Abstract
Ovarian granulosa cells play a central role in steroidogenesis, which is critical for female reproduction. Follicle-stimulating hormone (FSH) promotes cyclic adenosine monophosphate (cAMP)-mediated signaling to regulate granulosa cell steroidogenesis. We have shown previously that 2,2-bis-(p-hydroxyphenyl)-1,1,1-trichloroethane (HPTE) inhibits FSH- and dibutyryl cAMP-stimulated steroidogenesis and affects the messenger RNA levels of steroidogenic pathway enzymes in rat granulosa cells. However, HPTE showed a differential effect in FSH- and cAMP-stimulated cells in that HPTE more completely blocked FSH- when compared to cAMP-driven steroidogenesis. The objective of this study was to analyze the effects of HPTE on global gene expression profiles in untreated granulosa cells and those challenged with FSH or cAMP. Granulosa cells from immature rats were cultured with 0, 1, 5, or 10μM HPTE in the presence or absence of either 3 ng FSH/ml or 1mM cAMP for 48 h. Total RNA was isolated for real-time quantitative PCR and microarray analysis using the GeneChip Rat Genome 230 2.0 and ArrayAssist Microarray Suite. An investigation of changes in gene expression across all HPTE treatments showed that HPTE altered more genes in FSH- (∼670 genes) than in cAMP-stimulated cells (∼366 genes). Analysis confirmed that HPTE more effectively inhibited FSH- than cAMP-induced steroid pathway gene expression and steroidogenesis. Furthermore, expression patterns of novel genes regulating signal transduction, transport, cell cycle, adhesion, differentiation, motility and growth, apoptosis, development, and metabolism were all altered by HPTE. This study further established that HPTE exerts differential effects within the granulosa cell steroidogenic pathway and revealed that these effects include broader changes in gene expression.
Keywords: ovary, endocrine disruptors, gene microarray, steroidogenesis, follicle-stimulating hormone, cAMP
Methoxychlor (MXC) is an organochloride pesticide with estrogenic activities (Hall et al., 1997); this compound was used in the United States as a replacement for dichlorodiphenyltrichloroethane until 2004 (Stuchal et al., 2006). MXC is considered to be an endocrine-disrupting compound (EDC) based on the adverse reproductive effects of MXC observed in many species, including rats (Gray et al., 1989), mice (Eroschenko et al., 1995), and nonhuman primates (Gupta et al., 2007). This toxicity has been linked to the metabolism of MXC into the more potent compounds 2-(p-hydroxyphenyl)-2-(p-methoxyphenyl)-1,1,1-trichloroethane, also known as mono OH-MXC, and 2,2-bis-(p-hydroxyphenyl)-1,1,1-trichloroethane (HPTE) via cytochrome P450 activity in the liver (Hu and Kupfer, 2002). HPTE, e.g. can interact with the estrogen receptor (ER) subtypes and androgen receptor (AR) (Gaido et al., 2000), acting as an ERα agonist and an antagonist to both ERβ and AR (Gaido et al., 2000). Since other prevalent EDCs show similar activities, MXC and its metabolites are good model compounds that can be used to elucidate the actions of EDCs within the reproductive system.
The ultimate function of the ovary is ovulation. The process of folliculogenesis precedes ovulation and is exquisitely controlled by ovarian steroid hormones (Findlay et al., 2001) and growth factors (Skinner, 2005). Folliculogenesis and ovulation are intimately linked to multidirectional communication between the oocyte, granulosa cells, and theca cells (Uzumcu and Zachow, 2007).
The first somatic cell type that interacts with germ cells is granulosa cells or precursor granulosa cells (Pepling and Spradling, 2001). The nature of this interaction is bidirectionally established at the time of follicle formation and remains active throughout the life of a follicle (Matzuk et al., 2002). A second population of somatic cells, the theca cells, is recruited during later stages of folliculogenesis (Skinner, 2005). Granulosa cells and theca cells begin to interact with each other during the early stages of folliculogenesis through cell-to-cell signaling, which is primarily mediated by local paracrine factors and juxtacrine mechanisms (Skinner, 2005). Although the involvement of growth factors is still maintained, successful progression to the later stages of folliculogenesis requires actions of the gonadotropins, follicle-stimulating hormone (FSH), and luteinizing hormone (LH) (Eppig and O'Brien, 1996). Prior to the preovulatory follicular stage, LH targets theca cells, while FSH stimulates granulosa cells to induce steroidogenic differentiation, which enables the production of aromatizable androgens and estradiol-17β (E2) (Palermo, 2007). Within granulosa cells, FSH mobilizes numerous signaling motifs; noteworthy is the activation of the cyclic adenosine monophosphate (cAMP)-dependent signaling pathway, which exerts profound effects on granulosa cell steroidogenesis (Hsueh et al., 1984).
Previous studies from our laboratory have shown that HPTE inhibits FSH- and dibutyryl cAMP-stimulated steroid hormone production in granulosa cells isolated from the ovaries of immature rats (Zachow and Uzumcu, 2006). In addition, these studies showed that HPTE had a greater inhibitory effect on FSH-induced granulosa cell steroidogenic enzyme messenger RNA (mRNA) levels than on cAMP-induced mRNA levels. Specifically, HPTE inhibited levels of mRNAs encoding FSH-induced cytochrome P450 side-chain cleavage (CYP11A1), 3β-hydroxysteroid dehydrogenase type 1 (3β-HSD), and cytochrome P450 aromatase (CYP19A1), while having no effect on the level of steroidogenic acute regulatory protein (StAR) mRNA. In contrast, HPTE caused an increased accumulation of mRNA levels of cAMP-induced StAR, CYP11A1, and 3β-HSD, with a moderate reduction in CYP19A1 (Zachow and Uzumcu, 2006). These differential effects suggested that at the molecular level, HPTE acts somewhere between binding of FSH to its receptor and cAMP production. Although our previous report demonstrated that HPTE regulates steroidogenesis, with changes in gene expression as a plausible target (Zachow and Uzumcu, 2006), the potential mechanism of action for this regulation is unknown. Therefore, the objective of this study was to use a genomic approach to better understand the effect of HPTE in the ovary by examining its effect on FSH- and cAMP-induced gene expression in immature rat granulosa cells.
METHODS AND MATERIALS
Chemicals.
HPTE was generously supplied by Dr Stephen Safe, Texas A&M University. FSH (oFSH-20; 4453 IU/mg) was purchased from the National Hormone and Pituitary Program of the National Institute of Diabetes and Digestive and Kidney Diseases (Dr A.F. Parlow, Harbor-UCLA, Torrance, CA). N6,2′-O-dibutyryladenosine 3′,5′-cyclic monophosphate (cAMP) was purchased from Sigma (St Louis, MO).
Animals.
Sprague-Dawley rats were used. The animals were maintained in a room with controlled illumination (lights on 0700 h–2100 h), temperature (26°C–28°C), and humidity (30–70%) and were given free access to regular rat diet and water. All the procedures were carried out according to guidelines provided by Rutgers University Animal Care and Facilities Committee.
Granulosa cell culture.
Granulosa cells were prepared as described previously with some modifications (Zachow and Uzumcu, 2006). Briefly, intact, nonprimed, immature female rats (21–27 days old) were killed by cervical dislocation. Ovaries were removed from the animals, cleaned free of associated fat, oviduct, and bursa ovary, and then placed in ice-cold Dulbecco's Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F-12). Granulosa cells were isolated using a nonenzymatic needle puncture method with a sterile bundle of beading needles to release the cells from follicles. Cell viability was determined by the trypan blue exclusion method. The cells were plated and cultured (24-well plates) at approximately 3–4 × 105 viable cells/ml/well and incubated at 37°C for 24 h in DMEM/F-12 containing fetal bovine serum (5%) in an atmosphere of 5% CO2 in air. Following the 24-h acclimation period, the medium was replaced with serum-free DMEM/F-12 containing androstenedione (0.1M) as a substrate for aromatization.
