Abstract
Hepatitis C virus (HCV) is a human pathogen associated with chronic liver disease. Recently, the cell culture systems supporting complete replication and production of HCV genotype 2a (JFH1) have been established. This study investigated the effect of low-speed centrifugation on HCV JFH1 infection of human hepatocytes (Huh7.5.1). Higher levels of HCV RNA expression were observed in Huh7.5.1 cells infected with centrifugal inoculation of HCV JFH1 than those in the control cells. This increased HCV RNA expression was associated with the elevated expression of HCV NS3 protein in the hepatocytes. The centrifugal enhancement of HCV infection was time and speed dependent. However, the enhancement was not observed when centrifugation was performed before or after HCV infection. In addition, there was no association between centrifugal enhancement and the expression of HCV entry receptors (CD81 and claudin-1) and intracellular IFN-α in the hepatocytes. These data indicate that centrifugal inoculation is a useful tool for increasing the efficiency of HCV infection and replication in the target cells in vitro.
Keywords: Hepatitis C virus, centrifugal inoculation, infection
1. Introduction
Hepatitis C virus (HCV) infection is a major cause of chronic hepatitis, liver cirrhosis and hepatocellular carcinoma worldwide. HCV is a small, enveloped, and positive-sense RNA virus belonging to family Flaviviridae, genus Hepaivirus, which was first identified in 1989 (Choo et al., 1989). However, a protective vaccine is not yet available and the therapeutic options are still limited (Lindenbach et al., 2006). The elucidation of the viral lifecycle and the development of specific antiviral agents and preventive vaccine were long hampered by the poor replication of HCV in cultured cells. Although HCV replication in vivo is extremely robust (Neumann et al., 1998), the growth of HCV in cell cultures in vitro is very difficult. Several groups recently reported the establishment of efficient in vitro cell systems for complete and infectious HCV replication based on the hepatic cell lines and HCV genotype 2a JFH1 clone (Cai et al., 2005; Lindenbach et al., 2006; Wakita et al., 2005; Zhong et al., 2005).
Centrifugal inoculation has been used for many years to enhance infections of a variety of different types of viruses. The centrifugal enhancement of infection has been described for human and murine cytomegaloviruses (Ho et al., 1993; Hodgkin et al., 1988; Hudson et al., 1976; Osborn and Walker, 1968), retroviruses (Bahnson et al., 1995; Chuck et al., 1996; Ho et al., 1993; Kotani et al., 1994; Pietroboni et al., 1989), bluetongue virus (Sundin and Mecham, 1989), adenoviruses (Nyberg-Hoffman et al., 1997), human herpesvirus type 6 (Pietroboni et al., 1989) and influenza A virus (Reading et al., 2001). This study evaluated the impact of centrifugation on HCV JFH1 infection of human hepatocytes.
2. Materials and Methods
2.1. Reagents
Mouse monoclonal anti-HCV NS3 antibody was generated by Dr. Guangxiang Luo (Department of Microbiology, Immunology, and Molecular Genetics, University of Kentucky College of Medicine, Lexington, Kentucky). Rabbit polyclonal anti-CD81 antibody was purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Mouse monoclonal anti-claudin-1 was purchased from Invitrogen Co. (Carlsbad, CA). Rabbit polyclonal anti-actin antibody was purchased from Sigma-Aldrich Co. (St. Louis, MO). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG and HRP-conjugated goat anti-mouse IgG were purchased from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA).
2.2. Human hepatic cell lines and HCV plasmid
Hepatic cell line (Huh7.5.1) was kindly provided by Dr. Charles M. Rice (Laboratory of Virology and Infectious Diseases, the Rockefeller University, New York). Cells were maintained in the conditioned DMEM supplemented with 10% FCS, 10 mM Hepes, 100 units/ml penicillin, 100 mg/ml streptomycin, and 2 mM L-glutamine at 5% CO2. The plasmid pJFH1 that contains the full-length HCV genotype 2a JFH1 cDNA downstream of the T7 RNA promoter, was kindly provided by Dr. Takaji Wakita (Department of Microbiology, Tokyo Metropolitan Institute for Neuroscience, Tokyo, Japan).
