Summary
To date most antibiotics are targeted at intracellular processes, and must be able to penetrate the bacterial cell envelope. In particular, the outer membrane of Gram-negative bacteria provides a formidable barrier that must be overcome. There are essentially two pathways that antibiotics can take through the outer membrane: a lipid-mediated pathway for hydrophobic antibiotics, and general diffusion porins for hydrophilic antibiotics. The lipid and protein compositions of the outer membrane have a strong impact on the sensitivity of bacteria to many types of antibiotics, and drug resistance involving modifications of these macromolecules is common. This review will describe the molecular mechanisms for permeation of antibiotics through the outer membrane, and the strategies that bacteria have deployed to resist antibiotics by modifications of these pathways.
Keywords: outer membrane, antibiotic, porin, LPS, resistance
The outer membrane (OM) of Gram-negative bacteria performs the crucial role of providing an extra layer of protection to the organism without compromising the exchange of material required for sustaining life. In this dual capacity, the OM emerges as a sophisticated macromolecular assembly, whose complexity has been unraveled only in recent years. By combining a highly hydrophobic lipid bilayer with pore-forming proteins of specific size-exclusion properties, the OM acts as a selective barrier. The permeability properties of this barrier, therefore, have a major impact on the susceptibility of the microorganism to antibiotics, which, to date, are essentially targeted at intracellular processes. Small hydrophilic drugs, such as β-lactams, use the pore-forming porins to gain access to the cell interior, while macrolides and other hydrophobic drugs diffuse across the lipid bilayer. The existence of drug-resistant strains in a large number of bacterial species due to modifications in the lipid or protein composition of the OM indeed highlights the importance of the OM barrier in antibiotic sensitivity. This review will summarize the properties of the OM lipid barrier and porin-mediated permeability, and highlight the antibiotic resistance mechanisms that involve modifications of these properties.
It is important to note that many of the alterations in outer membrane permeability described below are often associated with increased levels of antibiotic efflux. Even intrinsic antibiotic resistance is likely to reflect the synergistic action of the outer membrane acting as a permeability barrier, and of the diverse and widely distributed efflux pumps. The review below essentially focuses on the permeability changes per se, as the roles of efflux pathways in antibiotic resistance are treated by others. Whether changes in outer membrane lipid or porin composition also mechanistically influences the efflux systems remains to be determined.
1. Organization of the OM
In most Gram-negative bacteria, the OM is an asymmetric bilayer of phospholipid and lipopolysaccharides (LPS), the latter exclusively found in the outer leaflet. A typical LPS molecule consists of three parts (Figure 1): 1) lipid A, a glucosamine-based phospholipid, 2) a relatively short core oligosaccharide, and 3) a distal polysaccharide (O-antigen) [1]. Since part of the core oligosaccharide and the O-antigen are not required for the growth of Escherichia coli, strains can exhibit varying length of these structures. The phospholipid composition of the inner leaflet of the OM is similar to that of the cytoplasmic membrane, i.e. about 80 % phosphatidylethanolamine, 15% phosphatidylglycerol and 5 % cardiolipin [2]. In mutants with altered LPS structure, phospholipids have also been detected in the outer leaflet of the OM, possibly due to consequent decrease in OM protein levels [3].
Figure 1.

Overall organization of LPS and structure of Kdo2-Lipid A. The left hand side shows the organization of LPS in 3 regions: Lipid A, core oligosaccharide (itself subdivided into inner core and outer core), and O-antigen. Abbreviations are: Kdo, 3-deoxy-D-manno-oct-2-ulosonic acid; Hep, L-glycero-D-manno-heptose; Glc, D-glucose; Gal, D-galactose; R, a variety of different substituents (see reference 13 for details). The right hand side shows the structure of Kdo2-Lipid A, the minimal entity required for E. coli growth.
