Summary
Increases in protein levels of XIAP in cancer cells have been associated with resistance to apoptosis induced by cellular stress. Herein we demonstrate that the upregulation of XIAP protein levels is regulated by MDM2 at the translational level. MDM2 was found to physically interact with the IRES of the XIAP 5′-UTR, and to positively regulate XIAP IRES activity. This XIAP IRES-dependent translation was significantly increased in MDM2-transfected cells where MDM2 accumulated in the cytoplasm. Cellular stress and DNA damage triggered by irradiation induced the dephosphorylation and cytoplasmic localization of MDM2, which also led to an increase in IRES-dependent XIAP translation. Upregulation of XIAP in MDM2-overexpressing cancer cells in response to irradiation resulted in resistance of these cells to radiation-induced apoptosis.
Significance
Overexpression of the MDM2 oncoprotein, which is often found in cancer cells, is associated with resistance to chemo-radiation therapy and poor prognosis of cancer patients. MDM2 is well characterized as an inhibitor of the tumor suppressor p53, and overexpression of MDM2 contributes to a growth advantage for cancer cells. However, the mechanism by which overexpression of MDM2 confers resistance to DNA damage induced by irradiation and chemotherapy are not fully elucidated. We report here that MDM2 was able to bind to XIAP mRNA and positively regulate the IRES-dependent XIAP translation during cellular stress triggered by irradiation. These results identify a p53-independent function for MDM2 in mediating XIAP translation, which is critical in effecting cancer cell response to chemo-radiation therapy.
Introduction
Inhibitor-of-Apoptosis Proteins (IAP) form a family of caspase inhibitors that block the downstream portion of the apoptosis pathway and inhibit cell death in response to multiple stimuli. There are currently eight known human IAP family members and XIAP is a very important inhibitor of apoptosis among the members (Schimmer, 2004; Eckelman et al., 2006). XIAP has been shown to bind specifically to caspase-3, -7, and -9, inhibiting them; but not the remaining caspases (Riedl et al., 2001; Shiozaki et al., 2003). The distinct cascades of caspase activation regulate different apoptosis pathways that can be triggered via intrinsic and extrinsic signals (Green, 2005). For example, activation of caspase-9 initiates the intrinsic (mitochondrial) pathway of apoptosis, which is the major mechanism involved in cell death induced by cellular stress and DNA damage, such as from exposure to UV and ionizing radiation (IR) (Kuida et al., 1998). Activation of caspase-3 and -7 are distal steps in this intrinsic apoptosis pathway (Lakhani et al., 2006). The specific inhibition of these caspases by XIAP suggests that XIAP is a molecule that is critical for regulating sensitivity to apoptosis induced by cellular stress and DNA damage.
Previous studies have demonstrated that the expression of XIAP mRNA in most tissues and cells is fairly consistent (Liston et al., 1996). Cellular stresses such as exposure to IR will induce alteration of XIAP protein expression, without concomitant changes in XIAP mRNA levels, in a variety of cancer cells (Holcik et al., 1999). These observations suggest that the expression of XIAP is mainly regulated at the translational level. Initiation of translation can occur by two distinct mechanisms, cap-dependent scanning and internal ribosome entry. The latter mechanism requires an internal ribosome entry segment (IRES) located in the 5′-UTR of the mRNA and the interaction of IRES trans-acting factor (ITAF)/ribonucleoprotein (RNP) (Stoneley et al., 2004). So far, IRES elements have been found mainly in mRNAs involved in regulating gene expression during development, differentiation, cell growth and survival (Bonnal et al., 2003). In particular, IRES of certain anti-apoptotic genes become activated under conditions where cap-dependent protein synthesis is greatly reduced, such as upon cellular stress and DNA damage; thus, activated IRES initiates translation of proteins that can protect cells from stress (Komar et al., 2005). It is known that XIAP translation is uniquely regulated by the IRES mechanism. There is a 162-nucleotide (nt) IRES sequence in the 5′-UTR of XIAP mRNA (Holcik et al., 1999). Three ITAFs/RNPs, namely La, hnRNP C1/C2, and hnRNP A1, have been identified as able to regulate XIAP IRES activity (Holcik et al., 2000a; Holcik et al., 2003; Lewis et al., 2007). Reports show that IRES-regulated XIAP translation is activated in cancer cells in response to IR, and that upregulation of XIAP results in increased resistance to apoptosis induced by this stress stimulation (Holcik et al., 2000b; Lewis et al., 2005).
The MDM2 protein is a multifunctional oncoprotein, and its ability to inactivate the p53 function through interaction with this tumor suppressor has been well characterized. In addition to interacting with and regulating p53, it has been demonstrated that MDM2 interacts with other molecules including specific protein and RNA, which may play a p53-independent role in oncogenesis. For example, MDM2 was shown to bind to and ubiquitinate Rb, resulting in Rb degradation and release of the E2F1 that promotes cell cycle progression (Xiao et al., 1995). MDM2 was also found to bind E2F1 directly, and to enhance E2F1 stability (Zhang et al., 2005). The C-terminal RING finger domain of MDM2 was found to exhibit specific RNA binding ability. A SELEX (systematic evolution of ligands by exponential enrichment) procedure yielded a subset of RNA molecules that bind efficiently to MDM2 in vitro (Elenbaas et al., 1996).
Aside from regulating other cellular molecules, MDM2 itself is modulated by various cellular signals. Phosphorylation and subcellular distribution of MDM2 is regulated by the PI3K/Akt pathway (Mayo et al., 2001). Cell growth/survival factor-induced activation of PI3K and its downstream target Akt will phosphorylate cytoplasmic MDM2 on serines 166 and 186. Phosphorylation of these sites is required for translocation of MDM2 from the cytoplasm into the nucleus. In contrast to survival signals that induce MDM2 phosphorylation, cellular stress and DNA damage invoke dephosphorylation of MDM2 (Meek et al., 2003; Blattner et al., 2002). It has been shown that dephosphorylation of the central acidic domain of MDM2 is essential for accumulation and stabilization of p53 in stressed wild-type (wt)-p53 cells. Stress also induces signaling for dephosphorylation of MDM2 at serine 166 (Okamoto et al., 2002), which may lead to inhibition of nuclear entry of MDM2 or an increase in its translocation from the nucleus to the cytoplasm. Once MDM2 is released from p53 and is localized in the cytoplasm, it could play a p53-independent role. Since MDM2 is able to bind RNA and shuttles between the nucleus and the cytoplasm, which are the properties of most ITAFs/RNPs, we hypothesize that the dephosphorylated cytoplasmic MDM2 may act as an ITAF/RNP to exhibit a p53-independent role in regulating translation through binding of its C-terminus to specific RNA.
We have previously demonstrated a link between MDM2 and XIAP. In a study utilizing transfection of MDM2 into a p53-null leukemia cell line, we found that the expression of XIAP protein is upregulated in the MDM2-transfected cells (Gu et al., 2002). Because the expression of XIAP is primarily regulated at the translational level though the IRES-dependent mechanism, we have now investigated the possible regulation of XIAP translation by MDM2 and the mechanism by which MDM2 acts as an ITAF/RNP to regulate XIAP IRES activity. Furthermore, we investigated the impact of MDM2-mediated XIAP expression on irradiation-induced apoptosis in human cancer cells.
Results
IR induces MDM2 modulation and XIAP translation
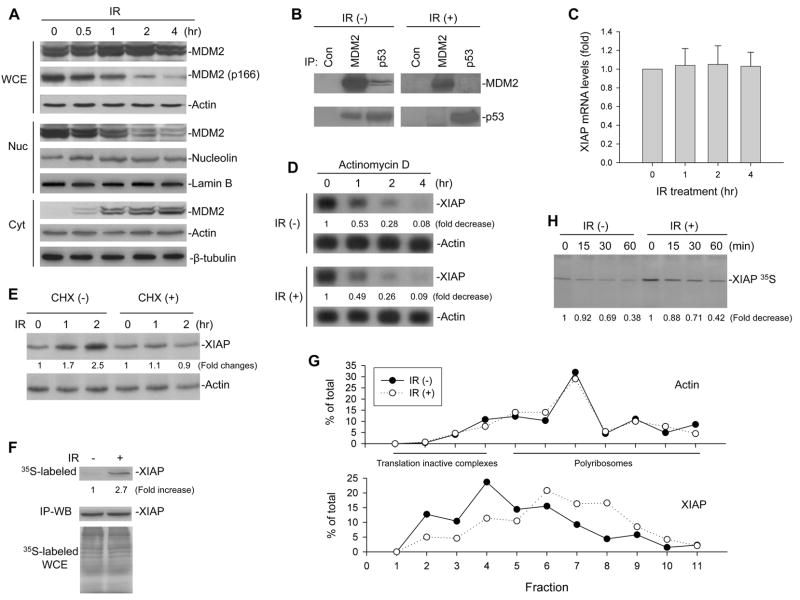
It is known that IR treatment dephosphorylates MDM2. To test whether IR-induced dephosphorylated MDM2 becomes dissociated from the MDM2-p53 complex and translocated from the nucleus to the cytoplasm, we examined the cellular redistribution of MDM2 as well as its association with p53 following IR treatment in an acute lymphoblastic leukemia (ALL) cell line EU-1. This cell line was chosen for the test because EU-1 cells have a wt-p53 and overexpress MDM2 (Figure S1A), and MDM2 is predominantly localized in the nucleus of cultured/untreated cells (Zhou et al., 2003). As is shown in Figure 1A, IR treatment decreased nuclear MDM2 and increased cytoplasmic MDM2 expression, as well as induced downregulation of MDM2 at serine 166 in EU-1 cells. IP-western blot analysis demonstrated that after IR treatment MDM2 dissociated from p53, as shown in Figure 1B, in which the levels of both MDM2 and p53 in immunocomplexed form precipitated with p53 and MDM2 antibodies, respectively, were significantly reduced in the IR-treated cells as compared to untreated cells.
