Abstract
Cathepsin P is a member of a family of placentally expressed cathepsins (PECs). The closest human homolog of cathepsin P is cathepsin L, a broad specificity enzyme that has functions in many tissues in addition to placenta. The gene duplications that gave rise to the PECs provide a rare opportunity to define proteolytic functions in placenta, a transient organ unique to mammals. Peptidyl substrate and inhibitor libraries have shown that cathepsin P has evolved an unusually restricted preference for substrates containing hydrophobic amino acids. Proteomic techniques were used to probe for substrates of this enzyme. Recombinant cathepsin P was incubated with rat choriocarcinoma (Rcho-1) cell proteins to identify substrates using two-dimensional difference gel electrophoresis. Substrate proteins were excised from gels and characterized by trypsin digestion and MALDI MS/MS. Two endoplasmic reticulum (ER) proteins, gp96 and calreticulin, emerged as potential substrates, and western blotting showed that these proteins are processed by cathepsin P from their C-terminus, removing the KDEL ER retention signal. Immunohistochemistry showed that a portion of cathepsin P co-localizes with calreticulin in Rcho-1 cells. Extracellular calreticulin induces differentiation of Rcho-1 cells, indicating a potential role of cathepsin P in processing and secretion of calreticulin during differentiation of trophoblast giant cells.
Keywords: placenta, cathepsin, proteolysis, processing
Introduction
Proteolysis by cathepsins plays critical roles in processes related to early development, including fertilization, embryonic and fetal development, tissue remodeling, antigen presentation, cell-cycle regulation, angiogenesis, apoptosis and inflammatory responses (Turk et al., 2001; Puente et al., 2003). Pharmacological targeting of cysteine proteases with broad-based inhibitors such as E-64 and leupeptin indicates a critical role for cathepsins in blastocyst hatching, implantation and normal embryonic development in mouse, rat and hamster (Babiarz et al., 1992; Afonso et al., 1997; Sireesha et al., 2008). Originally, cathepsins L and B were proposed to be the primary targets of these inhibitors, but genetic deletion of both of these enzymes in mice has no obvious effect on normal fetal development (Deussing et al., 1998; Roth et al., 2000; Felbor et al., 2002). Subsequently, a family of placentally expressed cathepsins (PECs) were discovered in rodent genomes (Sol-Church et al., 1999; Hemberger et al., 2000; Deussing et al., 2002; Sol-Church et al., 2002; Mason, 2008). This family of proteases are well conserved in rodent genomes; orthologs of cathepsins 6, M, P, Q and R are found in both old world and new world mice (Sol-Church et al., 2002; Glenn et al., 2008). The closest human homolog of these genes is cathepsin L, an enzyme that is expressed in all tissues.
Cathepsin P was the first PEC to be discovered and mRNA for this gene can be detected as early as Day 7.5 in the ectoplacental cone of the mouse conceptus and even earlier in the hamster blastocyst (Sol-Church et al., 1999; Hemberger et al., 2001; Deussing et al., 2002; Sireesha et al., 2008). Cathepsin P mRNA is up-regulated during gestation in a developmentally dependent manner in mouse (Sol-Church et al., 1999; Deussing et al., 2002). Later in gestation, it is localized to the labyrinth layer of the definitive placenta (Nakajima et al., 2000; Hemberger et al., 2001).
Recombinant pro-cathepsin P autoactivates at neutral pH, which is a unique property for a lysosomal cysteine protease (Mason et al., 2004; Puzer et al., 2005). The recombinant enzyme is optimally active at neutral pH (6.5–7.5) and preferentially hydrolyzes small synthetic substrates containing hydrophobic amino acids (Mason et al., 2004; Puzer et al., 2005). A screen with a combinatorial library of small peptidyl inhibitors confirmed the preferred specificity of cathepsin P for hydrophobic aromatic amino acids in six contiguous-binding sites spanning the enzyme’s active site cleft (Hassanein et al., 2007). Like other cathepsins, cathepsin P is found in the lysosome, partially co-localizing with cathepsin B in the rat placental cell line, Rcho-1 (Hassanein et al., 2007). Cathepsin P immunoreactivity was also detected in cellular compartments devoid of cathepsin B, indicating that the enzyme might also function outside the lysosome. mRNA and enzyme activity of cathepsin P increase during differentiation of Rcho-1 trophoblast stem cells to a trophoblast giant cells phenotype, indicating a potential role in these invasive cells (Hassanein et al., 2007).
Most studies of the role of lysosomal proteases in placental function have focused on general proteolysis of extracellular matrix proteins to facilitate invasive implantation (Afonso et al., 1997; Cheon et al., 2004). Such general proteolysis could be performed by several broad specificity proteases, such as the matrix metalloproteases and serine proteases (Lamarca et al., 2005). The tissue-specific expression, narrow specificity and unusual pH activity profile of cathepsin P indicate that this enzyme may fulfill a much more specific placental proteolytic role. The preferred peptide sequence for binding to cathepsin P is Trp-Trp-Phe-Val-Trp-Phe (Puzer et al., 2005; Hassanein et al., 2007). A bioinformatics search of the NCBI protein database reveals no exact matches with this peptide. Biological substrates are defined not only by peptide sequence but also by conformational access of enzyme to hydrolysable sequences within proteins. We therefore utilized the resolving power, quantitative capability and sensitivity of two-dimensional difference gel electrophoresis (2D-DIGE) to identify protein substrates of cathepsin P.
Materials and Methods
Materials
NCTC-135 medium was purchased from Sigma Aldrich, (St Louis, MO, USA). Fetal bovine serum (FBS), horse serum, trypsin and HBSS were purchased from Mediatech Inc. (Herndon, VA, USA). Rcho-1 cells were a kind gift from Michael J. Soares, Kansas Medical Center, Kansas City, KS, USA. Sheep anti-calreticulin antibody to aa 270–390 for immunohistochemistry was from BD Biosciences (San Jose, CA, USA). Polyclonal rabbit anti-calreticulin raised to aa 405–417, polyclonal goat anti-calreticulin raised to the N-terminus of the protein and polyclonal rabbit anti-Gp96 raised to the N-terminus of the protein were from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA). Antibody to cathepsin P was prepared as described previously (Mason et al., 2004). Texas Red-labeled mouse anti-rabbit IgG antibody, FITC-labeled donkey anti-sheep IgG antibody and horseradish peroxidase secondary antibodies were from Jackson Immunoresearch Inc. (West Grove, PA, USA). Recombinant mouse cathepsin P was expressed in Pichia pastoris, purified and activated as described previously (Mason et al., 2004). Cy dyes, ECL kits and IPG strips were purchased from GE Healthcare (Piscataway, NJ, USA). Gel Code Blue stain was from Pierce Biotechnology Inc. (Rockford, IL, USA). Sequencing grade modified trypsin was from Promega (Madison, WI, USA). C-18 Zip Tips were from Millipore (Billerica, MA, USA). Other reagents used in this study were analytical grade and were obtained from Sigma Aldrich.