Cells were treated with increasing doses of HPTE (0, 1, 5, and 10μM; in a final concentration of 0.1% dimethylsulfoxide) in the absence (basal) or presence of 3 ng FSH/ml or 1mM cAMP for 48 h. The doses of FSH and cAMP used in this study were chosen following a preliminary dose-response curve experiment, and FSH or cAMP concentrations that caused a submaximal stimulation for E2 production were selected (data not shown). HPTE concentrations were chosen based upon those previously used (Zachow and Uzumcu, 2006). Cultures were terminated at 48 h following the addition of treatments. At this time, cell-conditioned media were collected for E2 radioimmunoassay (RIA), and cell lysate was prepared for RNA isolation and microarray analysis.
E2 radioimmunoassay.
E2 was measured using a commercially available RIA kit, COAT-A-COUNT (Diagnostic Products Corp., Los Angeles, CA) via manufacturer's protocol. Hormone levels are expressed in picogram per milliliter of culture medium.
Oligonucleotide microarray.
Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA) followed by DNase I treatment. RNA quality was assessed by electrophoresis using the Agilent Bioanalyzer 2100 and spectrophotometric analysis prior to complementary DNA (cDNA) synthesis (data not shown). Forty nanograms of total RNA from each sample was used to generate a high-fidelity cDNA for array hybridization using the NuGen Ovation Biotin RNA Amplification and Labeling system (NuGen, San Carlos, CA). After fragmentation and biotin labeling, the samples were hybridized to Affymetrix Rat Genome 230 2.0 arrays, which have 11 pairs of oligonucleotide probes per chip. Washing and staining of all arrays were carried out in the Affymetrix fluidics module using the manufacturer's protocol. The detection and quantification of target hybridization were performed with an Affymetrix GeneChip Scanner. Three separate samples were analyzed for each of the treatment groups. A total of 36 separate arrays were used, and the data sets in their entirety are available through NCBI via the Gene Expression Omnibus (GEO) data repository (http://www.ncbi.nih.gov/geo/), GEO accession number GSE13883.
Microarray data analysis and statistics.
Raw data from three independent experiments were initially analyzed using Robust Multichip Analysis with a logarithmic base 2 conversion utilizing the Affymetrix ArrayAssist Suite (Affymetrix, Inc., Santa Clara, CA). One-way ANOVA of each group (basal, FSH, or cAMP) was used to determine genes that are significantly affected by HPTE. This was followed by an unpaired t-test to determine changes in global gene expression between HPTE-treated groups and the control groups to determine the levels of fold change. All statistical analyses for microarray analysis were performed using p ≤ 0.005 as the level of statistical significance. This is a level of probability consistently used in microarray analysis literature in order to minimize false positives (Moreira et al., 2008). Two-way completely randomized ANOVA was used to analyze the effect of HPTE on E2 production. The results showing changes in steroidogenic enzyme level were analyzed using one-way ANOVA followed by Tukey's multiple comparison test. Statistically significant differences were confirmed at p ≤ 0.05 for the last two parameters.
Validation of data consistency.
DNA-Chip Analyzer (dCHIP), a Windows-based software package developed by the Wing Wong Laboratory (www.dchip.org), was used to determine if any outliers were present in the oligonucleotide array repeats as described (Li and Wong, 2001). The arrays were shown to have no significant outliers (data not shown).
Real-time quantitative reverse transcription-PCR.
Gene expression was examined using Taqman chemistry with probes and primers designed using the Roche Universal Probe Library (UPL; www.universalprobelibrary.com). All cDNAs were measured in a 10-μl PCR reactions containing 5 μl of ABI 2× Universal Master Mix, 1.25 μl of each forward and reverse primers (final concentrations ranging from 200 to 900nM depending on the primer set), 1 μl of the corresponding UPL probe, and RNAase/DNAase-free water. All quantitative PCR (QPCR) reactions were performed in triplicate on triplicate biologic replicates leading to nine QPCR data points per condition measured. The cycling parameters for ABI 7900HT were 1 cycle of 50°C (2 min) followed by 95°C (10 min) and 40 cycles of 95°C (15 s) followed by 60°C (1 min). Data were collected at every temperature phase during every cycle. Raw data were analyzed using the Sequence Detection Software (ABI, Foster City, CA), while relative quantitation using the comparative threshold cycle (CT) method was performed in Microsoft Excel (ABI Technote #2: Relative Gene Expression Quantitation). Twelve differentially expressed genes were chosen as validation targets, those that exhibited gene expression downregulation (LHCGR, CYP11A1, CYP19A1, INHBA, and INHA) or upregulation (IGF1, CASP12, TGFB2, TGFB3, IGFBP1, IGFBP5, and CASP4). A gene that showed no changes in expression in the microarray data set (StAR) was used as the normalizer in the ΔΔCT analysis.
Enrichment analysis.
Enrichment analysis was conducted using two analysis tools: the U.S. Federal Drug Administration-supported ARRAY TRACK and the Medical College of Wisconsin’s (MCW) APROPOS. ARRAY TRACK software is both a management and an analysis tool designed to collect gene expression data from microarray and xenobiotic-genomic studies (Tong et al., 2003). It is equipped with pathway and gene expression analysis libraries linked to the KYOTO Encyclopedia of Genes and Genomes and Ingenuity Systems Library. ARRAY TRACK has been shown effective for examining pathways enriched by different treatment groups and also for conducting statistical analyses of microarray data (Guo et al., 2006).
APROPOS is an open source software package developed at the MCW (http://apropos.mcw.edu/). APROPOS, a MySQL database, is built on the top of the rat, mouse, and human International Protein Index data sets (Kersey et al., 2004). The system cross-references to the Rat Genome Database, Mouse Genome Database, Uniprot, Entrez gene, and other groups to facilitate annotations of each record.
RESULTS
Effect of HPTE on E2 Accumulation
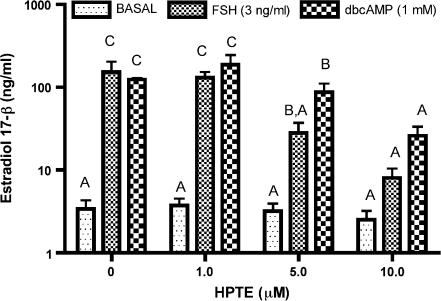
Basal E2 accumulation was not altered by HPTE (Fig. 1). In contrast, 5 and 10μM HPTE inhibited both FSH- and cAMP-stimulated E2 secretion, but the effect on FSH group was much greater as compared to that on cAMP group. These data corroborated that HPTE was more effective in inhibiting FSH- than cAMP-stimulated steroidogenesis in the present experiments. We then utilized the mRNA collected from the same granulosa cells for oligonucleotide microarray analysis. During the treatment period, microscopic evaluation suggested that cell viability was comparable between the groups.
FIG. 1.
The effect of HPTE on E2 accumulation in immature rat granulosa cells in vitro. Granulosa cells were exposed to increasing doses of HPTE (0, 1, 5, or 10μM) in the absence (basal) or presence of 3 ng FSH/ml or 1mM cAMP (dbcAMP). After a 48-h treatment, media were collected and E2 was measured by RIA as described in Materials and Methods. Different letters on the bars indicate statistically significant differences.
Effect of HPTE on FSH-Stimulated Steroidogenic Pathway Gene Expression
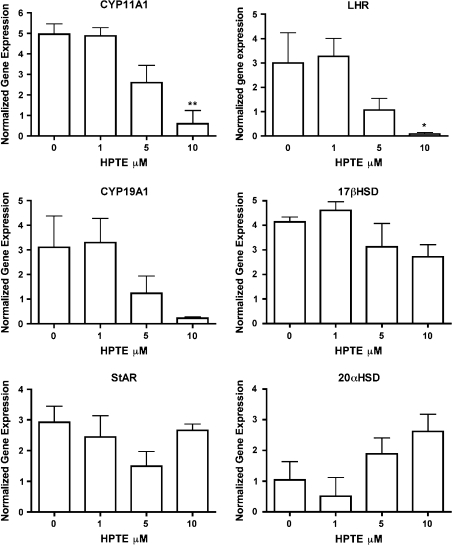
Since our laboratory and others have previously shown that HPTE and MXC affected steroidogenesis (Akgul et al., 2008; Chedrese and Feyles, 2001; Zachow and Uzumcu, 2006), we analyzed the microarray data to determine the effect of HPTE on the key steroidogenic enzymes. Figure 2 shows that HPTE (10μM) significantly lowered the FSH-dependent expression of CYP11A1 and LH receptor (LHR; p ≤ 0.05). In addition, 10μM HPTE showed a tendency to inhibit FSH-induced CYP19A1 mRNA expression. In contrast, HPTE did not significantly alter the expression of 17β-HSD or StAR (Fig. 2). Although it was not statistically significant, HPTE (5 and 10μM) caused an upregulation in 20α-HSD mRNA in the presence of FSH.