2.3. Preparation of infectious HCV JFH1 and HCV infection of hepatocytes
For generating infectious HCV JFH1, in vitro transcribed JFH1 genomic RNA was transfected into Huh7.5.1 cells as described (Wakita et al., 2005). Cell culture media collected at day 10 post transfection were centrifuged and passed through 0.22 μm filter. The cell-free media were used as JFH1 stocks, which were aliquoted and stored at −80°C.
2.4. Centrifugation
Huh7.5.1 cells were seeded into 12-well plates (1 × 105 cells/well) and cultured for 24 h and then incubated with HCV JFH1 as described (Wakita et al., 2005). The incubated plates were immediately centrifuged at 500×g or 1000×g for 30–120 min at room temperature following the addition of the JFH1 stock. After centrifugation, the inoculums were removed and the cells were washed 5 times with plain DMEM. The cells were refed with the conditioned DMEM and maintained at 5% CO2. Total cellular RNA samples were collected at day 2 and day 6 post infection.
2.5. Real Time Reverse Transcriptase (RT) PCR
Total cellular RNA was extracted from Huh7.5.1 cells using Tri-Reagent (Molecular Research Center, Cincinnati, OH) as described (Li et al., 2003). Total RNA (1μg) was then subjected to reverse transcription using the reverse transcription system from Promega (Madison, WI). The real time RT PCR for the quantification of HCV, CD81, claudin-1, IFN-α, Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was performed with the iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA) as previously described (Li et al., 2004; Zhang et al., 2005). The amplification results were visualized and analyzed using the software MyiQ provided with the thermocycler (iCycler iQ real time PCR detection system, Bio-Rad Laboratories). The levels of GAPDH mRNA were used as an endogenous reference to normalize the quantities of target mRNA. The special oligonucleotide primers used in this study were listed as follows: HCV: 5′-RAY CACTCC CCT GTG AGG AAC -3′ (sense) and 5′-TGR TGC ACG GTC TAC GAG ACC TC -3′ (anti-sense). CD81: 5′-CGC CAA GGA TGT GAA GCA GTT C -3′ (sense) and 5′-TCC CGG AGA AGA GGT CAT CGA T-3′ (anti-sense). Claudin-1: 5′-GGC AGA TCC AGT GCA AAG TC-3′ (sense) and 5′-TCT TCT GCA CCT CAT CGT CTT-3′ (anti-sense). IFN-α: 5′-TTT CTC CTG CCT GAA GGA CAG-3′ (sense) and 5′-GCT CAT GAT TTC TGC TCT GAC A-3′ (anti-sense). GAPDH: 5′-GGT GGT CTC CTC TGA CTT CAA CA-3′ (sense) and 5′-GTT GCT GTA GCC AAA TTC GTT GT-3′ (anti-sense). The oligonucleotide primers were synthesized by Integrated DNA Technologies Inc. (Coralville, IA).
2.6. Western Blot
Total cell lysates from the hepatic cells were prepared using a radioimmune precipitation assay (RIPA) buffer (Promega, Madison, WI) with 1% protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). Protein concentrations were determined by DC protein assay kit (Bio-Rad, Hercules, CA). Western blot were carried out as previous described (Li et al., 2003). HCV NS3, CD81, claudin-1 and actin proteins were detected using antibodies against HCV NS3 (1:500), CD81 (1:1000), claudin-1 (1:400), and actin (1:3000), respectively. HRP-conjugated goat anti-rabbit antibody (1:10 000) or goat anti-mouse antibody (1:5000) was used as the second antibody. The bound antibodies were recognized by using SuperSignal West Pico Chemiluminescent Substrate Kit (Pierce, Rockford, IL) according to the manufacturer’s instruction. Prestained molecular markers (Bio-Rad, Hercules, CA) were used to determine the molecular weight of immuno-reactive bands.