Proteolysis of Rcho-1 cell proteins by cathepsin P
Rcho-1 trophoblast stem cells were maintained in a proliferative state by culturing in hormone and growth-factor-rich medium (NCTC-135 containing 20% FBS, 50 µM β-mercaptoethanol and 1% pyruvic acid). Cells were split every 2–3 days as described (Faria and Soares, 1991; Sahgal et al., 2006). Confluent cells were scraped and washed three times with ice-cold PBS to remove serum and other media components. Cells were then lysed in cathepsin P activity buffer I (20 mM Tris, 1 mM EDTA, 1 mM dithiothreitol and 4% CHAPS, pH 7.4) by freeze-thawing (Bredemeyer et al., 2004). After removal of insoluble material by centrifugation at 12 000g for 5 min, supernatant protein (50 µg) was incubated with or without cathepsin P (0.2 µg) for 6 h at 37°C. The enzymatic reaction was stopped by placing the sample immediately on ice and by adding a 3-fold volume of denaturing isoelectric focusing buffer (7 M urea, 2 M thiourea, 4% CHAPS and 30 mM Tris buffer, pH 8.5).
For western blotting, a similar digestion protocol was used: Rcho-1 protein extracts were incubated with cathepsin P for 1–20 h, digestion was stopped by placing samples on ice and they were then boiled in SDS–PAGE sample buffer.
Two-dimensional electrophoresis and DIGE labeling
Equal amounts of control and treated samples were trace-labeled with Cy-3-red or Cy-5-green according to standard protocols (GE Healthcare). A pool of equal amounts of all control and treated samples were labeled with a third dye, Cy-2-blue, to be used as a standard for each gel. Four separate samples of both treated and controls were prepared to determine significance of quantitative differences in levels of individual proteins due to cathepsin P treatment. Pairs of control and treated samples were combined with a portion of the standard sample and proteins were separated on 24-cm immobilized gradient gels IPG (3–10) strips. Strips were focused for 50 000 Vh and then proteins were reduced and alkylated with iodoacetamide and equilibrated for SDS–PAGE separation (12% polyacrylamide gels, 200 × 230 × 1.0 mm).
Image analysis and differential display of cathepsin P digested proteins
2D SDS–PAGE gels were scanned using a Typhoon Trio imager (GE Healthcare) to identify proteins labeled by the three dyes. Images were analyzed using Decyder software to map protein spots and compare levels of proteins in digested samples with undigested samples (Alban et al., 2003). Statistical analysis using Student’s t-test was applied to identify the biologically significance of differentially processed proteins. Protein spots that were reduced or increased on cathepsin P treatment and received P-value of <0.05 were considered significant and included in a potential substrate list (products of hydrolysis may appear as new spots). A separate picking gel was run simultaneously using aliquots from the standard samples and was stained with Gel Code Blue stain.
In-gel trypsin digestion and mass spectrometric identification
Selected spots were excised from gels using a 1.5 mm Spot Picker Plus (The Gel Company, San Francisco, CA, USA). Picked spots were destained by washing in 50% acetonitrile. Gel plugs were sequentially treated with 10 mM dithiothreitol (45 min, 56°C), 50 mM iodoacetamide (30 min, 22°C), 100 mM NH4HCO3 (10 min) and 100% acetonitrile (10 min). Plugs were dried using speed vacuum and protein was digested with sequencing grade modified trypsin (200 ng in NH4HCO3, 16 h, 37°C). Peptides were purified with C-18 Zip Tips prior to loading onto 192 spot MALDI plates in 1 µl matrix solution (cyano-4-hydroxycinamic acid in 70% acetonitrile, 0.1% trifluoroacetic acid, 5 mg/ml). MS measurements were carried out on a 4700 proteomic analyzer, a MALDI-TOF-TOF-MS spectrometer (Applied Biosystems, Foster City, CA, USA). Mass spectra were obtained on a mass range of 800–3200 Da, using a laser beam (337 nm, 200 Hz) as ionization source. The instrument was used in reflector-positive mode with an acceleration voltage of 20 kV. The TOF-TOF mass spectra were acquired by the Data Dependent Acquisition method with the five strongest precursor ions selected from one MS scan. MS accuracy was externally calibrated with trypsin-digested peptides of horse myoglobin. The database searches were performed at a peptide mass tolerance of ± 0.3 Da and a fragment mass tolerance of ± 0.4 Da. All data analysis and database searching were performed by the GPS Explorer software using the NCBInr rodent subset of protein sequences (Applied Biosystems). Molecular weights and pIs of identified proteins were calculated from primary sequence using the protein sequence analysis tools on the ExPASy proteomics server (http://ca.expasy.org/cgi-bin/pi_tool).
Western blot analysis
Equal amounts (20 µg) of cathepsin P treated or control protein extracts from Rcho-1 cells were separated by 10% SDS–PAGE and then transferred onto PVDF membranes. Primary antibodies were: polyclonal rabbit anti-calreticulin raised to aa 405–417 (1:2000 in blocking solution); polyclonal goat anti-calreticulin raised to the N-terminus of the protein (1:2000 in blocking solution) and polyclonal rabbit anti-Gp96 raised to the N-terminus of the protein (1:1000 in blocking solution). Secondary antibodies were horseradish peroxidase-conjugated anti-rabbit or anti-goat IgG. ECL kits were used to detect horseradish peroxidase.
Immunohistochemistry
Rcho-1 cells were grown to confluence in four-chambered glass slides. Cells were washed with PBS, fixed in methanol/acetone (1:1) at −20°C for 5 min, permeabilized with 0.25% Triton 100-X for 15 min and then washed with PBS to remove detergent (Zheng et al., 2001). Rabbit anti-mouse cathepsin P antibody (1:300 in blocking solution) (Mason et al., 2004) and sheep anti-calreticulin antibody to aa 270–390 (1:200 in blocking solution) were then added and incubated at 4°C for 24 h. Texas Red-labeled anti-rabbit IgG and FITC-labeled anti-sheep IgG (each 1:500 dilution) were used to visualize the primary antibodies. 4,6-Diamidino-2-phenylindole (Dapi) was used as a nuclear marker. Cells incubated without primary antibodies or with pre-immune serum were used to control for non-specific signals.
