Abstract
The ubiquitin-proteasome pathway and autophagy are the two major mechanisms responsible for the clearance of cellular proteins. We have used the yeast Saccharomyces cerevisiae as a model system and the cystic fibrosis transmembrane conductance regulator (CFTR) as a model substrate to study the interactive function of these two pathways in the degradation of misfolded proteins. EGFP-tagged human CFTR was introduced into yeast and expressed under a copper-inducible promoter. The localization and degradation of EGFP-CFTR in live cells were monitored by time-lapse imaging following its de novo synthesis. EGFP-CFTR first appears within the perinuclear and sub-cortical ER and is mobile within the plane of the membrane as assessed by fluorescence recovery after photobleaching (FRAP). This pool of EGFP-CFTR is subsequently degraded through a proteasome-dependent pathway that is inhibited in the pre1-1 yeast strain defective in proteasomal degradation. Prolonged expression of EGFP-CFTR leads to the sequestration of EGFP-CFTR molecules into ER structures called ER-associated complexes (ERACs). The sequestration of EGFP-CFTR into ERACs appears to be driven by aggregation since EGFP-CFTR molecules present within ERACs are immobile as measured by FRAP. Individual ERACs are cleared from cells through the autophagic pathway that is blocked in the atg6Δ and atg1Δ yeast strains defective in autophagy. Our results suggest that the proteasomal and the autophagic pathways function together to clear misfolded proteins from the ER.
Keywords: Proteasome, Autophagy, ER-associated degradation, ER-associated complexes, Yeast, Live-cell imaging, FRAP, CFTR
Introduction
The ubiquitin-proteasomal pathway (UPP) and the autophagic pathway are the two major mechanisms responsible for the clearance of aberrant proteins and organelles (Rubinsztein et al., 1996). The UPP degrades short-lived proteins, in which the misfolded molecules are linked to a ladder of ubiquitin and targeted to proteasomes for degradation. The proteasomes contain multiple protein subunits that form a barrel-shaped cylinder. The ubiquitinated protein substrates are inserted into the narrow pore of the cylinder and chopped into peptides, which are further degraded to amino acids by cytosolic and nuclear peptidases (Ciechanover, 2006). In contrast, the autophagic pathway mediates the delivery of protein aggregates and organelles for degradation within the lysosomes/vacuolar compartments. The molecular players in the autophagic pathway have been identified primarily by genetic screens of the yeast S. cerevisiae (Harding et al., 1996; Thumm et al., 1994; Tsukada and Ohsumi, 1993). Genes encoding autophagy components are named ATG, for “autophagy-related” (Klionsky et al., 2003). They function in the four discrete steps of autophagy: 1) selection and packaging of cargos, 2) formation of autophagic vesicles called autophagosomes, 3) targeting and fusion of autophagosomes with vacuoles/lysosomes, and 4) protein hydrolysis inside the vacuoles/lysosomes (Wang and Klionsky, 2003). Despite the mechanistic differences between UPP and autophagic degradative pathways, crosstalk between proteasomal and autophagic degradation has been reported (Kruse et al., 2006; Pandey et al., 2007). For example, both pathways have been shown to be involved in the degradation of the Z variant of human alpha-1 proteinase inhibitor (A1PiZ) (Kruse et al., 2006) and polyglutamine-containing proteins (Rubinsztein, 2006). It has been proposed that autophagy plays a compensatory role when proteasomal function is compromised (Pandey et al., 2007). The details of how proteasomal and autophagic pathways intersect on the molecular and cellular level remain to be elucidated.
CFTR is an excellent model substrate to study cellular degradative mechanisms. CFTR is a cAMP-dependent chloride channel present on the apical surface of epithelial cells lining the respiratory, intestinal and exocrine tissues (Bertrand and Frizzell, 2003). A single amino acid deletion termed ΔF508-CFTR (Kerem et al., 1989) causes the protein to be misfolded and prematurely degraded through the ER-associated proteasomal degradation (ERAD). ERAD is known to degrade misfolded CFTR, and the proteasomal degradation of CFTR has been extensively studied (Amaral, 2005; Kopito, 1999; Younger et al., 2006). Newly synthesized CFTR molecules are first monitored for folding status by molecular chaperones such as calnexin (Okiyoneda et al., 2004), BiP (Sullivan et al., 2003), Hsp70 (Choo-Kang and Zeitlin, 2001; Zhang et al., 2001), Hsp40 and Hsp90 (Youker et al., 2004). The chaperone-mediated selection is coupled with ubiquitination of misfolded CFTR, which is sequentially regulated by the E3 RMA1/E2 Ubc6e/Derlin-1 complex and the E3 CHIP/E2 Ubc5/Hdj2 complex (Meacham et al., 2001; Younger et al., 2004, 2005). Once selected for degradation, misfolded CFTR is extracted from the ER membrane by the Cdc48p AAA-ATPase and degraded by the 26S proteasome in the cytoplasm (Romisch, 2005). Importantly, additional non-UPP mechanisms have been proposed to participate in CFTR degradation, but these have not been carefully examined (Jensen et al., 1995; Lukacs et al., 1994; Ward et al., 1995).
To understand the pathways that participate in the degradation of cellular substrates, we used novel real-time imaging approaches to track the clearance of newly synthesized EGFP-CFTR. We uncovered that EGFP-CFTR is targeted to distinct ER locations and is degraded differentially through both proteasomal and autophagic pathways. First, newly synthesized EGFP-CFTR localizes within the ER, is freely mobile within that compartment and is degraded through the proteasomal pathway. Second, EGFP-CFTR can accumulate in sub-ER compartments called ER-associated complexes (ERACs). ERACs co-localize with pre-autophagosomal marker and are adjacent to vacuolar degradative compartments. EGFP-CFTR molecules localized to ERACs are largely immobile and are degraded through an autophagy-dependent pathway. Our findings support a model in which the proteasomal and autophagic pathways function in tandem to clear misfolded proteins.