FIG. 2.
The effect of HPTE on FSH-stimulated steroidogenic pathway gene expression in granulosa cells in vitro. Granulosa cells were treated with HPTE (0, 1, 5, or 10μM) in the presence of 3 ng FSH/ml. After a 48-h treatment, cell lysate was collected for mRNA isolation and microarray analysis as described in Material and Methods. . **p ≤ 0.01 and *p ≤ 0.05.
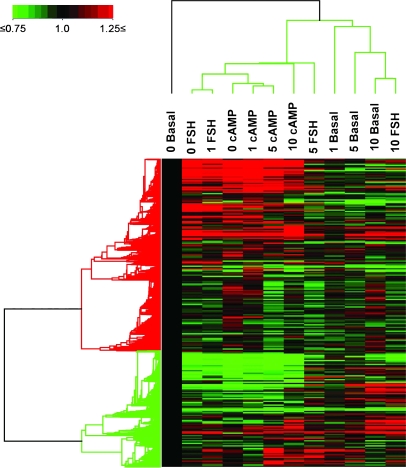
Hierarchical Clustering Analysis Reveals Three Transcriptome Patterns
Hierarchical clustering was used to compare global gene expression profiles and to identify similar gene expression patterns between the individual treatment groups. Before analysis, the data were normalized using values from the basal (0μM HPTE) group. Three vertical cluster patterns were observed. The highest dose of HPTE (10μM) in the FSH-stimulated and basal groups exhibited the same pattern (Fig. 3). The cAMP groups showed the least change across the HPTE doses used in this study as they clustered in the same node as the cells given FSH with 1μM HPTE and FSH alone (0μM HPTE). Granulosa cells concomitantly incubated with FSH, and 5μM HPTE displayed an intermediary pattern. A one-way ANOVA was used in order to determine if any consistent patterns of gene expression were observed between groups. Results are presented in the next section.
FIG. 3.
Hierarchical cluster analysis of the effect of HPTE in granulosa cells. Three vertical patterns of expression were observed among the 12 groups delineating the effect of HPTE (0, 1, 5, or 10μM) with and without FSH or cAMP. The legend indicates that data in 0μM HPTE in the basal group (0 basal) are set to 1 (black color) and used to normalize the data in the other groups. In addition, normalization values between 0.75 and 1.25 were excluded to reduce the background noise.
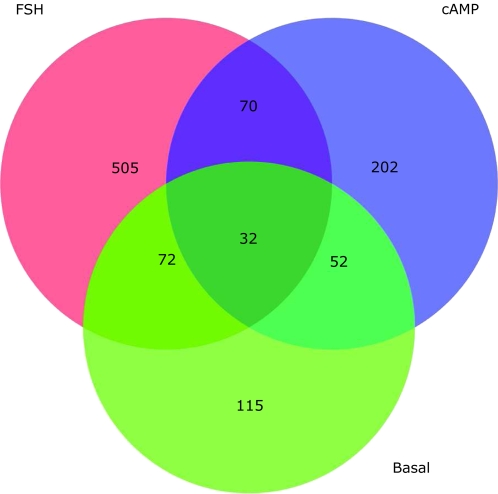
Genes That Are Common in Basal, FSH, and cAMP Groups Were Altered by HPTE
Across the basal, FSH, and cAMP groups, the number of genes that showed significant changes in response to HPTE treatment (1, 5, and 10μM) was determined (Fig. 4). In the FSH group, 679 genes showed significant changes in response to HPTE. This is compared to the cAMP group with 356 genes, and in the basal group, 271 genes were significantly affected by HPTE. Thirty-two genes that were in common between the three groups (i.e., basal, FSH, and cAMP) were affected by HPTE (Fig. 4). Using the APROPOS program, the commonly affected genes in granulosa cells that were untreated or treated with FSH or cAMP were categorized according to biologic function. Two or more genes were affected in cell cycle, multicellular organismal development, transport, amino acid transport, regulation of transcription, Wnt receptor signaling, and cell division (Table 1). Approximately 70 additional genes were commonly regulated between the FSH- and cAMP-treated granulosa cells (Fig. 4; Supplementary Table 1).
FIG. 4.
Venn diagrams of the genes under HPTE regulation in granulosa cells. Genes that were affected by HPTE in three groups were analyzed by one-way ANOVA. Between the cAMP and FSH groups, 102 common genes were regulated in granulosa cells (p ≤ 0.005).
TABLE 1.
Genes That Are Commonly Altered by HPTE in Untreated and Treated (FSH or cAMP) Granulosa Cells (p ≤ 0.005)
| Biologic processes | Genes |
| Multicellular organismal development | RGD1308165_predicted RGD1564008_predicted Cd24 Dixdc1 Itgb1 Angptl4 |
| Signal transduction | Angptl4 Ank2 Fcgr2b Hivep Igfbp5 Tas1r2 |
| Metabolic process | Ank2 Asns LOC3040860 Man1a_predicted Tas1r2 |
| Regulation of transcription, DNA dependent | E2f8 Bhlhb2 Atf5 Hivep2 Irf6_predicted |
| Cell cycle | E2F8 CDCa1 Kif11a IPI00768806 Mad2l1_predicted |
| Transport | Tcn2 Slc7a3 Slc6a9 Kcnd2 Cp |
| Cell differentiation | Angptl4 Cd24 Ctgf RGD1308697 |
| Cell adhesion | Cd24 Ctgf Itgb1 Sdc1 |
| Mitosis | Kif11 Kif15 Mad2l1_predicted Cdca1 |
| Regulation of progression through cell cycle | Atf5 E2f8 Itgb1 |
| Regulation of cell growth | Ctgf Htra1 Igfbp5 |
| Cell division | Cdca1 Madl1_predicted Kif11 |
| Ion transport | Cp Kcnd2 Tcn2 |
| Cell matrix adhesion | Ctgf Itgb1 |
| Humoral immune response | Cd24 Fcgr2b |
Confirmation of the Limited HPTE Effect Within Untreated and cAMP-Stimulated Granulosa Cells
In order to determine which genes exhibited the most changes in the level of expression relative to the baseline, a comparative analysis was performed. A twofold change was established in all groups as the cutoff criteria to filter out relatively small changes in gene expression. The result from this analysis confirmed the previous analysis, i.e., the greatest numbers of genes were affected in the FSH group (669 total, 159 downregulated, and 420 upregulated). In the basal group, 90 genes showed changes in expression; specifically, 52 genes were downregulated and 38 genes were upregulated. HPTE affected the least number of genes in the cAMP group, with the expression of 76 genes significantly altered (16 genes downregulated and 60 genes upregulated) (Tables 2 and 3). These results do not include expressed sequence tags.
TABLE 2.
Distribution of the Downregulatory Effect of HPTE on Gene Expression in Untreated (Basal) or Treated (FSH or cAMP) Granulosa Cells In Vitro. Only Those Genes Exhibiting a Twofold or Greater Level of Change Are Shown (p ≤ 0.005).
| HPTE (μM) |
Total | |||
| 1 | 5 | 10 | ||
| Basal | 1 | 6 | 45 | 52 |
| FSH | 0 | 64 | 95 | 159 |
| cAMP | 0 | 0 | 16 | 16 |
TABLE 3.