A large number of different types of proteins reside in the OM. Some of them are extremely abundant. For example, murein lipoprotein (Lpp), OmpA and general diffusion porins are present at > 105 copies per cell [4]. Lpp carries a fatty acid moiety that anchors it into the OM, while about a third of the Lpp population is also covalently attached to the peptidoglycan layer. Thus, Lpp is thought to play a role in providing OM-peptidoglycan interactions and in maintaining OM integrity. Indeed, mutants lacking Lpp produce OM vesicles and leak periplasmic enzymes [5]. Another abundant OM protein is OmpA. The protein is believed to have a structural role and the absence of OmpA and Lpp compromises the shape of the cell [6]. Along with the Pseudomonas aeruginosa homolog OprF, OmpA has pore-forming properties as well, but with extremely low permeation efficiency. Recent experimental evidence suggests that these proteins exhibit two different conformations, an abundant closed form that exists as a monomeric 8-stranded β-barrel with a C-terminal periplasmic domain, and a rare oligomeric form, that comprises large open β-barrels similar to the general diffusion porin OmpF [7, 8].
Other than general diffusion porins, which will be described in detail below, the OM also contains specialized protein channels and receptors used for the uptake of specific substrates (for example LamB and BtuB for maltodextrins and vitamin B12 transport, respectively), proteins involved in OM and surface appendages biogenesis (for example, Omp85 for membrane protein insertion, and a large array of translocators used in the assembly of adhesins, pili and flagella), translocons allowing release of secreted substrates (for example, translocon of the Type II secretion system involved in toxin release), various enzymes (such as the E. coli OmpT protease) and proteins involved in LPS assembly. The reader is referred to recent reviews for more information on these proteins [9–12].
2. The OM lipid barrier
2.1. Molecular description
The asymmetric presence of LPS is a salient and unique feature of the OM. LPS is composed of the hydrophobic, fatty acid chain bearing lipid A, a core oligosaccharide and the O-antigen (Figure 1). The O antigen is an immunogenic oligosaccharide of considerable variability among Gram-negative bacteria, consisting of 1 to 40 repeating units. The core oligosaccharide is branched and contains 6 to 10 sugars in addition to two Kdo’s (3-deoxy-D-manno-oct-2-ulosonic acid) linked to lipid A. This core region is also heterogeneous due to the variable presence and nature of additional substituents. Lipid A is a glucosamine disaccharide, phosphorylated at the 1 and 4′ positions, and acetylated at the 2, 2′, 3 and 3′ positions with 3-hydroxymyristic acid. It differs from a typical phospholipid by having six saturated fatty acid chains rather than two saturated or unsaturated chains. These characteristics make the asymmetric OM bilayer much more hydrophobic than a typical phospholipid bilayer, due to strong lateral interactions between LPS molecules and low fluidity [4]. The glucosamine backbone of lipid A and the core region bear multiple anionic groups, and LPS is known to bind strongly divalent cations, which compensates for the electrostatic repulsion between neighboring LPS molecules. Only the inner part of LPS, consisting of lipid A and Kdo, is required to sustain growth in E. coli [1]. Thus, many mutants (R or “rough” mutants, due to colony appearance) exist with varying length of core oligosaccharide, and have been classified as Ra to Re chemotypes [4, 13]. “Deep rough” mutants have the most truncated core, and show high sensitivity to lipophylic agents such as detergents, some antibiotics, bile salts, etc. “Smooth” strains have an intact O-antigen, of varying length, and are found among clinical isolates of Enterobacteriaceae. Excellent descriptions of LPS structure and biogenesis can be found in earlier reviews [1, 13].
2. 2 Lipid-mediated antibiotic resistance
Hydrophobic antibiotics that appear to gain access to the cell interior by permeating through the OM bilayer per se are aminoglycosides (gentamycin, kanamycin), macrolides (erythromycin), rifamycins, novobiocin, fusidic acid and cationic peptides [11, 14]. Tetracylcine and quinolones use both a lipid-mediated and a porin-mediated pathway (see below). The core region of LPS plays a major role in providing a barrier to hydrophobic antibiotics and other compounds, and the strains which express full length LPS have an intrinsic resistance to these. On the other hand, membrane permeabilizers, such as Tris/EDTA, polymyxin B and polymyxin B nonapeptide (PMBN), have the ability to increase the sensitivity of E. coli and Salmonella typhimurium to the hydrophobic antibiotics mentioned above by tens- to hundreds fold, depending on the treatment and the particular antibiotics [14]. The achieved sensitivities become similar to those of deep rough mutants [14]. Treatment by Tris/EDTA leads to massive release of LPS in the medium, and it is believed that the reduced amount of LPS in the OM outer leaflet is compensated by glycerophospholipids, essentially creating patches of phospholipid bilayer, which are much more permeable to lipophilic compounds [11, 14]. A similar situation may also be found in deep rough mutants, where there is a decrease in OM membrane protein incorporation, leaving a void which is also filled by phospholipids [11].