3. Results
3.1. The effect of centrifugation on HCV JFH1 infection
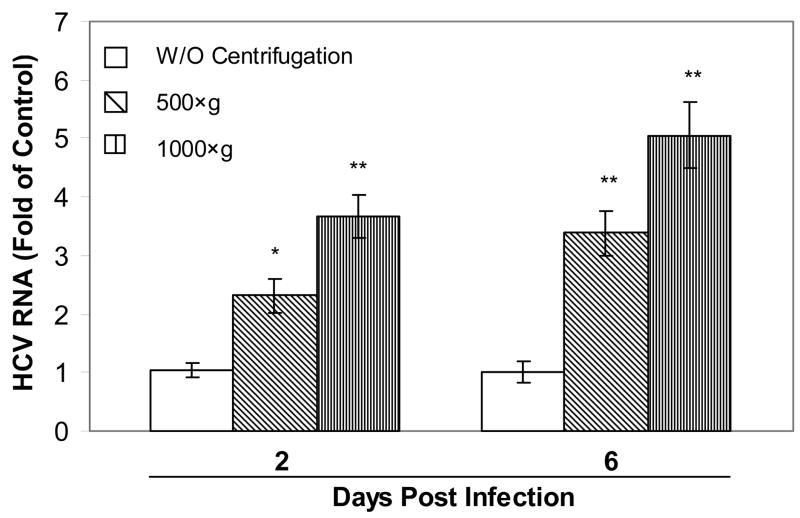
HCV JFH1 infection of Huh7.5.1 cells was greatly enhanced by centrifugal inoculation as determined by HCV RNA real time RT PCR (Fig. 1). Centrifugation resulted in increased expression of HCV RNA in Huh7.5.1 cells (Fig. 1). The centrifuged enhancement of HCV RNA expression was positively related to the time and speed of centrifugation, as the longer time (120 min) and higher speed (1000×g) resulted in the higher expression of HCV RNA in the hepatocytes (Figs. 1 and 2). The increase of HCV infection by centrifugation was also confirmed by Western blot assay, showing that the levels of HCV NS3 protein were elevated in Huh7.5.1 cells infected by centrifugal inoculation (500×g or 1000×g; 120 min). (Fig. 3).
Fig. 1.
Effect of centrifugation on HCV JFH1 infection. Huh7.5.1 cells were incubated with HCV JFH1 for 90 min with or without centrifugation (500×g; 1000×g). Total cellular RNA was collected at the indicated time points and subjected to real time RT PCR for HCV and GAPDH mRNA. Data are expressed as HCV RNA levels relative (fold) to control (without centrifugation), which is defined as 1.0. The results shown are the mean ± SD of three independent experiments (*, P<0.05; **, P<0.01, centrifugation vs. control).
Fig. 2.
Effect of centrifugation time on HCV JFH1 infection. Huh7.5.1 cells were incubated with HCV JFH1 with or without centrifugation (1000×g) for the indicated times. Total cellular RNA was collected at day 2 and day 6 post infection and subjected to real time RT PCR for HCV and GAPDH mRNA. Data are expressed as HCV RNA levels relative (fold) to control (without centrifugation), which is defined as 1.0. The results shown are the mean ± SD of three independent experiments (*, P<0.05; **, P<0.01, centrifugation vs. control).
Fig. 3.
Western blot analysis of HCV NS3 protein expression in HCV JFH1-infected Huh7.5.1 cells. Huh7.5.1 cells were incubated with HCV JFH1 for 120 min with or without centrifugation (500×g; 1000×g). At day 6 post infection, equal amount of proteins extracted from infected cells with or without centrifugation were subjected to Western blot assay using antibodies against HCV NS3 and actin, respectively. The numbers in the right panel are the signal intensities of protein bands of the Western blot expressed as densitometry scanning units (DSUs). One representative of three experiments with similar results is shown.
3.2. The effect of centrifugation on CD81, claudin-1 and IFN-α expression in Huh7.5.1 cells
In order to explore the mechanism(s) involved in the centrifugal enhancement of HCV infectivity, the effect of centrifugation on the expression of CD81 or claudin-1, two recognized receptors for HCV entry into human hepatocytes (Cormier et al., 2004; Evans et al., 2007), was investigated. The centrifugation had little effect on CD81 or claudin-1 expression in Huh7.5.1 cells at both RNA and protein levels (Fig. 4A and 4B). In addition, the impact of centrifugation on the endogenous IFN-α expressionαin Huh7.5.1 cell was examined. As demonstrated in Fig. 4A, the centrifugation had little effect on the expression of intracellular IFN-α in the hepatocytes.