Distribution of the Upregulatory Effect of HPTE on Gene Expression in Untreated (Basal) or Treated (FSH or cAMP) Granulosa Cells In Vitro. Only Those Genes Exhibiting a Twofold or Greater Level of Change Are Shown (p ≤ 0.005)
| HPTE (μM) |
Total | |||
| 1 | 5 | 10 | ||
| Basal | 0 | 1 | 37 | 38 |
| FSH | 0 | 163 | 257 | 420 |
| cAMP | 2 | 4 | 54 | 60 |
Analysis of Genes That Were Affected by 10μM HPTE
The expression of the greatest number of genes was affected by 10μM HPTE; therefore, we focused on this dose for further analysis. A list of the upregulated and downregulated genes was compiled, and an enrichment analysis was conducted to profile the targeted genes. Analysis revealed that 257 genes were upregulated and 95 genes were downregulated in the FSH group. Fifty-four genes were upregulated and 16 genes were downregulated in the cAMP group, whereas HPTE upregulated the expression of 37 genes and downregulated 45 genes in basal group. ARRAY TRACK and APROPOS software were used in order to determine the functional groups of the genes regulated by HPTE, and these are listed in Tables 4 and 5. Upregulation was observed in genes associated with signal transduction, cell adhesion, and various transport functions. Downregulation was observed in genes associated with signal transduction, transport, and cell division. In FSH-stimulated granulosa cells, HPTE induced the largest fold changes in the expression of several genes previously linked with ovarian function, and these data are shown in Table 6.
TABLE 4.
Biologic Function of Genes That Are Downregulated by HPTE (10μM) in Untreated (Basal) or Treated (FSH or cAMP) Granulosa Cells. Note that Some Genes Are Listed in Multiple Functional Groups
| Functional group | Basal | FSH | cAMP |
| Signal transduction | 3 | 15 | 4 |
| Transport | 3 | 11 | 8 |
| Ion transport | 8 | 3 | |
| Amino acid transport | 3 | ||
| Cell adhesion | 4 | 14 | |
| Cell differentiation | 6 | ||
| Cell-cell signaling | 2 | 5 | |
| Cell motility | 4 | ||
| Regulation of cell growth | 3 | 6 | |
| Regulation of progression through cell cycle | 3 | ||
| Antiapoptosis | 5 | ||
| Regulation of apoptosis | 4 | 2 | |
| G-protein-coupled receptor signaling pathway | 6 | ||
| Multicellular organismal development | 4 | 5 | |
| Skeletal development | 5 | ||
| Proteolysis | 4 | ||
| Metabolism | 3 | ||
| Glucose metabolism | 3 |
TABLE 5.
Biologic Function of Genes That Are Upregulated by HPTE (10μM) in Untreated (Basal) or Treated (FSH or cAMP) Granulosa Cells. Note that Some Genes Are Listed in Multiple Biologic Functional Groups
| Functional group | Basal | FSH | cAMP |
| Signal transduction | 2 | 4 | |
| Transport | 2 | ||
| Cell adhesion | 3 | ||
| Cell division | 6 | 1 | |
| Cell motility | 1 | ||
| Regulation of progression through cell cycle | 4 | ||
| Cell cycle | 7 | ||
| Apoptosis | 2 | ||
| Multicellular organismal development | 3 | ||
| Metabolic process | 3 | ||
| Lipid metabolism | 2 | ||
| Fatty acid metabolism | 2 |
TABLE 6.
Genes Associated with Ovarian Function, Which Were Significantly Affected by HPTE (10μM) in FSH-Stimulated Granulosa Cells. Fold Change and Summary of Function Are Included
| Gene symbol | Gene name | Summary of the functiona | Fold change |
| Lhcgr (LHR) | Luteinizing hormone/chorionic gonadotropin receptor | Receptor for both LH and chorionic gonadotropin; involved in reproductive development and function (RGD) | −5.3 |
| Prlr | Prolactin receptor | Expression changed during estrous cycle; may be regulated by prolactin receptor or steroid hormones (RGD) | −4.7 |
| Cyp11a1 | Cytochrome P450, family 11, subfamily a, polypeptide 1 | Monooxygenase that catalyzes synthesis of cholesterol and steroids (RGD) | −4.3 |
| Acsbg1 | Acyl-CoA synthetase bubblegum family member 1 | May play a role in fatty acid metabolism and biosynthesis of steroid hormone precursors (RGD) | −4.1 |
| Cyp19a1 | Cytochrome P450, family 19, subfamily a, polypeptide 1 | Catalyzes end-step estrogen formation from androgens; aromatization of testosterone to E2 (RGD) | −3.7 |
| Inhba | Inhibin β-A | Subunit that can homodimerize or heterodimerize with Inhbb to form activins or heterodimerize with α-subunit to form inhibin A to regulate FSH secretion [RGD] | −3.5 |
| Ddit4 | DNA-damage–inducible transcript 4 | HIF-1–responsive gene that may protect some types of cells from hypoxia and H(2)O(2)-triggered apoptosis (RGD) | −2.9 |
| Fabp6 | Fatty acid–binding protein 6, ileal (gastrotropin) | May mediate metabolism of steroids in steroid hormone–producing tissues (RGD) | −2.7 |
| Nr5a1(SF-1) | Nuclear receptor subfamily 5, group A, member 1 | Transcription factor; essential for adrenal and gonadal development (RGD) | −2.7 |
| Kitl | Kit ligand | Stimulates proliferation of both myeloid and lymphoid hematopoietic progenitors in bone marrow cultures and interacts with the basic fibroblast growth factor to promote the primordial to primary follicle transition in rat ovaries (RGD) | −2.5 |
| Rasd1 | RAS, dexamethasone-induced 1 | Interacts with neuronal NO synthase adaptor protein CAPON and is involved in nitric oxide-mediated signaling (RGD) | −2.5 |
| Fdxr | Ferredoxin reductase | Component of electron transfer system for mitochondrial cytochrome P450; mediates production of steroid hormones and bile acids; activates vitamin D3 in steroidogenic tissues, liver, and kidney (RGD) | −2.4 |
| Stmn4 | Stathmin-like 4 | Controls cell proliferation and activities for stathmin (RGD) | −2.4 |
| Mrap_predicted | Melanocortin 2 receptor accessory protein (predicted) | Homologous to the mouse gene that interacts with ACTH receptor (IHOP) | −2.4 |
| Pla2g1b | Phospholipase A2, group IB | Putative phospholipase A2 enzyme (RGD) | −2.4 |
| Inha | Inhibin α | α-subunit of protein hormone that heterodimerizes with one of the two alternative of β-subunits to form inhibin A and B (RGD) | −2.2 |
| Nr5a2 | Nuclear receptor subfamily 5, group A, member 2 | An orphan nuclear receptor that may bind DNA and activate gene transcription (RGD) | −2.2 |
| Cebpa | CCAAT/enhancer-binding protein (C/EBP), α | Binds the CCAAT promoter motif as well as a core enhancer homology; activates transcription (RGD) | −2.2 |
| Hsd3b1_predicted | Hydroxysteroid dehydrogenase-1, delta<5>-3-β (predicted) | A member of the C-21 steroid pathway and progesterone metabolism pathway (RGD) | −2.0 |
| Egfr | Epidermal growth factor receptor | Promotes cell proliferation and differentiation; mediates G protein-coupled receptor-regulated induction of protein synthesis, plays role in ovulation (RGD) | −2.0 |
| Snf1lk | SNF1-like kinase | Inhibits CREB binding and transcription of cAMP-responsive element-dependent genes Cyp11A and StAR (RGD) | −1.9 |
| Pgr | Progesterone receptor | Receptor for progesterone; regulates glutamic acid decarboxylase expression in the hypothalamus during proestrus; may mediate production of the GnRH and LH surge, critical for ovulation (RGD) | −1.9 |
| Nr0b1 | Nuclear receptor subfamily 0, group B, member 1 | Human homolog is an orphan nuclear hormone receptor (RGD) | −1.7 |
| Hsd17b7 | Hydroxysteroid (17β) dehydrogenase 7 | A phosphoprotein that associates with the short form of the prolactin receptor [RGD] | −1.6 |
| Akr1c18(20α-HSD) | Aldo-keto reductase family 1, member C18 | Catalyzes the covariant of progesterone into 20α-dihydroprogesterone; activity is regulated by prolactin (RGD) | 1.6 |
| Igf1 | Insulin-like growth factor 1 | Growth factor; plays a major role in mammalian growth (RGD) | 1.6 |
| Casp12 | Caspase 12 | Cysteine-aspartic acid protease (caspase); involved with the terminal stage of apoptosis (RGD) | 1.6 |
| Pdcd4 | Programmed cell death 4 | Acts as an inhibitor of apoptosis; has similarity to eukaryotic translation initiation factor (eIF)4G (RGD) | 1.6 |
| Hsd11b1 | Hydroxysteroid 11β dehydrogenase 1 | Catalyzes the interconversion of cortisol and cortisone; plays a role in glucocorticoid metabolism; may regulate blood pressure (RGD) | 1.7 |
| Ca3 | Carbonic anhydrase 3 | Catalyzes hydration of carbon dioxide; may be involved in cellular response to oxidative stress (RGD) | 1.7 |
| Akr1cl1_predicted | Aldo-keto reductase family 1, member C-like 1 (predicted) | Unknown function | 1.8 |