The molecular mechanism for permeabilization by polymyxin B and PMBN is thought to involve the competition for binding to LPS of these cations with the divalent cations that normally cross-bridge neighboring LPS molecules. The displacement of these stabilizing interactions leads to enhanced lateral diffusion of LPS. The resulting destabilization of the LPS layer allows the penetration of polymyxin B into the periplasm, providing essentially a “self-promoted uptake pathway” for polymyxin B to reach its target, the cytoplasmic membrane. Then, the fatty acid tail on polymyxin B allows it to permeabilize the inner membrane, thus leading to its antibacterial action. PMBN lacks the fatty acid chain, and is less bactericidal, but the fact that it sensitizes cells to hydrophobic antibiotics demonstrates that it retains OM permeabilizing properties [14]. To our knowledge there is no evidence that the cationic peptides induce phospholipid patches.
Polymyxin-resistant mutants have been isolated in S. typhimurium and E. coli [15, 16]. The polymyxin-resistant mutants of S. typhimurium bind only 25% of the amount of polymyxin bound by the parent strain, and tolerate up to 100 times higher concentrations of polymyxin B [17]. LPS isolated from these mutants also binds less polymyxin B [18, 19], and contains 4 to 6 times more 4-aminoarabinose and also more phosphoethanolamine [18], due to esterification of the lipid A phosphates by these moieties. These substitutions effectively lower the negative charge of the LPS molecule, and possibly decrease the repulsion between adjacent LPS molecules [11]. The resulting more closely packed LPS layer and decreased negative charge lead to a reduced sensitivity of the mutants not only to polymyxin B, but also to PMBN, EDTA and other cationic agents [14]. It was later found that the addition of 4-aminoarabinose and phosphoethanolamine to the 1- and 4′-phosphates of lipid A is operative in wildtype cells, creating a family of variant LPS molecules in S. typhimurium [20]. It is now known that the regulation of these modifications in wildtype and under antibiotic stress is under the control of the two component system PmrA/PmrB, itself regulated by the PhoP/PhoQ system [21]. Indeed the constitutive expression of pmrA confers a polymyxin-resistant phenotype [21, 22] and is associated with a larger amount of lipid A bearing 4-aminoarabinose modifications than in wildtype cells [21]. In addition, neither 4-aminoarabinose nor the ethanolamine substitutions occur in a pmrA null mutant [20], and genes controlled by the PmrAB system are involved in the aminoarabinose modification [23].
The PhoP/PhoQ and PmrA/PmrB two-component systems play important roles in the adaptation of S. typhimurium to cationic antimicrobial peptides and survival inside macrophages [24]. This adaptation is crucial for virulence as the bacteria need to be protected from the host innate immune system, which comprises numerous cationic peptides found at mucosal surfaces and in the phagosome. PhoQ is a membrane-bound protein with a periplasmic sensor domain, and a cytoplasmic kinase domain. It has been shown to be directly activated by cationic peptides that are thought to bind the acidic surface of the periplasmic domain of PhoQ [25]. The resulting autophosphorylation of PhoQ and subsequent phosphotransfer to PhoP lead to activation of PhoP, which itself negatively or positively controls the expression of specific genes, including the activation of the pmrAB operon [21]. In addition, a PhoP activated gene, pagP, is also required for resistance to a cationic antimicrobial peptide [26]. pagP codes for a palmitate acyl transferase, which links an additional palmitate to lipid A, creating a heptaacylated form [26]. The palmitoylation of lipid A allows for increased hydrophobic interactions between neighboring LPS molecules. Besides the addition of aminoarabinose, phosphoethanolamine and palmitate, the remodeling of LPS in response to antibiotic stress also includes the conversion to 2-hydroxymyristic acid of the “piggyback” fatty acid chain linked to the hydroxymyristic acid at the 3′-position, and the deacylation of the 3-hydroxymyristic acid at the 3-position [11, 24]. Altogether, these modifications lead to stabilization of the LPS leaflet and decreased electrostatic interactions with cations, and have been shown to play an important role in mediating resistance to lipophylic agents, including cationic antimicrobial peptides [24].