Figure 1.
Modulation of MDM2 and XIAP translation by IR. (A) EU-1 cells were treated with 10 Gy IR for different times as indicated, and cell extracts were tested for the nuclear (Nuc) and cytoplasmic (Cyt) expression of MDM2 as well as for total MDM2 and MDM2 phosphorylation at site 166 in whole cell extract (WCE) in western blot assays. (B) Cell lysates from EU-1 treated with or without 10 Gy IR for 4 hours were IP with antibodies as indicated. Normal mouse antibody served as control (Con). Proteins in immune complexes were detected by western blotting. (C) mRNA expression of XIAP after treatment with 10 Gy IR for the indicated time in EU-1 cells was determined by quantitative RT-PCR. Data represents the mean (±SD) levels of XIAP mRNA of three independent experiments. (D) EU-1 cells were treated with 10 Gy IR for 2 hours followed by addition of 5 mg/ml actinomycin D. At different times after actinomycin D addition, the cells were harvested, and total RNA was isolated. The amount of XIAP mRNA remaining was determined by Northern blotting and quantified by densitometric analysis. Labels under bands in the blot represent XIAP mRNA levels after normalization to actin, compared with samples (0, defined as 1 unit). (E) EU-1 cells were treated with and without 10μg/ml CHX for 10 min, before 10 Gy IR. Extracts from cells that were unirradiated (0) or harvested 1 or 2 h after IR were assessed for expression of XIAP, to show that CHX blocked IR-induced XIAP. (F) XIAP protein was IP from EU-1 cells that had been labeled for 5 min with [35S]-methionine, 30 min after 0 or 10 Gy IR, and then assessed by autoradiography (upper panel). Cells that had been pretreated with 50 μM MG132 and analyzed by western blot showed equivalent amounts of XIAP in the IP as a control for total XIAP protein that was not turnover in unirradiated versus irradiated cells (middle panel). Controls include analysis of whole cell extracts (WCE), where equal amounts of [35S]-methionine incorporated into the irradiated and unirradiated cells (bottom panel). (G) IR enhances association of the XIAP mRNA with translating polyribosomes. EU-1 cells were treated with or without 10 Gy IR for 12 h, and cytoplasmic lysates were fractionated on sucrose gradient. RNA was extracted from each of the fractions and subjected to quantitative RT-PCR for quantitative analysis of the distribution of XIAP and Actin mRNAs. Data represent percentage of the total amount of corresponding mRNA on each fraction. (H) Turnover of XIAP protein in IR-treated and control cells, as detected by pulse-chase assay. EU-1 cells were treated with or without 10 Gy IR for 2 hours followed by [35S]methionine labeling. At selected times as indicated after labeling cell lysates were collected for determination of the XIAP 35S protein levels by SDS-PAGE analysis.
We next evaluated whether there was concomitant upregulation of XIAP expression that would occur at a translational level, following the IR-induced cytoplasmic translocation of MDM2. We began by testing whether IR directly induces XIAP mRNA expression. Consistent with previous observation in other cancer cells (Holcik et al., 1999), XIAP mRNA was not induced by IR in EU-1 cell line (Figure 1C). The stability of XIAP mRNA was also not affected by IR treatment. As shown in Figure 1D, there was no difference in degradation rate of XIAP mRNA between IR-treated and untreated EU-1 cells. We next evaluated the effect of IR on XIAP translation in the MDM2-overexpressing cells. We treated EU-1 cells with IR in the presence or absence of the protein synthesis inhibitor cycloheximide (CHX). In the absence of CHX, IR induced XIAP, whereas XIAP was not induced by the same exposure of IR in the presence of CHX (Figure 1E). In order to directly demonstrate an increase in XIAP translation after IR in EU-1 cells, we further performed metabolic [35S]-methionine labeling and IP analysis. As shown in Figure 1F, a significant increase in metabolically labeled, newly synthesized XIAP was apparent in the IR-treated cells, while the total XIAP protein levels were found to be equivalent in untreated versus IR-treated cells in the presence of the proteasome inhibitor MG132. In addition, we performed linear sucrose gradient fractionation to assess the polyribosome association of the XIAP mRNA in EU-1 cells subjected to IR and mock-treatment. We found that XIAP mRNA was clearly shifted from fractions containing translation dormant complexes including mRNPs, ribosome subunits, and monosomes (Figure 1G, bottom, fractions 1–4) to fractions enriched of translating polyribosomes (Figure 1G, bottom, fractions 5–11), indicative of enhanced translation. Consistent with previous observation (Lü et al., 2006), IR treatment had no effect on the polyribosome profile of the Actin mRNA (Figure 1G, top). Furthermore, there was no appreciable difference in XIAP protein stability between EU-1 cells exposed to IR and those without IR exposure (Figure 1H); thus demonstrating that induction of XIAP by IR occurs at the translational level.
Cytoplasmic MDM2 does not regulate XIAP transcription and post-translational modification
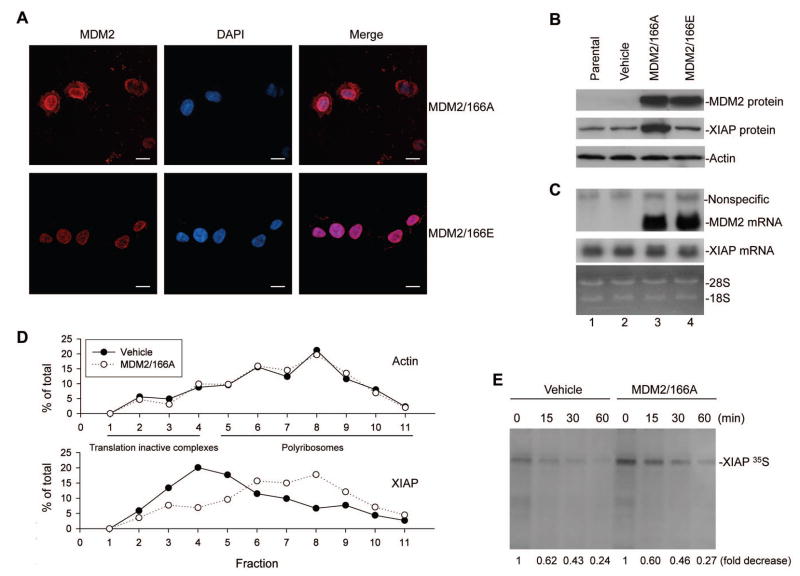
We considered the possibility that the cytoplasmic translocation of MDM2 was responsible for induction of XIAP. Due to the fact that MDM2 in cultured/unstressed cells is phosphorylated and located in the nucleus if the cells have a high level of PI3k/Akt activation, we designed an experiment using gene transfection of an MDM2 plasmid producing an MDM2 with a mutation at serine 166 substituted with alanine (MDM2/166A). This MDM2 with a serine-to-alanine substitution cannot be phosphorylated by PI3K/Akt; thus, it remains in the cytoplasm (Mayo et al., 2001). In this manner we were able to test the effect of cytoplasmic MDM2 on XIAP expression. First, we checked the subcellular distribution of the mutant MDM2 protein in a p53-null ALL cell line, EU-4 (Figure S1A), by transfecting it with the MDM2/166A plasmid tagged by red fluorescence protein (pDsRed1-C1). The red fluorophores indicated that approximately 50% of MDM2/166A was expressed in the cytoplasm (Figure 2A). A control, the mutant MDM2 protein in EU-4 cells transfected with plasmid MDM2/166E where serine 166 is mutated to glutamic acid that mimics phosphorylated MDM2 (Mayo et al., 2001), was predominantly localized in the nucleus.
Figure 2.
Effect of cytoplasmic MDM2 on XIAP transcription, translation, and post-translational modification. (A) Subcellular distribution of transfected MDM2/166A and MDM2/166E, as demonstrated by the red fluorophores in EU-4 cells characterized by confocal microscopy. Scale bars = 10μm. (B) and (C) Expression of transfected MDM2 and XIAP in both MDM2-transfected and control cells were detected by western blot assay (B), and their mRNA levels were detected by Northern blot assay (C). (D) Quantitative RT-PCR analysis of the MDM2/166A-induced translation enhancement of XIAP mRNA. SH-SY5Y cells were transfected with MDM2/166A and control plasmid (Vehicle), and cytoplasmic extracts were subjected to linear sucrose gradient fractionation. RNA was extracted from each of the fractions and quantitatively analyzed to evaluate polyribosome association by the XIAP and Actin mRNA. (E) Turnover of XIAP protein in MDM2/166A-transfected and control cells, as detected by pulse-chase assay. The radioactive labeled XIAP protein levels were quantified by densitometric analysis. All values were corrected relative to the load control and labeled under bands in the blot compared with samples (0, defined as 1 unit).