Calreticulin treatment of Rcho-1 cells
Rcho-1 cells were cultured in proliferative medium in the presence or absence of purified calreticulin (1 or 2 µg/ml) for 7 days. For comparison, a group of cells were cultured in differentiation medium for 7 days. Cells were evaluated morphologically using a Motic AE31 phase contrast light microscope with a ×20 objective.
Identification of mRNA in Rcho-1 cells
Expression of actin and markers of differentiation were determined in control cells and cells treated with calreticulin (2 µg/ml in proliferative media for 6 days, with fresh media added at Day 3). RT–PCR was performed as described previously (Hassanein et al., 2007). Primers used were: GCTCTCTTCCAGCCTTCCTT and CTTCTGCATCCTGTCAGCAA for β- and γ-actin, 168 bp product; TGAAGAGTTGAGTCTGTGGAGGACC and GCCAGTTTTTGAGAACATCTGACC for cathepsin P, 526 bp product; CTCTGAAACACTTGGTCGGC and CGGCACAGGTTACAAATGGC for placental lactogen, 247 bp product; CTTCAAGAAGTCCGCAGGTC and ACCAAAGAGGAAGGGTTCGT for Hand I, 252 bp product; and GGAGTGGAAGAGGAACAATGCG and TGGGACAACAAAAAAGCGGG for cathepsin 1, 592 bp product.
Cell counting
Cells were harvested, mixed with an equal volume of counting solution (0.4% Trypan Blue) and counted using a hemocytometer (Hausser Scientific, Horsham, PA, USA).
Results
Identification of cathepsin P substrates by 2D-DIGE proteomics
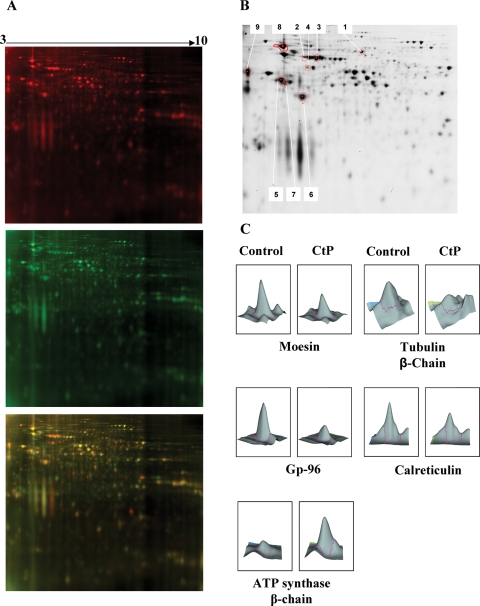
In this study, the rat choriocarcinoma cell line Rcho-1 was chosen as a reproducible source of proteins for digestion by recombinant rat cathepsin P. 2D-DIGE analysis revealed a number of potential substrates and products, although most proteins are not affected by cathepsin P treatment (Fig. 1A). Not all of the spots seen to change in individual gels proved to be altered significantly on batch analysis of multiple gels using the biological variation analysis program of the Decyder software package. After background subtraction, image cropping to remove poorly resolved areas of gel images and batch analysis, only 25 spots changed in abundance due to cathepsin treatment with a P-value of <0.05. A separate picking gel loaded with 1 mg Rcho-1 cell protein was stained with Gel Code Blue. Sixteen of the identified spots were matched with spots on the picking gel. After picking, enzymatic digestion and MALDI MS/MS, nine of these spots were characterized based on a Mascot score >100 and a close match of predicted molecular weight and pI with experimental values. Spots that did not reach these criteria were either limited in abundance or contained more than one protein so they could not be accurately identified or quantified. Identified spots are shown in Fig. 1B and Table I. ATP synthase was identified as a digestion product and the picked spot appeared to be smaller than the calculated size of the intact protein. The identified proteins are typically found in the cytoplasm, mitochondria or endoplasmic reticulum (ER) of cells (Table I). Three-dimensional representations of spots from digested and undigested samples show reduced volumes of digested spots (Fig. 1C). These results show that cathepsin P can degrade a limited number of cellular proteins in vitro. Cathepsin P is packaged into vesicular compartments of cells so it is not likely to encounter the cytoplasmic and mitochondrial proteins under normal cellular conditions. This leaves two ER proteins Gp-96 and calreticulin, as the only potentially physiologically relevant substrates for cathepsin P discovered in this study.
Figure 1.
2D-DIGE analysis of cathepsin P-mediated proteolysis of rat trophoblast extracts.
One example of the 2D gels is shown in (A). Control Rcho-1 proteins incubated for 6 h at 37°C in the absence (upper panel) or presence of cathepsin P (middle panel) labeled with Cy 3 (red) and Cy 5 (green), respectively. The merged images (lower panel) show protein spots that are reduced (red) or increased (green) in relative abundance due to cathepsin P proteolysis. Yellow spots indicate proteins that are not significantly altered. Red spots are potential substrates, whereas green spots are potential products. A picking gel stained with Gel Code Blue (B) shows the nine spots that were identified as in vitro substrates for cathepsin P (Table I). Three-dimensional representations of spots of control and digested proteins are shown in (C).
Table I.
Trophoblast proteins processed by cathepsin P in vitro
| ID | Protein name2 | Protein ID2 | MW3 | PI3 | t-test4 | Fold change5 | Subcellular distribution6 | Mascot score7 | Coverage (%)8 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Moesin | MOES_RAT | 67 565.7 | 6.2 | 1.6E−05 | ↓1.85 | Cytoplasm | 146 | 14 |
| 2 | Heat shock-related 70 kDa | HSP7C_RAT | 69 485.6 | 5.4 | 6.3E−07 | ↓2.74 | Cytoplasm | 400 | 37 |
| 3 | Stress-70 protein | GRP75_RAT | 73 813.8 | 6.0 | 7.6E−06 | ↓3.05 | Mitochondria | 291 | 27 |
| 4 | 60 kDa heat shock protein | CH60_RAT | 60 917.4 | 5.9 | 1.1E−05 | ↓3.70 | Mitochondria | 439 | 31 |
| 5 | Tubulin β-5 chain | TBB2A_RAT | 49 639.0 | 4.8 | 1.2E−04 | ↓1.87 | Cytoplasm | 125 | 22 |
| 6 | Actin, α skeletal muscle | ACTG_RAT | 42 023.9 | 5.2 | 1.2E−03 | ↓1.63 | Cytoplasm | 295 | 31 |
| 7 | ATP synthase β chain | ATPB_RAT | 56 318.5 | 5.2 | 1.8E−05 | ↑2.08 | Mitochondria | 329 | 33 |
| 8 | Gp96 | EDM17052 | 92 418.2 | 4.7 | 7.5E−06 | ↓2.18 | ER | 453 | 30 |
| 9 | Calreticulin | CALR_RAT | 48 137.0 | 4.4 | 1.1E−01 | ↓1.27 | ER | 138 | 32 |
2Substrate proteins are identified by name and Uniprot ID.