| Ddit3 | DNA damage–inducible transcript 3 | Plays a role in the endoplasmic reticulum stress response (RGD) | 2.1 |
| Tgfb2 | Transforming growth factor, β-2 | Binds the transforming growth factor-β receptor; plays a role in regulation of cell growth and proliferation; may be involved in mesenchymal-epithelial cell interactions during development (RGD) | 2.2 |
| Adamts5 | A disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 5 (aggrecanase-2) | Human homolog catalyzes the degradation of aggrecan, a component of cartilage extracellular matrix, plays role in ovulation (RGD) | 2.3 |
| Tgfb3 | Transforming growth factor, β-3 | Involved in epithelial and endothelial cell proliferation and differentiation during development (RGD) | 2.4 |
| Igfbp1 | Insulin-like growth factor–binding protein 1 | A modulator of a insulin growth factor bioavailability (RGD) | 2.4 |
| Ptgis (CYP 8A) | Prostaglandin I2 (prostacyclin) synthase | Catalyzes the conversion of prostaglandin H2 to prostacyclin in prostaglandin biosynthesis and appears to be important to implantation (RGD) | 2.4 |
| Igfbp5 | Insulin-like growth factor–binding protein 5 | May be involved in ovarian folliculogenesis and may play a modulatory role in type I fiber-dominated muscles (RGD) | 2.5 |
| Casp4, Casp11 | Caspase 4, apoptosis-related cysteine peptidase | Cysteine-aspartic acid protease (caspase); involved with the terminal stage of apoptosis; may also be involved with response to inflammation (RGD) | 2.9 |
| Grem1 | Gremlin 1 homolog, cysteine knot superfamily (Xenopus laevis) | Protein may play a role in cellular growth control, viability, and differentiation; high expression levels may induce apoptosis (RGD) | 3.3 |
| Tnfrsf11b | Tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) | Functions in the negative regulation of bone resorption by inhibiting formation of osteoclasts (RGD) | 4.2 |
The function of the genes as described in Rat Genome Database (RGD) or Information Hyperlinked over Proteins (IHOP).
Validation of Microarray Results for Select Transcripts by QPCR
Validation of microarray results was performed by examining the expression levels of 12 genes using QPCR. Similar gene expression patterns were observed for all targets measured by QPCR when compared to the results of the microarray gene expression study (Supplementary Fig. 1); specifically, the overall stimulatory and inhibitory effects of HPTE on the expression for target genes were the same. As expected, the magnitudes of change were greater for QPCR because this approach not only is more quantitative but also represents a different probe sequence than that used to conduct array measurements. In addition to StAR, other normalizers were evaluated and demonstrated similar results (data not shown).
DISCUSSION
We examined the effect of the potent MXC metabolite HPTE on global gene expression in cultured immature rat granulosa cells. This study shows that HPTE exerts a stronger inhibition on FSH-induced gene expression when compared to that regulated by exogenous cAMP. In addition, these data reveal for the first time the effects of HPTE on the expression of a plethora of genes which are important to numerous vital pathways in granulosa cells, including signal transduction, transport, cell differentiation, growth, survival, and apoptosis. We also analyzed the effect of HPTE on the expression of genes associated with ovarian function, showing that HPTE affects multiple genes previously linked with the processes of folliculogenesis, steroidogenesis, and/or ovulation.
In vivo exposure to MXC reduces serum progesterone levels and disrupts female reproductive parameters and ovarian morphology (Chapin et al., 1997; Gray et al., 1989). Transient developmental exposure to MXC can affect the level of key ovarian regulators including certain steroidogenic regulatory proteins (e.g., LHR and CYP11A1) (Armenti et al., 2008). Direct inhibition of CYP11A1 enzyme activity by HPTE, leading to reduced progesterone production in cultured granulosa cells, was previously observed, although HPTE did not alter protein or mRNA levels of this enzyme (Akgul et al., 2008). We have previously shown that HPTE inhibits FSH- and cAMP-induced steroid hormone production. Furthermore, we demonstrated HPTE-directed changes in the mRNA levels of several steroidogenic pathway proteins/enzymes in granulosa cells (Zachow and Uzumcu, 2006). While HPTE completely abrogated CYP11A11, 3β-HSD, and CYP19A1 mRNA levels in FSH-treated granulosa cells, HPTE reduced CYP19A1 mRNA levels by only 50% in the presence of cAMP (Zachow and Uzumcu, 2006), indicating that FSH-stimulated cells were more sensitive to the inhibitory effects of HPTE. The current study confirmed that FSH-induced steroidogenesis is more sensitive to downregulation by HPTE. This action was maintained on a global scale as shown by the hierarchical clustering analyses. Hierarchical clustering profiles suggest that in the presence of FSH, effects manifest above 1μM HPTE, while granulosa cells given exogenous cAMP are more resistant to HPTE. The expression profile of FSH groups that were treated with 5 and 10μM of HPTE resembled the gene expression profile of the unstimulated (basal) groups. However, the expression profile of the cAMP-stimulated cells remained distinct. These data suggest that HPTE targets the cAMP-dependent cascade at one or more loci preceding cAMP production (e.g., FSH receptor, G proteins, and/or adenylyl cyclase) in immature rat granulosa cells. This is in contrast to a study by Chedrese and Feyles, where MXC had no observable effect on the levels of FSH-induced cAMP but still inhibited FSH-dependent steroid accumulation in porcine granulosa cells (Chedrese and Feyles, 2001). This suggests that MXC exerts its effects distal to cAMP production. These apparently disparate results may be due to differences in action of MXC and HPTE in vitro, specific culture conditions, and/or species-specific effects.
In the present report, FSH-treated granulosa cells were more sensitive to the effects of HPTE, and FSH represents a more physiological stimulus. Therefore, the FSH group was further analyzed to determine the HPTE-dependent alterations in gene expression. We examined those genes that exhibited the largest fold changes in expression and detected that many of these genes are associated with ovarian function (Tables 6). HPTE suppressed the expression of several genes encoding mediators of steroidogenesis and/or ovulation (LHR, CYP11A1, HSD17B7, CYP19A1, FABP6, ACSBG1, PGR, and EGFR), transcription (SF-1, NR5A2, and CEBPA), and other aspects of ovarian function (KITL, INHβA, INHα, and PRLR). In contrast, HPTE upregulated the expression of several genes encoding mediators of apoptosis (CASP11, CASP12, and PDCD4), cell differentiation and growth regulation (GREM1, TGFβ2, TGFβ3, IGF-1, IGFBP1, and IGFBP5), ovulation/tissue remodeling (ADAMTS5), and stress response (DDIT3 and CA3).
Some genes of interest from this list are GREM1, INHα (α subunit of inhibin), INHβA (βA subunit of inhibin), TGFB2, TGFB3, ACSBG11, IGFBP5, CASP11, and CASP12. Gremlin 1 (GREM1) expression was upregulated by HPTE. GREM1 is an antagonist of bone morphogenetic protein (BMP) signaling (Merino et al., 1999) and is spatiotemporally expressed in the ovary (Pangas et al., 2004). It is primarily expressed in granulosa cells within preantral and antral follicles. In large antral follicles, GREM1 mRNA is detected in cumulus but not in mural granulosa cells (Pangas et al., 2004). While its expression is stimulated by BMPs and growth and differentiation factor-9 (GDF-9), GREM1 inhibits BMP signaling with no effect on GDF-9 signaling (Pangas et al., 2004). Thus, it has been speculated that this regulated expression of GREM1 may inhibit the actions of theca cell-derived BMP on granulosa cell luteinization, while allowing GDF-9 of oocytic origin to mediate cumulus expansion (Pangas et al., 2004). The fact that the expression of some of the luteinization markers (LHR and PGR EGFR) was inhibited by HPTE in the present study supports the above notion. Therefore, it maybe worth exploring the effect of HPTE on theca cell-derived BMPs and BMP-coupled signaling molecules.