Transfection of MDM2/166A in EU-4 cells upregulated endogenous XIAP protein expression (Figure 2B). In contrast, transfection of MDM2/166E did not upregulate XIAP. To investigate whether the enhanced XIAP expression seen in MDM2/166A-transfected cells is due to increased transcription of the XIAP gene, we performed a northern blot assay. Expression of XIAP mRNA was not increased in the MDM2/166A-transfected cells (Figure 2C). We next performed polyribosome profiling assay to directly evaluate the influence of MDM2/166A on translation of XIAP in a NB cell line SH-SY5Y that express low level of MDM2 (Figure S1A). Transfection of MDM2/166A resulted in a shift of XIAP mRNA from translationally inactive complexes to polyribosomes engaged in active translation (Figure 2D), similar to that observed in IR-treated EU-1 cells (Figure 1G). Furthermore, we performed pulse-chase experiments using metabolic [35S]-methionine labeling and IP assay, to evaluate whether the observed induced XIAP expression in MDM2/166A-transfected cells may also involve a post-translational mechanism. As shown in Figure 2E, the half-life of the XIAP protein in MDM2/166A-transfected cells did not change as compared with that of control cells. These results suggest that MDM2/166A-upregulated expression of XIAP most likely occurs only at the translational level.
Cytoplasmic MDM2 induces XIAP IRES activity
To test the possible role of cytoplasmic MDM2 in regulating XIAP translation via an IRES-dependent mechanism, the status of the 5′-UTR of XIAP mRNA containing IRES in cancer cells must first be characterized. The 5′-UTR of XIAP mRNA has been controversial in previous studies. Liston et al (Liston et al., 1996) reported that the 5′-UTR of XIAP mRNA is short (128-bp), whereas the 5′-UTR of XIAP mRNA reported by Holcik et al (Holcik et al., 1999) is long (1.6 or 1.7 kb) containing a 162 bp IRES sequence (Figure S2A). To clarify this discrepancy regarding the 5′-UTR of XIAP mRNA, we searched the Human Genome to obtain the entire human XIAP gene sequence (Accession number: AY886519). We have identified a DNA sequence for the putative promoter region of the XIAP gene located immediately upstream of the short 5′-UTR (Figure S2B). Importantly, we found that the long 5′-UTR of XIAP mRNA is a variant that retains a part of intron 1 as a cryptic exon (Figure S2C). We performed RT-PCR on both ALL and NB cells, searching for the presence of the short and long versions of the 5′-UTR of XIAP, and found that all the cancer cells studied expressed both the short and long 5′-UTR of XIAP mRNA (Figure S2D). Further RT-PCR analysis with various controls (DNAse digestion of RNA and absence of reverse transcriptase) confirmed that the long 5′-UTR of XIAP containing IRES was indeed an mRNA (exon), but not genomic DNA (intron) (Figure S2E).
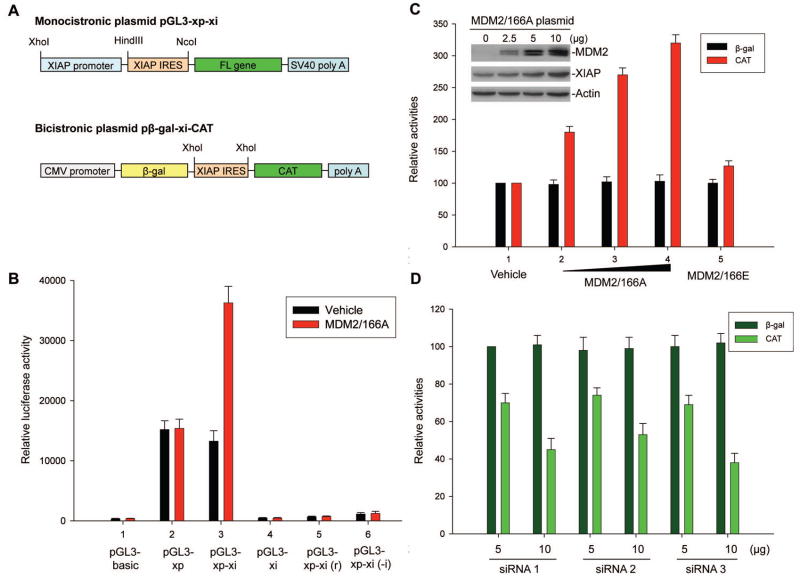
We performed gene transfection and reporter assays using EU-4 cells and both monocistronic and bicistronic plasmids containing the 162 bp XIAP IRES from the long 5′-UTR of XIAP. Results from experiments using the monocistronic plasmids (Figure 3A), which is driven by the XIAP promoter (xp), showed that insertion of the XIAP IRES (xi) into a pGL3-xp plasmid regulated luciferase translation, as demonstrated in Figure 3B, where the luciferase activity of pGL3-xp-xi was found to be only slightly reduced as compared to that of pGL3-xp (comparison of black bars in bar 3 versus bar 2). As is also shown in Fig. 3B, insertion of the xi directly into the pGL3 basic vector [pGL3-xi] resulted in no luciferase activity (bar 4), suggesting that a cryptic promoter is not present in the XIAP IRES. In addition, both insertion of the xi into pGL3-xp in a reverse orientation [pGL3-xp-xi(r)] and insertion of the xi with a deletion of the core RNP binding sequence [pGL3-xp-xi(-i)] abolished XIAP promoter-driven luciferase activity (bars 5 and 6), suggesting that the 162 bp XIAP IRES is indeed critical for proper regulation of luciferase translation. Insertion of the 128 bp short 5′-UTR of XIAP into the monocistronic plasmid did not regulate luciferase activity (data not shown).
Figure 3.
Effect of cytoplasmic MDM2 on XIAP IRES activity. (A) Schematic illustration of the monocistronic plasmids and bicistronic plasmids used for the reporter assays. (B) Transfected MDM2/166A induces XIAP IRES (xi) but not XIAP promoter (xp) activity. Here, 5μg of pGL3-xp or pGL3-xp-xi plasmids were co-transfected with 10μg MDM2/166A, respectively, into EU-4 cells; then luciferase activities were detected. Cells were co-transfected with the pGL3-basic vector and MDM2/166A as controls. Other controls include co-transfection of MDM2/166A with plasmids pGL3-xp-xi(r): insertion of the xi into pGL3-xp in a reverse orientation; and pGL3-xp-xi(-i): insertion of the xi with deletion of the core RNP binding sequence. In addition, the controls were co-transfected with each luciferase plasmid and with the pcDNA empty vector (vehicle). Data represent the mean ±SD of three independent experiments normalized to Renilla luciferase activity (co-transfection of pRL-SV40 vector as control). (C) Dose-dependent induction of xi-mediated translation of CAT by MDM2/166A in transfection using the bicistronic plasmid pβgal-xi-CAT. EU-4 cells were co-transfected with 5 μg pβgal-xi-CAT and with increasing amounts (2.5, 5, and 10 μg) of the MDM2/166A (bars 2–4) or vehicle only (bar 1), and 10 μg MDM2-166E (bar 5). After 24 h, cell extracts were prepared and the βgal and CAT activities were determined. The βgal and CAT activities in the control reactions were set at 100%. Bars represent the mean ±SD of three independent experiments. The titrations of MDM2 in cellular extracts from MDM2/166A-transfected cells and expression of the endogenous XIAP in these cells were detected by western blot assay (insert). (D) Co-transfection and IRES activity assay for the effect of MDM2 siRNA on xi-mediated translation. LA1-55N cells were co-transfected with 5μg pβgal-xi-CAT and different amounts (5, and 10 μg) of three pSUPER MDM2 plasmids (siRNA 1–3). Transfection and CAT/βgal activity assays were performed as described in (C). CAT (IRES) activity represents the mean percentage (±SD) of control (co-transfection of βgal-xi-CAT with vehicle, not shown) in at least three independent experiments.
Co-transfection of either wt-MDM2 (data not shown) or MDM2/166A did not induce XIAP promoter activity (comparison of red bar with black bar in bar 2 of Figure 3B), which is consistent with the finding in Fig. 2C that shows there was no XIAP mRNA induction in MDM2/166A- transfected cells. Interestingly, co-transfection of MDM2/166A significantly enhanced the XIAP IRES-mediated luciferase activity (comparison of red bar with black bar in bar 3). This MDM2-stimulated XIAP IRES function was confirmed in a reporter assay using the bicistronic plasmid pβgal-xi-CAT (Figure 3A). As shown in Fig. 3C, MDM2/166A significantly increased the activity of CAT (XIAP IRES) but not the activity of βgal, a control cistron, translation of which is initiated by cap-dependent mechanism. In addition, transfected MDM2/166A also increased a dose-dependent expression of endogenous XIAP (Figure 3C, insert). We also found that the MDM2/166E mutation slightly activated XIAP IRES (Figure 3C), whereas wt-MDM2 induced XIAP IRES activity depending on the status of PI3K/Akt activation in the cells used for transfection assays (data not shown).
To further confirm that MDM2 acts as an activator to regulate XIAP IRES activity, we performed experiments to knockdown endogenous MDM2 by siRNA to evaluate whether inhibition of MDM2 results in decreased XIAP IRES activity. We generated three pSUPER/MDM2 plasmids containing different 19 nt siRNA sequences specific for targeting MDM2. Transfection of these plasmids into LA1-55N, a NB cell line with MDM2 overexpression (Figure S1A), significantly suppressed the endogenous MDM2 expression (see below in Figure 6F). Co-transfection of these MDM2 siRNA plasmids with the pβgal-xi-CAT reporter plasmid into LA1-55N cells remarkably decreased the activity of CAT (Figure 3D).