3MW and pI were calculated from the primary amino acid sequence found in the NCBI database.
4t-tests were used to determine significantly altered spots on 2D-DIGE by analysis of eight separate samples. Only spots that changed significantly and closely matched predicted MW and pI (with the exception of ATP synthase which is a potential product that migrated faster than predicted for the full-length protein in the SDS dimension) are shown.
5Average fold decrease (or increase) in proteins are indicated.
6Cellular locations of each protein reported in the Swiss Atlas protein database are listed.
7Mascot scores for MS data are listed.
8Percent total protein sequence identified are also listed.
Validation of Gp-96 as a substrate for cathepsin P
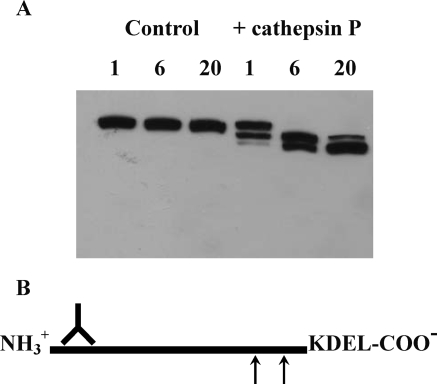
Western blotting was used to show processing of Gp-96 by cathepsin P in Rcho-1 cell extracts (Fig. 2). Under cathepsin P assay conditions at pH 6.5 (or 7.5), Gp-96 was not processed by endogenous proteases. After 1 h treatment with cathepsin P, an initial cleavage product of 84 kDa was observed. This band increased in intensity after 6 h and decreased after 20 h incubation. After 6 h of digestion, a new band of 72 kDa appeared and the intensity of this band increased at 20 h. No other bands appeared and the combined intensity due to all three bands did not change appreciably during this time course. These results indicate that Gp-96 is sequentially processed by cathepsin P at two sites toward the C-terminus of the protein. Analysis of the primary amino acid sequence of rat Gp-96 reveals two hydrophobic regions, Asn-Tyr-Tyr-Ala-Ser and Leu-Ala-Val-Val-Leu-Phe, cleavage of which would yield products of ∼72 and 84 kDa, respectively (Fig. 2B).
Figure 2.
Gp-96 proteolysis by cathepsin P.
Western blotting of time-dependent hydrolysis of Rcho-1 proteins by cathepsin P using an antibody to Gp-96 shows sequential processing to a 72 kDa product via a 84 kDa intermediate after digestion for 1, 6 and 20 h (A). No processing is observed in the control samples. Analysis of the primary sequence of rat Gp-96 reveals two hydrophobic domains (arrows) that are potential cleavage sites for cathepsin P that would yield products of the observed molecular weights (B). The antibody can recognize C-terminally processed products.
Validation of calreticulin as a substrate for cathepsin P
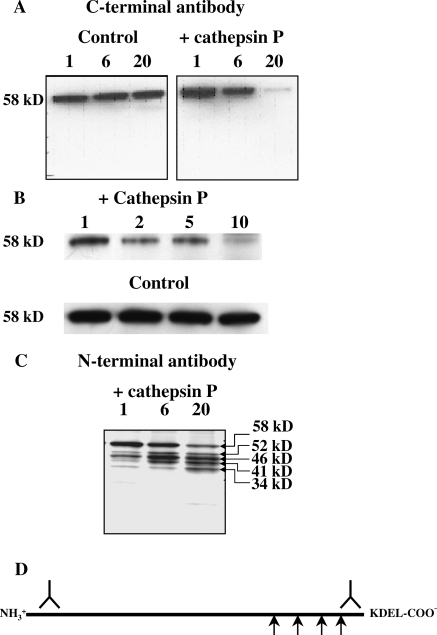
Western blot analysis of cathepsin P digestion of protein extracts of rat Rcho-1 cells using an antibody directed to the C-terminus of calreticulin showed a time-dependent reduction of the intact 58 kDa protein (Fig. 3A). Increasing the concentration of cathepsin P also increased loss of the intact protein (Fig. 3B). Control incubations without added cathepsin P show no loss of immunoreactive protein, demonstrating that endogenous enzymes do not significantly degrade calreticulin in the time frame and assay conditions of these experiments. The time and enzyme concentration-dependent effect on the hydrolysis of the rat protein confirms calreticulin as a substrate for cathepsin P. A lack of appearance of any intermediates indicates that either cathepsin P completely digests calreticulin down to small peptides and amino acids or that cathepsin P processes calreticulin from its C-terminus. The latter process was revealed by western blotting using an antibody directed to the N-terminus of calreticulin (Fig. 3C). The time-dependent experiments revealed four distinct proteolytic products of sizes ranging from 52 to 34 kDa. Total immune-reactive calreticulin was not significantly diminished over 24 h. Native calreticulin consists of three distinctive domains: a globular N-domain, a proline-rich central P-domain and a Glu/Asp-rich C-domain (Ellgaard et al., 2001; Hojrup et al., 2001). The N- and P-domains constitute the chaperone functions of calreticulin, whereas the C-domain is a high-capacity Ca2+-binding domain (Michalak et al., 1999). At the C-terminus, there is a KDEL ER retention signal. The sizes of the cleavage products indicate that processing occurs in the P-domain and at the start of the C-domain. Cleavage sites are difficult to predict from primary amino acid sequence but there is one hydrophobic region at the beginning of the C-domain, FAVLGL, that may be cleaved by cathepsin P. However, there are no long stretches of hydrophobic amino acids at other sites in the C-terminus of calreticulin, indicating that the structure of this protein must allow alternative less specific cleavage by cathepsin P. Nevertheless, as seen for Gp-96, calreticulin undergoes limited processing by cathepsin P from its C-terminus (Fig. 3D).
Figure 3.
Calreticulin proteolysis by cathepsin P.
Western blotting of time-dependent (1, 6 and 20 h) hydrolysis of Rcho-1 proteins by cathepsin P using an antibody to the C-terminus of calreticulin showed a time-dependent reduction of full-length calreticulin (A). No detectable proteolysis of calreticulin was observed in the absence of cathepsin P. The amount of full-length calreticulin was also reduced by increasing amounts of cathepsin P (B). Relative amounts of cathepsin P added to a constant amount of cellular protein are indicated above each lane, 1 representing a protein/enzyme ratio of 250:1. Probing of similar blots using an antibody to the N-terminus of calreticulin shows time-dependent (1, 6 and 20 h) appearance of discreet proteolytic fragments with of 34, 41, 46 and 52 kDa (C). A schematic view of processing of calreticulin is shown in (D). Representative gels of at least three separate experiments are shown.