Materials and methods
Antibodies and plasmid
The polyclonal rabbit anti-Kar2p polyclonal antibody was a gift from Dr. Jeffrey Brodsky (University of Pittsburgh). The monoclonal antibody against yeast Pdi1p (38H8) was purchased from EnCor Biotechnology Inc. and used at 1:500 dilution. FM4-64, Goat anti-rabbit and goat anti-mouse Alexa Fluor 594 were purchased from Molecular Probes (Invitrogen). The expression plasmid CFP-Ape1(414) was received from Daniel J. Klionsky (University of Michigan). The expression plasmid EGFP-CFTR with a copper-inducible promoter was described previously (Fu and Sztul, 2003).
Yeast strains
Yeast strain atg6Δ, atg1Δ and its wild-type counterpart BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) were obtained from Open Biosystems. YHI29/W (MATa, ura3, leu2-3, 112, his3-11, 15, GAL1) and YHI29/1 (MATa, pre1-1, ura3, leu2-3, 112, his3-11, 15, cans, GAL1) were a gift from Dr. D. Wolf (Universität Stuttgart).
Yeast media were prepared as described (Rose et al., 1990). YP and synthetic media (SM) were supplemented with 2% glucose (YPD or SMD). In all experiment unless specified, cultures were grown for a minimum of 5-6 generations to an A600 of no more than 1.0.
Microscopy
The subcellular localization of EGFP-CFTR was examined by light and electron microscopy as described previously (Fu and Sztul, 2003). Briefly, yeast containing the EGFP-CFTR construct was grown in SMD medium lacking uracil to an OD600 of no more than 1.0. The yeast was then induced to express EGFP-CFTR in the presence of 100 μM CuSO4 for 2 h and fixed in 3% formaldehyde for 30 min. Cells were harvested and washed twice with 0.1 M potassium phosphate, pH 6.5, and once with potassium phosphate buffer containing 1.2 M sorbitol. The cell walls were digested in 100 mM potassium phosphate buffer, pH 6.5, containing 1.2 M sorbitol, 0.1% β-mercaptoethanol and 20 μg/ml zymolase 20T (U.S. Biological). Cells were permeabilized with 0.1% Triton X-100 for 10 min and washed twice for 5 min each with blocking buffer (phosphate-buffered saline (PBS) containing 0.2% Tween 20 and 1 mg/ml bovine serum albumin). Cells were stained with primary and secondary antibodies followed by fluorescence microscopy imaging. The vacuolar membrane was stained with FM 4-64 as described previously (Vida and Emr, 1995). Images were obtained and processed using IpLab Spectrum software (Signal Analytics).
The relationship of ERACs containing EGFP-CFTR and pre-autophagosomal structures was analyzed by confocal microscopy. Wild-type yeast containing both the EGFP-CFTR and the CFP-APE1 plasmids were induced to express EGFP-CFTR in the presence of 100 μM CuSO4 for 2 h and fixed in 3% formaldehyde for 30 min. Imaging was performed on a Leica DMXRE epifluorescence/Nomarski microscope equipped with Leica TCS NT SP2 laser confocal optics (Leica Inc., Exton, PA). Precise control of fluorochrome excitation and emission, respectively, was afforded by an acousto-optical tunable filter and a TCS SP2 prism spectrophotometer. For spectral separation of CFP from EGFP during image acquisition, we used the 457-nm excitation line of the Argon laser, the TK 465-nm dichroic mirror, and a tight emission band pass between 465 nm – 485 nm. For spectral separation of EGFP from CFP, we used the 488-nm excitation line of the Argon laser, the RSP 500-nm dichroic mirror, and a tight emission band pass of 510 nm - 535 nm. To enhance the visualization, the cyan of CFP images was digitally converted to red. Electron microscopy of yeast cells expressing EGFP-CFTR was performed as described previously (Fu and Sztul, 2003).
Time-lapse imaging and FRAP analyses
Yeast cells containing the EGFP-CFTR plasmid were grown in SMD growth medium lacking uracil to log-phase. Two A600 units of cells were quickly harvested and washed once with growth medium containing 100 μM CuSO4. The cells were then resuspended in 10 μl copper-containing medium and placed onto a glass-bottom dish (Warner Instruments). The cells were then covered with a 1 mm thick agar pad made from the copper-containing medium to retain the cells stabilized in position and moisturized with growth medium. Green fluorescence images were acquired every 2 min for up to 3 h with an Olympus IX70 inverted microscope equipped with a 100×/1.35 NA objective lens and a cooled charge-coupled device camera. IpLab Spectrum software was used to control image acquisition and manipulation. The time period for the first image to be taken from the addition of copper induction was about 20 min. The exposure time for each image was limited to 2 s to limit photobleaching and possible photodamage to the cells.
To monitor the degradation of EGFP-CFTR in ERACs, yeast cells containing the EGFP-CFTR expression plasmid were grown to log-phase and incubated with copper-containing medium for 2 h to induce the formation of ERACs. The cells were then harvested, washed and resuspended in medium without copper followed by time-lapse imaging as described above. The lag time from harvesting the cells to taking the first image was typically 20 to 30 min.
FRAP was conducted as described previously (Fu et al., 2005). Yeast cells were incubated with copper-containing medium for 30 min or 2 h to induce the expression of EGFP-CFTR in the ER or ERACs, respectively. Cells were harvested, washed and placed onto a glass-bottom dish as described above. FRAP was performed at room temperature using an argon laser in conjunction with a 100× objective for optimum resolution. The outlined box was photobleached at full laser power, and recovery of fluorescence was monitored by scanning the entire cell at lower laser power (about 30% power) over 15-s intervals. The scanning laser intensity was set to minimal photobleach the specimen over the time course of the experiment. Fluorescence recovery was calculated by comparing the intensity ratio in regions of interest within the bleached area before the bleach and after recovery. Postbleach intensities were normalized upward to correct for total loss of fluorescence due to the photobleach by comparing the fluorescence intensities outside the bleached area using IpLab Spectrum software.
Results
EGFP-CFTR is degraded from distinct ER compartments
We used real-time fluorescent microscopy imaging to study the dynamics of synthesis, accumulation and degradation of EGFP-CFTR. The advantage of this approach is its ability to analyze behaviors of individual cells. Other biochemical approaches such as pulse-chase analyses monitor changes in large pools of cells and are unlikely to uncover cellular behaviors that might be masked by population analysis. GFP from the jellyfish Aequorea victoria has been fused to many cellular proteins to trace their synthesis, localization, movement, and degradation in living cells (Lippincott-Schwartz and Patterson, 2003). For our studies, we generated a construct containing EGFP fused in-frame to the N-terminus of human CFTR in a yeast expression vector. The expression of EGFP-CFTR in our system is controlled by a copper-inducible promoter and confirmed by immunoprecipitation experiments using antibodies against either GFP or CFTR protein (data not shown, (Fu and Sztul, 2003)).