A pmr mutant of E. coli has also been shown to be somewhat resistant to the aminoglycoside antibiotics gentamycin and kanamycin [27]. Like polymyxin B, aminoglycosides are thought to use a self-promoted uptake pathway to penetrate the OM [28]. Indeed, they carry three to six net positive charges, and bind to isolated LPS [27]. These antibiotics increase the permeability of the OM to fluorescent hydrophobic probes [27], and thus can been considered as OM permeabilizers. However, this effect is relatively weak, when one compares the ability of the aminoglycoside streptomycin to sensitize S. typhimurium to the hydrophobic antibiotic novobiocin to that of PMBN [29].
3. Porin-mediated OM permeability
3.1. Structural and functional properties of general diffusion porins
3.1.1. Structure
Except for the capsular polysaccharide translocon Wza [30], all OM proteins crystallized to date are built on a β-barrel structural motif. The E. coli general diffusion porins OmpF, PhoE and OmpC are trimers of 16-stranded β-barrels [31, 32] (Figure 2). The large number and configuration of the β-strands allow for the formation of a central hydrophilic pore in each β-barrel. The pore of some other OM proteins, such as the enterobactin transporter FepA [33] or the adhesin translocator FhaC [34], is essentially obstructed by a globular plug domain. But this is not the case for general diffusion porins. The pore is, however, somewhat constricted by the inwardly folded extracellular loop L3 (shown in orange in Figure 2). This loop, together with the opposite barrel wall, form the so-called eyelet or constriction zone, which determines the size exclusion limit and other permeation properties of the barrel (see below). At this level, the pore size of OmpF is 7 × 10 Å [32]. A conserved set of charged residues decorates the eyelet: negatively charged residues (in red in Figure 2) are typically found on the L3 loop itself, and positive charges (in blue in Figure 2) often form a cluster on the opposite barrel wall. These residues have been shown to play an important role in ionic movement and in ionic selectivity (see below). The β-strands are connected to each other by short turns on the periplasmic side and long loops on the extracellular side. This protruding extracellular domain provides a site for interactions with specific colicins and phages that use porins as surface receptors [11].
Figure 2.

Structure of an OmpF monomer. A) Side view of a single β-barrel of the OmpF trimer to highlight the location of the protein in the membrane bilayer (“EC” refers to the extracellular side, and “Peri” refers to the periplasmic side); note that some of the protein structure has been cut out of view in order to better visualize the constricting L3 loop (orange). B) View of the OmpF monomer from the periplasmic side, highlighting the configuration of the eyelet or constriction zone. Important residues of the eyelet are acidic residues of the L3 loop (in red) and a cluster of basic amino acids of the opposite barrel wall (in blue).
3.1.2. Pore properties and permeation
The functional properties of porins have been the subject of investigation for over 30 years. Initial work established the size exclusion cutoffs of porins by measuring the transport of various size sugars using liposome swelling assays [35]. A value of about 600 Daltons was determined for OmpF [36], which implies that ions, amino acids, and small sugars use general diffusion porins for gaining access to the periplasm. Disaccharides, larger sugars and other molecules need to use dedicated pathways for OM transport [11]. These early studies established the molecular sieving properties of porins, and provided an explanation for the high diffusion rates of these compounds through the OM [37].
The application of electrophysiology to the study of porins, along with computational studies, has permitted a better understanding of porin permeation at the molecular level. The traditional electrophysiological approach is the study of porin-mediated ion currents in planar lipid bilayers (also known as “black lipid membranes” or “BLM”). A lipid bilayer is formed over an aperture pierced through a Teflon film separating two chambers. Each chamber contains a buffered ionic solution and an electrode used to measure electric current due to the flow of ions across the bilayer and to clamp the transmembrane potential required to promote ion movement. Purified detergent-solubilized channel proteins or proteoliposomes are added to one chamber (the so-called cis side), and spontaneously insert in the bilayer over time. The sequential insertions of open channels in the membrane lead to discrete current jumps due to ion movement through the open channels. The conductance (i.e. the amount of current per unit voltage) of a channel can be obtained from measuring the size of these current jumps. In the case of porins, this would represent the trimeric conductance, since porins typically purify and insert in the bilayer as trimers. By manipulating the protein concentration, it is possible to ensure that either many or only one porin trimer inserts, and investigations can be performed on single channels or on populations of channels. After insertion, the channel activity can be studied in various conditions and membrane potentials.