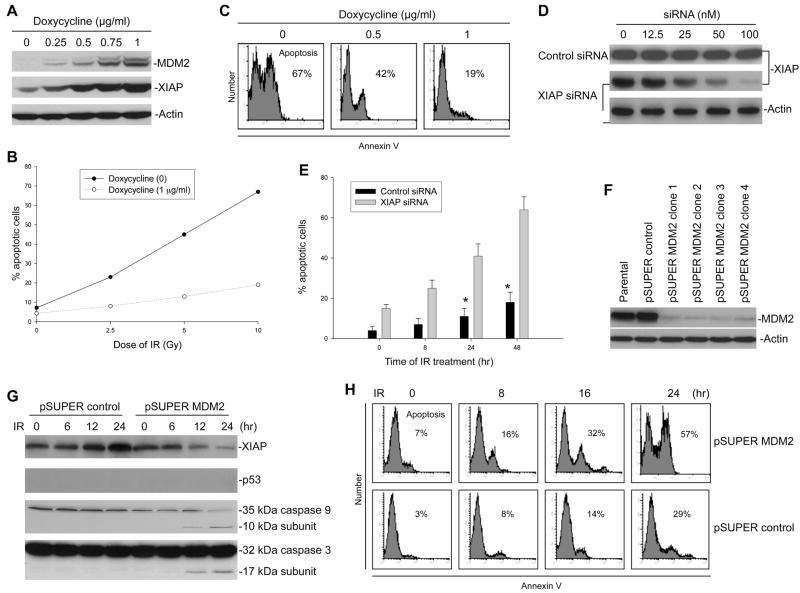
Figure 6.
Effect of MDM2-mediated XIAP translation on the sensitivity of cells to IR. (A) Dose-dependent induction of MDM2 by doxycycline in EU-4 cells transfected with Tet-controlled MDM2 plasmids followed by XIAP upregulation, detected by Western blot assay. (B) Dose-dependent response to IR-induced apoptosis of EU-4/Tet-on/MDM2 cells in cultures with or without 1 μg/ml of doxycycline. Cells were exposed to different dose of IR for 48 h, and apoptotic cells (Annexin V positive) were detected by flow cytometry. (C) Representative histograph of flow cytometry for detection of apoptosis in EU-4/Tet-on/MDM2 cells with different concentration of doxycycline and 10 Gy IR for 48 hours. (D) EU-4/Tet-on/MDM2 cells cultured with 1 μg/ml doxycycline were transfected with different concentrations of either XIAP siRNA or control siRNA for 48 h. Expression of endogenous XIAP was detected by western blotting. (E) Time-course of apoptosis induced by IR in combination with either XIAP siRNA or control siRNA in EU-4/Tet-on/MDM2 cells with 1 μg/ml doxycycline. Cells were treated with 10 Gy IR plus siRNA (100 nM) for the indicated time points, and apoptotic cells were detected by annexin-V staining. Data represent the mean percentage of annexin-V positive cells from three independent experiments; bars, ± SD. *p<0.01. (F) Stable suppression of MDM2 in p53-null NB cell line LA1-55N by transfection of pSUPER MDM2 siRNA plasmid. The expression of MDM2 in LA1-55N cells transfected with the pSUPER vector containing scrambled 19-nt (control) and in 4 clones transfected with pSUPER/MDM2 was detected by western blotting. (G) Kinetic analyses of XIAP protein expression and concomitant activation of caspases 9 and 3 in LA1-55N cells transfected with either control siRNA or MDM2 siRNA, following 10 Gy IR. (H) Time-course of apoptosis induced by IR in LA1-55N/MDM2 siRNA and control cells. Cells were treated with 10 Gy IR for the indicated time points, and apoptotic cells were detected by annexin-V staining using flow cytometry.
In addition, we performed translation assay to evaluate the effect of MDM2 on XIAP IRES activity using the dicistronic plasmid pFL-xi-FL. Previous studies have shown that the presence of XIAP IRES in pRL-FL vector decreases RL activity in cells transfected with this plasmid due to spurious splicing within the XIAP IRES, resulting in a relative increase of FL activity (Van Eden et al., 2004; Holcik et al., 2005). In agreement with this observation, transfection of pRL-xi-FL in EU-4 cells decreased the RL activity as compared to the transfection of pRL-FL (Figure S3A). However the presence of MDM2 did not affect the RL activity as compared to the transfection in the absence of MDM2, and MDM2 increased FL (XIAP IRES) activity (Figure S3A). Consistent with these results, an in vitro translation assay using pRL-xi-FL plasmid (with the T7 promoter) and TnT® T7 Quick Coupled Transcription/Translation System demonstrated that MDM2 induced FL (XIAP IRES) activity, but not affected RL activity (Figure S3B).
MDM2 physically interacts with XIAP IRES both in vivo and in vitro
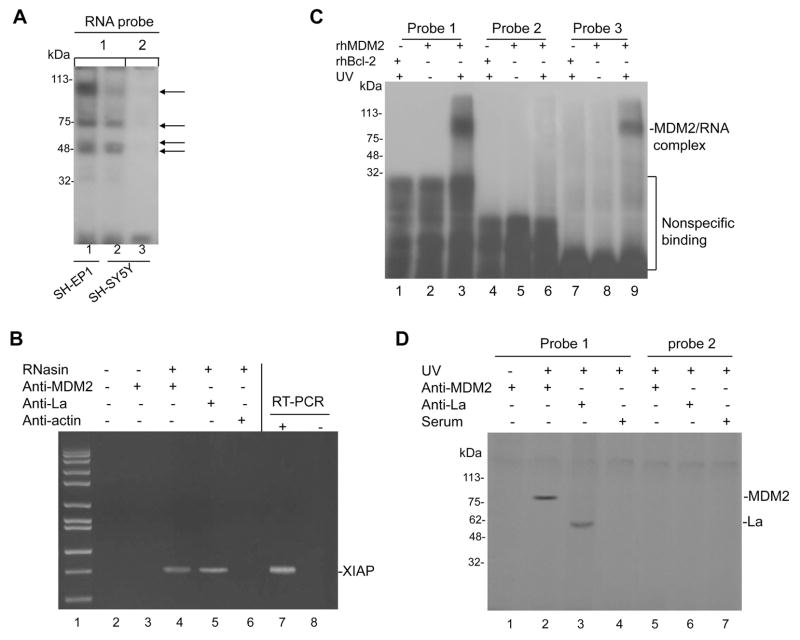
As MDM2 could induce XIAP IRES activity, we evaluated whether MDM2 interacts physically with the XIAP IRES. We performed UV cross-linking of 32P-labeled XIAP IRES probes and cell extracts from MDM2-overexpressing NB line SH-EP1 (Figure S1A) and SH-SY5Y to evaluate whether RNP was bound to the XIAP IRES. Similar results to previous experiments (Holcik et al., 2000) were obtained, as shown in Figure 4A, in which 4 major sizes of RNP (indicated by arrows) bound to the RNA probe of XIAP IRES. Interestingly, we observed a great reduction in the density of the high molecular size band in the non-MDM2-overexpressing SH-SY5Y cells as compared to MDM2-overexpressing SH-EP1 cells (see lanes 2 and 1 in Figure 4A), which implied that MDM2 might be a component of the RNP complex formed on the XIAP IRES. Next, we performed IP and RT-PCR to search for a possible association in vivo between the MDM2 protein and the XIAP mRNA. As shown in Figure 4B, the MDM2 protein was able to bind XIAP mRNA under these conditions as well. In addition, UV cross-linking and gel electrophoresis showed that the MDM2 protein strongly bound to the XIAP IRES RNA in vitro (Figure 4C, lane 3). MDM2 did not bind to the non-IRES upstream 5′-UTR of XIAP (Figure 4C, lane 6). Consistent with a previous report (Elenbaas et al., 1996), MDM2 bound to a RNA molecule (clone A) from the SELEX procedure (Figure 4C, lane 9). As a control, the non-related recombinant protein Bcl-2 did not bind to XIAP IRES. The binding of MDM2 to XIAP IRES was further confirmed by similar UV cross-linking and gel electrophoresis using cellular extract of SH-EP1 cells and IP with anti-MDM2 antibody (Figure 4D).
Figure 4.
MDM2 protein binds to XIAP IRES. (A) Cell extracts from SH-EP1 and SH-SY5Y were incubated with 32P-labeled RNA probes corresponding to the complete XIAP IRES element (probe 1) and non-IRES upstream 5′-UTR segment (probe 2) as a control; then UV cross-linked, run on an SDS-PAGE gel and imaged by autoradiography. RNP-RNA complexes are indicated by arrows. (B) Cell extracts from SH-EP1 were prepared in RNA-binding buffer in the presence of RNase inhibitor (RNasin). Following co-IP with anti-MDM2, anti-La (positive control), and anti-actin (negative control), the XIAP mRNA was detected by RT-PCR analysis. IP without addition of RNasin and antibody and with anti-MDM2 but no RNasin were done as additional controls. The positive (SH-EP1 RNA as template, lane 7) and negative (no template, lane 8) controls for RT-PCR are also shown. (C) Recombinant human MDM2 (rhMDM2) protein binds to XIAP IRES RNA in vitro. The rhMDM2 and rhBcl-2 as an unrelated recombinant control protein were incubated, respectively, with 32P-labeled RNA probes 1 and 2 (as described in A), and with a RNA probe (probe 3), clone A from the SELEX procedure carried out by Elenbaas et al, serving as positive control. The protein/RNA complexes were UV cross-linked, run on an SDS-PAGE gel, and imaged by autoradiography. The rhMDM2 were also incubated with probes without UV exposure as additional controls. (D) Cell extracts from SH-EP1 were incubated with probe 1 and probe 2, UV cross-linked, and then immunoprecipitated with antibodies (anti-La and serum as controls) as indicated. Controls also included the absence of UV cross-linking with MDM2 antibody.
