Abstract
Recent progress in sequencing the genomes of several Leishmania species, causative agents of cutaneous, mucocutaneous and visceral leishmaniasis, is revealing unusual features of potential relevance to parasite virulence and pathogenesis in the host. While the genomes of Leishmania major, Leishmania braziliensis and Leishmania infantum are highly similar in content and organisation, species-specific genes and mechanisms distinguish one from another. In particular, the presence of retrotransposons and the components of a putative RNA interference machinery in L. braziliensis suggest the potential for both greater diversity and more tractable experimentation in this Leishmania Viannia species.
Keywords: Leishmania, Comparative genomics, Disease phenotype, Retrotransposons, RNAi
1. Introduction
Species of the genus Leishmania cause a diverse spectrum of infectious diseases, the leishmaniases, in tropical and subtropical regions of the world (reviewed in Murray et al., 2005). Belonging to the order Kinetoplastida and the family Trypanosomatidae, mammalian-infective species of the genus can be divided into two subgenera, Leishmania (Leishmania) and Leishmania (Viannia), which differ principally in the site of parasite development within the female sandfly vector (reviewed in Sacks and Kamhawi, 2001). Once flagellated metacyclic promastigote stages are transmitted from vector to mammalian host, parasites enter resident tissue macrophages and transform into aflagellated replicative amastigotes, often inducing immuno-inflammatory responses and persistent tissue infection. The fate of these intracellular parasites determines disease type, which can range from cutaneous or mucocutaneous infection to diffuse cutaneous or visceral disease. The principal factors contributing to disease outcome are the infecting parasite species and immunological status of the host, including a well-established role for host genetic variation (reviewed in Lipoldova and Demant, 2006). Attributing the relative importance of these factors to disease outcome in humans is a complex area, however, particularly given the overlap in pathology associated with infection with different Leishmania species (Guerbouj et al., 2001;McMahon-Pratt and Alexander, 2004; Wilson et al., 2005; BenSaid et al., 2006). While multiple species cause cutaneous disease (see Table 1) and only three are usually considered as visceralising types (Leishmania donovani, Leishmania infantum and Leishmania chagasi), cutaneous lesions attributed to these latter species have also been reported (Gramiccia et al., 1995; Noyes et al., 1997; Gumy et al., 2004; BenSaid et al., 2006), while isolated cases of visceral disease due to cutaneous species have been recorded as well (Barral et al., 1991). Variable disease phenotype has also been observed in naturally and experimentally infected animals. Teasing out the relative importance of all contributing factors to clinical disease is a challenging objective.
Table 1.
Human-infective species of the Leishmania genus
| Old World species | New World species | Disease type |
|---|---|---|
| L. major complex | L. mexicana complex | Cutaneous |
| L. (L.) major | L. (L.) mexicana | |
| L. (L.) tropica | L. (L.) amazonensis | |
| L. (L.) aethiopica | L. (L.) pifanoi | |
| L. (L.) venezuelensis | ||
| L. (Viannia) subgenus | ||
| L. (V.) braziliensis | ||
| L. (V.) panamensis | ||
| L. (V.) guyanensis | ||
| L. (V.) peruviana | ||
| L. (V.) lansoni | ||
| L. (V.) braziliensis | Mucocutaneous | |
| L. (L.) aethiopica | L. (L.) amazonensis | Diffuse cutaneous |
| L. (L.) pifanoi | ||
| L. donovani complex | Visceral | |
| L. (L.) donovani | ||
| L. (L.) infantum⁎ | L. (L.) chagasi⁎ |
The main species complexes and subgenus are shown in bold; the species with complete genome sequences either available or being generated are underlined.
Species that can also be associated with cutaneous leishmaniasis.
A number of parasite factors affecting pathogenesis in the host have already been well-characterised. These include the surface zinc metalloprotease GP63 (or leishmanolysin) which protects promastigotes against host lysis by cleavage of complement, enhances phagocytosis and has recently been shown to protect against antimicrobial peptide-induced apoptotic killing (reviewed in Yao et al., 2005; Kulkarni et al., 2006); several parasite-derived cysteine proteases which can function as virulence factors (Mottram et al., 2004) and the complex glycoconjugates, lipophosphoglycan and the proteophosphoglycans, which promote parasite survival in the vector and host by a range of mechanisms (reviewed by Beverley and Turco, 1998; Ilg, 2000; Sacks and Kamhawi, 2001; Turco et al., 2001; Descoteaux and Turco, 2002; Lodge and Descoteaux, 2005).
Sequencing the genomes of representative parasite species provides a global framework within which to investigate the contribution of these and other parasite factors to the diverse forms of leishmaniasis and address the question: what is the contribution of parasite genotype to disease phenotype?
2. Sequencing the Leishmania genomes: which species?
The decision to initiate sequencing programmes for three kinetoplastid organisms, Leishmania, Trypanosoma brucei and Trypanosoma cruzi (the Tritryp genome projects), was made in principal at a Oswaldo Cruz Foundation (FIOCRUZ) – World Health Organization Special Programme for Research and Training in Tropical Diseases (WHO/TDR) Parasite Genome Network Planning Meeting in Rio de Janeiro in 1994. Although the available technology for completion of such large genomes was lacking at that time, these projects were considered feasible given the organisation of a committed consortium of participating laboratories, sufficient funding and anticipated advances in sequencing technology. Thirteen years later, three completed Leishmania genomes (Leishmania major, L. infantum, Leishmania braziliensis) are in the public domain (Ivens et al., 2005; Peacock et al., 2007), with sequencing of a fourth (Leishmania mexicana) in progress (http://www.genedb.org/). These first four species were chosen to represent the main species complexes of the L. Leishmania subgenus together with the best-characterised species of the L. Viannia subgenus. The strains selected were all infective, allowing study of host responses in suitable rodent models in vivo, and adapted for maintenance and manipulation in the laboratory (Laurentino et al., 2004; Ivens et al., 2005; Denise et al., 2006). Importantly, the three completed genomes represent species that usually give rise to distinct disease types (Table 1).
Leishmania major was the first species to be sequenced and has provided the model for subsequent genomic analyses. L. major is a causative agent of cutaneous leishmaniasis (CL), the most common disease form that is usually self-healing in humans but leaves unsightly scars in those affected (Table 1). Widespread in Africa and Asia, CL caused by L. major is predominantly a zoonotic infection, with parasites maintained in rodent reservoir hosts following transmission by phlebotomine sandflies. In the laboratory, L. major has been one of the principal experimental species used to dissect the roles of TH1/TH2 cells in response to infection in susceptible and resistant hosts (reviewed in Sacks and Noben-Trauth, 2002).
The case for sequencing a visceralising species of Leishmania as the second genome to be analysed was overwhelming: visceral leishmaniasis (VL) is the most serious disease form and frequently fatal if left untreated. Closely related species of the L. donovani complex are found in different geographical regions: L. donovani is the primary cause of VL in the Indian subcontinent and East Africa, L. infantum in the Mediterranean region and L. chagasi in the New World (Table 1). Humans are the only known reservoir of L. donovani while canines, especially domestic and stray dogs, provide the reservoir hosts for L. infantum and L. chagasi. While the last two species are generally considered to be genetically identical (Mauricio et al., 2000), all three species are very similar as demonstrated in recent comprehensive phylogenetic studies using both non-coding and protein-coding sequences (Kuhls et al., 2005, 2007; Mauricio et al., 2006; Lukes et al., 2007). The choice of a prevalent L. infantum strain for genome sequencing was made on the basis of its virulence in animals, transmissibility in sandflies and adaptability to laboratory experimentation (Denise et al., 2006).
The New World species L. braziliensis, within the subgenus L. (Viannia), is the third and most divergent species sequenced to date – and the principal cause of CL and mucocutaneous leishmaniasis (MCL) in tropical America. Originally associated with close contact with the forest environment, it is now accepted that L. braziliensis transmission may occur in peri-domestic and domestic habitats. More recently, studies on the reservoir hosts strongly suggest that a number of rodent species are involved, including domestic rats (Grimaldi and Tesh, 1993; Brandao-Filho et al., 1994; Oliveira et al., 2005).
The most severe pathology associated with this species is the disease affecting mucous membranes, mainly the anterior nasal region and occasionally the pharynx and larynx, which can result in destruction of the cartilagenous tissue. Most frequently, mucosal disease occurs after the appearance of cutaneous lesions and its diagnosis may happen months or years after treatment of the primary lesion (Zajtchuk et al., 1989). However, recent clinical investigation indicates that mucosal infection may be present, but not diagnosed, concomitantly with the primary cutaneous lesion (Boaventura et al., 2006). This is a relevant issue because the patient usually does not recognise MCL until partial destruction of the nose mucosa and/or cartilage occurs. Treatment of such cases is often difficult and death may occur as a result of uncontrollable secondary infections.
3. The L. major genome
The L. major genome is composed of both nuclear and independently replicating mitochondrial or kinetoplast DNA. The structure and function of kinetoplastid DNA have been reviewed recently (Lukes et al., 2005) and will not be discussed further here.
Prior to publication of the Tritryp kinetoplastid genomes in 2005, relatively little was known about the overall physical architecture of kinetoplastid chromosomes. However, earlier studies on antigenic variation in T. brucei had defined telomeric expression sites for variant surface glycoprotein (VSG) as polycistronic transcription units (reviewed in Pays et al., 1994), while work on RNA processing mechanisms in T. brucei and Leishmania had demonstrated linkage between 3′-polyadenylation sites and 5′-trans-splicing of adjacent genes, suggesting a coupled mechanism that could be processive along a chromosome (LeBowitz et al., 1993; Ullu et al., 1993; Matthews et al., 1994). The first whole chromosome sequence of L. major (Myler et al., 1999) revealed an unusual pattern of gene distribution, consistent with the polycistronic transcription model. Of the 79 identified genes, 29 were present as a contiguous unit on one DNA strand with the remaining 50 as a second unit on the opposite strand. Further chromosome sequencing revealed that the genes on other L. major chromosomes are similarly aligned in long arrays that appear to be transcribed as a single unit (Martinez-Calvillo et al., 2003; see Fig. 1) prior to trans-splicing and polyadenylation. These directional gene clusters (DGCs) range in size from a few to hundreds of genes stretching over 1 Mb of DNA. DGCs are separated by AT-rich strand switch regions (SSRs) believed to contain sites for transcription initiation and termination (Martinez-Calvillo et al., 2004). Whole genome microarray experiments have recently confirmed that most genes are constitutively transcribed in Leishmania (Holzer et al., 2006; Leifso et al., 2007). Differential gene expression between different life cycle stages is subsequently mediated by downstream processing events affecting mRNA stability and translation (Boucher et al., 2002; Myung et al., 2002).
Fig. 1.