The patch-clamp technique has also been applied to the study of purified porins reconstituted in artificial liposomes. Here a small patch of liposome membrane is drawn at the tip of a 1 μM-diameter glass pipette, and the current flowing through this patch is recorded at a fixed membrane potential. Because of the small area of membrane under investigation, the patch clamp technique typically offers a better signal-to-noise ratio than BLM. This technique permitted the discovery that porins flicker between multiple states, whose kinetics and conductance can be affected in mutants and in the presence of modulators (see below).
Studies performed by the Benz and the Rosenbusch groups in the 70’s and 80’s established some of the hallmark properties of the general diffusion porins, such as high ionic current due to the relatively large pore size, low ionic selectivity (although some porins show preference for cations (OmpC) or anions (PhoE)), and high open probability, in standard bilayer electrophysiology conditions of low voltage, neutral pH and high ionic strength (but see below) [38–41]. Computational modeling studies have suggested that the paths taken by anions and cations are divergent at the eyelet, as cations are drawn close to the negative charges of the L3 loop, and anions flow near the positively charged cluster of the opposite barrel wall [42]. This type of work emphasizes the notion that the permeating ions interact with the wall of the channel and that ion movement does not follow simple diffusion. This was demonstrated experimentally by measuring the conductance and selectivity of various general diffusion porins in solutions of varying ionic strength or pH, and in variants with mutations at specific pore exposed residues [43–48].
Bezrukov’s group showed that the selectivity of OmpF for cations relative to anions increases sharply in solutions of low ionic strength [43]. The channel reaches nearly ideal cation-selectivity in solutions of < 100 mM KCl. Furthermore, at pH’s < 4, the channel reverses its selectivity from preferring cations to preferring anions. The authors combined these experimental observations with calculations of the distribution of charged residues in the pore lumen and concluded that electrostatic interactions exist between the permeating ions and the charges of ionizable residues over the entire channel length [43]. However, shifts in selectivity are detected upon mutations of single residues. Substitution at the pore-exposed D113 residue in OmpF [48] and its homologs in OmpC [46] and the Vibrio cholerae porin OmpU [45] decreases cation-selectivity. Opposite effects are seen upon charge removal at arginines of the constriction zone [48].
The mutations also affect conductance, although there is not strict correlation between an apparent increase in pore size due to removal of a bulky side chain and increase conductance [49]. The results highlight the notion that conductance is a reflection not only of pore size but also interaction of permeating ions with channels walls, and strengthen the argument that the derivation of pore size from conductance measurements should be avoided [50]. In addition, an increase in conductance is not always a good predictor of an increased permeation of larger substrates or antibiotic susceptibility, as was shown for OmpF [44] and V. cholerae porin OmpU [45].
3.1.3. Functional modulation of porins
The fact that the activity of porins can be quickly modulated by ligand binding and a variety of physico-chemical parameters is an important - but relatively unappreciated - aspect of outer membrane permeability. Porins are thought of as permanently open pores, and for years, the only documented mechanism to reduce outer membrane permeability was through a lower porin expression due to environmental factors or mutations. The knowledge of which parameters lead to rapid closure of porins is important, since the resulting tightening of the OM will decrease the efficacy of penetration of antibiotics using the porin-mediated pathway. The first rapid modulation of porin function to be described was transmembrane voltage [41], but the significance of this phenomenon is often dismissed because the OM is believed to be without a transmembrane potential (but see below). Still, the voltage-dependent inactivation of porins is a robust phenomenon, shown to differ among different porins species, and affected by mutations at specific pore residues [48, 51–58]. The voltage sensitivity of porins is typically quantified by the so-called “threshold” potential, i.e. the minimum membrane potential at which porins start to close. When the membrane potential is above this value, porin monomers close, often sequentially, in a typical stepwise fashion. The protein appears to reach a deep inactivated state, as it is reluctant to re-opening, even at lower voltages, and hysteresis is observed when voltages are slowly ramped up and down in bilayers containing many channels [52]. The threshold potential is typically quite high (~150 mV for OmpF [48] and ~ 200 mV for OmpC [55]), but some porins are more voltage-sensitive (V. cholerae OmpT has a threshold potential of ~ 90 mV [57]). Nikaido demonstrated that Donnan potentials established by accumulation of periplasmic negatively charged membrane-derived oligosaccharides (MDO’s) are unable to decrease porin-mediated permeability to β-lactams [59]. However, it is possible that this negative result stems from the asymmetric voltage dependence of porins [60]. OmpF might close in vivo upon the opposite membrane potential (more positive on the periplasmic side relative to the outside); this potential could be established in vivo by a concentration gradient of potassium ions, for example, if the absolute ionic strength of the periplasmic and external solutions is relatively low (<100 mM), i.e. in the range where OmpF becomes a highly selective cation channel [43]. This still needs to be demonstrated experimentally.