The C-terminal RING domain of MDM2 binds to the core RNP binding sequence of XIAP IRES
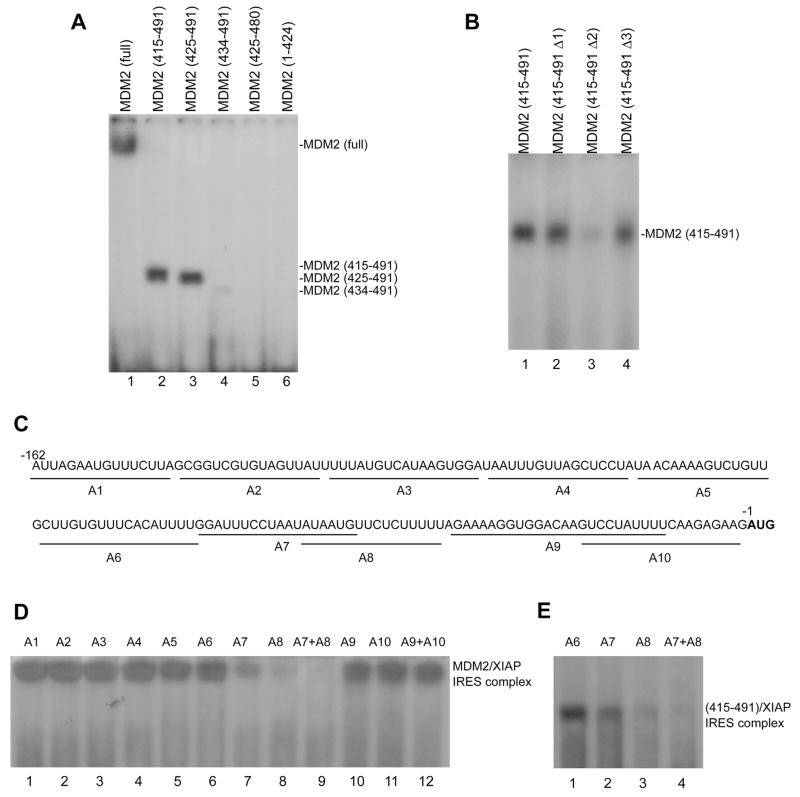
We performed UV cross-linking and gel electrophoresis to screen for the domain of MDM2 required for XIAP IRES binding. Fusion proteins of GST and full-length MDM2, or MDM2 with different deletions (415–491), (425–491), (434–491) and (425–480), as well as (1–424) were constructed and expressed in Escherichia coli. A subsequent RNA binding assay revealed that the C-terminal RING domain (425–491) but not other parts of MDM2 protein (1–424) was able to bind to XIAP IRES (Figure 5A). We found that the fragments (415–491) and (425–491) were similar to the full-length MDM2 binding to XIAP IRES. However, the binding of fragment (434–491) to the XIAP IRES was greatly reduced, suggesting that 10 amino acids (425–434) are critical for proper binding. In addition, when the last 11 hydrophobic amino acids were deleted, the binding of this fragment (425–480) to XIAP IRES was abolished, suggesting that these last 11 amino acids are also important for binding to the XIAP IRES.
Figure 5.
The C-terminal RING domain of MDM2 binds to the core RNP binding sequence of XIAP IRES. (A) GST-fused full-length MDM2 (lane 1) and different GST-fused MDM2 fragments of the C-terminal RING domain (lanes 2–5) and the fragment with deletion of C-terminal RING domain (lane 6) were incubated with 32P-labeled XIAP IRES (probe 1 in Figure 5); the formation of MDM2 XIAP IRES complexes was detected as described in Figure 5C. (B) UV cross-linking and binding assay similar to (A) for the interaction between XIAP IRES and GST-fused wt (lane 1) and different mutant (lanes 2–4) C-terminal RING domains of MDM2: MDM2 (415–491 Δ1) with a S428G mutation, MDM2 (415–491 Δ2) with a G448S mutation, MDM2 (415–491 Δ3) with a L487S mutation. (C) Mapping the MDM2-binding site within XIAP IRES. Nucleotide sequence of the 162-nt XIAP IRES (probe 1 as described in Fig. 5A) and the positions of antisense (A1–A10) used for binding site mapping are shown under the sequence. The first codon (AUG) is bold. (D) Antisense mapping of the XIAP IRES/MDM2 binding site. Indicated antisense oligonucleotides were annealed to probe 1, then incubated with full-length rhMDM2 and UV cross-linked. The MDM2/XIAP IRES complex was analyzed as described in Figure 5C. (E) Similar antisense mapping of binding within XIAP IRES to the 415–491 fragment containing the C-terminal RING domain of MDM2 protein.
Other similar UV cross-linking and gel electrophoresis experiments were performed using point mutagenesis of the MDM2 C-terminal domain to further characterize the binding of MDM2 to XIAP IRES. The G448S (G446S in mouse form, conserved as G448S in human) mutation has been previously demonstrated to abolish the RNA binding of MDM2 (Elenbaas et al., 1996). Thus, we generated this mutation in the 415–491 fragment (415–491 Δ2), and another two mutations 415–491 Δ1 with a S428G and 415–491 Δ3 with a L487S located in the first and last 10 amino acids of the C-terminal RING domain (425–491), respectively. These three mutant 415–491 fragments were fused with GST and purified. Using the three mutants, RNA binding assays showed that the G448S mutation greatly reduced the specific XIAP IRES binding of MDM2, while the 415–491 fragments with mutations S428G and L487S were still able to bind XIAP IRES (Figure 5B).
Because we found that the C-terminal RING domain of MDM2 binds to XIAP IRES, we also wished to determine what region on the XIAP IRES is critical for MDM2 binding. We performed a detailed study of the MDM2 binding site within XIAP IRES using antisense mapping. The antisenses spanning the entire region of the 162-nt XIAP IRES for mapping were shown in Figure 5C. We observed that antisense A8 (−34 to −49) containing the critical polypyrimidine tract that is essential for XIAP IRES function (Holcik et al., 2000) significantly disrupted the MDM2/XIAP IRES complex formation (Figure 5D lane 8). Antisense A7 (−44 to −62), to a lesser extent, inhibited the binding of MDM2 to XIAP IRES (Figure 5D lane 7). A combination of A8 and A7 completely blocked the binding between MDM2 and XIAP IRES (Figure 5D, lane 9). In concordance with these results, antisense mapping for testing the binding between XIAP IRES and the MDM2 fragment (415–491), resulted in binding to the same sequence from −34 to −62 (Figure 5E), a region that has been previously identified as RNP core binding site of XIAP IRES (Holcik et al., 2000).
Overexpression or inhibition of MDM2 increases or decreases resistance to IR-induced apoptosis, respectively, through regulation of XIAP
It is noted that in cancer cells treated with IR, XIAP is found to be upregulated in the resistant cells, but comparatively downregulated in the sensitive cells (Holcik et al., 2000). We further demonstrate here that the fluctuation of XIAP levels in response to IR is associated with MDM2. We found that following IR, XIAP protein levels were upregulated in MDM2-overexpressing SH-EP1 cell line resulting in resistance to apoptosis, but downregulated in non-MDM2-overexpressing line SH-SY5Y leading to apoptosis (Figure S1).
We have further evaluated the possible impact of MDM2-mediated upregulation of XIAP on IR resistance in isogenic cells. In order to rule out the p53-dependent role of MDM2 in regulating apoptosis, we used p53-null ALL cell line EU-4 (expressing very low level of MDM2) and NB cell line LA1-55N (expressing high level of MDM2) as models (Figure S1A). We first transfected MDM2 in EU-4 cells under control of Tet-on, and MDM2 expression was induced by adding doxycycline to the cell culture, accompanied by an increase in XIAP expression (Figure 6A). We tested the response to IR-induced apoptosis of cells with different levels of MDM2 expression. As shown in Figure 6B and C, cells cultured with 1 μg/ml doxycycline, which have a maximum expression of MDM2 and XIAP, become significantly resistant to IR as compared with the cells without doxycycline (p<0.01). To further confirm the role of upregulated XIAP following MDM2 induction in IR resistance, we tested the effect of IR treatment on resistant cells (with 1 μg/ml doxycycline) in the presence of XIAP siRNA. Treatment of XIAP siRNA inhibits over 90% of endogenous XIAP (Figure 6D). Apoptosis resistance in these cells is significantly reversed in the presence of XIAP siRNA but not control siRNA during the treatment with IR (Figure 6E).