In addition, GREM1 has been shown to affect other signaling pathways that may interact with the FSH/cAMP-dependent protein kinase A (PKA) signaling pathway. Overexpression of GREM1 has been observed to inhibit the activity of Wnt signaling through its connection with β-catenin (Gazzerro et al., 2007). Recently, β-catenin was proven to be critical for gonadotropin-directed signal transduction (specifically FSH) through the coordination of SF-1 and β-catenin (Parakh et al., 2006). Parakh et al. (2006) also showed that β-catenin selectively modified the FSH-driven production of CYP19A1 and CYP11A1 in granulosa cells. In addition, β-catenin, in conjunction with SF-1, regulated the activity of CYP19A1 and was necessary for the FSH- and cAMP-mediated regulation of CYP19A1. In the present study, HPTE showed an inhibitory effect on the mRNAs encoding CYP19A1 and CYP11A1. This may be a result of a downstream effect on Wnt signaling via GREM1-induced β-catenin inhibition or other possible upstream effectors of β-catenin (Hino et al., 2005). The Wnt pathway, and other signaling pathways that may interact with FSH-PKA signaling, appears to be one of many novel interactions that may be affected by HPTE in granulosa cells (Fig. 5).
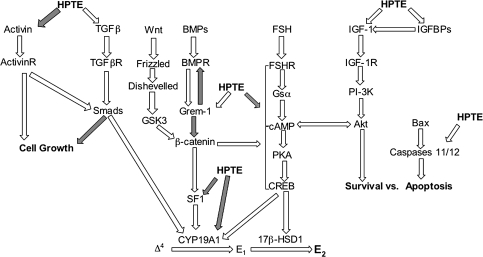
FIG. 5.
Proposed model for the effect of HPTE in the granulosa cell. FSH binding to the FSH receptor initiates the PKA signaling pathway that promotes E2 production in granulosa cells. HPTE appears to inhibit this cascade through an unknown mechanism, but effects in multiple pathways are implicated. HPTE affected the expression of several genes known to control granulosa cell function, as described in the text. Among these, activin, TGFβs, GREM1, SF1, IGF-1, IGFBPs, and caspases all have distinct effects in controlling granulosa cell growth, differentiation, survival, and steroidogenesis. White arrows indicate induction. Gray arrows indicate inhibition. Gsα, α subunit of stimulatory guanine nucleotide–binding protein; CREB, cAMP-response element–binding protein; SF-1, steroidogenic factor 1; ▵4, androstenedione; E1, estrone; PI-3K, phosphoinositide 3-kinase; TFGβ, transforming growth factor-β; GSK3, glycogen synthase kinase-3.
Downregulation of the expression of both INHα and INHβA by HPTE may have significant consequences for the production of inhibins and activins in granulosa cells. Inhibins are dimers of an α subunit combined with either a βA (inhibin A) or a βB subunit (inhibin B) (Knight and Glister, 2006). Thus, HPTE inhibition of INHα could affect the level of both inhibin isoforms. In addition, activins are homodimers (βAβA, activin A, or βBβB, activin B) or a heterodimer (activin AB) of β subunits (Knight and Glister, 2006). Therefore, inhibition of INHβA would also prevent formation of activins A and AB. Collectively, activins and inhibins have important roles in granulosa cell proliferation and differentiation. For example, activin stimulates basal and FSH-induced granulosa cell proliferation (Miro and Hillier, 1996). Activin also regulates basal and gonadotropin-induced steroid production in rat granulosa cells (Miro et al., 1991). A modulatory role of inhibin on follicular steroidogenesis has also been reported (Smyth et al., 1994). As a result, inhibition of INHβA and INHα may be causal to the reduced steroidogenesis in HPTE-treated granulosa cells (Zachow and Uzumcu, 2006).
In addition to inhibin and activin, the expression of the mRNA encoding TGFβ2 and TGFβ3 was increased by HPTE. The TGFβs have a synergistic effect on FSH-stimulated proliferation in granulosa cells, and TGFβ-directed changes in granulosa cell steroidogenesis are well documented (Knight and Glister, 2006). Since E2 has been shown to impair TGFβ expression (Kleuser et al., 2008), a possible mechanism that explains the upregulation of TGFβs is the HPTE-dependent decrease in E2 and/or a direct effect of HPTE on TGFβ expression. However, further experiments are needed to determine this.
The long-chain fatty acid synthase known as Acyl-CoA synthase bubblegum 1 (ACSBG1) was downregulated in granulosa cells challenged with HPTE. ACSBG1 is normally located in theca cells and testicular Leydig cells and has been linked to spermatogenesis (Pei et al., 2003). The role of ACSBG1 in folliculogenesis has not been defined, but it may function as a survival factor, which mediates the synthesis of steroid precursors (Pei et al., 2003). By inhibiting ACSBG1, HPTE may reduce steroidogenesis by directly reducing steroid precursors and/or energy availability.
Numerous reports have shown the importance of the insulin-like growth factor (IGF) system within granulosa cells. In conjunction with FSH, IGF-I stimulates cell proliferation and steroidogenesis in granulosa cells of various species (deMoura et al., 1997; Mazerbourg et al., 2003). In contrast, insulin-like growth factor–binding proteins (IGFBPs) can suppress FSH-induced follicular growth and differentiation, leading to atresia by possibly sequestering IGF-I protein and inhibiting its activity (Cataldo et al., 1993; Ui et al., 1989). Interestingly, FSH reduces IGFBP activity by stimulating proteolytic mechanisms that degrade IGFBPs (Fielder et al., 1993). In the present study, 10μM HPTE stimulated the genes encoding IGF-I and IGFBPs (i.e., IGFBP1 and IGFBP5) in FSH-stimulated granulosa cells. This appears to be controversial; however, the increase in IGF-I may counteract the HPTE-directed increase in the level of IGFBPs. In addition, E2 has been shown to inhibit IGFBP5 in granulosa cells from large follicles (Voge et al., 2004); thus, the apparent upregulation of IGFBP5 by HPTE could be a consequence of the HPTE-dependent reduction in E2 production. In general, some of the effects on gene expression that were observed in HPTE-treated cells may be an indirect effect due to reduced E2 secretion as well as due to the direct effects of HPTE.
Others have noted that HPTE inhibits growth and induces atresia in antral follicles (Gupta et al., 2006). HPTE upregulated caspases 11 and 12 mRNAs in granulosa cells, which may provide a mechanism that explains HPTE-induced atresia. Caspase 11 is classified as caspase 4 in humans and appears to be essential for activation of interleukin 1β–converting enzyme (caspase 1) (Wang et al., 1998). Caspase 12 mediates apoptosis driven by the endoplasmic reticulum (Liu and Baliga, 2005). Both caspases are upstream initiators of apoptosis through caspase 3 (Kang et al., 2000; Liu and Baliga, 2005). Whether HPTE affects caspases that regulate survival in granulosa cells is currently unknown but should be investigated.
Combining these observations, we have composed a working model that attempts to connect the observed gene expression changes in granulosa cells to some promising signaling mechanisms (Fig. 5). Evidence suggests that HPTE acts on the FSH-PKA signaling pathway (Zachow and Uzumcu, 2006). Whether this interaction is direct or through still vaguely defined cross talk of parallel signaling cascades (Hunzicker-Dunn and Maizels, 2006) is unknown. Multiple signaling complexes exist within granulosa cells, and each has some role in directing normal cellular function (Knight and Glister, 2006). Our model attempts to link these pathways with inhibition of E2 production and steroidogenic pathway proteins/enzymes. Induction of GREM1 by HPTE is an example of multiple pathways that can be perturbed by HPTE. GREM1 is known to inhibit β-catenin; β-catenin is a proven stimulator of SF-1 leading to CYP19A1 activity (Gazzerro et al., 2005; Michos et al., 2007). Also, GREM1 binds to BMP and therefore would block BMP activity. This provides another potential mechanism for the inhibitory effects of HPTE in granulosa cells. Finally, Wnt functions through a pathway leading to β-catenin; so it is plausible that the effect of HPTE is also manifested through that cascade (Michos et al., 2007). Since HPTE induced changes in the level of expression of several genes linked to granulosa cell signaling pathways, further efforts should be placed on determining the functional effects of the HPTE-stimulated alterations in gene expression.