The structure of a typical Leishmania chromosome: chromosome 2 of Leishmania major. The location and coding strands of the 74 protein-coding genes are shown as coloured boxes, coded according to the categories indicated below. The majority are genes conserved with other eukaryotes; distribution of the Leishmania-specific and scβ-galactosyltransferase (SCG) repeat family genes are also shown, together with three pseudogenes. Other chromosomal features include the telomeric hexamer repeats, the spliced leader tandem array and the ribosomal mobile element (RIME) degenerate LTR transposons.
The DGC type of chromosomal organization is remarkably well-conserved in all the kinetoplastids sequenced to date, with most of the L. major chromosomes being completely syntenic with corresponding regions in the larger T. brucei chromosomes (El-Sayed et al., 2005a), despite separation of these two species by at least 200 million years of evolution (Stevens et al., 2001). DGCs do not contain clusters of genes of related function similar to the organisation of prokaryotic operons, however.
In contrast to the high conservation of DGC gene order shared by L. major and the other Tritryps, organization of the L. major chromosome ends is distinctive, with short and highly conserved subtelomeric regions bordering distinctive repeat sequences that precede heterogeneous telomeres (Ivens et al., 2005). Interestingly, there are no large subtelomeric multigene families encoding proteins involved in host immune evasion – these are features not only of the T. brucei and T. cruzi genomes but also of other protozoan pathogen genomes such as those of Plasmodium spp. (Hall et al., 2002). Associated with this lack of telomeric repeated genes is a lack of transposable elements in the L. major genome (Bringaud et al., 2006) – these mobile sequence units are associated with gene re-arrangements in the subtelomeric regions of Trypanosoma spp. (Berriman et al., 2005).
4. The L. major genome – gene content
The 33 Mb genome of L. major is predicted to contain ∼8,300 genes, a figure likely to be accurate given the distinct codon bias in the coding regions and the rarity of cis-splicing in this species, features that make gene prediction reliable (Ivens et al., 2005; Peacock et al., 2007). Within this gene complement are a number of tandemly arrayed gene families, a kinetoplastid feature that may overcome the limitations of polycistronic transcription by increasing gene dosage (see below). Properties of the L. major genome, in comparison with the L. infantum, L. braziliensis and T. brucei genomes, are listed in Table 2.
Table 2.
Genome facts: the Leishmanias versus Trypanosoma brucei
| Leishmania major | Leishmania infantum | Leishmania braziliensis | Trypanosoma brucei | |
|---|---|---|---|---|
| Total size (Mb) | 32.8 | 32.1 | 32.0 | 26.1 |
| Contigs | 36 | 562 | 1,041 | 30 |
| No. of chromosomes | 36 | 36 | 35 | 11∗ |
| Chromosome size range (Mb) | 0.3–2.8 | 0.3–2.8 | 0.3–2.8 | 1–5.2 |
| Overall G + C content % | 59.7 | 59.3 | 60.4 | 46.4 |
| No. of genes | 8,298 | 8,154 | 8,153 | 9,068 |
| No. of pseudogenes | 97 | 41 | 161 | 904 |
| Average gene size (bp) | 1,894 | 1,868 | 1,873 | 1,592 |
| Gene density (per Mb) | 252 | 235 | 258 | 317 |
| Coding percentage | 48.0 | 44.0 | 48.5 | 50.5 |
| Coding G + C content % | 62.5 | 62.4 | 60.4 | 50.9 |
| No. of DGCs | 133 | 133 | n/a | 127 |
| Average DGC length (kb/genes) | 240/61 | n/a | n/a | 204/71 |
| No. of tRNAs | 83 | 62 | 66 | 65 |
| No. of snoRNAs | 693 | n/a | n/a | 353 |
| No. of snRNAs | 6 | n/a | n/a | 5 |
| No. of rRNAs | 63 | n/a | n/a | 56 |
| Average intergenic size | 1,939 | 2,049 | 1,976 | 1,279 |
| Active mobile elements | None (degenerate RIME/DIRE) | None (degenerate RIME/DIRE) | TATEs, SLACS | ingi, RIME, DIRE, SLACs, SIRE, VIPER |
Data included in this Table are correct as of February 2007 (for Leishmania species) and July 2005 (for T. brucei). *, only the megabase chromosome are included, not the intermediate or mini-chromosomes. DGC, directional gene cluster; RIME, ribosomal mobile element; DIRE, degenerate ingi/L1Tc-related element; TATE, telomere-associated transposable element; SLACS, Spliced Leader Associated Conserved Sequence; SIRE, short interspersed repetitive element; VIPER, LTR retroelement related to SIRE.
The L. major genes can be divided into three broad categories. Around 85% encode the core proteome common to all kinetoplastids. Many of these function in metabolism and can be described as house-keeping genes, similar to those found in higher eukaryotes. A second category contains genes restricted to kinetoplastids, with no orthologues in higher eukaryotes. These include sequences encoding cytoskeletal proteins, RNA editing components and other metabolic enzymes. A third category appears to be restricted to Leishmania: these number ∼1,000, ∼12% of the genome (El-Sayed et al., 2005a). Many of these genus-specific genes have no conserved or recognisable functional domains and it is reasonable to predict that some of them function in host/pathogen interactions.
Annotation of the L. major genome benefited considerably from the comparative genomics approach developed in the Tritryp projects: based on chromosomal position and sequence similarity, orthologues of genes experimentally characterised in the trypanosomes could be identified in Leishmania and vice versa. This approach was used to great effect in the identification of L. major metabolic pathways, which are more extensive than in either T. brucei or T. cruzi (Berriman et al., 2005). Leishmania metabolism reflects parasite lifestyle in the vector and the host and is influenced by the availability of nutrients in both locations (reviewed in Zilberstein, 1993; Tielens and Van Hellemond, 1998). As with the other Tritryps, Leishmania lacks the biosynthetic pathways for essential amino acids which must therefore be host-derived, explaining the presence of the large amino acid transporter gene families found in L. major (Ivens et al., 2005). Similarly, while all the Tritryps encode the full machinery for uptake and degradation of glucose (Berriman et al., 2005), L. major alone appears to have the capability to hydrolyse disaccharide sugars, probably as a consequence of sandfly nectar feeding that provides an alternative source of energy for the promastigote stage of the parasite (Muller and Schlein, 2004). L. major also encodes enzymes required for haem synthesis, which are lacking from the other Tritryps (Ivens et al., 2005). Haem is an important Leishmania growth factor that is required in both extracellular and intracellular stages of the life cycle (Chang and Chang, 1985). The last three enzymes in the biosynthetic pathway, encoded solely by Leishmania, share a high similarity to genes in prokaryotic pathways, like many of the apparent kinetoplastid gene acquisitions that lead to new metabolic activities – in the Tritryp analysis, as many as 50 genes were identified as likely candidates for lateral gene transfer from prokaryotic sources (Berriman et al., 2005). It should be noted, however, that synthesis of haem has not been demonstrated biochemically in Leishmania and genes coding for enzymes required for the rest of the pathway have yet to be identified.
While L. major lacks the highly expanded and divergent large subtelomeric immune evasion gene families common to other protozoan pathogens, the sub-telomeric regions are not totally devoid of genes, with some sequence families found in these locations (see Fig. 1). The largest and best studied gene family codes for a family of phosphoglycan scβ-galactosyltransferases (SCGs). These enzymes are involved in side chain modifications of the phosphoglycan repeats of the major promastigote glycoconjugate, LPG (Dobson et al., 2003). There are 14 members spread across three subfamilies (SCG, SCGR and SCGL) but seven of the “core” SCG genes are found exclusively at distinct subtelomeric sites (Dobson et al., 2006). Interestingly, SCG expression does not appear to be restricted to a single location that might support an antigenic variation model in Leishmania, similar to that operating with the T. brucei VSGs (reviewed in Barry and McCulloch, 2001). Further analysis is required to rigorously test this “single-cell exclusive-expression” model (Dobson et al., 2006). It is clear, however, that these sub-telomeric locations can facilitate gene conversion events that may ultimately support the strain-specific diversity in LPG galactosylation known to be critical for successful sandfly transmission of different Leishmania strains and species (reviewed in Sacks and Kamhawi, 2001; Kamhawi, 2006).
In contrast to the sub-telomeric SCG genes, the tandemly arrayed gene families coding for Leishmania surface proteins are found at internal chromosome sites. A good example is the GP63 gene family which encodes glycoproteins critical for host invasion and virulence (Joshi et al., 2002; Yao et al., 2003). These zinc metalloproteases are not confined to Leishmania species however: a single tandem GP63 gene array is present in all three Tritryp species (El-Sayed et al., 2005a), while recent studies suggest that GP63-type proteins are conserved and functional in insect and plant trypanosomatid species (Masini Masini d’Avila-Levy et al., 2006) as well as the more distant Trichomonas vaginalis (Carlton et al., 2007).
In Leishmania, the GP63 locus is limited to four to six copies in L. major and L. infantum but expanded approximately eightfold in the L. braziliensis genome (Peacock et al., 2007). Studies with multiple isolates of L. braziliensis and Leishmania peruviana (both L. Viannia spp., see Table 1) have shown extensive size variation and evidence of gene re-arrangement at this locus during long-term maintenance in the laboratory, suggesting that the GP63 locus is susceptible to dynamic change in this sub-genus (Victoir et al., 1995). This does not seem to be the case in a more limited study in L. major (Sadlova et al., 2006). Recent analysis of sequence diversity within a single GP63 gene copy in multiple L. donovani complex strains has confirmed earlier speculation that evolution of this family is influenced by mosaic gene conversion and provides strong genetic evidence for GP63 as an important marker for host preference in infection (Mauricio et al., 2006).
Another family of leucine-rich membrane surface proteins, the PSA-2 or GP46 family, is restricted to Leishmania spp.; analysis at both the DNA and protein levels failed to detect GP46 in the L. braziliensis complex (McMahon-Pratt et al., 1992). Confirming this earlier study, only one PSA-2-like gene has been annotated in the L. braziliensis genome. This is in contrast to the genome strains of L. major (with 25 PSA-2 genes) and L. infantum (also with a large gene complement, although the precise copy number has been difficult to determine accurately). The PSA-2s are known to bind to host cell macrophages and protect the parasite from complement-mediated lysis during infection with L. chagasi and L. infantum (Kedzierski et al., 2004; Lincoln et al., 2004). In L. infantum, different strains that cause either visceral or cutaneous disease show considerable correlative variation in the size of the repeat regions containing the PSA-2 genes (Guerbouj et al., 2001).
