Abstract
The CYP1A family of cytochrome P450s (CYPs), comprising CYP1A1, CYP1A2, and CYP1B1, plays a role in bioactivation of several procarcinogens to carcinogenic derivatives, and also in detoxification of several xenobiotic compounds. Resveratrol (3,4,5-trihydroxystelbine) is a naturally occurring compound that has been shown in a number of studies to inhibit the induction of CYP1A1 and CYP1B1 by dioxin (2,3,7,8-tetrachloro-dibenzo-p-dioxin), but the mechanism(s) of resveratrol inhibition is controversial. In the current study, 100nM dioxin treatment for 24, 48, and 72 h induced CYP1A1, CYP1A2, and CYP1B1 mRNA levels in the human breast cancer cell line MCF-7, and CYP1A1 and CYP1A2 mRNA levels in the human hepatocellular carcinoma cell line, HepG2. Simultaneous treatment with 10μM resveratrol significantly inhibited dioxin-induced mRNA expression levels of these genes in both cell lines. Our studies are novel in that we used the chromatin immunoprecipitation assay to assay dioxin-induced recruitment of the aryl hydrocarbon receptor (AHR), and aryl hydrocarbon nuclear translocator (ARNT) to the enhancer regions and recruitment of RNA polymerase II to the promoter regions, of the CYP1A1 and CYP1B1 genes in their natural chromosomal settings. These recruitments were significantly inhibited in cells cotreated with resveratrol. Our studies thus indicate that resveratrol inhibits dioxin induction of the CYP1 family members either by directly or indirectly inhibiting the recruitment of the transcription factors AHR and ARNT to the xenobiotic response elements of the corresponding genes. The reduced transcriptional factor binding at their enhancers then results in reduced pol II recruitment at the promoters of these genes.
Keywords: dioxin, resveratrol, CYP1A1, CYP1B1, and ChIP assay
The cytochrome P450 (CYP) CYP1 family includes three proteins, CYP1A1, CYP1A2, and CYP1B1, which are involved in the bioactivation of numerous procarcinogens compounds to mutagenic and carcinogenic derivatives (Mikstacka et al., 2008; Rendic, 2002). The transcriptional regulation of CYP1 family genes are regulated through the aryl hydrocarbon receptor (AHR) pathway. AHR is a ligand activated transcriptional factor. Its ligands include polycyclic aromatic hydrocarbons (PAHs), aromatic amines and halogenated aromatic hydrocarbons (HAHs, including 2,3,7,8-tetrachloro-dibenzo-p-dioxin (also commonly referred to as dioxin). After binding ligand the AHR translocates to the nucleus where it binds to its heterodimeric partner, the aryl hydrocarbon nuclear translocator (ARNT), thereby forming an active transcription factor complex, termed the aryl hydrocarbon receptor complex (AHRC) (Hankinson, 1995). The AHRC binds to the consensus regulatory sequences referred as xenobiotic response elements (XREs), located near its target genes, including CYP1A1, CYP1A2, and CYP1B1, resulting in their transcriptional upregulation (Hankinson, 1995).
Resveratrol is a naturally occurring polyphenolic compound that occurs in grapes, peanuts, berries, and a number of plants used in traditional Asian medicine (Aziz et al., 2003; Jayatilake et al., 1993). This compound displays several useful properties applicable to human health, including cardioprotective activity and inhibitory activity toward the ageing process. Resveratrol has also been reported to exhibit chemopreventive activity toward the development of several cancers, at all three stages of carcinogenesis: that is, initiation, promotion, and progression. Although resveratrol acts as a strong antioxidant and a free radical scavenger, much of its mechanism of action are not clearly understood (Aziz et al., 2003; Chen et al., 2004; Jang et al., 1997; Khan et al., 2008; Le Corre et al., 2005; Potter et al., 2002; Signorelli and Ghidoni, 2005; Stewart et al., 2003; Whitsett and Lamartiniere, 2006). Several studies have reported that resveratrol inhibits the expression of a number of cytochrome P450 genes, including CYP1A1, CYP1B1, CYP1A2, CYP2E1, CYP3A, and aromatase (CYP19) in cancer cell lines of different tissue origin in humans and other mammals, and also inhibits the catalytic activities of several of these CYPs, and it has been suggested that these inhibitions may underlie some of the cancer chemopreventive activity of this compound (Aluru and Vijayan, 2006; Casper et al., 1999; Chang et al., 2000, 2001; Chen et al., 2004; Chun et al., 1999; Ciolino and Yeh, 1999, 2001; Ciolino et al., 1998; Lee and Safe, 2001; Liu et al., 2004; Mikstacka et al., 2007, 2008; Mollerup et al., 2001; Piver et al., 2001; Wu et al., 2005).
Although a number of studies have demonstrated that resveratrol inhibits dioxin induction of CYP1A1 and CYP1B1 mRNAs in a dose dependent manner, several contradictory observations were made concerning the mechanism of this inhibitory effect, and no unifying model has emerged (Casper et al., 1999; Chen et al., 2004; Ciolino et al., 1998; Lee and Safe, 2001). Certain studies reported that resveratrol triggered AHR (in concert with ARNT) to bind an XRE-containing double-stranded oligonucleotide in an in vitro electromobility shift assay (EMSA), and also reported that resveratrol did not inhibit the dioxin-stimulated binding of AHR to the XRE sequence (Casper et al., 1999; Lee and Safe, 2001). In contrast, other investigators, also using the EMSA assay, reported that resveratrol did not induce binding of AHR to an XRE-containing oligonucleotide, but did inhibit dioxin-induced binding of AHR to the XRE (Chen et al., 2004; Ciolino et al., 1998). We address the mechanism of resveratrol's inhibitory activity toward CYP1A1 and CYP1B1 induction in this paper. Our studies differ from those previously reported in that we investigate the effect of resveratrol on the recruitment of the AHRC to the CYP1A1 and CYP1B1 genes in their natural chromosomal configurations in vivo. We confirm in this paper that resveratrol inhibits dioxin induction of the CYP1A1 and CYP1B1 genes. We further demonstrate using the in vivo ChIP assay, that resveratrol on its own does not induce binding of AHR/ARNT to the enhancer regions of the CYP1A1 or CYP1B1 genes, but does inhibit dioxin's ability to induce this binding, subsequently inhibiting the pol II recruitment to the promoter regions of these genes. Our observations therefore clarify the mechanism of resveratrol's inhibitory action, and enhance our understanding of this potentially important protective nutrient.