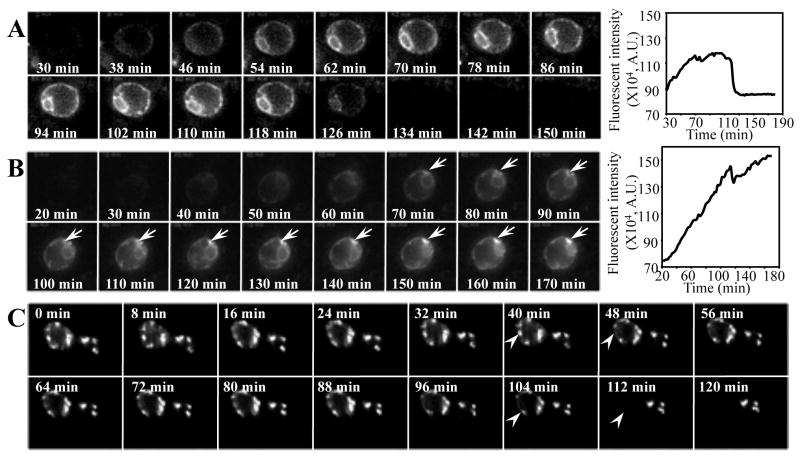
In the first set of experiments, we monitored the de novo expression, localization and degradation of EGFP-CFTR in different yeast cells by time-lapse imaging after inducing EGFP-CFTR synthesis by copper addition. Interestingly, yeast cells show distinctive EGFP-CFTR localization and degradation patterns. The first pattern (Fig. 1A) shows EGFP-CFTR localized to the ER ring (the characteristic ER localization in yeast, see Fig. 2 below) after approximately 30 to 45 min of induction (the first image was taken approximately 20 min after induction). EGFP-CFTR continues to accumulate in the ER during the next 80 to 90 min and then disappears from the ER over the following 10 to 15 min (graph in Fig. 1A). The rapid disappearance of the ER-localized EGFP-CFTR suggests that at times, the degradative efficiency of the ERAD exceeds the synthetic rate of inserting newly synthesized EGFP-CFTR into the ER. The disappearance of the fluorescent signal is not due to the movement of cells out of the focal field (all cells remained in place as detected by bright-field imaging after the time-lapse imaging, data not shown). The second pattern (Fig. 1B) shows that EGFP-CFTR appears within the ER at the early (about 30 min) time points. However, instead of being subsequently cleared, EGFP-CFTR expression increases until the end of imaging, approximately 3 h after induction (graph in Fig. 1B).
Fig. 1.
EGFP-CFTR is degraded from ER or ER-associated foci. (A, B) Wild-type yeast cells containing EGFP-CFTR construct were imaged for EGFP-CFTR immediately after the addition of copper to induce expression. The first image was captured ∼20 min after induction. Subsequent images were taken every 2 min by time-lapse imaging. Movie montages at the indicated time points are shown. The fluorescence intensity over time was quantified using IpLab software and is shown in the graphics (A.U.: arbitrary unit). (C) Wild-type yeast cells were induced with copper for 2 h to form EGFP-CFTR foci. Cells were then harvested by centrifugation and the first time-lapse image was taken ∼20 min afterwards (0 min). Subsequent images were taken every 2 min. EGFP-CFTR is degraded from the ER (A), or forms foci (B, arrows) that are degraded later (C, arrowheads). Movies (e-components 1-3) showing the different behaviors of EGFP-CFTR are presented as Supplementary online material.
Fig. 2.
EGFP-CFTR localized to ER and ER foci has distinct properties. The properties of EGFP-CFTR localized to the ER or the ER-associated foci were analyzed by FRAP. (A) Wild-type yeast was induced for 30 min to express EGFP-CFTR in the ER followed by FRAP in the region marked. (B) Wild-type yeast was induced for 2 h to form EGFP-CFTR foci followed by FRAP. Relative fluorescence recovery is plotted and shown in (C). (D, E) Yeast cells containing EGFP-CFTR plasmid were induced to form EGFP-CFTR foci followed by immunofluorescence using antibodies against yeast Kar2p (D) or Pdi1p (E). The EGFP-CFTR foci colocalize with Kar2p (D, arrows) but not Pdi1p. (F). Nuclear membrane and limited sub-plasmamembrane ER elements are visible in yeast cells not expressing EGFP-CFTR. (G) Extensive accumulations of tubules continuous with the ER are visible in cells expressing EGFP-CFTR.
Noteworthy, EGFP-CFTR starts to form foci after approximately 1 h of induction (Fig. 1B, arrow). The formation of the foci continues for another 2 to 4 h until numerous large foci of EGFP-CFTR are present within one single cell (see below). Quantifications of the fluorescence intensity over time suggest that the different patterns of EGFP-CFTR might be due to the different levels of protein expression in the cells (compare the graphs in Fig. 1A and B). In a typical experiment, about 50 to 100 yeast cells were imaged simultaneously in one focal field. Among them, ∼ 20% of the cells have the first pattern and ∼ 30% to 40% of the cells show the second pattern. It remains to be determined what might cause the discrepancy of EGFP-CFTR expressions in different yeast cells. The expression of EGFP-CFTR could be cell age or cell cycle dependent.
We explored whether EGFP-CFTR accumulated in the ER-associated foci can be degraded. Yeast cells containing the EGFP-CFTR construct were incubated in the presence of copper for 2 h to allow the formation of EGFP-CFTR foci. Copper was then removed from the medium to arrest further synthesis of EGFP-CFTR, and the behavior of EGFP-CFTR foci was examined by real-time imaging. As shown in Figure 1C, numerous foci containing EGFP-CFTR are detected after 2 h of induction. During the subsequent imaging, the EGFP-CFTR foci persist for different time periods and then rapidly (within 5 to 10 min) disappear (Fig. 1C, arrowheads), indicating that EGFP-CFTR can be degraded from the ER foci.