The amastin genes, the largest family in the Leishmania genome, encode small surface-expressed glycoproteins of as yet unknown function that are conserved in T. cruzi. Importantly, a majority of these genes are expressed specifically in intracellular amastigotes in L. major and therefore presumably contribute to survival in the host (Rochette et al., 2005). Unlike most other large Leishmania gene families, the amastin genes are not present in a single array but found on seven different chromosomes in the L. major genome (Ivens et al., 2005). Most of these sites contain small groups of genes (one to four copies) but two large arrays are also present, with the largest containing amastin genes alternating with the highly conserved tuzin genes; a similar arrangement is found in T. cruzi but is absent from T. brucei (El-Sayed et al., 2005b). The second large gene array, not present in T. cruzi, contains amastin genes separated by degenerate tuzin genes. A likely explanation for this gene complexity is that a common kinetoplastid ancestor possessed a single amastin/tuzin array, which underwent duplication, re-arrangement and diversification after separation of the Leishmania and Trypanosoma lineages. With the exception of a lone copy on L. major chromosome 36, L. infantum and L. braziliensis have amastin gene arrays at the same locations, although sequence read depth suggests that the large amastin array with intact tuzin genes is shorter in L. braziliensis.
Other Leishmania-specific proteins that are not necessarily surface-exposed may also be important regulators of host–parasite interactions. One interesting example is an apparent homologue of the human cytokine macrophage inhibitory factor (MIF). The first cytokine to be identified, MIF is also one of the most pleiotropic, implicated in a wide range of cellular activities that could influence parasite survival (Calandra and Roger, 2003). Addition of purified recombinant human MIF to L. major-infected macrophages activates parasite killing by inducible nitric oxide synthase and TNF-α (Juttner et al., 1998). In malaria, variations in the expression of MIF have been associated with susceptibility to high density parasitemia, while reduced MIF production may lead to increased severity of disease (Awandare et al., 2006). MIF-deficient mice also show increased susceptibility to L. major (Satoskar et al., 2001), highlighting the important role of this cytokine in mounting an effective immune response to infection.
MIF sequences are not confined to the host, however. All three Leishmania spp. sequenced to date have two similar MIF gene copies located at a strand switch region (see http://www.genedb.org/). Functional analysis of these sequences may be of considerable interest, given that MIF expression in the filarial worm Brugia malayi influences the progression of the CD4+ T cell response to infection towards a TH2 pathway (Falcone et al., 2001). The Leishmania MIF genes show greatest similarity to bacterial orthologues, suggesting that they too may have been acquired by lateral gene transfer following diversification from the trypanosome lineage.
5. Comparison of the L. major, L. infantum and L. braziliensis genomes
From an evolutionary perspective, phylogenetic analyses have suggested a neotropical origin for the Leishmania genus (Stevens et al., 2001) and, while there has been some controversy in this designation (see Kerr, 2000), this has been largely resolved in a recent multifactorial genetic study (Lukes et al., 2007). Irrespective of this debate, L. braziliensis is the most genetically and biologically divergent of the three sequenced species. Divergence between the Leishmania species complexes is estimated to have occurred 15–50 million years ago (Lukes et al., 2007), within the same range as two potential host species, mouse and human. Given this period of isolation, it was expected that there would be significant differences in both genome architecture and gene repertoire between L. braziliensis, L. infantum and L. major. Indeed, while the genomes have a similar DNA content of around 33 Mb, karyotypic differences had already been identified by linkage group analysis (Britto et al., 1998): L. major and L. infantum, in common with other Old World species, have a haploid content of 36 chromosomes, while the New World species have either 35 (L. braziliensis complex) or 34 (L. mexicana complex). These differences were shown to be due to fusion of pairs of chromosomes (chromosomes 20 + 34 in L. braziliensis; chromosomes 8 + 29 and 20 + 36 in L. mexicana), with the former observation now confirmed in the sequencing project.
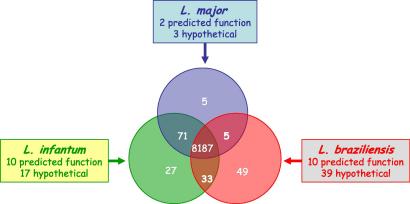
Surprisingly, comparison of the respective orthologous chromosomes has revealed remarkable conservation of both gene content and gene order in all three genome species. Despite the differences in gene copy number within some of the major protein-coding families described above, not a single chromosomal re-arrangement has been identified between L. major and L. infantum across the whole genome, while L. braziliensis has only a few possible sequence re-arrangements (Peacock et al., 2007). Equally surprising, from the total content of ∼8,300 genes in each species, only ∼200 can be identified as differentially distributed between the three genomes. The most divergent, L. braziliensis, possesses 47 genes that are absent from the other two species. In comparison, L. infantum has 27 species-specific genes while L. major has only five (see Fig. 2). A number of the other differentially distributed genes are found in two out of the three species. Some of these species-specific sequences have already been analysed at the molecular level. Examples include the L. infantum A2 gene that encodes an amastigote-specific repeat-containing protein previously characterised in L. donovani, the only Leishmania sequence to date that confers a change in virulence phenotype when introduced into L. major by genetic transfection (Zhang et al., 2003); and the HASP and SHERP genes, expressed from a single locus (absent in L. braziliensis) in infective stages of L. major and L. infantum, with their protein products localizing to the plasma membrane and intracellular membranes, respectively, in these species (Denny et al., 2000; Knuepfer et al., 2001).
Fig. 2.
Leishmania species-specific genes. The Venn diagram shows how many of the 8,187 protein-coding genes are species-specific or shared between two of the three sequenced Leishmania species. These genes are subdivided into those that have a predicted function (mostly through sequence identity) and those that are of unknown function at the time of publication (the “hypotheticals”).
In the Tritryp genome analyses, most genes specific to each of the representative species were found either at the ends of the DGCs or in the subtelomeric regions of the chromosomes, regions that appear to be more tolerant to genome re-arrangement. However, comparison of the three Leishmania genomes has revealed that gene variation is not predominantly restricted to the subtelomeric regions or even the SSRs but is evenly distributed across the genome (Peacock et al., 2007).
Leishmania is also distinctive from other eukaryotes in the apparent mechanism by which species-specific gene variation occurs. Whereas insertions/deletions and sequence re-arrangements play major roles in gene diversification in most other eukaryotes characterised to date, degeneration of existing genes (leading to probable loss of function) accounts for ∼80% of the species differences in Leishmania. These degenerate sequences have in-frame stop codons and frame shifts, generating truncated open reading frames that are presumably not translated. One example is the gene encoding cysteine peptidase Pfp1, which is present as an intact gene and translated in L. major (Eschenlauer et al., 2006). However, there are five in-frame stop codons and a frame shift in the L. infantum orthologue, while the syntenic region in L. braziliensis is even more degenerate.
Pfp1 like some of the other species-specific genes, appears to be another candidate for lateral gene transfer from bacteria. Of the remaining species-specific sequences not caused by loss of function, many also fall into this category. One example is the cyclopropane fatty acyl synthase (CFAS) gene, present in L. infantum and L. braziliensis but absent from L. major. Acquisition of novel genes in this way may be a mechanism for environmental adaptation to promote survival; similar adaptations to stress or other stimuli may lead to the redundancy of other sequences clearly identified as pseudogenes in the Leishmania genomes (Peacock et al., 2007). In the case of CFAS, acquisition of this gene may have an impact on parasite survival in the host, since the CFAS orthologue in Mycobacterium tuberculosis is associated with increased virulence and persistence, functions that apparently require cyclopropanation of a mycolic acid substrate in the bacterial cell wall (Rao et al., 2005).
Despite its chromosomal plasticity (Martinez-Calvillo et al., 2005), the incredible conservation of synteny revealed by comparative genomic analyses of these three species suggests that the Leishmania genome is highly stable and has not undergone major genomic re-arrangements during speciation. One contributing factor to this stability could be a lack of mobile DNA elements, as originally demonstrated in the L. major genome and verified in L. infantum. The comparative sequencing project has revealed some surprising observations, however, one of the most striking being the presence of transposable elements in L. braziliensis.
6. Retrotransposons in L. braziliensis
Repeat sequences are a common feature of both prokaryotic and eukaryotic genomes, with different categories contributing 50% or more to some eukaryotic genomes. Among these groups are the mobile genetic elements which, by virtue of their ability to move to different chromosomal locations, are considered important in shaping the course of genome evolution. Mobile elements are widespread in the biological kingdom. When actively proliferating (Wickstead et al., 2003), they can create and destroy or modify existing genes and regulatory elements, thus reshaping the structure and function of their host genomes. When no longer able to proliferate (“dead repeats”), they are still important markers for evolutionary events, a palaeontological record.
Although some protozoan parasites carry no mobile elements or any vestige of them (Katinka et al., 2001; Carlton et al., 2002; Gardner et al., 2002; Xu et al., 2004), different types of transposable elements have been found in kinetoplastids. The genomes of T. brucei and T.cruzi contain both highly reiterated interspersed elements (Bhattacharya et al., 2002; Wickstead et al., 2003; El-Sayed et al., 2005b) and retroelements, transposons that require reverse transcription from an RNA intermediate. Depending on their mechanism of transposition, these retroelements are classified either as long terminal repeat (LTR) retroelements (or retrotransposons) or non-LTR retroelements (the retroposons), which include the short and long interspersed nuclear elements (Singer, 1982).
In some kinetoplastids, retrotransposons (VIPER), site-specific retroposons (SLACS, CZAR, CRE) and non-site-specific retroposons (Ingi and L1Tc) have been described (Kimmel et al., 1987; Murphy et al., 1987; Martin et al., 1995; Teng et al., 1995; Wickstead et al., 2003). Trypanosoma cruzi bears the highest number of repetitive sequences, which occupy at least 50% of the parasite genome. Long terminal repeat (LTR) and non-LTR retroelements account for ∼5% and 2% of the haploid T. cruzi and T. brucei genomes, respectively (El-Sayed et al., 2005b). These mobile elements probably played an important role in shaping these parasites’ genome organization and evolution.
In contrast, analysis of several Leishmania species originally indicated a lack of active retroelements (Bhattacharya et al., 2002). In the L. major and L. infantum genomes, it was possible to detect remnants of retroposons, named DIREs (degenerate ingi/L1Tc-related elements), which had been previously described in trypanosomes (Ghedin et al., 2004; Bringaud et al., 2006). Other relics of retroelements, distinct from ingi/L1Tc, were also detected in L. major. These are conserved in some Old and New World Leishmania species, but absent from the T. cruzi and T. brucei genomes (Pedrosa et al., 2006).
Against this background, in addition to these degenerate retroelements, it was surprising to detect potentially active retroposons in the genome of L. braziliensis (Peacock et al., 2007). Similar to the organization described for the SLACS and CZAR elements in T. brucei and T. cruzi, these non-LTR retrotransposons are associated with the spliced leader array in L. braziliensis (Aksoy et al., 1987; Villanueva et al., 1991).