MATERIALS AND METHODS
Cell culture and reagents.
The human breast cancer cell line MCF-7, and human hepatic cancer cell line, HepG2, were grown as monolayers in α-minimal essential media and DMEM media respectively, containing 10% fetal bovine serum, 5% fungizone, 5% penicillin-streptomycin (Invitrogen, Carlsbad, CA) at 37°C and 5% C02. The tissue culture dishes used to grow HepG2 cells were coated with 5 ml of 50 μg/ml poly-l-lysine and dried before plating the cells. Cells were treated with 100nM dioxin (Wellington Laboratories, Guelph, Ontario, Canada) dissolved in dimethyl sulfoxide (DMSO), at a final concentration of 0.1% DMSO in the medium. Resveratrol (∼99% purity) was purchased from Sigma Chemical (St. Louis, MO). The antibodies used for ChIP analysis are from the indicated sources (Hsu et al., 2007; Probst et al., 1993) and SantaCruz Biotechnologies, CA, for AHR and pol II respectively.
Reverse transcription and real-time PCR.
he levels of the mRNAs for CYP1A1, CYP1B1, and the constitutively expressed ribosomal subunit 36B4 were determined by either Taqman or SYBR green quantitative polymerase chain reaction. Total RNA was isolated using RNeasy mini columns (Qiagen, Valencia, CA) according to the manufacture's protocol and quantified on a SmartSpec 3000 spectrophotometer (BioRad, Hercules, CA). Five micrograms of total RNA was used for complementary DNA (cDNA) synthesis in a 20-μl reaction using Superscript III reverse transcriptase (Invitrogen) and primed with random hexamers (Invitrogen) according to the manufacturer's instructions. cDNA synthesis was performed using incubations at 25°C for 10 min, 48°C for 30 min, and 95°C for 5 min, using an Icycler thermalcycler (BioRad). cDNAs were diluted 10-fold in autoclaved water. Standard curves were generated using the 72-h dioxin-treated cDNA sample and performing a 10-fold dilution series. The primers and probes for real-time PCR were designed using Primer Express software (Applied Biosystems, CA) and were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA) (see Table 1). Real-time PCR assays were performed using an Applied Biosystems 7500 machine. Real-time PCR reaction parameters were 50°C for 2 min, 95°C for 10 min, 92°C for 15 s, 60°C for 1 min, then back to the 92°C step 40 times. CYP1A1 and CYP1B1 mRNA quantities were normalized to the amount of 36B4 mRNA (Hsu et al., 2007). The relative expression levels of all genes measured were reported using the standard curve generated from MCF-7 cDNA treated for 72 h with dioxin, thus allowing us to report HepG2 mRNA expression levels relative to MCF-7 expression levels. In all real-time PCR analyses, three replicates were analyzed for each biological sample, and the standard deviations from those three replicates are reported.
TABLE 1.
List of the Real-Time PCR Primers
| S. no. | Primer name | Primer sequence |
| 1 | CYP1A1 forward | CAAGAGGAGCTAGACACAGTGATT |
| 2 | CYP1A1 reverse | AGCCTTTCAAACTTGTGTCTCTTGT |
| 3 | CYP1B1 forward | TTCGGCCACTACTCGGAGC |
| 4 | CYP1B1 reverse | AAGAAGTTGCGCATCATGCTG |
| 5 | CYP1A2 forward | TGGCCTCTGCCATCTTCTG |
| 6 | CYP1A2 reverse | GGACCCGAGGCCTCAAAC |
| 7 | AHR forward | CCTCCTGGGTTCAAGTGATTCT |
| 8 | AHR reverse | CACGCCACCATGCCTGTA |
| 9 | CYP1A1 Ehancer F | CAGGCTTACGCACGCTAGC |
| 10 | CYP1A1 Ehancer R | ACGCGAGACAGCAGGAGG |
| 11 | CYP1B1 Ehancer F | AGTCACGCAACCTCTCTGAACC |
| 12 | CYP1B1 Ehancer R | GCCCTTTCCTACATGCTGATG |
| 13 | CYP1A1 promoter F | CCCGCCTATAAAGGTGGCA |
| 14 | CYP1A1 promoter R | AGCAACTCACCTGAGGTACTG |
| 15 | CYP1B1 promoter F | GTTTGGCGCTGGGTTAC |
| 17 | CYP1B1 promoter R | AGGTCGGAGCTGACTCTCT |
Chromatin immunoprecipitation assay.