Topological co-localization of calreticulin and cathepsin P
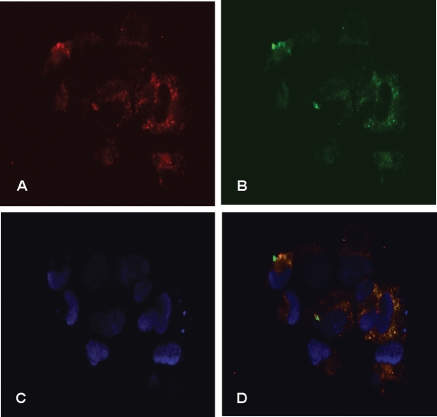
For calreticulin or Gp-96 to be physiologically significant substrates for cathepsin P, the enzymes and substrates must be physically in the same cellular compartment. We have previously shown that cathepsin P co-localizes with cathepsin B in lysosomes (Hassanein et al., 2007). This co-localization was not 100%, indicating that cathepsin P might also be in other cellular compartments. Immunohistochemistry shows that cathepsin P (red) and calreticulin (green) are largely co-localized in Rcho-1 cells (yellow, Fig. 4).
Figure 4.
Cellular co-localization of cathepsin P and calreticulin in Rcho-1 cells.
Rcho-1 cells were fixed, permeabilized and incubated with rabbit anti-cathepsin P and sheep anti-calreticulin-specific antibodies. Species-specific fluorescent antibodies (Texas Red-anti-rabbit IgG and FITC-anti-sheep IgG) were used to identify the primary IgGs. Calreticulin is shown as green (B) and cathepsin P as red (A). Dapi stain was used as a nuclear marker (C). Merged images are shown in (D).
Calreticulin promotes trophoblast differentiation to a giant cell phenotype
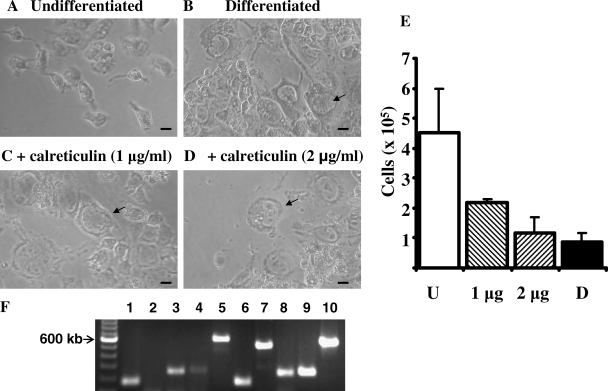
Calreticulin is well established as an ER chaperone but it is also proposed to have extracellular functions. Calreticulin is secreted by an Epstein–Barr virus immortalized human B cell line and the protein was shown to inhibit proliferation and adhesion of endothelial cells of the vasculature, both in vitro and in vivo (Pike et al., 1998; Yao et al., 2002). The mechanism by which the protein is secreted or how the protein inhibits proliferation and adhesion of the endothelial cells is not yet established, but the protein (also termed vasostatin) is a promising regulator of angiogenesis (Ma et al., 2007). The vasostatin properties of calreticulin reside in the N-terminal portion of the protein and both intact and truncated forms of the protein are active in vitro (Pike et al., 1999). We discovered that bovine calreticulin induces differentiation of Rcho-1 cells into a giant cell phenotype in the presence of media that normally maintains cells in a proliferative state (Fig. 5). Although the undifferentiated cells have an angular shape and proliferate in vitro, the calreticulin-treated cells are much larger and have an enlarged nuclei, multiple nucleoli and appear very granular. These cells are indistinguishable from cells differentiated by removal of growth-factor-rich media (Fig. 5B). Cell numbers continue to increase in undifferentiated cells, whereas cell growth is impaired in cells differentiated by calreticulin treatment or growth factor withdrawal (Fig. 5E). RT–PCR shows that expression of giant cell differentiation markers, cathepsin P, placental lactogen, hand1 and cathepsin 1 are all increased in the treated cells (Fig. 5F).
Figure 5.
Calreticulin-induced Rcho-1 differentiation.
Phase contrast images of undifferentiated Rcho-1 cells (A), differentiated cells (B) and cells treated with calreticulin (C and D) are shown. Size bars are 10 µm. Cells were seeded at 105 per well of 12-well plates. Cells were counted and shown as mean with error bars showing standard deviations (E) for undifferentiated cell (U), differentiated cells (D) and calreticulin-treated cells (1 and 2 µg). Cell numbers were significantly reduced (P < 0.05, Student's t-test) when treated with calreticulin or differentiation media compared with controls. This experiment is representative of three separate experiments that gave similar results. Expression of actin and markers of cell differentiation were determined for control cells and cells treated with 2 µg of calreticulin for 6 days (F). Lanes 1–5 show RT–PCR products from control cells and lanes 6–10 show products from treated cells. Lanes 1 and 6, actin; lanes 2 and 7, cathepsin P; lanes 3 and 8, placental lactogen; lanes 4 and 9, hand1; lanes 5 and 10, cathepsin 1. A 100 bp ladder is shown on the left.
Discussion
Cathepsin P is a cysteine protease that is expressed exclusively in placenta (Sol-Church et al., 1999). mRNA for this gene is detected in trophoblasts of the ectoplacental and expression levels increase in a developmentally dependent manner toward the end of fetal development (Sol-Church et al., 1999; Deussing et al., 2002). Rcho-1 cells are a well-established model for differentiation of trophoblast stem cells into a giant cell phenotype (Faria and Soares, 1991). Differentiation of these cells is associated with up-regulation of cellular levels of mRNA and enzymatic activity of cathepsin P, making these cells a valuable model system to dissect the biological function of cathepsin P (Hassanein et al., 2007).
Our first approach was to perform a proteomic analysis of the major proteins extracted from Rcho-1 cells that are sensitive to hydrolysis by cathepsin P. Although a total cell extract was hydrolyzed with cathepsin P under optimal conditions for enzyme activity, relatively few proteins were degraded during a 6 h treatment. Our earlier work with peptide libraries of substrates and inhibitors indicated that cathepsin P would have a restricted specificity (Puzer et al., 2005; Hassanein et al., 2007), but this proteomic study provides the first evidence that cathepsin P has a restricted specificity for cellular proteins. Despite the marked preference of cathepsin P for peptidyl substrates and inhibitors that contain hydrophobic amino acids, several of the cleavage sites in calreticulin are in the very hydrophilic C-terminal end of the protein. Cleavage specificity for linear peptides is not a good predictor for processing of native proteins and most hydrophobic regions of proteins are not accessible to proteases. In contrast to its closest human homolog, cathepsin L, it appears that the degradome of cathepsin P will be quite limited (Kirschke et al., 1982; Lopez-Otin and Overall, 2002). Although cathepsin L has a restricted activity against some native proteins (Johnson et al., 1986), it has a very broad specificity and can degrade proteins down to small peptides and amino acids (Barrett and Kirschke, 1981). Low abundance protein substrates are likely to be missed by proteomic screens and several substrates in total cell homogenates will not be physiologically relevant if they do not encounter the enzyme in living tissues. Nevertheless, this proteomic screen has shown that two potentially significant protein substrates undergo limited processing by cathepsin P.