In addition, the organization of the L. braziliensis telomeres is remarkably distinct from the Old World species. They accommodate the so called TATEs (Telomeric Associated Transposable Elements), a family of 20–30 novel DNA transposable elements, each including putative reverse transcriptase, phage integrase (site-specific recombinase) and DNA/RNA polymerase domains. These are highly conserved retroelements with a precise site of insertion within the telomeric hexamer repeats and a typical TT duplication flanking the transposon. The clusters of tandemly arrayed TATEs at the ends of chromosomes resemble ribosomal mobile element (RIME)/Ingi retroelements that are also organised in clusters within the subtelomeric regions of T. brucei chromosomes (Bringaud et al., 2002; Peacock et al., 2007). Based on current annotation of the L. braziliensis genome, TATEs are present on at least 12 chromosomes, precisely inserted in the middle of the GGGTTA repeats (Peacock et al., 2007). The TATE-containing L. braziliensis chromosome ends share longer stretches of sequence with a higher number of heterologous chromosomes, compared with the corresponding chromosomes in L. major (Brito, L.O. et al., unpublished data). On the other hand, only remnants of TATE elements are detected at L. braziliensis internal chromosomal sites. Besides the bias of abundance of these elements at the chromosome ends, selection against disruption of coding regions at chromosome central domains could explain TATE distribution patterns. To understand the role of the TATEs in shaping the L. braziliensis genome, it will be necessary to improve genome assembly and further analyse TATE distribution in different strains and species of the Viannia subgenus.
7. RNAi in L. braziliensis
Among different functional classes of non-coding RNAs, there is a large class of post-transcriptional regulators implicated in gene expression control in various organisms. Two classes of small RNAs, the short interfering RNAs (siRNAs) and the microRNAs (miRNAs) have been extensively studied in gene silencing. Initially described in Caenorhabditis elegans (Lee et al., 1993; Wightman et al., 1993), the term RNA interference (RNAi) was used to describe these events (Fire et al., 1998) and link them to similar post-transcriptional silencing phenomena in plants and fungi (Jorgensen, 1990; Romano and Macino, 1992).
Currently, RNAi is the best-known mechanism of gene silencing, an evolutionary conserved process in which double-stranded RNA induces the sequence-specific degradation of homologous mRNA (Fire et al., 1998). Besides its role in the control of endogenous mRNAs, the process seems to have an important defensive role against viruses and mobile element activity.
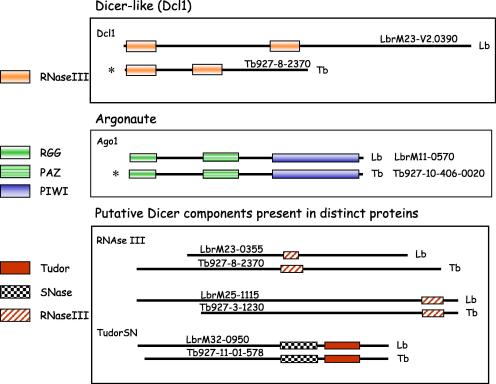
An RNAi mechanism was rapidly identified in T. brucei (Ngo et al., 1998) but has not been demonstrated in other kinetoplastids including L. major and T. cruzi (Robinson and Beverley, 2003; El-Sayed et al., 2005b). Therefore, one of the most unexpected differences between the sequenced Leishmania genomes is the identification of genes implicated in the RNAi pathway in L. braziliensis. One of the hallmarks of this pathway is Dicer activity, which converts double-stranded RNA (dsRNA) into small interfering RNA (siRNA). Annotation of the genomes of three Leishmania species and the trypanosomes did not reveal a Dicer orthologue (Berriman et al., 2005; El-Sayed et al., 2005b; Ivens et al., 2005; Peacock et al., 2007). Instead, it was speculated that a combination of independent proteins carrying the relevant domains DSRBD, DEAD/H box RNA helicase and RNase III could be responsible for Dicer activity in T. brucei. Orthologues of genes carrying these domains have also been annotated in the L. braziliensis genome (see Fig. 3 for an outline of the putative RNAi machinery in L. braziliensis, compared with T. brucei).
Fig. 3.
RNAi machinery: Leishmania braziliensis versus Trypanosoma brucei. Schematic representation of the main components of the RNAi pathways identified by sequence similarity and/or functional evaluation in T. brucei (Tb) and L. braziliensis (Lb). Comparisons of the gene products and their domains in each species are accompanied by their GeneDB gene IDs; Tb927, T. brucei genes; LbrM, L. braziliensis genes. Domain IDs are depicted on the left. RGG indicates a domain containing arginine-glycine-glycine repeats; SNase indicates the Staphylococcal nuclease domain present in TudorSN proteins. ∗Indicates genes confirmed to be functionally active in the T. brucei RNAi pathway.
Curiously, it has recently been shown that a T. brucei Dicer-like protein (TbDcl1), quite divergent from the typical Dicer proteins described for other eukaryotes, is required for the generation of siRNA-size molecules and for RNAi (Shi et al., 2006a). Interestingly, a TbDcl1 gene is also present in L. braziliensis (Peacock et al., 2007), although there are no orthologues in T. cruzi, L. major or L. infantum.
Another crucial component of the RNAi pathway is Argonaute, an endonuclease involved in the dsRNA-triggered cleavage of mRNA. Unlike L. major and L. infantum, L. braziliensis contains one orthologue for Ago1, the functional argonaute gene of T. brucei. TbAGO1 can be functionally replaced by the human gene Argonaute 2, which suggests that TbAGO1 has the endonuclease activity involved in mRNA target degradation in the trypanosome RNAi pathway (Shi et al., 2006b). The L. braziliensis gene contains the typical argonaute domains PAZ and PIWI, the latter containing conserved amino acid residues that are essential for functional TbAGO1 (Shi et al., 2004). In addition, an N-terminal RGG domain, present in TbAGO1 and shown to be essential for its association with polyribosomes, is also present in the L. braziliensis Ago1 gene (Shi et al., 2004). Thus, the L. braziliensis gene product may also participate in the control of translation.
It is an intriguing observation that RNAi machinery is present in only one of the three, highly syntenic Leishmania species sequenced to date. While this could indicate a lack of biological relevance for this pathway in the parasite, a more interesting possibility is that the availability of RNAi in L. braziliensis plays a role in the biological differences observed between the two subgenera. The demonstration of RNAi and its biological consequences in L. braziliensis awaits further experimentation.
Whether RNAi machinery has been lost or acquired across the kinetoplastid species and genera is an open question. Examination of the syntenic regions in L. major and L. infantum identified remnants of Argonaute in both, an observation that favours the proposal that RNAi was lost in the subgenus Leishmania. This speculation is reinforced by the conservation of key RNAi members between organisms as distant as T. brucei and humans (Shi et al., 2006a,b). It is relevant to stress that RNA viruses have been detected in species of the subgenus L. Viannia (Guilbride et al., 1992; Widmer and Dooley, 1995), suggesting that L. braziliensis could have retained RNAi as an antiviral defense mechanism (although there is also a single report of an RNAi virus in L. major; Scheffter et al., 1995). In addition, RNAi could have a role in regulating the function of the L. braziliensis-specific transposable elements described above. A role for RNAi as an endogenous mechanism to hamper expression of specific genes and defend cells from viruses and mobile elements has been demonstrated in a range of organisms, including T. brucei (Wickstead et al., 2003; Ullu et al., 2004).
The discovery of a Leishmania RNAi mechanism could also be of practical importance, given the wide and effective use of inducible RNAi methods to study gene function in other eukaryotes. An RNAi system for reducing gene expression would be especially relevant in Leishmania, a diploid pathogen that lacks a sexual cycle, and provide an interesting alternative to the classical two-step gene knockout strategy (Cruz et al., 1991). RNAi has been rigorously tested for in L. major and L. donovani with no success (Robinson and Beverley, 2003). Detection of an RNAi machinery so similar to that of T. brucei in L. braziliensis, however, suggests that this mechanism may be shown to be a useful tool in L. braziliensis. A word of caution however: the genetic plasticity of the Leishmania genome, which impedes the generation of null mutants for essential genes, may also interfere with the outcome of RNAi experiments. In fact, downregulation of T. brucei essential genes by RNAi led to selection of cell lines that escaped from the deleterious effect of RNA interference (Ullu et al., 2004). Notwithstanding this concern, the combination of RNAi and classical reverse genetic approaches promises to bring novel insights into the functional biology of Leishmania parasites.
8. How do the genome sequences impact on our understanding of parasite tropism?
It is premature to speculate on how knowledge of the genome sequences of different Leishmania species will impact on our global understanding of disease phenotype. However the primary comparative analysis (Peacock et al., 2007) supports some preliminary observations. Firstly, the genes that differ between the three species are good candidates for functional analysis to determine their roles in establishment of infection. With the A2 gene as a precedent (Zhang et al., 2003), it can be speculated that one or more of these sequences, expressed separately or together, may impact upon the course of disease. Testing this hypothesis will require the generation and functional testing of specific mutations in suitable models of infection. Murine models of CL are well-established, with experimental L. major infection reproducing distinct aspects of the clinical spectrum of human disease (reviewed in Gumy et al., 2004). The establishment of a dermal infection model, that introduces small numbers of infective L. major metacyclics into the host dermis via sandfly feeding, more closely mimics the natural course of parasite transmission (Belkaid et al., 1998). Murine models for L. infantum and L. braziliensis are not as well advanced, partly because these species are more difficult to grow and differentiate in vitro and partly because of the variable sensitivity of model mouse strains to infection with these parasites. Progress has recently been made, however, in the development of dermal models for both L. infantum and L. braziliensis infection (Ahmed et al., 2003; de Moura et al., 2005). For all species, more work is required to relate the immune correlates of protection in mice to those that are important in human and, as appropriate, canine infection.
Second, as most of the species-specific genes encode proteins sharing little identity with other eukaryotic sequences, the use of a combination of genetic, biochemical and immunological analyses will be essential to help delineate relevant biological functions. It will also be necessary to unravel the importance of those tandemly repeated genes that vary in copy number between species, given that this mechanism can support increased gene expression and potentially lead to detectable phenotypic effects. In this context, the recently published whole genome microarray analyses suggest that RNA expression is largely constitutive throughout the Leishmania life cycle with few genes showing increased expression at the RNA level in different parasite stages (Holzer et al., 2006; Leifso et al., 2007). Further studies are required to verify these observations in different parasite species and correlate RNA and protein expression levels, using high resolution proteomic methods (Foucher et al., 2006).
A third observation from the comparative genome analysis is that, despite the well-conserved synteny between the three model species, L. braziliensis may have the capacity for more extensive genomic reorganisation, given the putative retrotransposons and RNAi machinery found in this species. Clearly, whether RNAi actually operates as a functional mechanism in L. braziliensis awaits definitive experimental validation, while the functional relevance of the retroelements requires more extensive multiple strain analysis to determine possible levels of sequence variation. However, it cannot be discounted that retrotransposons play a role in shaping the L. braziliensis genome, impacting on the different disease outcomes associated with this species.
9. Future perspectives
Sequencing the genomes of three Leishmania species, with a fourth in progress, has provided a wealth of new information that is reshaping research initiatives for leishmaniasis. Extending this type of analysis to further species and virulent strains may reveal further surprises. Of most immediate practical application perhaps are the unlimited number of potential vaccine candidates that are now available and require robust evaluation, to identify the most favourable molecules to incorporate into clinical trials (Kedzierski et al., 2006). Similarly, the Leishmania-specific metabolic pathways revealed by comparative analysis are underpinning focused biochemical analyses to identify valid drug targets for development. New, more sensitive and specific diagnostic reagents for the leishmaniases should also emerge from exploring the different genome sequences.