Our results suggest that CFTR is targeted to and degraded from distinct ER sites. CFTR molecules are first synthesized and localized in the ER. After the initial synthesis, CFTR is either degraded from the ER or accumulates as ER-associated foci, from which it is subsequently removed.
EGFP-CFTR molecules localized to the ER and the ER foci have different properties
The real-time imaging experiments revealed that EGFP-CFTR can be targeted to either the ER or ER foci and can be degraded from both compartments. This result suggests that EGFP-CFTR molecules may be degraded from distinct ER domains through different mechanisms. To test this possibility, we analyzed the properties of EGFP-CFTR localized to the ER and to ER foci by FRAP. As shown in Figure 2A, photobleaching of a section of the ER membrane causes about 80% reduction in fluorescence intensity of EGFP-CFTR within the region. Fluorescence intensity starts to recover within a minute and recovers to 80% of that before photobleaching within 4 to 5 min. The half-life (t1/2) for maximum FRAP is about 2 min. This result suggests that EGFP-CFTR molecules localized to the ER are diffusible. In contrast, the FRAP of EGFP-CFTR targeted to the ER-associated foci is minimal. As shown in Figure 2B, photobleaching a single focus reduces the fluorescence intensity of EGFP-CFTR in that structure to about 10% of that before photobleaching. Two minutes after photobleaching, the fluorescence intensity reaches a plateau of approximately 15% of the initial fluorescence (Fig. 2C). Importantly, the fluorescence never recovers to higher levels, indicating that the EGFP-CFTR molecules present within the ER-associated foci exist as protein aggregates.
These real-time imaging experiments revealed that EGFP-CFTR targets to either the ER ring or to ER-associated foci. The relationship between the EGFP-CFTR foci and the ER was characterized further by analyzing the localization of two ER markers, the molecular chaperone Kar2p and the protein disulfide isomerase, Pdi1p. As shown in Figure 2D, the EGFP-CFTR foci often colocalize with Kar2p, in agreement with previous reports (Fu and Sztul, 2003; Zhang et al., 2001). In contrast, the EGFP-CFTR foci show limited colocalization with Pdi1p (Fig. 2E), suggesting that the EGFP-CFTR foci may represent specialized sub-ER domains.
The ultrastructural nature of the foci was examined by electron microscopy. As shown in Figure 2F, a normal nuclear membrane and sub-cortical ER are visible in cells not expressing EGFP-CFTR. In contrast, in cell expressing EGFP-CFTR (Fig. 2G, arrows), we observed accumulation of anastomozing tubules that are similar to ER extensions observed in yeast expressing CFTR (Zhang et al., 2001) or Ste6p* (Huyer et al., 2004). These structures are called ER-associated-complexes (ERACs).
The degradation of EGFP-CFTR from ERACs is proteasome independent
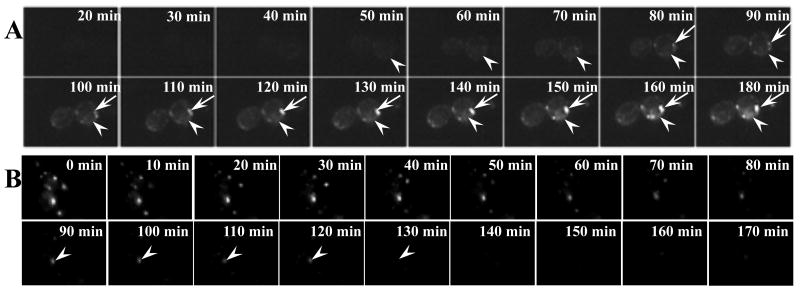
Misfolded CFTR has been documented to be degraded by proteasomes in a variety of model systems (Jensen et al., 1995; Lukacs et al., 1994; Ward et al., 1995). Because we found that EGFP-CFTR is targeted to either the ER or ERACs and can be cleared from both locations, we tested whether the degradations of EGFP-CFTR from the ER and the ERACs are mediated through the proteasome. To test the involvement of the proteasomal pathway, we introduced our copper-inducible EGFP-CFTR construct into the pre1-1 mutant yeast strain that lacks a functional α-subunit of the proteasome (Heinemeyer et al., 1991). The localization and behavior of EGFP-CFTR were monitored by real-time imaging immediately after copper induction. As shown in the cells in Figure 3A, EGFP-CFTR can be detected within the ER after 50 to 60 min of induction (arrowheads). Continuous monitoring for additional 2 h shows EGFP-CFTR is detectable in the ER after 3 h of induction (arrowheads). In addition, EGFP-CFTR starts to form ERACs approximately 80 min after induction and these ERACs enlarge around the ER 3 h after induction (arrow). This suggests that degradation of EGFP-CFTR from the ER requires a functional proteasomal system. When the proteasome function is compromised, EGFP-CFTR accumulates to form ERACs.
Fig. 3.
EGFP-CFTR localized within ERACs is degraded via a proteasome-independent pathway. (A) The proteasome-deficient pre1-1 yeast strain containing the EGFP-CFTR plasmid was imaged following induction of expression. The first image was captured ∼20 min after induction. Subsequent images were taken every 2 min. Movie montages at indicated time points are shown. CFTR in the pre1-1 strain forms ERACs (arrows). (B) The pre1-1 yeast strain was induced with copper for 2 h to form EGFP-CFTR foci followed by time-lapse imaging as described in Figure 1C. The arrowhead in (B) shows the rapid clearance of ERACs in the proteasome-deficient strain.
We also explored the degradation of EGFP-CFTR from ERACs in the pre1-1 strain. As shown in Figure 3B, EGFP-CFTR ERACs accumulated in the pre1-1 yeast strain after a 2-h induction may persist for 2 to 4 h after initial induction and are then cleared over the next 90 min (Fig. 3B, arrowheads). The kinetics of clearance was analogous to those seen in control cells. These results suggest that EGFP-CFTR degradation from the ER is proteasome dependent, while the degradation from ERACs does not require a functional UPP.