In contrast, we inhibited endogenous MDM2 expression using siRNA in the MDM2-overexpressing LA1-55N, to determine whether there were differences in XIAP regulation and sensitivity to IR in these cells, with or without overexpression of MDM2. For this, we used the pSUPER/MDM2 siRNA plasmid (pSUPER MDM2 (1) in Figure 3D) to silence MDM2. Transfection of this plasmid into the LA1-55N cells suppressed the endogenous MDM2 protein (Figure 6F). We then investigated the kinetic expression of the XIAP protein following IR treatment of LA1-55N cells, transfected with either control siRNA or MDM2 siRNA (clone 3). We found that IR induced upregulation of XIAP in the LA1-55N control cells and downregulation of this protein in the LA1-55N cells transfected with MDM2 siRNA. Consequently, activation of caspases 9 and 3 occurred in the MDM2-silenced LA1-55N, but not in control cells, following IR treatment (Figure 6G). Importantly, flow-cytometry assays showed that an increased percentage of the LA1-55N/MDM2 siRNA cells treated with IR were annexin-V positive, as compared to control cells (Figure 6H).
Blockade of MDM2/XIAP IRES interaction reduces resistance to IR-induced apoptosis
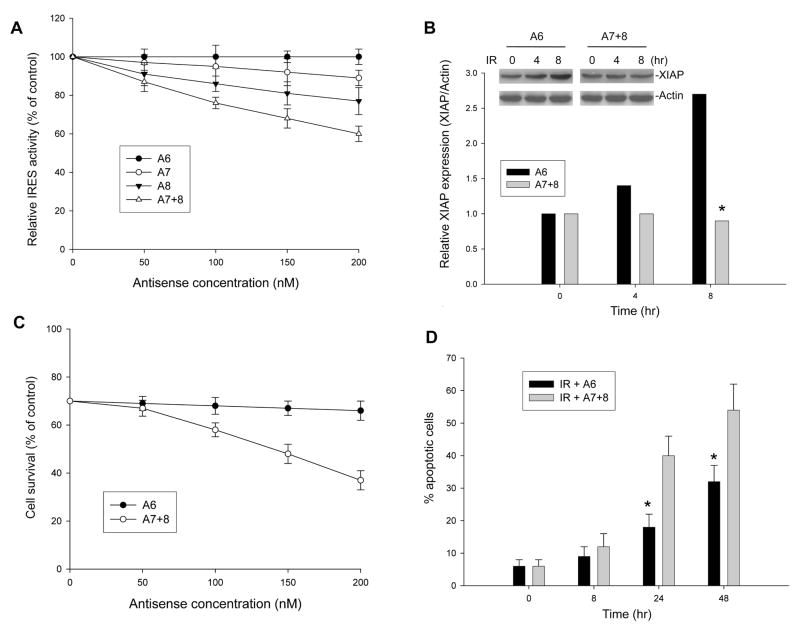
In order to convincingly demonstrate that it is the binding of MDM2 to XIAP IRES that upregulates XIAP translation leading to resistance to apoptosis, we evaluated whether block of the MDM2/XIAP IRES interaction could inhibit XIAP induction in MDM2-expressing cells following IR and reverse apoptosis resistance. The antisense A7 and A8 as shown in Figure 5C and D, which block the binding of MDM2 to XIAP IRES, were used for the assay. The antisense A6 without activity to the binding between MDM2 and XIAP IRES served as control. The antisense used for transfection is phosphorothioate form that is resistant to nuclease digestion in cells. Transfection and reporter assays were performed in p53-null/MDM2-overexpressing LA1-5S cells using pβgal-xi-CAT plasmid in the presence or absence of the antisenses. We found that A7 and A8 reduced XIAP IRES-mediated CAT translation by approximately 10% and 20%, respectively, if provided at a concentration of 200 nM. When the combination of A7 and A8 (A7+8), which completely blocks the binding between MDM2 and XIAP IRES (Figure 5D and E), was used, an approximately 40% reduction of CAT activity was achieved. At the same concentration, A6 did not inhibit XIAP IRES-mediated CAT activity (Figure 7A). In addition, we tested the effect of the combination of antisense A7+8 on expression of endogenous XIAP in response to IR treatment. When A7+8 were transfected into LA1-5S cells, they also significantly inhibited the IR-induced upregulation of endogenous XIAP, as compared to the control (A6) antisense (Figure 7B). Consequently, obvious reverse of resistance occurred in the LA1-5S cells treated with A7+8, but not in the same cells treated with A6, following IR exposure. As shown in Figure 7C, there was a significant reduced survival of LA1-5S cells following IR treatment in the presence of A7+8 at concentrations of 100nM and greater (p<0.01) compared to that in the presence of A6, as detected by WST assay. Consistent with these observations, a flow-cytometric apoptosis assay showed that a increased percentage of LA1-5S cells treated with IR plus A7+8 were annexin-V positive at 24-h post-treatment as compared to IR plus A6 (Figure 7D).
Figure 7.
Effect of blockage between MDM2 and XIAP IRES interaction on XIAP expression and IR-induced apoptosis. (A) LA1-5S cells were co-transfected with 5μg pβgal-xi-CAT plasmid and increasing amounts of antisense (A6, A7, A8 and A7+8 as shown in Fig. 5C). The % of CAT activity (mean ± SD, calculated by normalizing with βgal activity) was given relative to a translation reaction performed in the absence of antisense. (B) The effect of antisense (A6 and A7+8) on IR-induced upregulation of endogenous XIAP in LA1-5S cells. Cells transfected with 200 nM A6 or A7+8 were exposed to IR for the indicated time. The expression levels of XIAP, using Actin as a control, were analyzed by western blotting (insert). Data in the graph represent the relative expression of XIAP as compared to Actin after this western blot was densitometrically scanned. *p<0.01. (C) LA1-5S cells were treated with 10 Gy IR in the presence or absence of different dose of A6 or A7+8 as indicated. Cells were incubated for 48 h, and cell viability was determined by WST assay. Data represent the mean percentage (±SD) of cell survival from three independent experiments. (D) Time-course of apoptosis induced by IR in LA1-5S cells in the presence of A6 or A7+8 antisense (200 nM). Cells were treated with 10 Gy IR for the indicated time points, and apoptotic cells were detected by annexin-V staining using flow cytometry. Data represent the mean percentage of annexin-V positive cells from three independent experiments; bars, ± SD. *p<0.01.
Discussion
It is noted that overexpression of MDM2 independently of p53 is involved in chemoresistance and tumor progression. However, the mechanisms underlying all these activities are presently not well defined. For example, it is not clear why many alternatively spliced forms of MDM2 lacking the N-terminal p53 binding domain are overexpressed in a number of human cancers, nor exactly how transfection of p53-negative cells with these MDM2 variants results in tumor promotion (Bartel et al., 2002; Fridman et al., 2003). Several studies have noted that the C-terminal RING domain of MDM2 regulates cell proliferation via a p53-independent pathway (Argentini et al., 2000; Gu et al., 2003). Here we show that the RING finger domain of MDM2 interacted with the XIAP IRES mRNA. Importantly, cytoplasmic redistribution of MDM2, which is modulated by IR, causes a significant increase in XIAP protein levels in MDM2-overexpressing cancer cells, resulting in resistance to apoptosis. These results provide a mechanism by which MDM2 plays a p53-independent role in regulating expression of the anti-apoptotic factor XIAP, which may be involved in resistance to cancer treatment and disease progression.
A quite recent study showed that the RING finger domain of MDM2 binds to p53 mRNA in vivo, which increases p53 synthesis (Candeias et al., 2008). Our results from the present study indicate that MDM2 binds to the XIAP IRES mRNA in a manner similar to binding to p53 mRNA as well as the RNA molecule, clone A, from the SELEX procedure (Elenbaas et al., 1996): The amino acids 425–491 of the RING domain of MDM2 are definitely necessary and sufficient for XIAP IRES binding and the single point mutation G448S abolished that binding. We have further demonstrated that deletions of the first 10 amino acids or the last 11 amino acids of the RING domain of MDM2 disturb binding, suggesting that the entire RING domain of MDM2 is necessary for XIAP IRES binding to occur. This is also consistent with the results reported by Lai et al (Lai et al., 1998), showing that the first 10 amino acids and the last 11 amino acids of the RING domain of MDM2 are critical for binding to the clone A RNA and a fragment (328–406) of human ribosomal RNA. Interestingly, in comparing the nucleotide sequences between clone A RNA and the MDM2-binding region of XIAP IRES, no similar consensus sequences were identified in these two RNAs. However, the predicted secondary structure of the MDM2-binding region within XIAP IRES was similar to that of clone A RNA. The structure of clone A RNA consists of a 3′ stem-loop (6 bp) and a 5′ stem-loop (9 bp) that are connected with a large single-stranded region of 13 nt (Elenbaas et al., 1996). A recent study using enzymatic probing with RNase T1, RNase T2, and RNase V1 proposed the XIAP IRES RNA secondary structure model (Baird et al., 2007). An analogous structure to clone A RNA was found in the MDM2-binding region of XIAP IRES that consists of a 3′ stem-loop (6 bp) connecting with a large single-stranded region of 14 nt and a 5′ 9-bp incomplete stem-loop (2 bp mismatches). Binding of MDM2 to both clone A RNA and the XIAP IRES suggests that the specific secondary structure, but not the primary nucleotide sequence, is critical for MDM2 binding.