More fundamentally, genomic knowledge is revealing fascinating insights into Leishmania biology. In particular, the current comparative analyses are generating testable hypotheses relevant to understanding the importance of parasite genotype in pathogenesis. It is possible that just a few species-specific parasite genes are crucial to disease outcome and/or alternatively, that expression levels of key conserved parasite genes differ considerably both between and within species (perhaps as a consequence of variation in gene copy number). It is also possible that the parasite genome may play only a minor role in determining disease phenotype. In-depth functional analyses may ultimately unravel the relative contribution of parasite genes to disease progression in this spectrum of crippling infections.
Acknowledgements
Research in the laboratory of D.F.S. is supported by the Wellcome Trust and the BBSRC and in the laboratory of A.K.C., by FAPESP, CNPq and TDR/WHO. C.P. is also supported by the Wellcome Trust through core funding to the Pathogen Sequencing Unit, Sanger Institute. Space restraints require us to restrict the number of references for this review; we apologise to those colleagues whose work has not been cited here.
References
- Ahmed S., Colmenares M., Soong L., Goldsmith-Pestana K., Munstermann L., Molina R., McMahon-Pratt D. Intradermal infection model for pathogenesis and vaccine studies of murine visceral leishmaniasis. Infect. Immun. 2003;71:401–410. doi: 10.1128/IAI.71.1.401-410.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksoy S., Lalor T.M., Martin J., Van der Ploeg L.H., Richards F.F. Multiple copies of a retroposon interrupt spliced leader RNA genes in the African trypanosome, Trypanosoma gambiense. EMBO J. 1987;6:3819–3826. doi: 10.1002/j.1460-2075.1987.tb02718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awandare G.A., Ouma C., Keller C.C., Were T., Otieno R., Ouma Y., Davenport G.C., Hittner J.B., Ong’echa J.M., Ferrell R., Perkins D.J. A macrophage migration inhibitory factor promoter polymorphism is associated with high-density parasitemia in children with malaria. Genes Immun. 2006;7:568–575. doi: 10.1038/sj.gene.6364332. [DOI] [PubMed] [Google Scholar]
- Barral A., Pedral-Sampaio D., Grimaldi Junior G., Momen H., McMahon-Pratt D., Ribeiro de Jesus A., Almeida R., Badaro R., Barral-Netto M., Carvalho E.M. Leishmaniasis in Bahia, Brazil: evidence that Leishmania amazonensis produces a wide spectrum of clinical disease. Am. J. Trop Med. Hyg. 1991;44:536–546. doi: 10.4269/ajtmh.1991.44.536. [DOI] [PubMed] [Google Scholar]
- Barry J.D., McCulloch R. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 2001;49:1–70. doi: 10.1016/s0065-308x(01)49037-3. [DOI] [PubMed] [Google Scholar]
- Belkaid Y., Kamhawi S., Modi G., Valenzuela J., Noben-Trauth N., Rowton E., Ribeiro J., Sacks D.L. Development of a natural model of cutaneous leishmaniasis: powerful effects of vector saliva and saliva preexposure on the long-term outcome of Leishmania major infection in the mouse ear dermis. J. Exp. Med. 1998;188:1941–1953. doi: 10.1084/jem.188.10.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BenSaid M., Guerbouj S., Saghrouni F., Fathallah-Mili A., Guizani I. Occurrence of Leishmania infantum cutaneous leishmaniasis in central Tunisia. Trans. R. Soc. Trop. Med. Hyg. 2006;100:521–526. doi: 10.1016/j.trstmh.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Bohme U., Hannick L., Aslett M.A., Shallom J., Marcello L., Hou L., Wickstead B., Alsmark U.C., Arrowsmith C., Atkin R.J., Barron A.J., Bringaud F., Brooks K., Carrington M., Cherevach I., Chillingworth T.J., Churcher C., Clark L.N., Corton C.H., Cronin A., Davies R.M., Doggett J., Djikeng A., Feldblyum T., Field M.C., Fraser A., Goodhead I., Hance Z., Harper D., Harris B.R., Hauser H., Hostetler J., Ivens A., Jagels K., Johnson D., Johnson J., Jones K., Kerhornou A.X., Koo H., Larke N., Landfear S., Larkin C., Leech V., Line A., Lord A., Macleod A., Mooney P.J., Moule S., Martin D.M., Morgan G.W., Mungall K., Norbertczak H., Ormond D., Pai G., Peacock C.S., Peterson J., Quail M.A., Rabbinowitsch E., Rajandream M.A., Reitter C., Salzberg S.L., Sanders M., Schobel S., Sharp S., Simmonds M., Simpson A.J., Tallon L., Turner C.M., Tait A., Tivey A.R., Van Aken S., Walker D., Wanless D., Wang S., White B., White O., Whitehead S., Woodward J., Wortman J., Adams M.D., Embley T.M., Gull K., Ullu E., Barry J.D., Fairlamb A.H., Opperdoes F., Barrell B.G., Donelson J.E., Hall N., Fraser C.M., Melville S.E., El-Sayed N.M. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- Beverley S.M., Turco S.J. Lipophosphoglycan (LPG) and the identification of virulence genes in the protozoan parasite Leishmania. Trends Microbiol. 1998;6:35–40. doi: 10.1016/S0966-842X(97)01180-3. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S., Bakre A., Bhattacharya A. Mobile genetic elements in protozoan parasites. J. Genet. 2002;81:73–86. doi: 10.1007/BF02715903. [DOI] [PubMed] [Google Scholar]
- Boaventura V.S., Cafe V., Costa J., Oliveira F., Bafica A., Rosato A., de Freitas L.A., Brodskyn C., Barral-Netto M., Barral A. Concomitant early mucosal and cutaneous leishmaniasis in Brazil. Am. J. Trop Med. Hyg. 2006;75:267–269. [PubMed] [Google Scholar]
- Boucher N., Wu Y., Dumas C., Dube M., Sereno D., Breton M., Papadopoulou B. A common mechanism of stage-regulated gene expression in Leishmania mediated by a conserved 3′-untranslated region element. J. Biol. Chem. 2002;277:19511–19520. doi: 10.1074/jbc.M200500200. [DOI] [PubMed] [Google Scholar]
- Brandao-Filho S.P., de Carvalho F.G., de Brito M.E., Almeida Fde A., Nascimento L.A. American cutaneous leishmaniasis in Pernambuco, Brazil: eco-epidemiological aspects in ‘Zona da Mata’ region. Mem. Inst. Oswaldo Cruz. 1994;89:445–449. doi: 10.1590/s0074-02761994000300028. [DOI] [PubMed] [Google Scholar]
- Bringaud F., Biteau N., Melville S.E., Hez S., El-Sayed N.M., Leech V., Berriman M., Hall N., Donelson J.E., Baltz T. A new, expressed multigene family containing a hot spot for insertion of retroelements is associated with polymorphic subtelomeric regions of Trypanosoma brucei. Eukaryot Cell. 2002;1:137–151. doi: 10.1128/EC.1.1.137-151.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringaud F., Ghedin E., Blandin G., Bartholomeu D.C., Caler E., Levin M.J., Baltz T., El-Sayed N.M. Evolution of non-LTR retrotransposons in the trypanosomatid genomes: Leishmania major has lost the active elements. Mol. Biochem. Parasitol. 2006;145:158–170. doi: 10.1016/j.molbiopara.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Britto C., Ravel C., Bastien P., Blaineau C., Pages M., Dedet J.P., Wincker P. Conserved linkage groups associated with large-scale chromosomal rearrangements between Old World and New World Leishmania genomes. Gene. 1998;222:107–117. doi: 10.1016/s0378-1119(98)00472-7. [DOI] [PubMed] [Google Scholar]
- Calandra T., Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat. Rev. Immunol. 2003;3:791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J.M., Angiuoli S.V., Suh B.B., Kooij T.W., Pertea M., Silva J.C., Ermolaeva M.D., Allen J.E., Selengut J.D., Koo H.L., Peterson J.D., Pop M., Kosack D.S., Shumway M.F., Bidwell S.L., Shallom S.J., van Aken S.E., Riedmuller S.B., Feldblyum T.V., Cho J.K., Quackenbush J., Sedegah M., Shoaibi A., Cummings L.M., Florens L., Yates J.R., Raine J.D., Sinden R.E., Harris M.A., Cunningham D.A., Preiser P.R., Bergman L.W., Vaidya A.B., van Lin L.H., Janse C.J., Waters A.P., Smith H.O., White O.R., Salzberg S.L., Venter J.C., Fraser C.M., Hoffman S.L., Gardner M.J., Carucci D.J. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature. 2002;419:512–519. doi: 10.1038/nature01099. [DOI] [PubMed] [Google Scholar]
- Carlton J.M., Hirt R.P., Silva J.C., Delcher A.L., Schatz M., Zhao Q., Wortman J.R., Bidwell S.L., Alsmark U.C., Besteiro S., Sicheritz-Ponten T., Noel C.J., Dacks J.B., Foster P.G., Simillion C., Van de Peer Y., Miranda-Saavedra D., Barton G.J., Westrop G.D., Muller S., Dessi D., Fiori P.L., Ren Q., Paulsen I., Zhang H., Bastida-Corcuera F.D., Simoes-Barbosa A., Brown M.T., Hayes R.D., Mukherjee M., Okumura C.Y., Schneider R., Smith A.J., Vanacova S., Villalvazo M., Haas B.J., Pertea M., Feldblyum T.V., Utterback T.R., Shu C.L., Osoegawa K., de Jong P.J., Hrdy I., Horvathova L., Zubacova Z., Dolezal P., Malik S.B., Logsdon J.M., Jr., Henze K., Gupta A., Wang C.C., Dunne R.L., Upcroft J.A., Upcroft P., White O., Salzberg S.L., Tang P., Chiu C.H., Lee Y.S., Embley T.M., Coombs G.H., Mottram J.C., Tachezy J., Fraser-Liggett C.M., Johnson P.J. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.S., Chang K.P. Heme requirement and acquisition by extracellular and intracellular stages of Leishmania mexicana amazonensis. Mol. Biochem. Parasitol. 1985;16:267–276. doi: 10.1016/0166-6851(85)90069-6. [DOI] [PubMed] [Google Scholar]