The voltage-dependent inactivation of porins demonstrated that porins can exist in non-conducting, i.e. closed, forms, and set the stage for the discovery of other possible modulators of porin function. In particular, two other important forms of modulation lead to closing of porins: acidic pH and binding of polyamines. Besides effects on conductance and selectivity [43, 61], acid pH also promotes kinetic changes in porins. Fast flickering in the open channel noise drastically increases, in particular at pH’s < 4, and may be attributed to protonation-deprotonation events of key acidic residues in the pore [61, 62]. In addition, the channel increases its closing probability at acidic pH. In OmpF, this is often seen as a sequential stepwise closing of monomeric units after application of a transmembrane voltage. It is similar to the effect of voltage, but occurs at much lower potentials than at neutral pH. It is possible that it stems from an enhanced voltage-sensitivity, as documented [63]. In OmpU, channels are immediately stabilized in a closed configuration, and surprisingly, individual closures of three monomers are not observed, but rather closing events of an apparent single channel of increasingly large size as the pH is decreased [64]. In OmpF, extracellular loops L1, L7 and L8 have been implicated in the conformational changes that might lead to acidic pH-induced channel closures [62]. Altogether, these loops form a lid-type domain that might close up above the pore, as suggested by atomic force microscopy of OmpF surface at low pH [65].
E. coli OmpF and OmpC porins are inhibited by the polyamines spermine, spermidine and cadaverine [66, 67]. This is also true for the V. cholerae porin OmpU (Delcour, unpublished). These linear, highly charged, amine compounds are small enough to pass through porins, and indeed, the kinetics of the modulation observed in patch clamp experiments do not bear the hallmarks of open channel block. Rather, it appears that the compounds bind to an internal pore-exposed site and trigger channel closures. These effects are rather complex, with a greatly increase flickering activity to states of lower conductance than the monomeric conductance (subconductance states) [68], and prolonged monomeric closures [67]. Mutagenesis work defined a binding site involving the L3 loop acidic residues D113 and D121, and also Y294 for the case of spermine [69]. We envisage a model whereby polyamines would saddle over the L3 loop through ionic interactions involving the amine groups, and cause a destabilization or a possible movement of the L3 loop, leading to channel closure. Modulation of this kind by polyamines has a marked impact on the overall outer membrane permeability [70]. The porin closure induced by spermine might have some important therapeutic consequences in treatment of infections of tissues where spermine content is high, such as in the prostate.
Cadaverine is endogenously produced by E. coli and secreted in conditions of acidic pH. By manipulating the Cad operon, we have shown that the production and release of endogenous cadaverine decreases outer membrane permeability [71]. The cadaverine-dependent modulation of porin is part of adaptive response to a pH drop, since a cadaverine-resistant porin mutant is outcompeted by wildtype in acidic conditions [72]. These observations reinforce the notion that the rapid modulation of porin function can provide cells with an emergency mechanism to shut down OM permeability until slower mechanisms involving regulation of porin expression are put in place. Importantly, they suggest that the permeability of the OM to antibiotics, for example, might be changing for cells in different external conditions. Indeed polyamines were shown to inhibit the flux of cephaloridine through the porins OmpF and OmpC [70].