MCF-7 cells were treated with 100nM dioxin for 60 min. Cross-linking was achieved by the addition of a 1% formaldehyde solution for 10 min at 37°C. The cells were then rinsed twice with cold PBS and collected in 1 ml of ice-cold PBS + 1× protease inhibitor solution (Roche, Palo Alto, CA). Cells were collected by centrifugation at 600 × g for 5.5 min at 4°C in a Beckman tabletop centrifuge. The pellets were then resuspended in 300 μl of lysis buffer (1% sodium dodecyl sulfate [SDS], 10mM ethylenediaminetetraacetic acid [EDTA], 50mM Tris-HCl [pH 8.1] + 1 × protease inhibitor) and incubated on ice for 10 min. The cell lysates were then sonicated three times on 80% power for 10 s to shear DNA fragments to sizes between 200 and 900 base pairs using an ultra sonicator (Diagenode, NJ). Cellular debris was precipitated by centrifugation for 10 min at 7450 × g at 4°C. The supernatants were then diluted 1:5 in 1% Triton-X 100, 2mM EDTA, 150mM NaCl, 20mM Tris-HCl (pH 8.1), 1 × protease inhibitor solution. Immunoclearing was achieved by the addition of 30 μl of a 50% slurry of protein-A sepharose beads in Tris-EDTA/2.5 μg of sonicated salmon sperm DNA/bovine serum albumin solution, and incubated on a rotator at 4°C for 1 h. Supernatants were placed in a new tube and treated with 2 μg of antibodies of interest overnight on a rotator at 4°C. The solutions were then treated with 50 μl of a 50% slurry of protein-A sepharose beads in Tris EDTA/Sonicated Salmon sperm DNA/Bovine serum albumin solution and incubated for 2 h at 4°C on a rotator. The beads were then pelleted and sequentially washed in buffers A, B, LiCl, and two times in 10mM Tris-HCl (pH 8.1), 1mM EDTA (Tris EDTA). Buffer A: (0.1% SDS, 1% Triton-X 100, 2mM EDTA, 20mM Tris-HCl [pH 8.1], 150mM NaCl); buffer B: (0.1% SDS, 1% Triton-X 100, 2mM EDTA, 20mM Tris-HCl [pH 8.1], 500mM NaCl); LiCl buffer: (0.25M LiCl, 1% NP-40, 1% deoxycholate, 1mM EDTA, 10mM Tris-HCl [pH 8.1]). Chromatin complexes were eluted by the addition of 0.5 ml of freshly prepared elution buffer (1% SDS, 0.1M NaHCO3). The cross-linking was reversed by incubating samples in high salt conditions at 65°C water bath overnight for 18 h. The solutions were then digested with 40 μg of PCR-grade recombinant proteinase K solution (Roche, Palo Alto, CA) for 1 h at 45°C. DNA was extracted using DNeasy mini columns (Qiagen), and finally eluted in a volume of 50 μl using PCR-grade water. The primers used for housekeeping gene, 36B4, were reported previously (Hsu et al., 2007). The remainder primer sets used for real-time PCR analysis are listed in Table 1. All the ChIP analyses were carried out at least three times, and the data from representative experiments are reported in the manuscript. All statistical analyses were done using the Student's two-tailed t-test.
RESULTS
Dioxin Induction of CYP1A1, CYP1B1, CYP1A2, AHR, and ARNT mRNAs
The human breast cancer cell line MCF-7 and the human hepatic cancer cell line HepG2 were treated with 100nM dioxin for 0, 24, 48, and 72 h. CYP1A1, CYP1B1, CYP1A2, AHR, and ARNT mRNAs were then measured using SYBR green real-time PCR. 100nM dioxin was used to ensure maximal induction of the above mentioned genes. Our laboratory and several others have consistently showed that 100nM dioxin levels are not toxic to the human cancer cell lines used in these studies. The relative expression levels of all genes measured were reported using the standard curve generated from MCF-7 cDNA treated for 72 h with dioxin, thus allowing us to report HepG2 mRNA expression levels relative to MCF-7 expression levels. Dioxin strongly induced CYP1A1 mRNA levels in both cell lines by 24 h, and the induced levels were maintained or further increased over the rest of the treatment period (Fig. 1A). Dioxin induction of the mRNA for CYP1B1 was observed only in MCF-7 cells: no CYP1B1 mRNA levels were observed in HepG2 cells even after 72 h of dioxin treatment (Fig. 1C), similar to the results reported by others (Kress and Greenlee, 1997). Dioxin-induced CYP1A2 mRNA in both the cell lines by 24 h, although the induced levels were significantly greater in HepG2 cells than that in MCF-7 cells during the entire treatment period (Fig. 1B). We also investigated the time course of dioxin-induced mRNA expression levels of the CYP1A1, CYP1A2 and CYP1B1 genes at 2-h intervals up to 24 h after dioxin treatment, and the results indicated that significant dioxin-induction of these genes was observed by 2 h, though maximal induction was at 24 h (Beedanagari and Hankinson, submitted). We also investigated the mRNA expression levels of the transcription factors AHR and ARNT in both the cell lines. Expression of AHR and ARNT mRNAs were unaffected by dioxin treatment, and the expression levels of these mRNAs were very similar between the two cell lines (Figs. 1D and 1E).
FIG. 1.