- Cruz A., Coburn C.M., Beverley S.M. Double targeted gene replacement for creating null mutants. Proc. Natl. Acad. Sci. USA. 1991;88:7170–7174. doi: 10.1073/pnas.88.16.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denise H., Poot J., Jimenez M., Ambit A., Herrmann D.C., Vermeulen A.N., Coombs G.H., Mottram J.C. Studies on the CPA cysteine peptidase in the Leishmania infantum genome strain JPCM5. BMC Mol. Biol. 2006;7:42. doi: 10.1186/1471-2199-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moura T.R., Novais F.O., Oliveira F., Clarencio J., Noronha A., Barral A., Brodskyn C., de Oliveira C.I. Toward a novel experimental model of infection to study American cutaneous leishmaniasis caused by Leishmania braziliensis. Infect. Immun. 2005;73:5827–5834. doi: 10.1128/IAI.73.9.5827-5834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny P.W., Gokool S., Russell D.G., Field M.C., Smith D.F. Acylation-dependent protein export in Leishmania. J. Biol. Chem. 2000;275:11017–11025. doi: 10.1074/jbc.275.15.11017. [DOI] [PubMed] [Google Scholar]
- Descoteaux A., Turco S.J. Functional aspects of the Leishmania donovani lipophosphoglycan during macrophage infection. Microbes Infect. 2002;4:975–981. doi: 10.1016/s1286-4579(02)01624-6. [DOI] [PubMed] [Google Scholar]
- Dobson D.E., Scholtes L.D., Valdez K.E., Sullivan D.R., Mengeling B.J., Cilmi S., Turco S.J., Beverley S.M. Functional identification of galactosyltransferases (SCGs) required for species-specific modifications of the lipophosphoglycan adhesin controlling Leishmania major-sand fly interactions. J. Biol. Chem. 2003;278:15523–15531. doi: 10.1074/jbc.M301568200. [DOI] [PubMed] [Google Scholar]
- Dobson D.E., Scholtes L.D., Myler P.J., Turco S.J., Beverley S.M. Genomic organization and expression of the expanded SCG/L/R gene family of Leishmania major: internal clusters and telomeric localization of SCGs mediating species-specific LPG modifications. Mol. Biochem. Parasitol. 2006;146:231–241. doi: 10.1016/j.molbiopara.2005.12.012. [DOI] [PubMed] [Google Scholar]
- El-Sayed N.M., Myler P.J., Blandin G., Berriman M., Crabtree J., Aggarwal G., Caler E., Renauld H., Worthey E.A., Hertz-Fowler C., Ghedin E., Peacock C., Bartholomeu D.C., Haas B.J., Tran A.N., Wortman J.R., Alsmark U.C., Angiuoli S., Anupama A., Badger J., Bringaud F., Cadag E., Carlton J.M., Cerqueira G.C., Creasy T., Delcher A.L., Djikeng A., Embley T.M., Hauser C., Ivens A.C., Kummerfeld S.K., Pereira-Leal J.B., Nilsson D., Peterson J., Salzberg S.L., Shallom J., Silva J.C., Sundaram J., Westenberger S., White O., Melville S.E., Donelson J.E., Andersson B., Stuart K.D., Hall N. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309:404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- El-Sayed N.M., Myler P.J., Bartholomeu D.C., Nilsson D., Aggarwal G., Tran A.N., Ghedin E., Worthey E.A., Delcher A.L., Blandin G., Westenberger S.J., Caler E., Cerqueira G.C., Branche C., Haas B., Anupama A., Arner E., Aslund L., Attipoe P., Bontempi E., Bringaud F., Burton P., Cadag E., Campbell D.A., Carrington M., Crabtree J., Darban H., da Silveira J.F., de Jong P., Edwards K., Englund P.T., Fazelina G., Feldblyum T., Ferella M., Frasch A.C., Gull K., Horn D., Hou L., Huang Y., Kindlund E., Klingbeil M., Kluge S., Koo H., Lacerda D., Levin M.J., Lorenzi H., Louie T., Machado C.R., McCulloch R., McKenna A., Mizuno Y., Mottram J.C., Nelson S., Ochaya S., Osoegawa K., Pai G., Parsons M., Pentony M., Pettersson U., Pop M., Ramirez J.L., Rinta J., Robertson L., Salzberg S.L., Sanchez D.O., Seyler A., Sharma R., Shetty J., Simpson A.J., Sisk E., Tammi M.T., Tarleton R., Teixeira S., Van Aken S., Vogt C., Ward P.N., Wickstead B., Wortman J., White O., Fraser C.M., Stuart K.D., Andersson B. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- Eschenlauer S.C., Coombs G.H., Mottram J.C. PFPI-like genes are expressed in Leishmania major but are pseudogenes in other Leishmania species. FEMS Microbiol. Lett. 2006;260:47–54. doi: 10.1111/j.1574-6968.2006.00303.x. [DOI] [PubMed] [Google Scholar]
- Falcone F.H., Loke P., Zang X., MacDonald A.S., Maizels R.M., Allen J.E. A Brugia malayi homolog of macrophage migration inhibitory factor reveals an important link between macrophages and eosinophil recruitment during nematode infection. J. Immunol. 2001;167:5348–5354. doi: 10.4049/jimmunol.167.9.5348. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Foucher A.L., Papadopoulou B., Ouellette M. Prefractionation by digitonin extraction increases representation of the cytosolic and intracellular proteome of Leishmania infantum. J. Proteome Res. 2006;5:1741–1750. doi: 10.1021/pr060081j. [DOI] [PubMed] [Google Scholar]
- Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S., Paulsen I.T., James K., Eisen J.A., Rutherford K., Salzberg S.L., Craig A., Kyes S., Chan M.S., Nene V., Shallom S.J., Suh B., Peterson J., Angiuoli S., Pertea M., Allen J., Selengut J., Haft D., Mather M.W., Vaidya A.B., Martin D.M., Fairlamb A.H., Fraunholz M.J., Roos D.S., Ralph S.A., McFadden G.I., Cummings L.M., Subramanian G.M., Mungall C., Venter J.C., Carucci D.J., Hoffman S.L., Newbold C., Davis R.W., Fraser C.M., Barrell B. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghedin E., Bringaud F., Peterson J., Myler P., Berriman M., Ivens A., Andersson B., Bontempi E., Eisen J., Angiuoli S., Wanless D., Von Arx A., Murphy L., Lennard N., Salzberg S., Adams M.D., White O., Hall N., Stuart K., Fraser C.M., El-Sayed N.M. Gene synteny and evolution of genome architecture in trypanosomatids. Mol. Biochem. Parasitol. 2004;134:183–191. doi: 10.1016/j.molbiopara.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Gramiccia M., Gradoni L., Troiani M. Heterogeneity among zymodemes of Leishmania infantum from HIV-positive patients with visceral leishmaniasis in south Italy. FEMS Microbiol. Lett. 1995;128:33–38. doi: 10.1111/j.1574-6968.1995.tb07496.x. [DOI] [PubMed] [Google Scholar]
- Grimaldi G., Jr., Tesh R.B. Leishmaniases of the New World: current concepts and implications for future research. Clin. Microbiol. Rev. 1993;6:230–250. doi: 10.1128/cmr.6.3.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerbouj S., Guizani I., Speybroeck N., Le Ray D., Dujardin J.C. Genomic polymorphism of Leishmania infantum: a relationship with clinical pleomorphism? Infect. Genet. Evol. 2001;1:49–59. doi: 10.1016/s1567-1348(01)00008-9. [DOI] [PubMed] [Google Scholar]
- Guilbride L., Myler P.J., Stuart K. Distribution and sequence divergence of LRV1 viruses among different Leishmania species. Mol. Biochem. Parasitol. 1992;54:101–104. doi: 10.1016/0166-6851(92)90099-6. [DOI] [PubMed] [Google Scholar]
- Gumy A., Louis J.A., Launois P. The murine model of infection with Leishmania major and its importance for the deciphering of mechanisms underlying differences in Th cell differentiation in mice from different genetic backgrounds. Int. J. Parasitol. 2004;34:433–444. doi: 10.1016/j.ijpara.2003.11.021. [DOI] [PubMed] [Google Scholar]
- Hall, N., Pain, A., Berriman, M., Churcher, C., Harris, B., Harris, D., Mungall, K., Bowman, S., Atkin, R., Baker, S., Barron, A., Brooks, K., Buckee, C.O., Burrows, C., Cherevach, I., Chillingworth, C., Chillingworth, T., Christodoulou, Z., Clark, L., Clark, R., Corton, C., Cronin, A., Davies, R., Davis, P., Dear, P., Dearden, F., Doggett, J., Feltwell, T., Goble, A., Goodhead, I., Gwilliam, R., Hamlin, N., Hance, Z., Harper, D., Hauser, H., Hornsby, T., Holroyd, S., Horrocks, P., Humphray, S., Jagels, K., James, K.D., Johnson, D., Kerhornou, A., Knights, A., Konfortov, B., Kyes, S., Larke, N., Lawson, D., Lennard, N., Line, A., Maddison, M., McLean, J., Mooney, P., Moule, S., Murphy, L., Oliver, K., Ormond, D., Price, C., Quail, M.A., Rabbinowitsch, E., Rajandream, M.A., Rutter, S., Rutherford, K.M., Sanders, M., Simmonds, M., Seeger, K., Sharp, S., Smith, R., Squares, R., Squares, S., Stevens, K., Taylor, K., Tivey, A., Unwin, L., Whitehead, S., Woodward, J., Sulston, J.E., Craig, A., Newbold, C., Barrell, B.G., 2002. Sequence of Plasmodium falciparum chromosomes 1, 3–9 and 13. Nature 419, 527–531. [DOI] [PubMed]
- Holzer T.R., McMaster W.R., Forney J.D. Expression profiling by whole-genome interspecies microarray hybridization reveals differential gene expression in procyclic promastigotes, lesion-derived amastigotes, and axenic amastigotes in Leishmania mexicana. Mol. Biochem. Parasitol. 2006;146:198–218. doi: 10.1016/j.molbiopara.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Ilg T. Proteophosphoglycans of Leishmania. Parasitol. Today. 2000;16:489–497. doi: 10.1016/s0169-4758(00)01791-9. [DOI] [PubMed] [Google Scholar]
- Ivens A.C., Peacock C.S., Worthey E.A., Murphy L., Aggarwal G., Berriman M., Sisk E., Rajandream M.A., Adlem E., Aert R., Anupama A., Apostolou Z., Attipoe P., Bason N., Bauser C., Beck A., Beverley S.M., Bianchettin G., Borzym K., Bothe G., Bruschi C.V., Collins M., Cadag E., Ciarloni L., Clayton C., Coulson R.M., Cronin A., Cruz A.K., Davies R.M., De Gaudenzi J., Dobson D.E., Duesterhoeft A., Fazelina G., Fosker N., Frasch A.C., Fraser A., Fuchs M., Gabel C., Goble A., Goffeau A., Harris D., Hertz-Fowler C., Hilbert H., Horn D., Huang Y., Klages S., Knights A., Kube M., Larke N., Litvin L., Lord A., Louie T., Marra M., Masuy D., Matthews K., Michaeli S., Mottram J.C., Muller-Auer S., Munden H., Nelson S., Norbertczak H., Oliver K., O’Neil S., Pentony M., Pohl T.M., Price C., Purnelle B., Quail M.A., Rabbinowitsch E., Reinhardt R., Rieger M., Rinta J., Robben J., Robertson L., Ruiz J.C., Rutter S., Saunders D., Schafer M., Schein J., Schwartz D.C., Seeger K., Seyler A., Sharp S., Shin H., Sivam D., Squares R., Squares S., Tosato V., Vogt C., Volckaert G., Wambutt R., Warren T., Wedler H., Woodward J., Zhou S., Zimmermann W., Smith D.F., Blackwell J.M., Stuart K.D., Barrell B., Myler P.J. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R. Altered gene expression in plants due to trans interactions between homologous genes. Trends Biotechnol. 1990;8:340–344. doi: 10.1016/0167-7799(90)90220-r. [DOI] [PubMed] [Google Scholar]