3.2. Porin-mediated antibiotic permeability
The permeability of porins to β-lactam antibiotics has been demonstrated by various means. Evidence for a direct role of porins in mediating the diffusion of β-lactams was provided by purifying and reconstituting porins into liposomes and using either a liposome swelling assay [35], or measuring the antibiotic degradation rate by an entrapped β-lactamase [73]. Measurement of antibiotic flux in whole cells was originally developed by Zimmermann and Rosselet [74] and then extensively used by Nikaido’s group to characterize the permeability of cephaloridine and other cephalosporins in various cells types (wildtype and porin mutants), by taking advantage of the fast rate of cephalosporin degradation by periplasmic β-lactamase [75]. Rates of the order of ~ 10–50 10−5 cm/s were found for the permeation of zwitterionic drugs through OmpF, but were much reduced for anionic compounds.
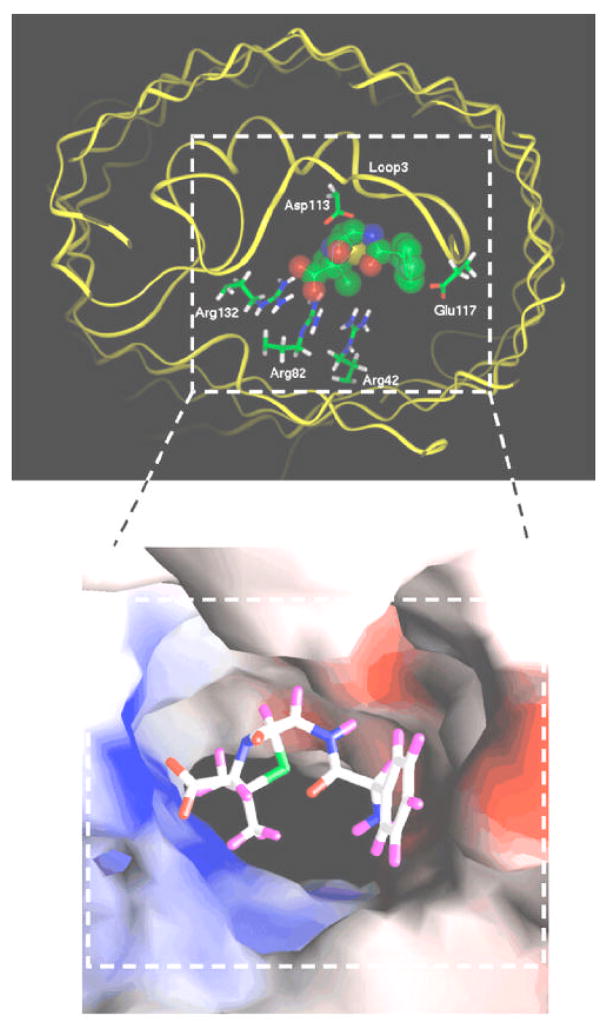
A molecular explanation for these findings has recently emerged from a more detailed view of the interactions of the permeating drugs with the porin channels, obtained from the combination of electrophysiology and computational studies. Bezrukov and colleagues demonstrated that ampicillin acts as a transient open channel blocker of the OmpF porin in a pH dependent manner, with a maximum block in a pH range where the ampicillin molecule is zwitterionic [76]. Molecular dynamics calculations explain this pH dependence, as they reveal that the drug molecule perfectly occludes the pore in the zwitterionic form, as it interacts simultaneously with negatively charged residues of L3 and positively charged residues of the barrel wall (Figure 3). Such complementation between the charge distributions on the drug molecule and the narrowest region of the OmpF pore has also been found for another zwitterionic β-lactam, amoxicillin [77]. On the contrary, poor interactions were delineated for the di-anionic carbenicillin and the mono-anionic β-lactams azlocillin and piperacillin. This negligible binding correlates with the poor diffusion rates measured from such compounds from liposome swelling assays [78]. On the other hand, high diffusion rates were obtained for ampicillin and amoxicillin. Thus, it appears that interactions at the OmpF constriction zone facilitate the drug translocation, and that the nature and position of specific charges on the antibiotic molecule and on OmpF play a major role in these interactions.
Figure 3.