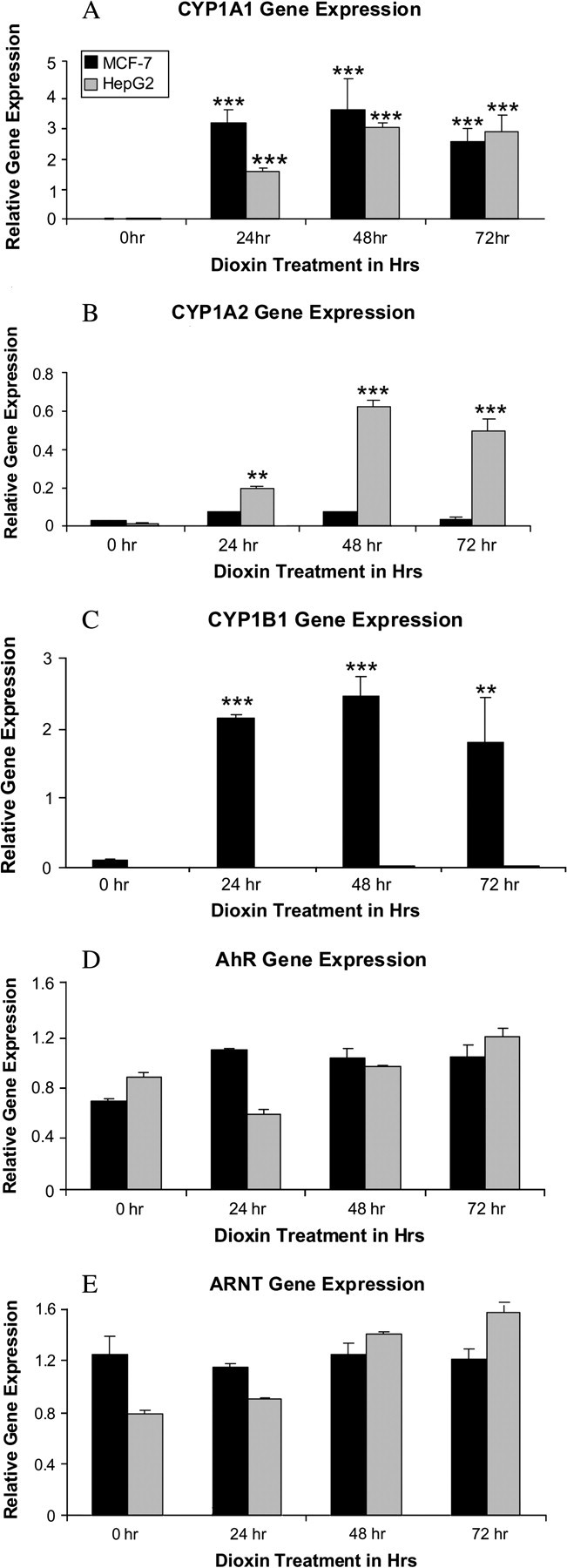
Dioxin-induction of the CYP1A1, CYP1B1, CYP1A2, AHR and ARNT genes expression. MCF-7 and HepG2 cells were treated with 100nM dioxin for 0, 24, 48, and 72 h. The relative amounts of the CYP1A1 (A), CYP1A2 (B), CYP1B1 (C), AHR (D), and ARNT (E) mRNAs were measured using SYBR green real-time PCR, and were normalized to the constitutively expressed ribosomal subunit 36B4 gene. The mRNA levels of the MCF-7 cells are indicated with black bars and HepG2 cells are indicated with gray bars. In this and subsequent figures, the relative expression levels of all genes were reported using standard curves generated from MCF-7 cDNA treated for 72 h with dioxin, thus allowing us to report HepG2 mRNA expression levels relative to MCF-7 expression levels. *p < 0.05, **p < 0.01, ***p < 0.001 compared with control DMSO or 0-h treatment.
Resveratrol Inhibits Dioxin-Induced Expression of CYP1A1, CYP1A2, and CYP1B1
MCF-7 and HepG2 cells were treated for 48 h with or without resveratrol (10 or 20μM) and with or without 100nM dioxin and then analyzed for mRNA levels of the cytochrome P450s. Resveratrol treatment on its own did not affect the expression of the CYP1A1, CYP1A2, and CYP1B1 mRNAs. However, resveratrol significantly inhibited dioxin induction of CYP1A1, CYP1A2 and CYP1B1 mRNAs in MCF-7 cells and CYP1A1, CYP1A2 in HepG2 cells (Fig. 2). Maximal inhibition of dioxin-induced mRNAs expression by resveratol was achieved at 10μM in a dose dependent manner (Fig. 3).