Fig. 4.
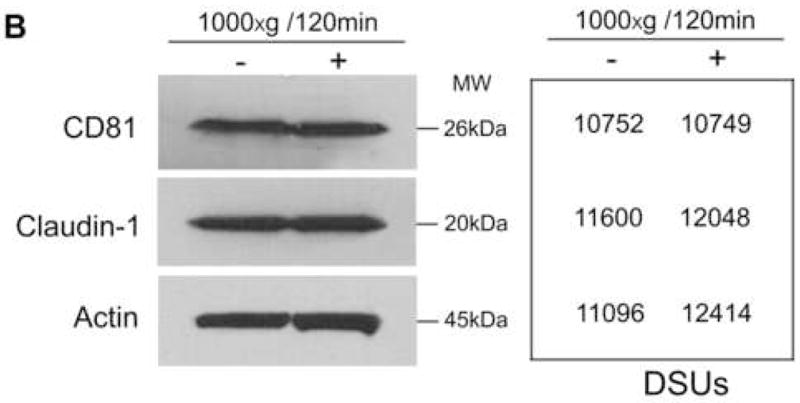
Effect of centrifugation on CD81, claudin-1 and IFN-α expression in Huh7.5.1 cells. (A) Total cellular RNA was collected from Huh7.5.1 cells with or without centrifugation (1000×g) for 120 min immediately after centrifugation and subjected to real time RT PCR for CD81, claudin-1, IFN-α and GAPDH mRNA. Data are expressed as mRNA levels relative (%) to control (without centrifugation), which is defined as 100. The results shown are the mean ± SD of three independent experiments. (B) Western blot analysis of CD81 and claudin-1 proteins in Huh7.5.1. Equal amount of proteins extracted from the cells with or without centrifugation immediately after centrifugation were subjected to Western blot assay using antibodies against CD81, claudin-1 and actin. The numbers in the right panel are the signal intensities of protein bands of the Western blot expressed as densitometry scanning units (DSUs). One representative of these experiments with similar results is shown.
4. Discussion
The technique of centrifugal inoculation has been used for many years to enhance infections of different types of viruses. However, some viruses such as herpes simplex virus type 1 and 2, bovine herpevirus type 1 and pseudorabies virus did not show enhanced infectivity by centrifugation (Sundin and Mecham, 1989). In addition, infection by some RNA viruses such as Sindbis and hepatic necrosis virus of fish failed to response to centrifugation (Hudson, 1988). Effect of centrifugation on positive-sense RNA viruses, however, has not been examined. This study for the first time demonstrated that low-speed centrifugation is effective on the enhancement of HCV JFH1 infection of human hepatocytes. There is a simultaneous requirement for both HCV and hepatocytes during the centrifugation, since enhancement effects were not observed when either HCV or hepatocytes were centrifuged alone (data not shown). These data indicate that the interaction between HCV and cell membrane of the hepatocytes during centrifugation is important in promoting HCV infection.
HCV isolates are classified into genotypes and subtypes, and there are 6 major genotypes that differ in their nucleotide sequence by 30–35% (Moradpour et al., 2007). The productive HCV infectious systems have been achieved mainly with genotype 2a (JFH1). A recent report described the production of HCV genotype 1a (H77-S strain) in human hepatocytes (Yi et al., 2006). However, the infectivity of HCV H77-S in the hepatocytes was significantly lower (~ 400-fold) than HCV genotype 2a (JFH1) (Yi et al., 2006). In this study, the investigation of the effect of centrifugation on HCV H77-S infection showed that although there was an increase (~5 fold) of viral RNA at 24h postinfection in the hepatocytes infected with centrifugal inoculation of HCV H77-S, the virus failed to persist in the hepatocytes (data not shown).