The degradation of EGFP-CFTR from ERACs is autophagy dependent
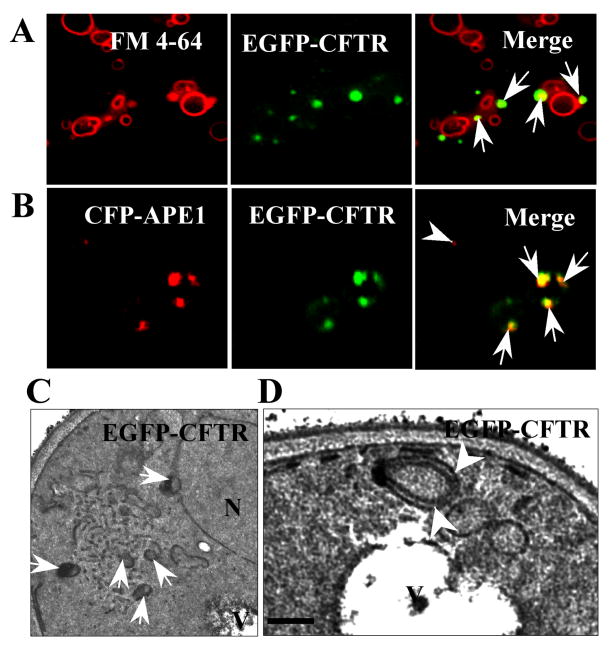
The results above show that only the ER-localized EGFP-CFTR molecules are degraded through the proteasomal pathway. In contrast, EGFP-CFTR molecules accumulated in ERACs are removed through proteasome-independent mechanisms. Besides the proteasomal pathway, autophagy is the other major pathway for the clearance of aberrant proteins (Rubinsztein et al., 1996). During autophagy, misfolded proteins are enclosed as autophagosomes and delivered to the acidic lysosomes/vacuoles for degradation (Mizushima, 2004). We used the red-fluorescent dye FM4-64 that labels acidic membrane compartments to explore the relationship between EGFP-CFTR ERACs and the vacuoles. As shown in Figure 4A, EGFP-CFTR foci are frequently adjacent to the surface of vacuoles (arrows).
Fig. 4.
Relationship of ERACs to autophagic structures. (A) The EGFP-CFTR ERACs (arrows) are located adjacent to vacuolar membranes stained with FM4-64. Wild-type yeast was induced to form EGFP-CFTR ERACs and stained with FM4-64 followed by fluorescence microscopy. (B) The EGFP-CFTR ERACs are enriched with pre-autophagosomal marker, Ape1p. Wild-type yeast cells were co-transformed with EGFP-CFTR and CFP-APE1 constructs. The cells were then induced with copper for 2 h to form EGFP-CFTR ERACs followed by confocal microscopy analysis. The CFP-Ape1p image was digitally converted to red to enhance visualization. (C, D) Ultrastructure of EGFP-CFTR ERACs. Wild-type yeast cells expressing the EGFP-CFTR were analyzed by electron microscopy. The membranous ERACs are often associated with autophagosome-like structures (C, arrows) and located close to the vacuole (D, arrowheads). Bar: ??? nm.
The approximate position of EGFP-CFTR-containing ERACs to the vacuolar membrane resembles the localization of the pre-autophagosomal structure (PAS) that is considered to be the site of autophagosome formation. The precursor of the vacuolar aminopeptidase I (prApe1p) is localized to the PAS before it is delivered to the vacuole to generate the mature form of Ape1p (Shintani et al., 2002). We tested if the EGFP-CFTR ERACs represent the PAS by analyzing the colocalization of Ape1p and EGFP-CFTR. Wild-type yeast was co-transformed with the EGFP-CFTR and CFP-APE1 constructs and induced with copper for 2 h to synthesize EGFP-CFTR. As shown in Figure 4B, the EGFP-CFTR-containing ERACs often co-localize with Ape1p, that appears to be induced by the formation of ERACs (arrows). In contrast, cells that lack EGFP-CFTR ERACs contain low levels of Ape1p (a single Ape1p-labeled PAS is visible in a cell marked with an arrowhead). These results indicate that the formation of ERACs may induce the sequestration of autophagic components to the sites.
The presence of autophagosomes is the hallmark of autophagy as detected by electron microscopy (Mizushima, 2004). We inspected the possible presence of autophagosomes in yeast cells expressing EGFP-CFTR by electron microscopy. As shown in Figure 4C, membranous ERACs induced by EGFP-CFTR expression are often associated with autophagosome-like structures. Specifically, doughnut-shaped and double-membrane organelles accumulate adjacent to ERACs (arrows). Significantly, these structures often adjoin the vacuolar membranes (Fig. 4D, arrowheads). Control cells without EGFP-CFTR expression do not contain ERACs and autophagosome-like structures (see Fig. 2F). This result raises the possibility that EGFP-CFTR-containing ERACs may be removed through autophagy.
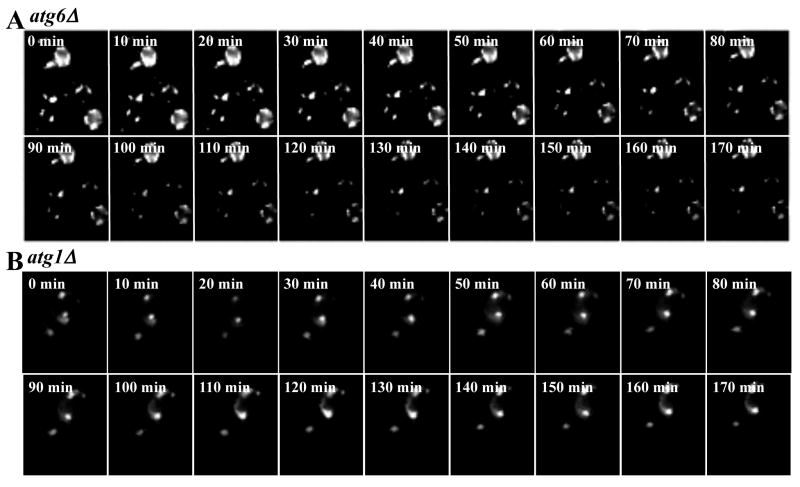
The involvement of autophagy was tested by monitoring EGFP-CFTR degradation from the ERACs in the atg6Δ mutant. Atg6p is a subunit of phosphatidylinositol 3-kinase (PI3-kinase) complexes that have been shown recently to participate in the degradation of the Z variant of human alpha-1 proteinase inhibitor (α1PiZ) (Kruse et al., 2006). As shown in Figure 5A, formation of EGFP-CFTR ERACs in atg6Δ cells is analogous to that seen in wild-type and pre1-1 cells. In contrast to wild-type and pre1-1 cells, EGFP-CFTR is not cleared from ERACs in atg6Δ cells 3 h post ERAC formation. This result strongly suggests that EGFP-CFTR degradation from ERACs is autophagy dependent. The autophagy-dependent degradation of EGFP-CFTR ERACs was also tested with another autophagic mutant lacking the yeast ATG1 gene (Figure 5B). ATG1 encodes a cytosolic serine-threonine kinase required for autophagosomal vesicle formation during the initial stage of autophagy (Abeliovich et al., 2003; Matsuura et al., 1997). As shown in Figure 5B, the atg1Δ cells are deficient in removing the EGFP-CFTR ERACs 3 h after their formation. Together, our findings suggest that autophagy functions to degrade aberrant proteins within ERACs. Our data appears to contradict to the results published by Kruse et al. (2006), in which it was shown by immunoblotting that Atg6p/Vps30p is not required for CFTR degradation. We assume that this seeming discrepancy is due to the methodological differences. In the immunoblotting and pulse-chase experiments, the results represent the average phenotypes from an entire population of cells within a large culture. Our data obtained by real-time imaging clearly demonstrate that different cells have different routes to degrade CFTR that would not be detected by traditional immunoblotting or pulse-chase experiments.