FIG. 2.
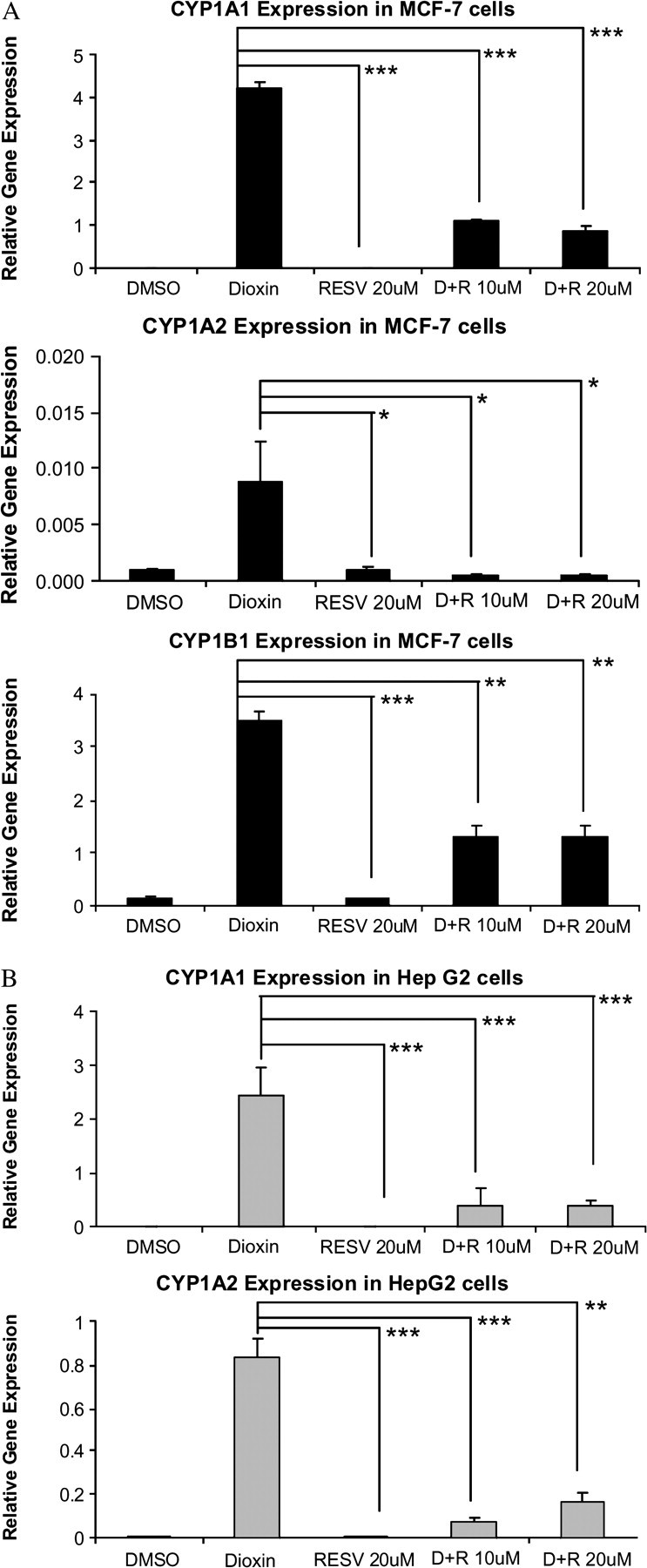
Resveratrol inhibits dioxin induction of CYP1A1, CYP1A2, and CYP1B1 mRNAs in MCF-7 and HepG2 cells. MCF-7 and HepG2 cells were treated with either 10 or 20μM resveratrol (R) or the vehicle, DMSO, for 48 h. The cells were also treated with 100 nM dioxin (D) or the vehicle, DMSO during this time. The cells were either treated with resveratrol alone or cotreated with dioxin and resveratrol for 48 h. DMSO was used as vehicle for resveratrol and dioxin and was therefore included in the negative controls. The mRNA levels of the MCF-7 cells are indicated with black bars (A) and HepG2 (B) cells are indicated with gray bars. *p < 0.05, **p < 0.01, ***p < 0.001 compared with dioxin alone.
FIG. 3.
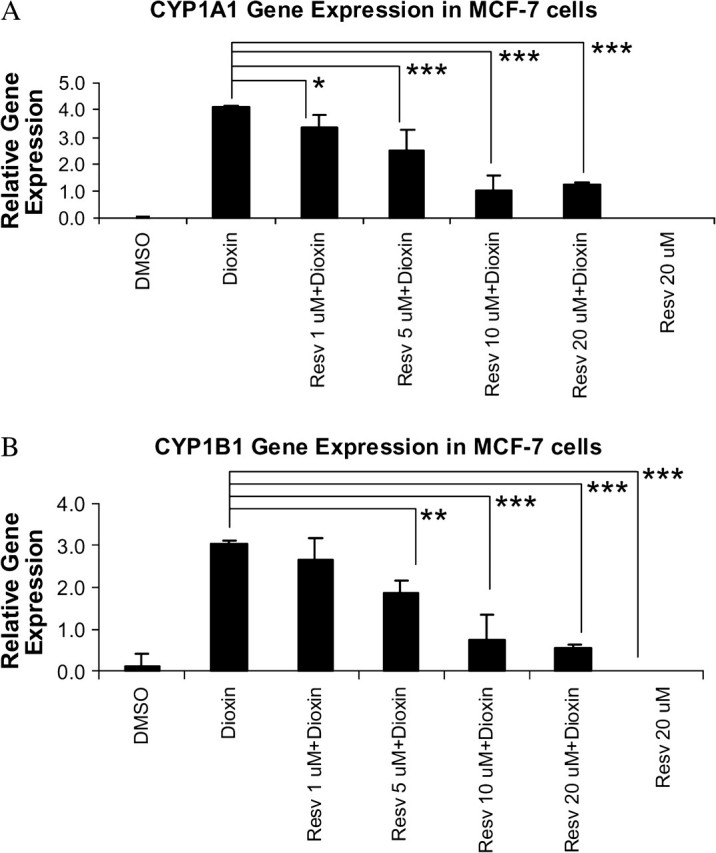
Resveratrol inhibits dioxin-induced expression of the CYP1A1 and CYP1B1 genes in a dose dependent manner. MCF-7 cells were treated with 1, 5, 10, or 20μM resveratrol or with DMSO (control) for 48 h. The cells were also treated with 100nM dioxin or DMSO (control) for 48 h. Resveratrol inhibited mRNA levels of the CYP1A1 (A) and CYP1B1 (B) genes were measured by real-time PCR. *p < 0.05, **p < 0.01, ***p < 0.001 compared with dioxin alone.