In summary, the current results show that HPTE differentially affects FSH- and cAMP-stimulated gene expression in granulosa cells. In addition, these results indicate that parallel pathways, besides FSH-cAMP-PKA, may be involved in the effects of HPTE on FSH-mediated steroidogenesis in granulosa cells. The discovery of the HPTE-directed involvement of these and other pathways, and perhaps cross talk among numerous cascades, is compelling and will no doubt provide a better understanding of the effect of HPTE and MXC in the ovary.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institute of Health (ES013854 to M.U.); National Institute of Environmental Health Sciences (NIEHS) training grant (ES07148 to C.N.H.); NIEHS Center grant (ES005022); Bristol-Myers Squibb (to C.N.H.); Rutgers/New Jersey Agricultural Experimental Station; funds from UMDNJ-RWJMS to R.Z.
Acknowledgments
Special thanks to Dr Stephen Safe (Texas A&M University, Texas) for his generous donation of HPTE.
References
- Akgul Y, Derk RC, Meighan T, Rao KM, Murono EP. The methoxychlor metabolite, HPTE, directly inhibits the catalytic activity of cholesterol side-chain cleavage (P450scc) in cultured rat ovarian cells. Reprod. Toxicol. 2008;25:67–75. doi: 10.1016/j.reprotox.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Armenti AE, Zama AM, Passantino L, Uzumcu M. Developmental methoxychlor exposure affects multiple reproductive parameters and ovarian folliculogenesis and gene expression in adult rats. Toxicol. Appl. Pharmacol. 2008;233:286–296. doi: 10.1016/j.taap.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo NA, Woodruff TK, Giudice LC. Regulation of insulin-like growth factor binding protein production by human luteinizing granulosa cells cultured in defined medium. J. Clin. Endocrinol. Metab. 1993;76:207–215. doi: 10.1210/jcem.76.1.7678423. [DOI] [PubMed] [Google Scholar]
- Chapin RE, Harris MW, Davis BJ, Ward SM, Wilson RE, Mauney MA, Lockhart AC, Smialowicz RJ, Moser VC, Burka LT, et al. The effects of perinatal/juvenile methoxychlor exposure on adult rat nervous, immune, and reproductive system function. Fundam. Appl. Toxicol. 1997;40:138–157. doi: 10.1006/faat.1997.2381. [DOI] [PubMed] [Google Scholar]
- Chedrese PJ, Feyles F. The diverse mechanism of action of dichlorodiphenyldichloroethylene (DDE) and methoxychlor in ovarian cells in vitro. Reprod. Toxicol. 2001;15:693–698. doi: 10.1016/s0890-6238(01)00172-1. [DOI] [PubMed] [Google Scholar]
- deMoura MD, Choi D, Adashi EY, Payne DW. Insulin-like growth factor-I-mediated amplification of follicle-stimulating hormone-supported progesterone accumulation by cultured rat granulosa cells: Enhancement of steroidogenic enzyme activity and expression. Biol. Reprod. 1997;56:946–953. doi: 10.1095/biolreprod56.4.946. [DOI] [PubMed] [Google Scholar]
- Eppig JJ, O'Brien MJ. Development in vitro of mouse oocytes from primordial follicles. Biol. Reprod. 1996;54:197–207. doi: 10.1095/biolreprod54.1.197. [DOI] [PubMed] [Google Scholar]
- Eroschenko VP, Abuel-Atta AA, Grober MS. Neonatal exposures to technical methoxychlor alters ovaries in adult mice. Reprod. Toxicol. 1995;9:379–387. doi: 10.1016/0890-6238(95)00025-6. [DOI] [PubMed] [Google Scholar]
- Fielder PJ, Pham H, Adashi EY, Rosenfeld RG. Insulin-like growth factors (IGFs) block FSH-induced proteolysis of IGF-binding protein-5 (BP-5) in cultured rat granulosa cells. Endocrinology. 1993;133:415–418. doi: 10.1210/endo.133.1.7686483. [DOI] [PubMed] [Google Scholar]
- Findlay JK, Britt K, Kerr JB, O'Donnell L, Jones ME, Drummond AE, Simpson ER. The road to ovulation: The role of oestrogens. Reprod. Fertil. Dev. 2001;13:543–547. doi: 10.1071/rd01071. [DOI] [PubMed] [Google Scholar]
- Gaido KW, Maness SC, McDonnell DP, Dehal SS, Kupfer D, Safe S. Interaction of methoxychlor and related compounds with estrogen receptor alpha and beta, and androgen receptor: Structure-activity studies. Mol. Pharmacol. 2000;58:852–858. [PubMed] [Google Scholar]
- Gazzerro E, Pereira RC, Jorgetti V, Olson S, Economides AN, Canalis E. Skeletal overexpression of gremlin impairs bone formation and causes osteopenia. Endocrinology. 2005;146:655–665. doi: 10.1210/en.2004-0766. [DOI] [PubMed] [Google Scholar]
- Gazzerro E, Smerdel-Ramoya A, Zanotti S, Stadmeyer L, Durant D, Economides AN, Canalis E. Conditional deletion of gremlin causes a transient increase in bone formation and bone mass. J. Biol. Chem. 2007;282:31549–31557. doi: 10.1074/jbc.M701317200. [DOI] [PubMed] [Google Scholar]
- Gray LE, Jr, Ostby J, Ferrell J, Rehnberg G, Linder R, Cooper R, Goldman J, Slott V, Laskey J. A dose-response analysis of methoxychlor-induced alterations of reproductive development and function in the rat. Fundam. Appl. Toxicol. 1989;12:92–108. doi: 10.1016/0272-0590(89)90065-1. [DOI] [PubMed] [Google Scholar]
- Guo L, Fang H, Collins J, Fan XH, Dial S, Wong A, Mehta K, Blann E, Shi L, Tong W, et al. Differential gene expression in mouse primary hepatocytes exposed to the peroxisome proliferator-activated receptor alpha agonists. BMC Bioinformatics. 2006;7(Suppl. 2):S18. doi: 10.1186/1471-2105-7-S2-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Aberdeen G, Babus JK, Albrecht ED, Flaws JA. Methoxychlor and its metabolites inhibit growth and induce atresia of baboon antral follicles. Toxicol. Pathol. 2007;35:649–656. doi: 10.1080/01926230701459960. [DOI] [PubMed] [Google Scholar]
- Gupta RK, Miller KP, Babus JK, Flaws JA. Methoxychlor inhibits growth and induces atresia of antral follicles through an oxidative stress pathway. Toxicol. Sci. 2006;93:382–389. doi: 10.1093/toxsci/kfl052. [DOI] [PubMed] [Google Scholar]
- Hall DL, Payne LA, Putnam JM, Huet-Hudson YM. Effect of methoxychlor on implantation and embryo development in the mouse. Reprod. Toxicol. 1997;11:703–708. doi: 10.1016/s0890-6238(97)00026-9. [DOI] [PubMed] [Google Scholar]
- Hino S, Tanji C, Nakayama KI, Kikuchi A. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase stabilizes beta-catenin through inhibition of its ubiquitination. Mol. Cell. Biol. 2005;25:9063–9072. doi: 10.1128/MCB.25.20.9063-9072.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh AJ, Adashi EY, Jones PB, Welsh TH., Jr Hormonal regulation of the differentiation of cultured ovarian granulosa cells. Endocr. Rev. 1984;5:76–127. doi: 10.1210/edrv-5-1-76. [DOI] [PubMed] [Google Scholar]
- Hu Y, Kupfer D. Metabolism of the endocrine disruptor pesticide-methoxychlor by human P450s: Pathways involving a novel catechol metabolite. Drug Metab. Dispos. 2002;30:1035–1042. doi: 10.1124/dmd.30.9.1035. [DOI] [PubMed] [Google Scholar]