- Joshi P.B., Kelly B.L., Kamhawi S., Sacks D.L., McMaster W.R. Targeted gene deletion in Leishmania major identifies leishmanolysin (GP63) as a virulence factor. Mol. Biochem. Parasitol. 2002;120:33–40. doi: 10.1016/s0166-6851(01)00432-7. [DOI] [PubMed] [Google Scholar]
- Juttner S., Bernhagen J., Metz C.N., Rollinghoff M., Bucala R., Gessner A. Migration inhibitory factor induces killing of Leishmania major by macrophages: dependence on reactive nitrogen intermediates and endogenous TNF-alpha. J. Immunol. 1998;161:2383–2390. [PubMed] [Google Scholar]
- Kamhawi S. Phlebotomine sand flies and Leishmania parasites: friends or foes? Trends Parasitol. 2006;22:439–445. doi: 10.1016/j.pt.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Katinka M.D., Duprat S., Cornillot E., Metenier G., Thomarat F., Prensier G., Barbe V., Peyretaillade E., Brottier P., Wincker P., Delbac F., El Alaoui H., Peyret P., Saurin W., Gouy M., Weissenbach J., Vivares C.P. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- Kedzierski L., Montgomery J., Bullen D., Curtis J., Gardiner E., Jimenez-Ruiz A., Handman E. A leucine-rich repeat motif of Leishmania parasite surface antigen 2 binds to macrophages through the complement receptor 3. J. Immunol. 2004;172:4902–4906. doi: 10.4049/jimmunol.172.8.4902. [DOI] [PubMed] [Google Scholar]
- Kedzierski L., Zhu Y., Handman E. Leishmania vaccines: progress and problems. Parasitology. 2006;133:S87–S112. doi: 10.1017/S0031182006001831. [DOI] [PubMed] [Google Scholar]
- Kerr S.F. Palaearctic origin of Leishmania. Mem. Inst. Oswaldo Cruz. 2000;95:75–80. doi: 10.1590/s0074-02762000000100011. [DOI] [PubMed] [Google Scholar]
- Kimmel B.E., ole-MoiYoi O.K., Young J.R. Ingi, a 5.2-kb dispersed sequence element from Trypanosoma brucei that carries half of a smaller mobile element at either end and has homology with mammalian LINEs. Mol. Cell Biol. 1987;7:1465–1475. doi: 10.1128/mcb.7.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuepfer E., Stierhof Y.D., McKean P.G., Smith D.F. Characterization of a differentially expressed protein that shows an unusual localization to intracellular membranes in Leishmania major. Biochem. J. 2001;356:335–344. doi: 10.1042/0264-6021:3560335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhls K., Mauricio I.L., Pratlong F., Presber W., Schonian G. Analysis of ribosomal DNA internal transcribed spacer sequences of the Leishmania donovani complex. Microbes Infect. 2005;7:1224–1234. doi: 10.1016/j.micinf.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Kuhls K., Keilonat L., Ochsenreither S., Schaar M., Schweynoch C., Presber W., Schonian G. Multilocus microsatellite typing (MLMT) reveals genetically isolated populations between and within the main endemic regions of visceral leishmaniasis. Microbes Infect. 2007;9:334–343. doi: 10.1016/j.micinf.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Kulkarni M.M., McMaster W.R., Kamysz E., Kamysz W., Engman D.M., McGwire B.S. The major surface-metalloprotease of the parasitic protozoan, Leishmania, protects against antimicrobial peptide-induced apoptotic killing. Mol. Microbiol. 2006;62:1484–1497. doi: 10.1111/j.1365-2958.2006.05459.x. [DOI] [PubMed] [Google Scholar]
- Laurentino E.C., Ruiz J.C., Fazelinia G., Myler P.J., Degrave W., Alves-Ferreira M., Ribeiro J.M., Cruz A.K. A survey of Leishmania braziliensis genome by shotgun sequencing. Mol. Biochem. Parasitol. 2004;137:81–86. doi: 10.1016/j.molbiopara.2004.05.001. [DOI] [PubMed] [Google Scholar]
- LeBowitz J.H., Smith H.Q., Rusche L., Beverley S.M. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Leifso K., Cohen-Freue G., Dogra N., Murray A., McMaster W.R. Genomic and proteomic expression analysis of Leishmania promastigote and amastigote life stages: the Leishmania genome is constitutively expressed. Mol. Biochem. Parasitol. 2007;152:35–46. doi: 10.1016/j.molbiopara.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Lincoln L.M., Ozaki M., Donelson J.E., Beetham J.K. Genetic complementation of Leishmania deficient in PSA (GP46) restores their resistance to lysis by complement. Mol. Biochem. Parasitol. 2004;137:185–189. doi: 10.1016/j.molbiopara.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Lipoldova M., Demant P. Genetic susceptibility to infectious disease: lessons from mouse models of leishmaniasis. Nat. Rev. Genet. 2006;7:294–305. doi: 10.1038/nrg1832. [DOI] [PubMed] [Google Scholar]
- Lodge R., Descoteaux A. Modulation of phagolysosome biogenesis by the lipophosphoglycan of Leishmania. Clin. Immunol. 2005;114:256–265. doi: 10.1016/j.clim.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Lukes J., Hashimi H., Zikova A. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr. Genet. 2005;48:277–299. doi: 10.1007/s00294-005-0027-0. [DOI] [PubMed] [Google Scholar]
- Lukes J., Mauricio I.L., Schonian G., Dujardin J.C., Soteriadou K., Dedet J.P., Kuhls K., Tintaya K.W., Jirku M., Chocholova E., Haralambous C., Pratlong F., Obornik M., Horak A., Ayala F.J., Miles M.A. Evolutionary and geographical history of the Leishmania donovani complex with a revision of current taxonomy. Proc. Natl. Acad. Sci. USA. 2007;104:9375–9380. doi: 10.1073/pnas.0703678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F., Maranon C., Olivares M., Alonso C., Lopez M.C. Characterization of a non-long terminal repeat retrotransposon cDNA (L1Tc) from Trypanosoma cruzi: homology of the first ORF with the ape family of DNA repair enzymes. J. Mol. Biol. 1995;247:49–59. doi: 10.1006/jmbi.1994.0121. [DOI] [PubMed] [Google Scholar]
- Martinez-Calvillo S., Yan S., Nguyen D., Fox M., Stuart K., Myler P.J. Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol. Cell. 2003;11:1291–1299. doi: 10.1016/s1097-2765(03)00143-6. [DOI] [PubMed] [Google Scholar]
- Martinez-Calvillo S., Nguyen D., Stuart K., Myler P.J. Transcription initiation and termination on Leishmania major chromosome 3. Eukaryot. Cell. 2004;3:506–517. doi: 10.1128/EC.3.2.506-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Calvillo S., Stuart K., Myler P.J. Ploidy changes associated with disruption of two adjacent genes on Leishmania major chromosome 1. Int. J. Parasitol. 2005;35:419–429. doi: 10.1016/j.ijpara.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Masini d’Avila-Levy C., de Almeida Dias F., Nogueira de Melo A.C., Martins J.L., De Carvalho Santos Lopes A.H., Souza Dos Santos A.L., Vermelho A.B., Branquinha M.H. Insights into the role of gp63-like proteins in lower trypanosomatids. FEMS Microbiol. Lett. 2006;254:149–156. doi: 10.1111/j.1574-6968.2005.00022.x. [DOI] [PubMed] [Google Scholar]
- Matthews K.R., Tschudi C., Ullu E. A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 1994;8:491–501. doi: 10.1101/gad.8.4.491. [DOI] [PubMed] [Google Scholar]
- Mauricio I.L., Stothard J.R., Miles M.A. The strange case of Leishmania chagasi. Parasitol. Today. 2000;16:188–189. doi: 10.1016/s0169-4758(00)01637-9. [DOI] [PubMed] [Google Scholar]
- Mauricio I.L., Gaunt M.W., Stothard J.R., Miles M.A. Glycoprotein 63 (gp63) genes show gene conversion and reveal the evolution of Old World Leishmania. Int. J. Parasitol. 2006;27:27. doi: 10.1016/j.ijpara.2006.11.020. [DOI] [PubMed] [Google Scholar]
- McMahon-Pratt D., Traub-Cseko Y., Lohman K.L., Rogers D.D., Beverley S.M. Loss of the GP46/M-2 surface membrane glycoprotein gene family in the Leishmania braziliensis complex. Mol. Biochem. Parasitol. 1992;50:151–160. doi: 10.1016/0166-6851(92)90252-f. [DOI] [PubMed] [Google Scholar]
- McMahon-Pratt D., Alexander J. Does the Leishmania major paradigm of pathogenesis and protection hold for New World cutaneous leishmaniases or the visceral disease? Immunol. Rev. 2004;201:206–224. doi: 10.1111/j.0105-2896.2004.00190.x. [DOI] [PubMed] [Google Scholar]
- Mottram J.C., Coombs G.H., Alexander J. Cysteine peptidases as virulence factors of Leishmania. Curr. Opin. Microbiol. 2004;7:375–381. doi: 10.1016/j.mib.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Muller G., Schlein Y. Nectar and honeydew feeding of Phlebotomus papatasi in a focus of Leishmania major in Neot Hakikar oasis. J. Vector Ecol. 2004;29:154–158. [PubMed] [Google Scholar]
- Murphy N.B., Pays A., Tebabi P., Coquelet H., Guyaux M., Steinert M., Pays E. Trypanosoma brucei repeated element with unusual structural and transcriptional properties. J. Mol. Biol. 1987;195:855–871. doi: 10.1016/0022-2836(87)90490-6. [DOI] [PubMed] [Google Scholar]
- Murray H.W., Berman J.D., Davies C.R., Saravia N.G. Advances in leishmaniasis. Lancet. 2005;366:1561–1577. doi: 10.1016/S0140-6736(05)67629-5. [DOI] [PubMed] [Google Scholar]
- Myler P.J., Audleman L., deVos T., Hixson G., Kiser P., Lemley C., Magness C., Rickel E., Sisk E., Sunkin S., Swartzell S., Westlake T., Bastien P., Fu G., Ivens A., Stuart K. Leishmania major Friedlin chromosome 1 has an unusual distribution of protein-coding genes. Proc. Natl. Acad. Sci. USA. 1999;96:2902–2906. doi: 10.1073/pnas.96.6.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K.S., Beetham J.K., Wilson M.E., Donelson J.E. Comparison of the post-transcriptional regulation of the mRNAs for the surface proteins PSA (GP46) and MSP (GP63) of Leishmania chagasi. J. Biol. Chem. 2002;277:16489–16497. doi: 10.1074/jbc.M200174200. [DOI] [PubMed] [Google Scholar]
