adenovirus fiber, protein IX, and hexon capsomeres as scaffolds for vector purification and cell targeting. Virology. 2006;349:453–62. doi: 10.1016/j.virol.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 90.Corjon S, Wortmann A, Engler T, van Rooijen N, Kochanek S, Kreppel F. Targeting of adenovirus vectors to the LRP receptor family with the high-affinity ligand RAP via combined genetic and chemical modification of the pIX capsomere. Mol Ther. 2008;16:1813–24. doi: 10.1038/mt.2008.174. [DOI] [PubMed] [Google Scholar]
- 91.Uil TG, Seki T, Dmitriev I, Kashentseva E, Douglas JT, Rots MG, Middeldorp JM, Curiel DT. Generation of an adenoviral vector containing an addition of a heterologous ligand to the serotype 3 fiber knob. Cancer Gene Ther. 2003;10:121–4. doi: 10.1038/sj.cgt.7700543. [DOI] [PubMed] [Google Scholar]
- 92.Hesse A, Kosmides D, Kontermann RE, Nettelbeck DM. Tropism modification of adenovirus vectors by peptide ligand insertion into various positions of the adenovirus serotype 41 short-fiber knob domain. J Virol. 2007;81:2688–99. doi: 10.1128/JVI.02722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nettelbeck DM, Rivera AA, Kupsch J, Dieckmann D, Douglas JT, Kontermann RE, Alemany R, Curiel DT. Retargeting of adenoviral infection to melanoma: combining genetic ablation of native tropism with a recombinant bispecific single-chain diabody (scDb) adapter that binds to fiber knob and HMWMAA. Int J Cancer. 2004;108:136–45. doi: 10.1002/ijc.11563. [DOI] [PubMed] [Google Scholar]
- 94.Alemany R, Suzuki K, Curiel DT. Blood clearance rates of adenovirus type 5 in mice. J Gen Virol. 2000;81:2605–9. doi: 10.1099/0022-1317-81-11-2605. [DOI] [PubMed] [Google Scholar]
- 95.Parker AL, Waddington SN, Nicol CG, Shayakhmetov DM, Buckley SM, Denby L, Kemball-Cook G, Ni S, Lieber A, McVey JH, Nicklin SA, Baker AH. Multiple vitamin K-dependent coagulation zymogens promote adenovirus-mediated gene delivery to hepatocytes. Blood. 2006;108:2554–61. doi: 10.1182/blood-2006-04-008532. [DOI] [PubMed] [Google Scholar]
- 96.Roberts DM, Nanda A, Havenga MJ, Abbink P, Lynch DM, Ewald BA, Liu J, Thorner AR, Swanson PE, Gorgone DA, Lifton MA, Lemckert AA, Holterman L, Chen B, Dilraj A, Carville A, Mansfield KG, Goudsmit J, Barouch DH. Hexon-chimaeric adenovirus serotype 5 vectors circumvent preexisting anti-vector immunity. Nature. 2006;441:239–43. doi: 10.1038/nature04721. [DOI] [PubMed] [Google Scholar]
- 97.Krasnykh V, Dmitriev I, Mikheeva G, Miller CR, Belousova N, Curiel DT. Characterization of an adenovirus vector containing a heterologous peptide epitope in the HI loop of the fiber knob. J Virol. 1998;72:1844–52. doi: 10.1128/jvi.72.3.1844-1852.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chartier C, Degryse E, Gantzer M, Dieterle A, Pavirani A, Mehtali M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J Virol. 1996;70:4805–10. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95:2509–14. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Maizel JV, Jr, White DO, Scharff MD. The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology. 1968;36:115–25. doi: 10.1016/0042-6822(68)90121-9. [DOI] [PubMed] [Google Scholar]
- 101.Mittereder N, March KL, Trapnell BC. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J Virol. 1996;70:7498–509. doi: 10.1128/jvi.70.11.7498-7509.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zinn KR, Douglas JT, Smyth CA, Liu HG, Wu Q, Krasnykh VN, Mountz JD, Curiel DT, Mountz JM. Imaging and tissue biodistribution of 99mTc-labeled adenovirus knob (serotype 5) Gene Ther. 1998;5:798–808. doi: 10.1038/sj.gt.3300659. [DOI] [PubMed] [Google Scholar]