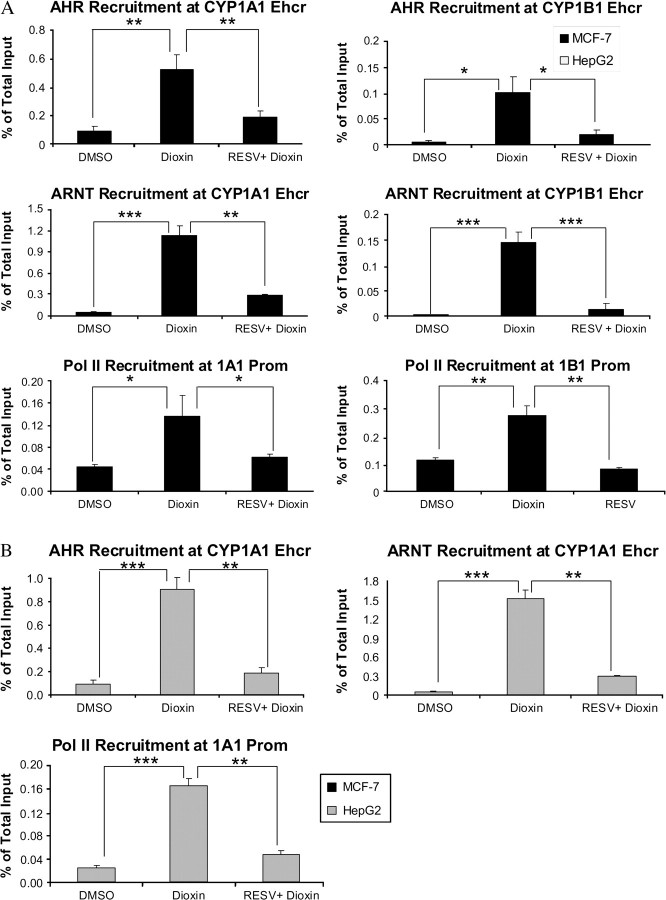
Resveratrol Inhibits Dioxin-Induced Recruitment of AHR, ARNT, and pol II to the Regulatory Regions of the CYP1A1 and CYP1B1 Genes In Vivo
To understand the mechanism(s) involved in the inhibition of dioxin-induced expression of the CYP1A1and CYP1B1 genes by resveratrol, we studied the recruitment of the transcription factors AHR and ARNT to the enhancer regions, and pol II to the promoter regions of these genes using the ChIP assay. The results showed that the recruitment of both the AHR and ARNT to the enhancer regions of the CYP1A1 and CYP1B1 genes were significantly reduced in the samples treated with 20μM resveratrol and 100nM dioxin, compared with the samples treated with dioxin alone (Fig. 4). We also studied the recruitment of the pol II to the promoter regions of these genes. The results showed that this recruitment was significantly inhibited in the samples treated with 20μM resveratrol and 100nM dioxin, compared with the samples treated with dioxin alone, in both the cell lines (Figs. 4A-3 and 4A-6 and 4B-3). (We did not examine recruitment of AHR/ARNT to the promoter or pol II to the enhancer regions because our laboratory has consistently demonstrated an absence of recruitment of these proteins to these regions [Taylor et al., 2009]). ChIP analyses were also performed at 24 h in MCF-7 cells, and the results are in total agreement with the 60-min ChIP analyses (Supplementary Fig. S1). Thus resveratrol prevents dioxin-induced association of the AHRC to the XREs in the enhancer regions of the CYP1A1 and CYP1B1 genes in their natural chromosomal configuration in vivo, resulting in inhibition of the subsequent recruitment of RNA polymerase to the promoter regions of the genes. The lack of this last recruitment would inhibit transcriptional initiation of these genes.
FIG. 4.
Resveratrol inhibits dioxin-induced recruitment of AHR and polII to the enhancer and promoter regions of the CYP1A1 and CYP1B1 genes. MCF-7 (A) and HepG2 (B) cells were treated with DMSO or with100nM dioxin, with or without 20μM resveratrol for 60 min and subjected to ChIP analysis. Sonicated whole cell lysates were probed with 2 μg of anti-human AHR, ARNT, or pol II antibodies. The recruitments of AHR and ARNT were measured using primers targeted to the enhancer regions, and pol II recruitment was performed using primers targeted to the promoter regions of the CYP1A1 and CYP1B1 genes. The results were plotted relative to those of the total input controls (untreated chromatin). The levels of recruitment in the MCF-7 cells are indicated with black bars and those for HepG2 cells are indicated with gray bars. *p < 0.05, **p < 0.01, ***p < 0.001 compared with dioxin alone.
DISCUSSION
A diet rich in certain vegetables has long been associated with cancer prevention (Köhle and Bock, 2006). Phytochemicals in plants are thought to have chemopreventive effects and prevent or disrupt carcinogenesis at one or more stages of the process including initiation, promotion and progression. The naturally occurring compound resveratrol has been reported to have a diverse spectrum of beneficial biological effects on humans, including cancer chemoprevention (Aziz et al., 2003). Resveratrol has previously been reported to inhibit the induction of CYP1A1 and CYP1B1 by dioxin (and presumably by PAHs) and this effect has been suggested to underlie its cancer chemopreventive effects, at least in part. However, conflicting results have been reported concerning resveratrol's mechanism of action in this regard. We endeavored to resolve these conflicts and to further delineate resveratrol's mechanism of action, because such information would be important for further development of this potentially promising cancer chemopreventive agent.
Some investigations reported that resveratrol triggered AHR (in concert with ARNT) to bind an XRE-containing double-stranded oligonucleotide in an in vitro EMSA, and also reported that resveratrol did not inhibit the dioxin-stimulated binding of AHR to the XRE sequence (Casper et al., 1999; Lee and Safe, 2001). In contrast, other investigators, also using the EMSA assay, reported that resveratrol did not induce binding of AHR to an XRE-containing oligonucleotide, but did inhibit dioxin-induced binding of AHR to the XRE (Chen et al., 2004; Ciolino et al., 1998). We demonstrate in this paper, using the ChIP assay, that resveratrol on its own does not induce binding of AHR/ARNT to the enhancers of the CYP1A1 or CYP1B1 genes, but does inhibit dioxin's ability to induce this binding. The results of our ChIP analysis are therefore consistent with the results reported by the last mentioned investigators. Unlike the EMSA assay, our ChIP data address the effect of resveratrol on the binding of the AHRC to the XRE sequences of the CYP1A1 and CYP1B1 enhancers in vivo, and also in their natural chromosomal settings, and are therefore more truly reflective of resveratrol's mechanism of action in vivo. Our observation that resveratrol inhibits dioxin induction of the recruitment of RNA polymerase II to the promoter regions of the CYP1A1 and CYP1B1 genes indicates that resveratrol inhibits transcriptional initiation of these genes. Although our results are compatible with the notion that resveratrol inhibits nuclear translocation of the dioxin-induced AHR, this is unlikely, because Casper et al. (1999) using a GFP-tagged AHR vector, demonstrated that resveratrol-induced AHR nuclear translocation in a similar manner to that of dioxin, suggesting that nucleolar translocation is not inhibited by resveratrol. Hence we propose that binding of AHR to the XREs represents the target of resveratrol in vivo.