Importantly, we have demonstrated that cytoplasmic relocalization of MDM2 is required to regulate XIAP translation. MDM2 is a nuclear phosphoprotein that is constitutively phosphorylated under normal growth conditions, but becomes hypophosphorylated in response to IR (Meek et al., 2003; Blattner et al., 2002; Okamoto et al., 2002). Our results indicate that IR-induced hypophosphorylation (dephosphorylation) of MDM2 led to the translocation of this protein from the nucleus to the cytoplasm. In a previous report, it has been shown that the basal levels of XIAP protein in growing/unstressed cells are similar (Holcik et al., 2000). Consistent with this observation, our results showed that 5 of 6 cultured cancer cell lines studied expressed similar level of XIAP, no matter whether the cells expressed high or low levels of MDM2. This suggested that phosphorylated MDM2 in the nucleus under normal growth conditions is unable to regulate XIAP translation; thus, that MDM2 overexpression in cultured or growing cells does not necessarily relate with high levels of XIAP. However, it was IR-induced dephosphorylation of MDM2 and its movement to the cytoplasm that influenced XIAP. We have not only demonstrated that upregulation of XIAP translation following IR occurs in MDM2-overexpressing cells, but have also shown that transfection of a mutant MDM2 that localizes in the cytoplasm significantly increased XIAP IRES activity. Our findings support the notion that the subcellular compartmentalization of ITAFs controls their trans-acting activity (Lewis et al., 2007).
The XIAP mRNA has both a short and long form of 5′-UTR, suggesting that XIAP translation is most likely regulated by both cap-dependent (use of short 5′-UTR) and cap-independent (use of long 5′-UTR containing IRES) mechanisms. Previous studies demonstrate that during induced cellular stresses such as exposure to IR, cap-dependent translation is rapidly inhibited (Sheikn et al., 1999; Spriggs et al., 2005); while cap-independent (IRES-dependent) translation can persist for certain anti-apoptotic genes, including XIAP (Stoneley et al., 2004; Hellen et al., 2001). Because the cap-independent translation of these anti-apoptotic genes under stress stimulation might delay apoptosis, allowing time for the cells to repair any damage that has incurred, it could lead to the development of resistance to apoptosis. Because MDM2 is a stress-responsive protein, the MDM2-mediated IRES-dependent translation of XIAP may be critical for resistance to radiation therapy.
It is well known that genotoxic stress such as exposure to IR activates p53, which regulates the transcription of many genes controlling cell cycle checkpoints and apoptosis. Less study has been done to evaluate genotoxic stress-regulated protein translation, which also forms an important component of the total cellular stress response (Kaufman et al., 1999). Previous studies have claimed that the IRES-mediated translation of certain genes (such as XIAP and HIAP2) following IR confers resistance of cells to this stimulation (Lewis et al., 2005). However, the mechanisms by which IRES-mediated translation contributes to IR-induced resistance are unknown. Many efforts have been made to discover the mediators involved in the modification of IRES-regulated translation in cells. In the case of XIAP, the ITAF/RNP (La, hnRNP C1/C2, and hnRNP A1) have been shown to modify XIAP IRES function (Holcik et al., 2000; Holcik et al., 2003; Lewis et al., 2007); however, the association of this modification with cell stress-induced signaling is as yet unclear. Results from the present study show that stress-induced signaling regulated cytoplasmic translocation of MDM2, which subsequently induced XIAP IRES activity, resulting in upregulation of XIAP protein, conferring resistance to apoptosis. The mechanistic insights generated in this study would suggest a functional role for MDM2 in the regulation of XIAP IRES-dependent translation during cellular stress and DNA damage, extending our current understanding of resistance to radiotherapy in cancer.
Experimental Procedures
Cells and plasmids
Four human NB cell lines (SH-EP1, SH-SY5Y, LA1-55N and LA1-5S) and two human leukemia cell lines (EU-1 and EU-4) were used in this study. The plasmids MDM2/166A or MDM2/166E were constructed by inserting the MDM2 cDNA with mutations at serine 166 to either alanine (166A) or glutamic acid (166E) into the pDsRed1-C1 vector. The wt and various C-terminal truncated and mutated GST-tagged MDM2 constructs were generated by PCR and cloned into the bacterial pGEX expression vector. The Tet-On gene expression systems including pTet-On and its response (pTRE2hyg) plasmids were purchased from Clontech (Palo Alto, CA). The human MDM2 cDNA was cloned into the pTRE2hyg at the Bam H1 restriction site. The pSUPER/MDM2 siRNA plasmids were constructed by inserting several specific 19 nt MDM2 sequence into an expression plasmid, pSUPER-neo, purchased from OligoEngine (Seattle, WA). The monocistronic plasmid pGL3-xp-xi was constructed by first inserting the XIAP promoter (xp), which is a 1 kb DNA sequence 5′-flank immediately upstream of XIAP mRNA (Figure S1B), into the pGL3-basic vector, then inserting a 670 bp cDNA fragment from the 5′UTR of XIAP containing the 162-bp XIAP IRES (xi) immediately downstream of xp. The bicistronic vector pβgal/CAT (containing the XIAP IRES in either sense or antisense orientations) was kindly provided by Dr. M. Holcik (University of Ottawa, Canada). The dicistronic vector pRL-xi-FL was kindly provided by Dr. Richard E. Lloyd (Baylor College of Medicine).
Gene transfection and reporter assay
To establish an MDM2-inducible model, both pTet-On and pTRE2hyg-MDM2 plasmids were stably transfected into EU-4 cells by electroporation to generate a cell line EU-4/Tet-On/MDM2, where MDM2 expression was induced by addition of doxycycline in the cell culture. For stable MDM2 siRNA transfection, LA1-55N cells were transfected with pSUPER/MDM2 or pSUPER containing a scrambled 19-nt (control) plasmid as described in detail online.
Transient transfection was performed to examine the effect of MDM2 on XIAP IRES activity. Cells were co-transfected with various MDM2 expression plasmids and either monocistronic or bicistronic and dicistronic XIAP IRES reporter plasmids. The subcellular distribution of transfected MDM2 in pDsRed1-C1 vector was detected by confocal microscopy. Luciferase activities were analysed by Luciferase Assay Reagent II (Promega) and a Microplate Luminometer (Turner Designs). In the transfection of cells with bicistronic plasmid, β-Galactosidase (βgal) enzymatic activity was determined by the chemiliminescent reporter gene assay system (Applied Biosystems, Bedford, Massachusetts). CAT levels were determined using the CAT ELISA kit, according to the protocol provided by the manufacturer (Roche Molecular Biochemicals, Indianapolis, IN).
UV cross-linking and RNA binding assays
UV cross-linking and immunoprecipitation assays were performed as described previously (Holcik et al., 2000). Briefly, the DNA templates for synthesis of the XIAP IRES RNA probe (probe 1) and a control RNA probe (probe 2) from non-IRES upstream 5′-UTR of XIAP mRNA were generated by PCR using XIAP IRES-specific primer pairs. A previously identified MDM2-binding RNA, clone A RNA (Elenbaas et al., 1996), (probe 3) was used as a positive control. Internally labeled RNA probes were synthesized by in vitro transcription with T7 polymerase (MAXIScript T7 RNA polymerase kit, Ambion) in the presence of [α-32P] UTP (Amersham). The cell extracts or purified MDM2 proteins were mixed with 32P-labeled RNA probes. UV cross-linking of the RNA-protein complexes was performed and then resolved by 10% SDS-PAGE gel and visualized by autoradiography.
XIAP mRNA was co-immunoprecipitated with MDM2 antibody from whole-cell extracts by using the modified method, as described previously (Seto et al., 1999) to detect the in vivo binding of MDM2 protein and XIAP mRNA (see detailed description in the supplemental data online).
Antisense mapping and treatment
Mapping of the MDM2 binding site within the XIAP IRES by antisense oligonucleotides was performed as described previously (Thomson et al., 1999). Briefly, the internally 32P-labeled 162-nt XIAP IRES probe (probe 1) was mixed with various antisense oligonucleotides spanning the entire 162-nt region. The mixtures were incubated at 80°C for 10 min, and allowed to cool gradually to room temperature. The RNA probe-oligonucleotide hybrids were then incubated either with rhMDM2 protein or with the GST-fused C-terminal RING domain of MDM2 and analyzed. For antisense treatment in cells, 50 to 200 nM phosphorothioate oligodenucleotides antisenses targeting the XIAP IRES were delivered in the form of complex with Lipofectamine (Invitrogen).
Metabolic labeling, pulse-chase assay and cycloheximide treatment
The protein synthesis and the half-life of XIAP were detected by pulse-chase experiments, metabolic labeling, immunoprecipitation and cycloheximide treatment as described in the supplemental data online.
Polyribosome profile analysis
Linear sucrose gradient fractionation was carried out as described previously (Feng et al., 1997). Briefly, cells treated with IR or transfected with MDM2-166A or mock-treated were lysed to isolate cytoplasmic extracts, followed by fractionation on a 15–45% (w/v) linear sucrose gradient. After centrifugation at 39,000rpm in a SW41Ti rotor for 1 hr, eleven 1 ml fractions were collected from each gradient by upward replacement and the RNA in each fraction was extracted and subjected to Real-Time PCR as described in detail in the supplemental data online.
Supplementary Material
Acknowledgments
We are most grateful to Dr. Martin Holcik and Dr. Richard E. Lloyd for providing us with the bicistronic plasmid pβgal-XIAP-CAT and dicistronic plasmid pRL-XIAP-FL, respectively. This work was supported by grants from NIH (R01 CA123490), The Leukemia and Lymphoma Society (6033-08), and CURE.