- Hunzicker-Dunn M, Maizels ET. FSH signaling pathways in immature granulosa cells that regulate target gene expression: Branching out from protein kinase A. Cell. Signal. 2006;18:1351–1359. doi: 10.1016/j.cellsig.2006.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SJ, Wang S, Hara H, Peterson EP, Namura S, Amin-Hanjani S, Huang Z, Srinivasan A, Tomaselli KJ, Thornberry NA, et al. Dual role of caspase-11 in mediating activation of caspase-1 and caspase-3 under pathological conditions. J. Cell Biol. 2000;149:613–622. doi: 10.1083/jcb.149.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. The International Protein Index: An integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- Kleuser B, Malek D, Gust R, Pertz HH, Potteck H. 17-{beta}-estradiol inhibits transforming growth factor-{beta} signaling and function in breast cancer cells via activation of extracellular signal-regulated kinase through the G protein-coupled receptor 30. Mol. Pharmacol. 2008;74:1533–1543. doi: 10.1124/mol.108.046854. [DOI] [PubMed] [Google Scholar]
- Knight PG, Glister C. TGF-beta superfamily members and ovarian follicle development. Reproduction. 2006;132:191–206. doi: 10.1530/rep.1.01074. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc. Natl. Acad. Sci. U. S. A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Baliga R. Endoplasmic reticulum stress-associated caspase 12 mediates cisplatin-induced LLC-PK1 cell apoptosis. J. Am. Soc. Nephrol. 2005;16:1985–1992. doi: 10.1681/ASN.2004090768. [DOI] [PubMed] [Google Scholar]
- Matzuk MM, Burns KH, Viveiros MM, Eppig JJ. Intercellular communication in the mammalian ovary: Oocytes carry the conversation. Science. 2002;296:2178–2180. doi: 10.1126/science.1071965. [DOI] [PubMed] [Google Scholar]
- Mazerbourg S, Bondy CA, Zhou J, Monget P. The insulin-like growth factor system: A key determinant role in the growth and selection of ovarian follicles? A comparative species study. Reprod. Domest. Anim. 2003;38:247–258. doi: 10.1046/j.1439-0531.2003.00440.x. [DOI] [PubMed] [Google Scholar]
- Merino R, Rodriguez-Leon J, Macias D, Ganan Y, Economides AN, Hurle JM. The BMP antagonist Gremlin regulates outgrowth, chondrogenesis and programmed cell death in the developing limb. Development. 1999;126:5515–5522. doi: 10.1242/dev.126.23.5515. [DOI] [PubMed] [Google Scholar]
- Michos O, Goncalves A, Lopez-Rios J, Tiecke E, Naillat F, Beier K, Galli A, Vainio S, Zeller R. Reduction of BMP4 activity by gremlin 1 enables ureteric bud outgrowth and GDNF/WNT11 feedback signalling during kidney branching morphogenesis. Development. 2007;134:2397–2405. doi: 10.1242/dev.02861. [DOI] [PubMed] [Google Scholar]
- Miro F, Hillier SG. Modulation of granulosa cell deoxyribonucleic acid synthesis and differentiation by activin. Endocrinology. 1996;137:464–468. doi: 10.1210/endo.137.2.8593790. [DOI] [PubMed] [Google Scholar]
- Miro F, Smyth CD, Hillier SG. Development-related effects of recombinant activin on steroid synthesis in rat granulosa cells. Endocrinology. 1991;129:3388–3394. doi: 10.1210/endo-129-6-3388. [DOI] [PubMed] [Google Scholar]
- Moreira DF, Strauss BE, Vannier E, Belizario JE. Genes up- and down-regulated by dermcidin in breast cancer: A microarray analysis. Genet. Mol. Res. 2008;7:925–932. doi: 10.4238/vol7-3x-meeting009. [DOI] [PubMed] [Google Scholar]
- Palermo R. Differential actions of FSH and LH during folliculogenesis. Reprod. Biomed. Online. 2007;15:326–337. doi: 10.1016/s1472-6483(10)60347-1. [DOI] [PubMed] [Google Scholar]
- Pangas SA, Jorgez CJ, Matzuk MM. Growth differentiation factor 9 regulates expression of the bone morphogenetic protein antagonist gremlin. J. Biol. Chem. 2004;279:32281–32286. doi: 10.1074/jbc.M403212200. [DOI] [PubMed] [Google Scholar]
- Parakh TN, Hernandez JA, Grammer JC, Weck J, Hunzicker-Dunn M, Zeleznik AJ, Nilson JH. Follicle-stimulating hormone/cAMP regulation of aromatase gene expression requires beta-catenin. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12435–12440. doi: 10.1073/pnas.0603006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z, Oey NA, Zuidervaart MM, Jia Z, Li Y, Steinberg SJ, Smith KD, Watkins PA. The acyl-CoA synthetase “bubblegum” (lipidosin): Further characterization and role in neuronal fatty acid beta-oxidation. J. Biol. Chem. 2003;278:47070–47078. doi: 10.1074/jbc.M310075200. [DOI] [PubMed] [Google Scholar]
- Pepling ME, Spradling AC. Mouse ovarian germ cell cysts undergo programmed breakdown to form primordial follicles. Dev. Biol. 2001;234:339–351. doi: 10.1006/dbio.2001.0269. [DOI] [PubMed] [Google Scholar]
- Skinner MK. Regulation of primordial follicle assembly and development. Hum. Reprod. Update. 2005;11:461–471. doi: 10.1093/humupd/dmi020. [DOI] [PubMed] [Google Scholar]
- Smyth CD, Gosden RG, McNeilly AS, Hillier SG. Effect of inhibin immunoneutralization on steroidogenesis in rat ovarian follicles in vitro. J. Endocrinol. 1994;140:437–443. doi: 10.1677/joe.0.1400437. [DOI] [PubMed] [Google Scholar]
- Stuchal LD, Kleinow KM, Stegeman JJ, James MO. Demethylation of the pesticide methoxychlor in liver and intestine from untreated, methoxychlor-treated, and 3-methylcholanthrene-treated channel catfish (Ictalurus punctatus): Evidence for roles of CYP1 and CYP3A family isozymes. Drug. Metab. Dispos. 2006;34:932–938. doi: 10.1124/dmd.105.009068. [DOI] [PubMed] [Google Scholar]
- Tong W, Cao X, Harris S, Sun H, Fang H, Fuscoe J, Harris A, Hong H, Xie Q, Perkins R, et al. ArrayTrack—supporting toxicogenomic research at the U.S. Food and Drug Administration National Center for Toxicological Research. Environ. Health Perspect. 2003;111:1819–1826. doi: 10.1289/ehp.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui M, Shimonaka M, Shimasaki S, Ling N. An insulin-like growth factor-binding protein in ovarian follicular fluid blocks follicle-stimulating hormone-stimulated steroid production by ovarian granulosa cells. Endocrinology. 1989;125:912–916. doi: 10.1210/endo-125-2-912. [DOI] [PubMed] [Google Scholar]
- Uzumcu M, Zachow R. Developmental exposure to environmental endocrine disruptors: Consequences within the ovary and on female reproductive function. Reprod. Toxicol. 2007;23:337–352. doi: 10.1016/j.reprotox.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voge JL, Santiago CA, Aad PY, Goad DW, Malayer JR, Spicer LJ. Quantification of insulin-like growth factor binding protein mRNA using real-time PCR in bovine granulosa and theca cells: Effect of estradiol, insulin, and gonadotropins. Domest. Anim. Endocrinol. 2004;26:241–258. doi: 10.1016/j.domaniend.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Wang S, Miura M, Jung YK, Zhu H, Li E, Yuan J. Murine caspase-11, an ICE-interacting protease, is essential for the activation of ICE. Cell. 1998;92:501–509. doi: 10.1016/s0092-8674(00)80943-5. [DOI] [PubMed] [Google Scholar]
- Zachow R, Uzumcu M. The methoxychlor metabolite, 2,2-bis-(p-hydroxyphenyl)-1,1,1-trichloroethane, inhibits steroidogenesis in rat ovarian granulosa cells in vitro. Reprod. Toxicol. 2006;22:659–665. doi: 10.1016/j.reprotox.2006.04.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.