It is important to note that we and all the above mentioned investigators studied the same or similar human cell lines (HepG2, MCF-7, or other breast cancer cell lines), and so the differences in results obtained are unlikely to result from differences in the cell types used. Interestingly, we found that resveratrol did not inhibit non-induced levels of the cytochrome P450s in MCF-7 cells, which contrasts with the observations of Ciolino et al. (1998) and Chen et al. (2004). In one study it was concluded that resveratrol competes with binding of dioxin to the AHR (Casper et al., 1999), whereas in another study it was reported that resveratrol is not a ligand for AHR (Ciolino et al., 1998). Our data does not allow us to distinguish between these two possibilities.
In conclusion, we demonstrate here that resveratrol on its own does not activate binding of AHR (or ARNT) to the enhancer regions of the CYP1A1 and CYP1B1 genes, but inhibits the dioxin induction of such binding, thereby preventing the subsequent recruitment of RNA polymerase II to the promoter regions of the genes, and their transcription.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institute of Health (R01CA28868) to O.H.; and University of California Toxic Substances Research and Training Program fellowships to S.R.B, I.B., and P.B.
Supplementary Material
Acknowledgments
We thank Kelly Joiner for her help in formatting this manuscript. We also thank Feng Wang, Erin Hsu, and Robert Taylor for their advice.
References
- Aluru N, Vijayan MM. Resveratrol affects CYP1A expression in rainbow trout hepatocytes. Aquat. Toxicol. 2006;77:291–297. doi: 10.1016/j.aquatox.2005.12.010. [DOI] [PubMed] [Google Scholar]
- Aziz MH, Kumar R, Ahmad N. Cancer chemoprevention by resveratrol: In vitro and in vivo studies and the underlying mechanisms (review) Int. J. Oncol. 2003;23:17–28. [PubMed] [Google Scholar]
- Casper RF, Quesne M, Rogers IM, Shirota T, Jolivet A, Milgrom E, Savouret JF. Resveratrol has antagonist activity on the aryl hydrocarbon receptor: Implications for prevention of dioxin toxicity. Mol. Pharmacol. 1999;56:784–790. [PubMed] [Google Scholar]
- Chang TK, Chen J, Lee WB. Differential inhibition and inactivation of human CYP1 enzymes by trans-resveratrol: Evidence for mechanism-based inactivation of CYP1A2. J. Pharmacol. Exp. Ther. 2001;299:874–882. [PubMed] [Google Scholar]
- Chang TK, Lee WB, Ko HH. Trans-resveratrol modulates the catalytic activity and mRNA expression of the procarcinogen-activating human cytochrome P450 1B1. Can. J. Physiol. Pharmacol. 2000;78:874–881. [PubMed] [Google Scholar]
- Chen ZH, Hurh YJ, Na HK, Kim JH, Chun YJ, Kim DH, Kang KS, Cho MH, Surh YJ. Resveratrol inhibits TCDD-induced expression of CYP1A1 and CYP1B1 and catechol estrogen-mediated oxidative DNA damage in cultured human mammary epithelial cells. Carcinogenesis. 2004;25:2005–2013. doi: 10.1093/carcin/bgh183. [DOI] [PubMed] [Google Scholar]
- Chun YJ, Kim MY, Guengerich FP. Resveratrol is a selective human cytochrome P450 1A1 inhibitor. Biochem. Biophys. Res. Commun. 1999;262:20–24. doi: 10.1006/bbrc.1999.1152. [DOI] [PubMed] [Google Scholar]
- Ciolino HP, Daschner PJ, Yeh GC. Resveratrol inhibits transcription of CYP1A1 in vitro by preventing activation of the aryl hydrocarbon receptor. Cancer Res. 1998;58:5707–5712. [PubMed] [Google Scholar]
- Ciolino HP, Yeh GC. Inhibition of aryl hydrocarbon-induced cytochrome P-450 1A1 enzyme activity and CYP1A1 expression by resveratrol. Mol. Pharmacol. 1999;56:760–767. [PubMed] [Google Scholar]
- Ciolino HP, Yeh GC. The effects of resveratrol on CYP1A1 expression and aryl hydrocarbon receptor function in vitro. Adv. Exp. Med. Biol. 2001;492:183–193. doi: 10.1007/978-1-4615-1283-7_14. [DOI] [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Hsu EL, Yoon D, Choi HH, Wang F, Taylor RT, Chen N, Zhang R, Hankinson O. A proposed mechanism for the protective effect of dioxin against breast cancer. Toxicol. Sci. 2007;98:436–444. doi: 10.1093/toxsci/kfm125. [DOI] [PubMed] [Google Scholar]
- Jang M, Cai L, Udeani GO, Slowing KV, Thomas CF, Beecher CW, Fong HH, Farnsworth NR, Kinghorn AD, Mehta RG, et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275:218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- Jayatilake GS, Jayasuriya H, Lee ES, Koonchanok NM, Geahlen RL, Ashendel CL, McLaughlin JL, Chang CJ. Kinase inhibitors from Polygonum cuspidatum. J. Nat. Prod. 1993;56:1805–1810. doi: 10.1021/np50100a021. [DOI] [PubMed] [Google Scholar]