Footnotes
Supplemental data
The supplemental data include three figures, supplemental experimental procedures and supplemental references, and can be found with this article online at http//www.cancercell.org
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Argentini M, Barboule N, Wasylyk B. The contribution of the RING finger domain of MDM2 to cell cycle progression. Oncogene. 2000;19:3849–3857. doi: 10.1038/sj.onc.1203737. [DOI] [PubMed] [Google Scholar]
- Bartel F, Taubert H, Harris LC. Alternative and aberrant splicing of MDM2 mRNA in human cancer. Cancer Cell. 2002;2:9–15. doi: 10.1016/s1535-6108(02)00091-0. [DOI] [PubMed] [Google Scholar]
- Blattner C, Hay T, Meek DW, Lane DP. Hypophosphorylation of Mdm2 augments p53 stability. Mol Cell Biol. 2002;22:6170–6182. doi: 10.1128/MCB.22.17.6170-6182.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnal S, Boutonnet C, Prado-Lourenco L, Vagner S. IRESdb: the Internal Ribosome Entry Site database. Nucleic Acids Res. 2003;31:427–428. doi: 10.1093/nar/gkg003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candeias MM, Malbert-Colas L, Powell DJ, Daskalogianni C, Maslon MM, Naski N, Bourougaa K, Calvo F, Fåhraeus R. p53 mRNA controls p53 activity by managing Mdm2 functions. Nat Cell Biol. 2008;10:1098–1105. doi: 10.1038/ncb1770. [DOI] [PubMed] [Google Scholar]
- Eckelman BP, Salvesen GS, Scott FL. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 2006;7:988–994. doi: 10.1038/sj.embor.7400795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elenbaas B, Dobbelstein M, Roth J, Shenk T, Levine AJ. The MDM2 oncoprotein binds specifically to RNA through its RING finger domain. Mol Med. 1996;2:439–451. [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- Fridman JS, Hernando E, Hemann MT, de Stanchina E, Cordon-Cardo C, Lowe SW. Tumor promotion by Mdm2 splice variants unable to bind p53. Cancer Res. 2003;63:5703–5706. [PubMed] [Google Scholar]
- Green DR. Apoptotic pathways: ten minutes to dead. Cell. 2005;121:671–674. doi: 10.1016/j.cell.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Gu L, Findley HW, Zhou M. MDM2 induces NF-kB/p65 expression transcriptionally through Sp1-binding sites: A novel, p53-independent role of MDM2 in adriamycin resistance in acute lymphoblastic leukemia. Blood. 2002;99:3367–3375. doi: 10.1182/blood.v99.9.3367. [DOI] [PubMed] [Google Scholar]
- Gu L, Ying H, Zheng H, Murray SA, Xiao ZX. The MDM2 RING finger is required for cell cycle-dependent regulation of its protein expression. FEBS Lett. 2003;544:218–222. doi: 10.1016/s0014-5793(03)00502-7. [DOI] [PubMed] [Google Scholar]
- Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Holcik M, Lefebvre C, Yeh C, Chow T, Korneluk RG. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat Cell Biol. 1999;1:190–192. doi: 10.1038/11109. [DOI] [PubMed] [Google Scholar]
- Holcik M, Korneluk RG. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol Cell Biol. 2000a;20:4648–4657. doi: 10.1128/mcb.20.13.4648-4657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M, Yeh C, Korneluk RG, Chow T. Translational upregulation of X-linked inhibitor of apoptosis (XIAP) increases resistance to radiation induced cell death. Oncogene. 2000b;19:4174–4177. doi: 10.1038/sj.onc.1203765. [DOI] [PubMed] [Google Scholar]
- Holcik M, Gordon BW, Korneluk RG. The internal ribosome entry site-mediated translation of antiapoptotic protein XIAP is modulated by the heterogeneous nuclear ribonucleoproteins C1 and C2. Mol Cell Biol. 2003;23:280–288. doi: 10.1128/MCB.23.1.280-288.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M, Graber T, Lewis SM, Lefebvre CA, Lacasse E, Baird S. Spurious splicing within the XIAP 5′ UTR occurs in the Rluc/Fluc but not the betagal/CAT bicistronic reporter system. RNA. 2005;11:1605–1609. doi: 10.1261/rna.2158605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- Komar AA, Hatzoglou M. Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J Biol Chem. 2005;280:23425–23428. doi: 10.1074/jbc.R400041200. [DOI] [PubMed] [Google Scholar]
- Kuida K, Haydar TF, Kuan CY, Gu Y, Taya C, Karasuyama H, Su MS, Rakic P, Flavell RA. Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell. 1998;94:325–337. doi: 10.1016/s0092-8674(00)81476-2. [DOI] [PubMed] [Google Scholar]
- Lai Z, Freedman DA, Levine AJ, McLendon GL. Metal and RNA binding properties of the hdm2 RING finger domain. Biochemistry. 1998;37:7005–7015. doi: 10.1021/bi980596r. [DOI] [PubMed] [Google Scholar]
- Lakhani SA, Masud A, Kuida K, Porter GA, Jr, Booth CJ, Mehal WZ, Inayat I, Flavell RA. Caspases 3 and 7: key mediators of mitochondrial events of apoptosis. Science. 2006;311:847–851. doi: 10.1126/science.1115035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SM, Holcik M. IRES in distress: translational regulation of the inhibitor of apoptosis proteins XIAP and HIAP2 during cell stress. Cell Death Differ. 2005;12:547–553. doi: 10.1038/sj.cdd.4401602. [DOI] [PubMed] [Google Scholar]
- Lewis SM, Veyrier A, Hosszu UN, Bonnal S, Vagner S, Holcik M. Subcellular relocalization of a trans-acting factor regulates XIAP IRES-dependent translation. Mol Biol Cell. 2007;18:1302–1311. doi: 10.1091/mbc.E06-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston P, Roy N, Tamai K, Lefebvre C, Baird S, Cherton-Horvat G, Farahani R, McLean M, Ikeda JE, MacKenzie A, Korneluk RG. Suppression of apoptosis in mammalian cells by NAIP and a related family of IAP genes. Nature. 1996;379:349–353. doi: 10.1038/379349a0. [DOI] [PubMed] [Google Scholar]
- Lü X, de la Peña L, Barker C, Camphausen K, Tofilon PJ. Radiation-induced changes in gene expression involve recruitment of existing messenger RNAs to and away from polysomes. Cancer Res. 2006;66:1052–1061. doi: 10.1158/0008-5472.CAN-05-3459. [DOI] [PubMed] [Google Scholar]
- Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci USA. 2001;98:11598–11603. doi: 10.1073/pnas.181181198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek DW, Knippschild U. Posttranslational modification of MDM2. Mol Cancer Res. 2003;1:1017–1026. [PubMed] [Google Scholar]
- Okamoto K, Li H, Jensen MR, Zhang T, Taya Y, Thorgeirsson SS, Prives C. Cyclin G recruits PP2A to dephosphorylate Mdm2. Mol Cell. 2002;9:761–771. doi: 10.1016/s1097-2765(02)00504-x. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Renatus M, Schwarzenbacher R, Zhou Q, Sun C, Fesik SW, Liddington RC, Salvesen GS. Structural basis for the inhibition of caspase-3 by XIAP. Cell. 2001;104:791–800. doi: 10.1016/s0092-8674(01)00274-4. [DOI] [PubMed] [Google Scholar]
- Schimmer AD. Inhibitor of apoptosis proteins: translating basic knowledge into clinical practice. Cancer Res. 2004;64:7183–7190. doi: 10.1158/0008-5472.CAN-04-1918. [DOI] [PubMed] [Google Scholar]
- Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- Sheikh MS, Fornace AJ., Jr Regulation of translation initiation following stress. Oncogene. 1999;18:6121–6128. doi: 10.1038/sj.onc.1203131. [DOI] [PubMed] [Google Scholar]
- Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, Alnemri ES, Fairman R, Shi Y. Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell. 2003;11:519–527. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Mitchell SA, Willis AE. Internal ribosome entry segment-mediated translation during apoptosis: the role of IRES-trans-acting factors. Cell Death Differ. 2005;12:585–591. doi: 10.1038/sj.cdd.4401642. [DOI] [PubMed] [Google Scholar]
- Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- Thomson AM, Rogers JT, Walker CE, Staton JM, Leedman PJ. Optimized RNA gel-shift and UV cross-linking assays for characterization of cytoplasmic RNA-protein interactions. Biotechniques. 1999;27:1032–1042. doi: 10.2144/99275rr03. [DOI] [PubMed] [Google Scholar]
- Van Eden ME, Byrd MP, Sherrill KW, Lloyd RE. Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA. 2004;10:720–730. doi: 10.1261/rna.5225204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao ZX, Chen J, Levine AJ, Modjtahedi N, Xing J, Sellers WR, Livingston DM. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature. 1995;375:694–698. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wang H, Li M, Rayburn ER, Agrawal S, Zhang R. Stabilization of E2F1 protein by MDM2 through the E2F1 ubiquitination pathway. Oncogene. 2005;24:7238–7247. doi: 10.1038/sj.onc.1208814. [DOI] [PubMed] [Google Scholar]
- Zhou M, Gu L, Findley HW, Jiang R, Woods WG. PTEN reverses MDM2-mediated chemotherapy resistance by interacting with p53 in acute lymphoblastic leukemia cells. Cancer Res. 2003;63:6357–6362. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.