Placental tissue contains many proteases and endogenous proteases, which cause rapid degradation of calreticulin (see Supplementary Fig. 1). Neither a cocktail of broad-based inhibitors nor a specific inhibitor of cathepsin P could arrest this proteolysis, indicating that many proteases can degrade this protein. The identity and physiological significance of this proteolysis is not known, but this precluded placental tissue as a source of protein to determine specificity of cathepsin P.
The co-localization of cathepsin P with calreticulin indicates that cathepsin P may function in the ER as a processing enzyme. Such a location for a lysosomal enzyme is unusual but not without precedent; cathepsin W is predominantly localized in the ER (Wex et al., 2001). Unlike other lysosomal cathepsins, the pro-peptide of cathepsin P is processed at neutral pH and the enzyme is optimally active at neutral pH, so it is ideally suited for an ER function (Mason et al., 2004).
Gp-96 (also known as GRP94) processing revealed only two cleavage products, indicating that cathepsin P processes this protein in a limited, sequential fashion. Endogenous cathepsins B and L rapidly degrade this protein down to small peptides and amino acids at pH 5.5 (M.H. and R.W.M., unpublished data). Processing of Gp-96 by cathepsin P is from the C-terminus, cleaving within the dimerization domain of this protein (Facciponte et al., 2006). This C-terminal processing would remove the KDEL ER retention signal, allowing release of Gp-96 from the cell. Extracellular Gp-96 acts as a potent adjuvant, eliciting anti-tumor immunity but a role in placental function is not known (Srivastava et al., 1986; Facciponte et al., 2006).
Calreticulin is expressed at its highest level in the definitive placenta and appears to play an important role in trophoblast maturation and embryonic development (Hershberger and Tuan, 1999; Su et al., 2004). The C-terminal domain of calreticulin is sensitive to proteolysis and can be degraded by a range of digestive serine proteases (Hojrup et al., 2001). Genetic deletion of the C-terminal domain and KDEL signal of calreticulin has been shown to stimulate secretion of this protein from transfected cells (Sonnichsen et al., 1994). Although intracellular processing of calreticulin would be expected to impair the chaperone and calcium-binding functions of the protein (Hershberger and Tuan, 1998; Wainwright et al., 1998), the function of the secreted processed calreticulin may be more significant.
Our data indicate that proteolytic processing by cathepsins may be an important mediator of calreticulin secretion by the placenta. Calreticulin released by removal of its KDEL ER retention signal could induce differentiation of trophoblasts in an autocrine fashion. This differentiation would further induce expression of cathepsin P (Hassanein et al., 2007) to create a feedback loop to enhance efficiency of differentiation of adjacent cells in a paracrine fashion to ensure coordinated differentiation of trophoblast cells invading the maternal uterus during placentation. Differentiation of primary trophoblast to highly invasive and phagocytic giant cells is critical during implantation into the maternal decidua during early gestation (Zybina et al., 2000).
The differentiated giant cells secrete a wide array of angiogenic factors and vasodilators (Cross et al., 2002) to promote continuous blood flow from maternal arteries throughout fetal development. During the latter half of pregnancy, endovascular trophoblast giant cells target the endothelial cells lining the maternal arteries, replacing them and enhancing vasodilatation. Calreticulin could contribute to this arterial remodeling as it disrupts endothelial cell attachment to laminin of basement membranes (Yao et al., 2002). The released endothelial cells can then be replaced by trophoblast giant cells to promote vasodilation and enhanced placental blood supply. This contrasts with the anti-angiogenic action of calreticulin that impairs endothelial cell proliferation (Pike et al., 1998). Calreticulin levels are increased 5-fold in plasma of pregnant women and are further elevated in plasma from women with pre-eclampsia, leading to a proposal that the anti-angiogenic function of this protein may be a cause of increased blood pressure in this disorder (Gu et al., 2008). A significant proportion of calreticulin isolated from human placenta is devoid of six C-terminal amino acids, indicating proteolytic removal of the KDEL ER retention signal (Hojrup et al., 2001). In rodents, cathepsin P may be the protease that regulates placental release of calreticulin to promote placental blood flow without elevating maternal blood pressure. Other members of the cathepsin family are also expressed in trophoblast giant cells, such as cathepsin 1 in this study, but these enzymes have not yet been characterized, so cathepsin P may not be the only enzyme that can process calreticulin in placenta. These or other uncharacterized proteases may contribute to calreticulin processing in the mature placenta (see Supplementary Fig. 1). There is no direct placenta-specific ortholog of cathepsin P in the human genome, but a functional ortholog may be a less specialized enzyme such as cathepsin L.
In summary, we have used a proteomic approach to identify Gp-96 and calreticulin as protein substrates for cathepsin P. The study indicates that cathepsin P has limited proteolytic activity and acts as a processing enzyme, unlike its closest human homolog, cathepsin L. Processing of Gp-96 and calreticulin by cathepsin P removes the C-terminal KDEL ER retention signal that will permit secretion of the proteins during differentiation of Rcho-1 cells into a giant cell phenotype. The secreted calreticulin may enhance this differentiation and promote placental vasodilation. Secreted calreticulin may also play a role in the maternal vasoconstriction that causes pre-eclampsia, indicating that proteolytic-mediated release of this protein must be tightly regulated. Detailed comparative studies are required to determine whether proteolytic-mediated release of calreticulin in this rodent system provides a model to study the role of calreticulin in human physiology and pathology.
Supplementary data
Supplementary data are available at http://molehr.oxfordjournals.org/.
Funding
This work was supported by Nemours Research Programs, and the National Institutes of Health (grant # P20RR20173 to the Center for Pediatric Research and grant # P20RR016472 to the Delaware INBRE).
Acknowledgements
We thank our colleagues, especially Bruce Korant, for many valuable discussions. Particular thanks go to Charlotte Mobarak of the UNM Mass Spectrometry Facility for mass analysis by MALDI MS/MS.