Fig. 5.
EGFP-CFTR degradation from ERACs is autophagy dependent. The atg6Δ and atg1Δ strains were induced with copper for 2 h to form EGFP-CFTR foci and were imaged every 2 min afterwards. Movie montages show that ERACs in the atg6Δ cells (A) and atg1Δ cells (B) are not degraded even after 170 min.
Discussion
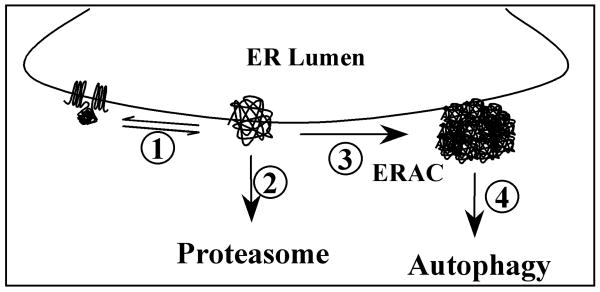
Using a novel real-time imaging approach and EGFP-CFTR as a model substrate, we monitored the temporal and spatial distribution and degradation of misfolded protein following its de novo synthesis. Our findings shed light on the functional relationship between the two major protein degradative pathways, the UPP and the autophagic pathway, in the removal of misfolded proteins such as CFTR. Based on our data, we propose a model in which the proteasome and autophagy function sequentially to remove misfolded proteins from distinct ER compartments (Fig. 6). Proteins synthesized in the ER are subject to ER-quality control in which correctly folded molecules are distinguished from misfolded ones (process 1). The misfolded molecules undergo a dynamic triage decision to be either degraded by the proteasome (process 2) or aggregate in sub-ER domains to form ERACs (process 3). ERACs are subsequently cleared by autophagy (process 4).
Fig. 6.
Model for the functional relationship of proteasomal degradation and autophagy in removing misfolded proteins from the ER. 1) The folding states of newly synthesized proteins are monitored by the ER quality-control machineries. 2) Misfolded proteins are initially subjected to proteasomal degradation. 3) Misfolded proteins form ER-associated complexes (ERACs). 4) ERACs are delivered for autophagic degradation.
The mechanisms that govern protein partitioning to proteasomal degradation or accumulation in ERACs are unknown. One of the determinants may reside in the intrinsic aggregation quotient of protein substrates. Proteins that are not correctly folded will be ubiquitinated and targeted to the proteasomes for degradation. Prior to degradation, proteasomal substrates need to be unfolded to be inserted to the narrow pore of the 26S cylinder of the proteasome. Some proteasomal substrates may not be easily unfolded and efficiently degraded, and therefore be prone to aggregation. For example, polyglutamine-containing proteins are prone to aggregation because they are not efficiently degraded by proteasomes (Holmberg et al., 2004; Venkatraman et al., 2004). By the same tokens, CFTR molecules may form intracellular aggregates under certain conditions (Kopito, 1999).
Here, we have characterized the formation and the degradation of ERACs in live cells. We report that ERACs form rapidly in response to EGFP-CFTR synthesis, and large ERAC structures are evident 50 min after induction of EGFP-CFTR synthesis (Fig. 1B). EGFP-CFTR-induced ERACs consist of a tubular network of ER membranes (Fig. 2G) and appear morphologically analogous to those induced by the overexpression of select mutants of Ste6p (Huyer et al., 2004) and the overexpression of the PM ATPase Pma2p or select mutants of Pma1p (Ferreira et al., 2002; Harris et al., 1994; Supply et al., 1993). Like CFTR, Ste6p is a member of the ATP-binding cassette (ABC) superfamily and consist of two homologous halves, each with six transmembrane domains and a cytoplasmic ATP-binding domain. The ultrastructural similarity of ERACs formed by these distinct ATPases suggests that the cellular response to the presence of aggregation-prone transmembrane proteins within the ER is common and that the formation of ERACs containing different proteins may occur through a common mechanism. The formation of ERACs might be driven by relatively weak protein-protein interactions between EGFP-CFTR molecules that synergize to form planar domains of tightly-bound proteins. This model is supported by our FRAP data suggesting extensive aggregation of EGFP-CFTR within ERACs. In this model the sorting of EGFP-CFTR into ERACs is due to the inherent aggregation-prone status of the protein.
The expansion observed in ERACs induced by EGFP-CFTR differs significantly from the ER expansion seen in response to unfolded protein responses (UPR) (Bernales et al., 2006). In those cells, the ER folds into tightly packed lamellar structures, which are similar to those seen in cells over-expressing HMG-CoA reductase (Koning et al., 1996). Irrespective of their ultimate morphology, the increase in ER membrane area seems logical as it increases the space allocated for protein folding. The details of how the ER membrane expands and what cellular mechanisms govern lamellar formation in the case of UPR and tubulation in response to EGFP-CFTR load remain to be defined. The formation of ERACs may function to spatially remove aggregated proteins from the remaining ER membrane, thus promoting ER homeostasis. It is likely that the sequestration of misfolded protein into ERACs is beneficial since cells containing ERACs show normal processing of secretory and plasma membrane proteins (Huyer et al., 2004), and ERACs do not cause UPR (Zhang et al., 2001), suggesting that the function of the ER is not compromised by the presence of ERACs.