- Khan N, Afaq F, Mukhtar H. Cancer chemoprevention through dietary antioxidants: Progress and promise. Antioxid. Redox. Signal. 2008;10:475–510. doi: 10.1089/ars.2007.1740. [DOI] [PubMed] [Google Scholar]
- Köhle C, Bock KW. Activation of coupled Ah receptor and Nrf2 gene batteries by dietary phytochemicals in relation to chemoprevention. Biochem. Pharmacol. 2006;72:795–805. doi: 10.1016/j.bcp.2006.04.017. [DOI] [PubMed] [Google Scholar]
- Kress S, Greenlee WF. Cell-specific regulation of human CYP1A1 and CYP1B1 genes. Cancer Res. 1997;57:1264–1269. [PubMed] [Google Scholar]
- Le Corre L, Chalabi N, Delort L, Bignon YJ, Bernard-Gallon DJ. Resveratrol and breast cancer chemoprevention: Molecular mechanisms. Mol. Nutr. Food Res. 2005;49:462–471. doi: 10.1002/mnfr.200400094. [DOI] [PubMed] [Google Scholar]
- Lee JE, Safe S. Involvement of a post-transcriptional mechanism in the inhibition of CYP1A1 expression by resveratrol in breast cancer cells. Biochem. Pharmacol. 2001;62:1113–1124. doi: 10.1016/s0006-2952(01)00763-8. [DOI] [PubMed] [Google Scholar]
- Liu J, Wang Q, Wu DC, Wang XW, Sun Y, Chen XY, Zhang KL, Li H. Differential regulation of CYP1A1 and CYP1B1 expression in resveratrol-treated human medulloblastoma cells. Neurosci. Lett. 2004;363:257–261. doi: 10.1016/j.neulet.2004.03.075. [DOI] [PubMed] [Google Scholar]
- Mikstacka R, Baer-Dubowska W, Wieczorek M, Sobiak S. Thiomethylstilbenes as inhibitors of CYP1A1, CYP1A2 and CYP1B1 activities. Mol. Nutr. Food Res. 2008;52:77–83. doi: 10.1002/mnfr.200700202. [DOI] [PubMed] [Google Scholar]
- Mikstacka R, Przybylska D, Rimando AM, Baer-Dubowska W. Inhibition of human recombinant cytochromes P450 CYP1A1 and CYP1B1 by trans-resveratrol methyl ethers. Mol. Nutr. Food Res. 2007;51:517–524. doi: 10.1002/mnfr.200600135. [DOI] [PubMed] [Google Scholar]
- Mollerup S, Ovrebo S, Haugen A. Lung carcinogenesis: Resveratrol modulates the expression of genes involved in the metabolism of PAH in human bronchial epithelial cells. Int. J. Cancer. 2001;92:18–25. [PubMed] [Google Scholar]
- Piver B, Berthou F, Dreano Y, Lucas D. Inhibition of CYP3A, CYP1A and CYP2E1 activities by resveratrol and other non volatile red wine components. Toxicol. Lett. 2001;125:83–91. doi: 10.1016/s0378-4274(01)00418-0. [DOI] [PubMed] [Google Scholar]
- Potter GA, Patterson LH, Wanogho E, Perry PJ, Butler PC, Ijaz T, Ruparelia KC, Lamb JH, Farmer PB, Stanley LA, et al. The cancer preventative agent resveratrol is converted to the anticancer agent piceatannol by the cytochrome P450 enzyme CYP1B1. Br. J. Cancer. 2002;86:774–778. doi: 10.1038/sj.bjc.6600197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst MR, Reisz-Porszasz S, Agbunag RV, Ong MS, Hankinson O. Role of the aryl hydrocarbon receptor nuclear translocator protein in aryl hydrocarbon (dioxin) receptor action. Mol. Pharmacol. 1993;44:511–518. [PubMed] [Google Scholar]
- Rendic S. Summary of information on human CYP enzymes: Human P450 metabolism data. Drug Metab. Rev. 2002;34:83–448. doi: 10.1081/dmr-120001392. [DOI] [PubMed] [Google Scholar]
- Signorelli P, Ghidoni R. Resveratrol as an anticancer nutrient: molecular basis, open questions and promises. J. Nutr. Biochem. 2005;16:449–466. doi: 10.1016/j.jnutbio.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Stewart JR, Artime MC, O'Brian CA. Resveratrol: A candidate nutritional substance for prostate cancer prevention. J. Nutr. 2003;133:2440S–2443S. doi: 10.1093/jn/133.7.2440S. [DOI] [PubMed] [Google Scholar]
- Taylor RT, Wang F, Hsu EL, Hankinson O. Roles of coactivator proteins in dioxin induction of CYP1A1 and CYP1B1 in human breast cancer cells. Toxicol. Sci. 2009;107:1–8. doi: 10.1093/toxsci/kfn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitsett TG, Jr, Lamartiniere CA. Genistein and resveratrol: Mammary cancer chemoprevention and mechanisms of action in the rat. Expert Rev. Anticancer Ther. 2006;6:1699–1706. doi: 10.1586/14737140.6.12.1699. [DOI] [PubMed] [Google Scholar]
- Wu ML, Li H, Wu DC, Wang XW, Chen XY, Kong QY, Ma JX, Gao Y, Liu J. CYP1A1 and CYP1B1 expressions in medulloblastoma cells are AhR-independent and have no direct link with resveratrol-induced differentiation and apoptosis. Neurosci. Lett. 2005;384:33–37. doi: 10.1016/j.neulet.2005.04.055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.