References
- Afonso S, Romagnano L, Babiarz B. The expression and function of cystatin C and cathepsin B and cathepsin L during mouse embryo implantation and placentation. Development. 1997;124:3415–3425. doi: 10.1242/dev.124.17.3415. [DOI] [PubMed] [Google Scholar]
- Alban A, David SO, Bjorkesten L, Andersson C, Sloge E, Lewis S, Currie I. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics. 2003;3:36–44. doi: 10.1002/pmic.200390006. [DOI] [PubMed] [Google Scholar]
- Babiarz BS, Romagnano LC, Kurilla GM. Interaction of mouse ectoplacental cone trophoblast and uterine decidua in vitro. In Vitro Cell Dev Biol. 1992;28A:500–508. doi: 10.1007/BF02634133. [DOI] [PubMed] [Google Scholar]
- Barrett AJ, Kirschke H. Cathepsin B, cathepsin H and cathepsin L. Methods Enzymol. 1981;80:535–561. doi: 10.1016/s0076-6879(81)80043-2. [DOI] [PubMed] [Google Scholar]
- Bredemeyer AJ, Lewis RM, Malone JP, Davis AE, Gross J, Townsend RR, Ley TJ. A proteomic approach for the discovery of protease substrates. Proc Natl Acad Sci USA. 2004;101:11785–11790. doi: 10.1073/pnas.0402353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon YP, DeMayo FJ, Bagchi MK, Bagchi IC. Induction of cytotoxic T-lymphocyte antigen-2beta, a cysteine protease inhibitor in decidua: a potential regulator of embryo implantation. J Biol Chem. 2004;279:10357–10363. doi: 10.1074/jbc.M309434200. [DOI] [PubMed] [Google Scholar]
- Cross JC, Hemberger M, Lu Y, Nozaki T, Whiteley K, Masutani M, Adamson SL. Trophoblast functions, angiogenesis and remodeling of the maternal vasculature in the placenta. Mol Cell Endocrinol. 2002;187:207–212. doi: 10.1016/s0303-7207(01)00703-1. [DOI] [PubMed] [Google Scholar]
- Deussing J, Roth W, Saftig P, Peters C, Ploegh HL, Villadangos JA. Cathepsins B and D are dispensable for major histocompatibility complex class II-mediated antigen presentation. Proc Natl Acad Sci USA. 1998;95:4516–4521. doi: 10.1073/pnas.95.8.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deussing J, Kouadio M, Rehman S, Werber I, Schwinde A, Peters C. Identification and characterization of a dense cluster of placenta-specific cysteine peptidase genes and related genes on mouse chromosome 13. Genomics. 2002;79:225–240. doi: 10.1006/geno.2002.6696. [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Riek R, Herrmann T, Guntert P, Braun D, Helenius A, Wuthrich K. NMR structure of the calreticulin P-domain. Proc Natl Acad Sci USA. 2001;98:3133–3138. doi: 10.1073/pnas.051630098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facciponte JG, Wang XY, MacDonald IJ, Park JE, Arnouk H, Grimm MJ, Li Y, Kim H, Manjili MH, Easton DP, et al. Heat shock proteins HSP70 and GP96: structural insights. Cancer Immunol Immunother. 2006;55:339–346. doi: 10.1007/s00262-005-0020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria TN, Soares MJ. Trophoblast cell differentiation: establishment, characterization, and modulation of a rat trophoblast cell line expressing members of the placental prolactin family. Endocrinology. 1991;129:2895–2906. doi: 10.1210/endo-129-6-2895. [DOI] [PubMed] [Google Scholar]
- Felbor U, Kessler B, Mothes W, Goebel HH, Ploegh HL, Bronson RT, Olsen BR. Neuronal loss and brain atrophy in mice lacking cathepsins B and L. Proc Natl Acad Sci USA. 2002;99:7883–7888. doi: 10.1073/pnas.112632299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn JL, Chen CF, Lewandowski A, Cheng CH, Ramsdell CM, Bullard-Dillard R, Chen J, Dewey MJ, Glenn TC. Expressed sequence tags from Peromyscus testis and placenta tissue: analysis, annotation, and utility for mapping. BMC Genomics. 2008;9:300. doi: 10.1186/1471-2164-9-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu VY, Wong MH, Stevenson JL, Crawford KE, Brennecke SP, Gude NM. Calreticulin in human pregnancy and pre-eclampsia. Mol Hum Reprod. 2008;14:309–315. doi: 10.1093/molehr/gan017. [DOI] [PubMed] [Google Scholar]
- Hassanein M, Korant BD, Lu G, Mason RW. Expression of cathepsin P mRNA, protein and activity in the rat choriocarcinoma cell line, Rcho-1, during giant cell transformation. Placenta. 2007;28:912–919. doi: 10.1016/j.placenta.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemberger M, Himmelbauer H, Ruschmann J, Zeitz C, Fundele R. cDNA subtraction cloning reveals novel genes whose temporal and spatial expression indicates association with trophoblast invasion. Dev Biol. 2000;222:158–169. doi: 10.1006/dbio.2000.9705. [DOI] [PubMed] [Google Scholar]
- Hemberger M, Cross JC, Ropers HH, Lehrach H, Fundele R, Himmelbauer H. UniGene cDNA array-based monitoring of transcriptome changes during mouse placental development. Proc Natl Acad Sci USA. 2001;98:13126–13131. doi: 10.1073/pnas.231396598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberger ME, Tuan RS. Placental 57-kDa Ca(2+)-binding protein: regulation of expression and function in trophoblast calcium transport. Dev Biol. 1998;199:80–92. doi: 10.1006/dbio.1998.8926. [DOI] [PubMed] [Google Scholar]
- Hershberger ME, Tuan RS. Functional analysis of placental 57-kDa Ca(2+)-binding protein: overexpression and downregulation in a trophoblastic cell line. Dev Biol. 1999;215:107–117. doi: 10.1006/dbio.1999.9439. [DOI] [PubMed] [Google Scholar]
- Hojrup P, Roepstorff P, Houen G. Human placental calreticulin characterization of domain structure and post-translational modifications. Eur J Biochem. 2001;268:2558–2565. doi: 10.1046/j.1432-1327.2001.02138.x. [DOI] [PubMed] [Google Scholar]
- Johnson DA, Barrett AJ, Mason RW. Cathepsin L inactivates alpha-1-proteinase inhibitor by cleavage in the reactive site region. J Biol Chem. 1986;261:14748–14751. [PubMed] [Google Scholar]