- Ngo H., Tschudi C., Gull K., Ullu E. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA. 1998;95:14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes H., Chance M., Ponce C., Ponce E., Maingon R. Leishmania chagasi: genotypically similar parasites from Honduras cause both visceral and cutaneous leishmaniasis in humans. Exp. Parasitol. 1997;85:264–273. doi: 10.1006/expr.1996.4133. [DOI] [PubMed] [Google Scholar]
- Oliveira F.S., Pirmez C., Pires M.Q., Brazil R.P., Pacheco R.S. PCR-based diagnosis for detection of Leishmania in skin and blood of rodents from an endemic area of cutaneous and visceral leishmaniasis in Brazil. Vet. Parasitol. 2005;129:219–227. doi: 10.1016/j.vetpar.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Pays E., Vanhamme L., Berberof M. Genetic controls for the expression of surface antigens in African trypanosomes. Annu. Rev. Microbiol. 1994;48:25–52. doi: 10.1146/annurev.mi.48.100194.000325. [DOI] [PubMed] [Google Scholar]
- Peacock C.S., Seeger K., Harris D., Murphy L., Ruiz J.C., Quail M.A., Peters N., Adlem E., Tivey A., Aslett M., Kerhornou A., Ivens A., Fraser A., Rajandream M.A., Carver T., Norbertczak H., Chillingworth T., Hance Z., Jagels K., Moule S., Ormond D., Rutter S., Squares R., Whitehead S., Rabbinowitsch E., Arrowsmith C., White B., Thurston S., Bringaud F., Baldauf S.L., Faulconbridge A., Jeffares D., Depledge D.P., Oyola S.O., Hilley J.D., Brito L.O., Tosi L.R., Barrell B., Cruz A.K., Mottram J.C., Smith D.F., Berriman M. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet. 2007;39:839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrosa A.L., Silva A.M., Ruiz J.C., Cruz A.K. Characterization of LST-R533: uncovering a novel repetitive element in Leishmania. Int. J. Parasitol. 2006;36:211–217. doi: 10.1016/j.ijpara.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Rao V., Fujiwara N., Porcelli S.A., Glickman M.S. Mycobacterium tuberculosis controls host innate immune activation through cyclopropane modification of a glycolipid effector molecule. J. Exp. Med. 2005;201:535–543. doi: 10.1084/jem.20041668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson K.A., Beverley S.M. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol. Biochem. Parasitol. 2003;128:217–228. doi: 10.1016/s0166-6851(03)00079-3. [DOI] [PubMed] [Google Scholar]
- Rochette A., McNicoll F., Girard J., Breton M., Leblanc E., Bergeron M.G., Papadopoulou B. Characterization and developmental gene regulation of a large gene family encoding amastin surface proteins in Leishmania spp. Mol. Biochem. Parasitol. 2005;140:205–220. doi: 10.1016/j.molbiopara.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Romano N., Macino G. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 1992;6:3343–3353. doi: 10.1111/j.1365-2958.1992.tb02202.x. [DOI] [PubMed] [Google Scholar]
- Sacks D., Kamhawi S. Molecular aspects of parasite–vector and vector–host interactions in leishmaniasis. Annu. Rev. Microbiol. 2001;55:453–483. doi: 10.1146/annurev.micro.55.1.453. [DOI] [PubMed] [Google Scholar]
- Sacks D., Noben-Trauth N. The immunology of susceptibility and resistance to Leishmania major in mice. Nat. Rev. Immunol. 2002;2:845–858. doi: 10.1038/nri933. [DOI] [PubMed] [Google Scholar]
- Sadlova J., Volf P., Victoir K., Dujardin J.C., Votypka J. Virulent and attenuated lines of Leishmania major: DNA karyotypes and differences in metalloproteinase GP63. Folia Parasitol. (Praha) 2006;53:81–90. [PubMed] [Google Scholar]
- Satoskar A.R., Bozza M., Rodriguez Sosa M., Lin G., David J.R. Migration-inhibitory factor gene-deficient mice are susceptible to cutaneous Leishmania major infection. Infect. Immun. 2001;69:906–911. doi: 10.1128/IAI.69.2.906-911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffter S.M., Ro Y.T., Chung I.K., Patterson J.L. The complete sequence of Leishmania RNA virus LRV2-1, a virus of an Old World parasite strain. Virology. 1995;212:84–90. doi: 10.1006/viro.1995.1456. [DOI] [PubMed] [Google Scholar]
- Shi H., Ullu E., Tschudi C. Function of the Trypanosome Argonaute 1 protein in RNA interference requires the N-terminal RGG domain and arginine 735 in the Piwi domain. J. Biol. Chem. 2004;279:49889–49893. doi: 10.1074/jbc.M409280200. [DOI] [PubMed] [Google Scholar]
- Shi H., Tschudi C., Ullu E. An unusual Dicer-like1 protein fuels the RNA interference pathway in Trypanosoma brucei. RNA. 2006;12:2063–2072. doi: 10.1261/rna.246906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Tschudi C., Ullu E. Functional replacement of Trypanosoma brucei Argonaute by the human slicer Argonaute2. RNA. 2006;12:943–947. doi: 10.1261/rna.20806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M.F. SINEs and LINEs: highly repeated short and long interspersed sequences in mammalian genomes. Cell. 1982;28:433–434. doi: 10.1016/0092-8674(82)90194-5. [DOI] [PubMed] [Google Scholar]
- Stevens J.R., Noyes H.A., Schofield C.J., Gibson W. The molecular evolution of Trypanosomatidae. Adv. Parasitol. 2001;48:1–56. doi: 10.1016/s0065-308x(01)48003-1. [DOI] [PubMed] [Google Scholar]
- Teng S.C., Wang S.X., Gabriel A. A new non-LTR retrotransposon provides evidence for multiple distinct site-specific elements in Crithidia fasciculata miniexon arrays. Nucleic Acids Res. 1995;23:2929–2936. doi: 10.1093/nar/23.15.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tielens A.G., Van Hellemond J.J. Differences in energy metabolism between trypanosomatidae. Parasitol. Today. 1998;14:265–272. doi: 10.1016/s0169-4758(98)01263-0. [DOI] [PubMed] [Google Scholar]
- Turco S.J., Spath G.F., Beverley S.M. Is lipophosphoglycan a virulence factor? A surprising diversity between Leishmania species. Trends Parasitol. 2001;17:223–226. doi: 10.1016/s1471-4922(01)01895-5. [DOI] [PubMed] [Google Scholar]
- Ullu E., Matthews K.R., Tschudi C. Temporal order of RNA-processing reactions in trypanosomes: rapid trans splicing precedes polyadenylation of newly synthesized tubulin transcripts. Mol. Cell Biol. 1993;13:720–725. doi: 10.1128/mcb.13.1.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E., Tschudi C., Chakraborty T. RNA interference in protozoan parasites. Cell Microbiol. 2004;6:509–519. doi: 10.1111/j.1462-5822.2004.00399.x. [DOI] [PubMed] [Google Scholar]
- Victoir K., Dujardin J.C., de Doncker S., Barker D.C., Arevalo J., Hamers R., Le Ray D. Plasticity of gp63 gene organization in Leishmania (Viannia) braziliensis and Leishmania (Viannia) peruviana. Parasitology. 1995;111:265–273. doi: 10.1017/s0031182000081828. [DOI] [PubMed] [Google Scholar]
- Villanueva M.S., Williams S.P., Beard C.B., Richards F.F., Aksoy S. A new member of a family of site-specific retrotransposons is present in the spliced leader RNA genes of Trypanosoma cruzi. Mol. Cell Biol. 1991;11:6139–6148. doi: 10.1128/mcb.11.12.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B., Ersfeld K., Gull K. Repetitive elements in genomes of parasitic protozoa. Microbiol. Mol. Biol. Rev. 2003;67:360–375. doi: 10.1128/MMBR.67.3.360-375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer G., Dooley S. Phylogenetic analysis of Leishmania RNA virus and Leishmania suggests ancient virus–parasite association. Nucleic Acids Res. 1995;23:2300–2304. doi: 10.1093/nar/23.12.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wilson M.E., Jeronimo S.M., Pearson R.D. Immunopathogenesis of infection with the visceralizing Leishmania species. Microb. Pathog. 2005;38:147–160. doi: 10.1016/j.micpath.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Xu P., Widmer G., Wang Y., Ozaki L.S., Alves J.M., Serrano M.G., Puiu D., Manque P., Akiyoshi D., Mackey A.J., Pearson W.R., Dear P.H., Bankier A.T., Peterson D.L., Abrahamsen M.S., Kapur V., Tzipori S., Buck G.A. The genome of Cryptosporidium hominis. Nature. 2004;431:1107–1112. doi: 10.1038/nature02977. [DOI] [PubMed] [Google Scholar]
- Yao C., Donelson J.E., Wilson M.E. The major surface protease (MSP or GP63) of Leishmania sp. Biosynthesis, regulation of expression, and functionc. Mol. Biochem. Parasitol. 2003;132:1–16. doi: 10.1016/s0166-6851(03)00211-1. [DOI] [PubMed] [Google Scholar]
- Yao C., Luo J., Hsiao C., Donelson J.E., Wilson M.E. Internal and surface subpopulations of the major surface protease (MSP) of Leishmania chagasi. Mol. Biochem. Parasitol. 2005;139:173–183. doi: 10.1016/j.molbiopara.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Zajtchuk J.T., Casler J.D., Netto E.M., Grogl M., Neafie R.C., Hessel C.R., de Magalhaes A.V., Marsden P.D. Mucosal leishmaniasis in Brazil. Laryngoscope. 1989;99:925–939. doi: 10.1288/00005537-198909000-00006. [DOI] [PubMed] [Google Scholar]
- Zhang W.W., Mendez S., Ghosh A., Myler P., Ivens A., Clos J., Sacks D.L., Matlashewski G. Comparison of the A2 gene locus in Leishmania donovani and Leishmania major and its control over cutaneous infection. J. Biol. Chem. 2003;278:35508–35515. doi: 10.1074/jbc.M305030200. [DOI] [PubMed] [Google Scholar]
- Zilberstein D. Transport of nutrients and ions across membranes of trypanosomatid parasites. Adv. Parasitol. 1993;32:261–291. doi: 10.1016/s0065-308x(08)60209-2. [DOI] [PubMed] [Google Scholar]