Previous reports suggest that ERACs represent temporary holding compartments for misfolded transmembrane proteins that are eventually degraded through ERAD (Huyer et al., 2004). When expressed in yeast cells, specific mutants of Ste6p are sequestered within ERACs. The degradation from those structures is dependent on Ubc7p (an E2 ubiquitin-conjugating enzyme), suggesting the involvement of the proteasomal pathway. It appears therefore, that mutant Ste6p sequestered within ERACs can be refolded sufficiently to allow translocation into the cytoplasm and the degradation through cytoplasmic proteasomes. Interestingly, different mutants of Ste6p that localize to ERACs have different rates of degradation. This difference could be due to differences in unfolding and extraction from the ER membrane or in engaging the ubiquitin/proteasome system. One may also speculate that proteins difficult to extract might require another mechanism for their clearance.
Our results suggest that aggregated proteins are sequestered into ERACs and that ERACs represent final destination depots that are cleared from cells by autophagy. This is based on multiple lines of evidence: the rapid kinetics of ERAC disappearance (an entire ERAC can be cleared within 8 min; Fig. 1C); the finding that EGFP-CFTR within ERACs is largely immobile (Fig. 2B) suggesting that it is aggregated; the co-localization of the Ape1p marker for PAS (Fig. 4B) and the requirement for a functional autophagic pathway (Fig. 5) for ERAC clearance. Autophagy provides a bulk method to eliminate large cytoplasmic protein aggregates in cell culture models of conformational diseases such as spinocerebellar ataxia 7 (Zander et al., 2001), Huntington's (Yamamoto et al., 2006) and Parkinson's (Stefanis et al., 2001) diseases. Here, we report the autophagic clearance of aggregated transmembrane EGFP-CFTR. Autophagy provides a general method to sequester protein aggregates into membranous compartments that ultimately fuse with the vacuole (or lysosomes in metazoan cells) for proteolytic degradation within that organelle. Thus, the formation of ERACs and their subsequent clearance by autophagy provide means to remove severely misfolded and aggregated proteins from the ER membrane. It appears that the removal of aggregated EGFP-CFTR from the ER occurs exclusively by autophagy. This is suggested by the finding that EGFP-CFTR sequestered into ERACs is not degraded when autophagy is inhibited (Fig. 5). In yeast strains lacking ATG1 or ATG6, ERACs remain for prolonged periods of time, indicating that EGFP-CFTR is not cleared through the proteasomal pathway. Interestingly, induction of UPR causes ER proliferation into closely packed lamellar structures that are cleared through autophagy (Bernales et al., 2006). These results suggest that autophagy is a common pathway to degrade the elongated ER structures under both conditions. Our studies document the novel autophagic clearance of aggregated protein from subdomains of the ER. This adds to the known repertoire of degradative pathways operational at the ER and suggests that the proteasomal and autophagic pathways participate in the effective clearance of misfolded protein from distinct domains of the ER.
Supplementary Material
Acknowledgments
The authors wish to express gratitude to Drs. J.L. Brodsky, K.B. Kruse, A.A McCracken, J.L. Hartman and D. Wolf for reagents and strains. We also thank A. Tousson, L. Millican and T. Szul for technique assistance and Drs. E. Sorscher, J. Collawn, Z. Bebok and D.M. Bedwell for helpful discussions. This work was supported by a NIH grant DK68074 (to L. Fu).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abeliovich H, Zhang C, Dunn WA, Jr, Shokat KM, Klionsky DJ. Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol Biol Cell. 2003;14:477–490. doi: 10.1091/mbc.E02-07-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral MD. Processing of CFTR: traversing the cellular maze – how much CFTR needs to go through to avoid cystic fibrosis? Pediatr Pulmonol. 2005;39:479–491. doi: 10.1002/ppul.20168. [DOI] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand CA, Frizzell RA. The role of regulated CFTR trafficking in epithelial secretion. Am J Physiol Cell Physiol. 2003;285:C1–18. doi: 10.1152/ajpcell.00554.2002. [DOI] [PubMed] [Google Scholar]
- Choo-Kang LR, Zeitlin PL. Induction of HSP70 promotes DeltaF508 CFTR trafficking. Am J Physiol Lung Cell Mol Physiol. 2001;281:L58–68. doi: 10.1152/ajplung.2001.281.1.L58. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Exp Biol Med (Maywood) 2006;231:1197–1211. doi: 10.1177/153537020623100705. [DOI] [PubMed] [Google Scholar]
- Ferreira T, Mason AB, Pypaert M, Allen KE, Slayman CW. Quality control in the yeast secretory pathway: a misfolded PMA1 H+-ATPase reveals two checkpoints. J Biol Chem. 2002;277:21027–21040. doi: 10.1074/jbc.M112281200. [DOI] [PubMed] [Google Scholar]
- Fu L, Sztul E. Traffic-independent function of the Sar1p/COPII machinery in proteasomal sorting of the cystic fibrosis transmembrane conductance regulator. J Cell Biol. 2003;160:157–163. doi: 10.1083/jcb.200210086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Gao YS, Tousson A, Shah A, Chen TL, Vertel BM, Sztul E. Nuclear aggresomes form by fusion of PML-associated aggregates. Mol Biol Cell. 2005;16:4905–4917. doi: 10.1091/mbc.E05-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J Biol Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- Harris SL, Na S, Zhu X, Seto-Young D, Perlin DS, Teem JH, Haber JE. Dominant lethal mutations in the plasma membrane H(+)-ATPase gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1994;91:10531–10535. doi: 10.1073/pnas.91.22.10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer W, Kleinschmidt JA, Saidowsky J, Escher C, Wolf DH. Proteinase yscE, the yeast proteasome/multicatalytic-multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J. 1991;10:555–562. doi: 10.1002/j.1460-2075.1991.tb07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg CI, Staniszewski KE, Mensah KN, Matouschek A, Morimoto RI. Inefficient degradation of truncated polyglutamine proteins by the proteasome. EMBO J. 2004;23:4307–4318. doi: 10.1038/sj.emboj.7600426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyer G, Longsworth GL, Mason DL, Mallampalli MP, McCaffery JM, Wright RL, Michaelis S. A striking quality control subcompartment in Saccharomyces cerevisiae: the endoplasmic reticulum-associated compartment. Mol Biol Cell. 2004;15:908–921. doi: 10.1091/mbc.E03-07-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TJ, Loo MA, Pind S, Williams DB, Goldberg AL, Riordan JR. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- Kerem BS, Rommens JM, Buchanan JA, Markiewitz D, Cox TK, Chakravarti A, Buchwald M, Tsui LC. Identification of the cystic fibrosis gene: Genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Cregg JM, Dunn WA, Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Koning AJ, Roberts CJ, Wright RL. Different subcellular localization of Saccharomyces cerevisiae HMG-CoA reductase isozymes at elevated levels corresponds to distinct endoplasmic reticulum membrane proliferations. Mol Biol Cell. 1996;7:769–789. doi: 10.1091/mbc.7.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito RR. Biosynthesis and degradation of CFTR. Physiol Rev. 1999;79:S167–173. doi: 10.1152/physrev.1999.79.1.S167. [DOI] [PubMed] [Google Scholar]
- Kruse KB, Brodsky JL, McCracken AA. Characterization of an ERAD gene as VPS30/ATG6 reveals two alternative and functionally distinct protein quality control pathways: one for soluble Z variant of human alpha-1 proteinase inhibitor (A1PiZ) and another for aggregates of A1PiZ. Mol Biol Cell. 2006;17:203–212. doi: 10.1091/mbc.E04-09-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Patterson GH. Development and use of fluorescent protein markers in living cells. Science. 2003;300:87–91. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- Lukacs GL, Mohamed A, Kartner N, Chang XB, Riordan JR, Grinstein S. Conformational maturation of CFTR but not its mutant counterpart (delta F508) occurs in the endoplasmic reticulum and requires ATP. EMBO J. 1994;13:6076–6086. doi: 10.1002/j.1460-2075.1994.tb06954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol. 2004;36:2491–2502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Okiyoneda T, Harada K, Takeya M, Yamahira K, Wada I, Shuto T, Suico MA, Hashimoto Y, Kai H. Delta F508 CFTR pool in the endoplasmic reticulum is increased by calnexin overexpression. Mol Biol Cell. 2004;15:563–574. doi: 10.1091/mbc.E03-06-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao TP, Baehrecke EH, Taylor JP. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- Romisch K. Endoplasmic reticulum-associated degradation. Annu Rev Cell Dev Biol. 2005;21:435–456. doi: 10.1146/annurev.cellbio.21.012704.133250. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1990. [Google Scholar]
- Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Leggo J, Crow TJ, DeLisi LE, Walsh C, Jain S, Paykel ES. Analysis of polyglutamine-coding repeats in the TATA-binding protein in different human populations and in patients with schizophrenia and bipolar affective disorder. Am J Med Genet. 1996;67:495–498. doi: 10.1002/(SICI)1096-8628(19960920)67:5<495::AID-AJMG12>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Shintani T, Huang WP, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanis L, Larsen KE, Rideout HJ, Sulzer D, Greene LA. Expression of A53T mutant but not wild-type alpha-synuclein in PC12 cells induces alterations of the ubiquitin-dependent degradation system, loss of dopamine release, and autophagic cell death. J Neurosci. 2001;21:9549–9560. doi: 10.1523/JNEUROSCI.21-24-09549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan ML, Youker RT, Watkins SC, Brodsky JL. Localization of the BiP molecular chaperone with respect to endoplasmic reticulum foci containing the cystic fibrosis transmembrane conductance regulator in yeast. J Histochem Cytochem. 2003;51:545–548. doi: 10.1177/002215540305100417. [DOI] [PubMed] [Google Scholar]
- Supply P, Wach A, Thines-Sempoux D, Goffeau A. Proliferation of intracellular structures upon overexpression of the PMA2 ATPase in Saccharomyces cerevisiae. J Biol Chem. 1993;268:19744–19752. [PubMed] [Google Scholar]
- Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf DH. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Venkatraman P, Wetzel R, Tanaka M, Nukina N, Goldberg AL. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol Cell. 2004;14:95–104. doi: 10.1016/s1097-2765(04)00151-0. [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CW, Klionsky DJ. The molecular mechanism of autophagy. Mol Med. 2003;9:65–76. [PMC free article] [PubMed] [Google Scholar]
- Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Cremona ML, Rothman JE. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol. 2006;172:719–731. doi: 10.1083/jcb.200510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youker RT, Walsh P, Beilharz T, Lithgow T, Brodsky JL. Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol Biol Cell. 2004;15:4787–4797. doi: 10.1091/mbc.E04-07-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger JM, Ren HY, Chen L, Fan CY, Fields A, Patterson C, Cyr DM. A foldable CFTR{Delta}F508 biogenic intermediate accumulates upon inhibition of the Hsc70-CHIP E3 ubiquitin ligase. J Cell Biol. 2004;167:1075–1085. doi: 10.1083/jcb.200410065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger JM, Fan CY, Chen L, Rosser MF, Patterson C, Cyr DM. Cystic fibrosis transmembrane conductance regulator as a model substrate to study endoplasmic reticulum protein quality control in mammalian cells. Methods Mol Biol. 2005;301:293–303. doi: 10.1385/1-59259-895-1:293. [DOI] [PubMed] [Google Scholar]
- Younger JM, Chen L, Ren HY, Rosser MF, Turnbull EL, Fan CY, Patterson C, Cyr DM. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- Zander C, Takahashi J, El Hachimi KH, Fujigasaki H, Albanese V, Lebre AS, Stevanin G, Duyckaerts C, Brice A. Similarities between spinocerebellar ataxia type 7 (SCA7) cell models and human brain: proteins recruited in inclusions and activation of caspase-3. Hum Mol Genet. 2001;10:2569–2579. doi: 10.1093/hmg/10.22.2569. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Nijbroek G, Sullivan ML, McCracken AA, Watkins SC, Michaelis S, Brodsky JL. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol Biol Cell. 2001;12:1303–1314. doi: 10.1091/mbc.12.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.