- Kirschke H, Kembhavi AA, Bohley P, Barrett AJ. Action of rat liver cathepsin L on collagen and other substrates. Biochem J. 1982;201:367–372. doi: 10.1042/bj2010367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarca HL, Ott CM, Honer Zu Bentrup K, Leblanc CL, Pierson DL, Nelson AB, Scandurro AB, Whitley GS, Nickerson CA, Morris CA. Three-dimensional growth of extravillous cytotrophoblasts promotes differentiation and invasion. Placenta. 2005;26:709–720. doi: 10.1016/j.placenta.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C, Overall CM. Protease degradomics: a new challenge for proteomics. Nat Rev Mol Cell Biol. 2002;3:509–519. doi: 10.1038/nrm858. [DOI] [PubMed] [Google Scholar]
- Ma L, Luo L, Qiao H, Dong X, Pan S, Jiang H, Krissansen GW, Sun X. Complete eradication of hepatocellular carcinomas by combined vasostatin gene therapy and B7H3-mediated immunotherapy. J Hepatol. 2007;46:98–106. doi: 10.1016/j.jhep.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Mason RW. Emerging functions of placental cathepsins. Placenta. 2008;29:385–390. doi: 10.1016/j.placenta.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Mason RW, Bergman CA, Lu G, Frenck Holbrook J, Sol-Church K. Expression and characterization of cathepsin P. Biochem J. 2004;378:657–663. doi: 10.1042/BJ20031548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak M, Corbett EF, Mesaeli N, Nakamura K, Opas M. Calreticulin: one protein, one gene, many functions. Biochem J. 1999;344:281–292. [PMC free article] [PubMed] [Google Scholar]
- Nakajima A, Kataoka K, Takata Y, Huh NH. Cathepsin-6, a novel cysteine proteinase showing homology with and co-localized expression with cathepsin J/P in the labyrinthine layer of mouse placenta. Biochem J. 2000;349:689–692. doi: 10.1042/bj3490689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike SE, Yao L, Jones KD, Cherney B, Appella E, Sakaguchi K, Nakhasi H, Teruya-Feldstein J, Wirth P, Gupta G, et al. Vasostatin, a calreticulin fragment, inhibits angiogenesis and suppresses tumor growth. J Exp Med. 1998;188:2349–2356. doi: 10.1084/jem.188.12.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike SE, Yao L, Setsuda J, Jones KD, Cherney B, Appella E, Sakaguchi K, Nakhasi H, Atreya CD, Teruya-Feldstein J, et al. Calreticulin and calreticulin fragments are endothelial cell inhibitors that suppress tumor growth. Blood. 1999;94:2461–2468. [PubMed] [Google Scholar]
- Puente XS, Sanchez LM, Overall CM, Lopez-Otin C. Human and mouse proteases: a comparative genomic approach. Nat Rev Genet. 2003;4:544–558. doi: 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- Puzer L, Barros NM, Oliveira V, Juliano MA, Lu G, Hassanein M, Juliano L, Mason RW, Carmona AK. Defining the substrate specificity of mouse cathepsin P. Arch Biochem Biophys. 2005;435:190–196. doi: 10.1016/j.abb.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Roth W, Deussing J, Botchkarev VA, Pauly-Evers M, Saftig P, Hafner A, Schmidt P, Schmahl W, Scherer J, Anton-Lamprecht I, et al. Cathepsin L deficiency as molecular defect of furless: hyperproliferation of keratinocytes and pertubation of hair follicle cycling. FASEB J. 2000;14:2075–2086. doi: 10.1096/fj.99-0970com. [DOI] [PubMed] [Google Scholar]
- Sahgal N, Canham LN, Canham B, Soares MJ. Rcho-1 trophoblast stem cells: a model system for studying trophoblast cell differentiation. Methods Mol Med. 2006;121:159–178. [PubMed] [Google Scholar]
- Sireesha GV, Mason RW, Hassanein M, Tonack S, Navarrete Santos A, Fischer B, Seshagiri PB. Role of cathepsins in blastocyst hatching in the golden hamster. Mol Hum Reprod. 2008;14:337–346. doi: 10.1093/molehr/gan026. [DOI] [PubMed] [Google Scholar]
- Sol-Church K, Frenck J, Troeber D, Mason RW. Cathepsin P, a novel protease in mouse placenta. Biochem J. 1999;343:307–309. [PMC free article] [PubMed] [Google Scholar]
- Sol-Church K, Picerno GN, Stabley DL, Frenck J, Xing S, Bertenshaw GP, Mason RW. Evolution of placentally expressed cathepsins. Biochem Biophys Res Commun. 2002;293:23–29. doi: 10.1016/S0006-291X(02)00167-5. [DOI] [PubMed] [Google Scholar]
- Sonnichsen B, Fullekrug J, Nguyen Van P, Diekmann W, Robinson DG, Mieskes G. Retention and retrieval: both mechanisms cooperate to maintain calreticulin in the endoplasmic reticulum. J Cell Sci. 1994;107:2705–2717. doi: 10.1242/jcs.107.10.2705. [DOI] [PubMed] [Google Scholar]
- Srivastava PK, DeLeo AB, Old LJ. Tumor rejection antigens of chemically induced sarcomas of inbred mice. Proc Natl Acad Sci USA. 1986;83:3407–3411. doi: 10.1073/pnas.83.10.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk V, Turk B, Turk D. Lysosomal cysteine proteases: facts and opportunities. EMBO J. 2001;20:4629–4633. doi: 10.1093/emboj/20.17.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainwright SD, Simpson KL, Holmes CH. Calreticulin associates with non-HLA-A,-B class I proteins in the human choriocarcinoma cell lines JEG-3 and BeWo. Immunology. 1998;93:437–445. doi: 10.1046/j.1365-2567.1998.00439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wex T, Buhling F, Wex H, Gunther D, Malfertheiner P, Weber E, Bromme D. Human cathepsin W, a cysteine protease predominantly expressed in NK cells, is mainly localized in the endoplasmic reticulum. J Immunol. 2001;167:2172–2178. doi: 10.4049/jimmunol.167.4.2172. [DOI] [PubMed] [Google Scholar]
- Yao L, Pike SE, Tosato G. Laminin binding to the calreticulin fragment vasostatin regulates endothelial cell function. J Leukoc Biol. 2002;71:47–53. [PubMed] [Google Scholar]
- Zheng JQ, Kelly TK, Chang B, Ryazantsev S, Rajasekaran AK, Martin KC, Twiss JL. A functional role for intra-axonal protein synthesis during axonal regeneration from adult sensory neurons. J Neurosci. 2001;21:9291–9303. doi: 10.1523/JNEUROSCI.21-23-09291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybina EV, Zybina TG, Stein GI. Trophoblast cell invasiveness and capability for the cell and genome reproduction in rat placenta. Early Pregnancy. 2000;4:39–57. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.