The mechanism(s) of centrifugation enhancement of infectivity remain unclear. One possible explanation is that most virus particles are too far from the cells layer to make the initial contact that are needed for the establishment of infection, and that this barrier can be somehow overcome by sedimenting virus particles to cell surface by centrifugal forces (Huyghe et al., 1995; Kotani et al., 1994; Nyberg-Hoffman et al., 1997; Reading et al., 2001). However, this explanation can not account for the viruses on which centrifugation has no effects (Sundin and Mecham, 1989). In addition, virus particles in suspension are too small to be precipitated by low-speed centrifugation. Since the enhancing effect of centrifugation for some viruses can be observed even at fields below 100×g (Hodgkin et al., 1988), while much higher forces are needed to bring other viruses out of suspension (e.g 10 000–12 000×g for 5–16 h) (Bahnson et al., 1995), it is likely that other factors are involved in the enhancing effect of centrifugation on viral infection. One possibility is that centrifugation influences the expression of cellular genes that favor or resist viral infection (Hodgkin et al., 1988). Thus, the impact of centrifugation on the known cellular factors that modulate HCV infection of human hepatocytes was examined. In addition to determine the effect of centrifugation on the expression of CD81 and claudin-1, the two recognized receptors used by HCV to entry into hepatocyts (Cormier et al., 2004; Evans et al., 2007), whether centrifugation inhibit the expression of IFN-α, a potent anti-HCV cytokine (Zhang et al., 2005), was also investigated. These cellular factors, however, were not affected by centrifugation (Fig. 4A and 4B). Therefore, it is likely that there are some common and nonspecific changes occurred in the cells during and after centrifugation, which facilitate certain step(s) of viral infection, for example, the virus entry or uncoating.
In conclusion, despite unknown mechanism of the centrifuged enhancement of HCV infection, centrifugation technique maybe a useful tool for the establishment of robust HCV infection in the in vitro cell systems. Future studies are needed to determine whether centrifugation enhances HCV infection of non-hepatic cells that serve as potential extrahepatic sites or a reservoir for HCV replication.
Acknowledgments
The authors thank Dr. Charles Rice (The Rockefeller University, New York, NY) for generously providing Huh7.5.1 cell line, Dr. Takaji Wakita (Tokyo Metropolitan Institute for Neuroscience, Tokyo, Japan) for providing HCV JFH1 molecular clone. The investigation was supported by the National Institute of Health grants DA 12815 and DA 22177 (to W.-Z. H) and Foerderer Murray Award (to W.-Z. H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bahnson AB, Dunigan JT, Baysal BE, Mohney T, Atchison RW, Nimgaonkar MT, Ball ED, Barranger JA. Centrifugal enhancement of retroviral mediated gene transfer. J Virol Methods. 1995;54:131–43. doi: 10.1016/0166-0934(95)00035-s. [DOI] [PubMed] [Google Scholar]
- Cai Z, Zhang C, Chang KS, Jiang J, Ahn BC, Wakita T, Liang TJ, Luo G. Robust production of infectious hepatitis C virus (HCV) from stably HCV cDNA-transfected human hepatoma cells. J Virol. 2005;79:13963–73. doi: 10.1128/JVI.79.22.13963-13973.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–62. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- Chuck AS, Clarke MF, Palsson BO. Retroviral infection is limited by Brownian motion. Hum Gene Ther. 1996;7:1527–34. doi: 10.1089/hum.1996.7.13-1527. [DOI] [PubMed] [Google Scholar]
- Cormier EG, Tsamis F, Kajumo F, Durso RJ, Gardner JP, Dragic T. CD81 is an entry coreceptor for hepatitis C virus. Proc Natl Acad Sci U S A. 2004;101:7270–4. doi: 10.1073/pnas.0402253101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–5. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- Ho WZ, Cherukuri R, Ge SD, Cutilli JR, Song L, Whitko S, Douglas SD. Centrifugal enhancement of human immunodeficiency virus type 1 infection and human cytomegalovirus gene expression in human primary monocyte/macrophages in vitro. J Leukoc Biol. 1993;53:208–12. doi: 10.1002/jlb.53.2.208. [DOI] [PubMed] [Google Scholar]
- Hodgkin PD, Scalzo AA, Swaminathan N, Price P, Shellam GR. Murine cytomegalovirus binds reversibly to mouse embryo fibroblasts: implications for quantitation and explanation of centrifugal enhancement. J Virol Methods. 1988;22:215–30. doi: 10.1016/0166-0934(88)90104-8. [DOI] [PubMed] [Google Scholar]
- Hudson JB. Further studies on the mechanism of centrifugal enhancement of cytomegalovirus infectivity. J Virol Methods. 1988;19:97–108. doi: 10.1016/0166-0934(88)90153-x. [DOI] [PubMed] [Google Scholar]
- Hudson JB, Misra V, Mosmann TR. Cytomegalovirus infectivity: analysis of the phenomenon of centrifugal enhancement of infectivity. Virology. 1976;72:235–43. doi: 10.1016/0042-6822(76)90326-3. [DOI] [PubMed] [Google Scholar]
- Huyghe BG, Liu X, Sutjipto S, Sugarman BJ, Horn MT, Shepard HM, Scandella CJ, Shabram P. Purification of a type 5 recombinant adenovirus encoding human p53 by column chromatography. Hum Gene Ther. 1995;6:1403–16. doi: 10.1089/hum.1995.6.11-1403. [DOI] [PubMed] [Google Scholar]
- Kotani H, Newton PB, 3rd, Zhang S, Chiang YL, Otto E, Weaver L, Blaese RM, Anderson WF, McGarrity GJ. Improved methods of retroviral vector transduction and production for gene therapy. Hum Gene Ther. 1994;5:19–28. doi: 10.1089/hum.1994.5.1-19. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang T, Douglas SD, Lai JP, Xiao WD, Pleasure DE, Ho WZ. Morphine enhances hepatitis C virus (HCV) replicon expression. Am J Pathol. 2003;163:1167–75. doi: 10.1016/S0002-9440(10)63476-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang T, Ho C, Orange JS, Douglas SD, Ho WZ. Natural killer cells inhibit hepatitis C virus expression. J Leukoc Biol. 2004;76:1171–9. doi: 10.1189/jlb.0604372. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Meuleman P, Ploss A, Vanwolleghem T, Syder AJ, McKeating JA, Lanford RE, Feinstone SM, Major ME, Leroux-Roels G, Rice CM. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc Natl Acad Sci U S A. 2006;103:3805–9. doi: 10.1073/pnas.0511218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–63. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, Perelson AS. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282:103–7. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- Nyberg-Hoffman C, Shabram P, Li W, Giroux D, Aguilar-Cordova E. Sensitivity and reproducibility in adenoviral infectious titer determination. Nat Med. 1997;3:808–11. doi: 10.1038/nm0797-808. [DOI] [PubMed] [Google Scholar]
- Osborn JE, Walker DL. Enhancement of infectivity of murine cytomegalovirus in vitro by centrifugal inoculation. J Virol. 1968;2:853–8. doi: 10.1128/jvi.2.9.853-858.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietroboni GR, Harnett GB, Bucens MR. Centrifugal enhancement of human immunodeficiency virus (HIV) and human herpesvirus type 6 (HHV-6) infection in vitro. J Virol Methods. 1989;24:85–90. doi: 10.1016/0166-0934(89)90010-4. [DOI] [PubMed] [Google Scholar]
- Reading SA, Edwards MJ, Dimmock NJ. Increasing the efficiency of virus infectivity assays: small inoculum volumes are as effective as centrifugal enhancement. J Virol Methods. 2001;98:167–9. doi: 10.1016/s0166-0934(01)00368-8. [DOI] [PubMed] [Google Scholar]
- Sundin DR, Mecham JO. Enhanced infectivity of bluetongue virus in cell culture by centrifugation. J Clin Microbiol. 1989;27:1659–60. doi: 10.1128/jcm.27.7.1659-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–6. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, Villanueva RA, Thomas DL, Wakita T, Lemon SM. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc Natl Acad Sci U S A. 2006;103:2310–5. doi: 10.1073/pnas.0510727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Lin RT, Li Y, Douglas SD, Maxcey C, Ho C, Lai JP, Wang YJ, Wan Q, Ho WZ. Hepatitis C virus inhibits intracellular interferon alpha expression in human hepatic cell lines. Hepatology. 2005;42:819–27. doi: 10.1002/hep.20854. [DOI] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102:9294–9. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]