Docking of an ampicillin molecule at the constriction zone of an OmpF monomer. The top panel shows the fit of the ampicillin molecule within the pore, with the carboxylate group attracted to the cluster of arginines in the OmpF barrel, and the ammonium group close to the acidic L3 loop residues. Colors of atoms in ampicillin are as follows: green for carbon, red for oxygen, blue for nitrogen and yellow for sulfur. Hydrogen atoms are not shown. The OmpF backbone is shown as a yellow ribbon. The lower panel shows the solvent accessible surface of the OmpF eyelet highlighting the electrostatic potential with blue color for positive potential and red color for negative potentials. Ampicillin is shown in a stick model with the following colors: white for carbon, red for oxygen, blue for nitrogen, green for sulfur and violet for hydrogen. Reproduced from reference 70 with permission (copyright © 1993–2008 by The National Academy of Sciences of the United States of America, all rights reserved).
Experimentally, site-directed mutations of many key charged residues of the porin constriction zone affect β-lactam flux and sensitivity [79–82]. The involvement of specific OmpF residues as anchorage points for several cephalosporins has been suggested from computational studies, as well [79]. Some mutations also involved uncharged residues. For example, the diffusion of radiolabeled cefepime was drastically decreased in the G119D and G119E mutants [83]. The X-ray structure of the G119D mutant OmpF shows that the introduced aspartate residue protrudes in the eyelet and constricts the diameter the pore [84]. Consequently, the channel conductance, diffusion rate of various sugars and sensitivity to cephalosporins are greatly reduced [83, 84]. On the other hand, mutations at the R132 residues lead to improved growth on maltodextrins relative to wildtype [48] and increased cefepime diffusion [83], possibly due to an increase in pore diameter [85].
Carbapenems, such as imipenem and merpenem, are β-lactam antibiotics with a high resistance to the action of β-lactamase. They have been particularly effective against P. aeruginosa, an organism which appears less susceptible to most antibiotics than Enterobacteriaceae, because of decreased OM permeability and an efficient drug efflux system. The low OM permeability stems from the lack of general diffusion porins, as P. aeruginosa acquires its nutrients through dedicated specific porins [11]. The isolation of an imipenem resistant strain pointed to the role of the OprD porin (previously known as protein D2) in the permeability to imipenem [86], and indeed this protein was later shown to allow the facilitated diffusion of carbapenems and penems through the OM [87]. When the purified protein is reconstituted into artificial bilayers, the formed channels have a very low conductance, but can be blocked by imipenem, indicating the presence of a specific binding site [88]. Additional studies demonstrated that, in fact, the OprD porin is used for the uptake of basic amino acids and peptides, which share structural similarity with the carbapenem molecule [89].
Quinolones are believed to use a dual pathway for entry into bacterial cells, because drug flux and susceptibility are both sensitive to the presence of porins (in particular OmpF) and to the manipulations that disrupt the outer membrane LPS barrier [90, 91]. The relative contribution of the two pathways correlates with the hydrophobicity and the protonation state of the quinolones, in the manners described below. Hydrophobic quinolones are more effective in LPS mutants [91]. There is a report that the quinolone fleroxacin induces the same perturbations of the OM as does gentamycin or EDTA, supporting the contention that quinolones might act as chelating agents and use a self-promoted pathway as aminoglycosides and cationic peptides do [90]. However, the sensitivity of cells to less hydrophobic quinolones, such as norfloxacin and ciprofloxacin and other drugs with similar hydrophobicity coefficient of less than 0.1, was not much affected in mutants in LPS structure [91] suggesting that they might use porin for access through the OM. Indeed a reduced accumulation of radiolabeled norfloxacin was observed in E. coli strains lacking OmpF [92]. Moreover, the flux of norfloxacin in Enterobacter cloacae was inhibited in the presence of spermine or cefepime, both known to use porins for permeation through the OM, thus confirming that norfloxacin diffuses through the porin lumen [93]. Nikaido and Thanassi have proposed that quinolones exist in an equilibrium of charged and uncharged species depending on the solution pH [94]. For example, they calculated that about 10% of norfloxacin exists as an uncharged species at pH 7.4, and this ratio is even higher (~ 40% at pH 6.5) for amifloxacin. These authors have argued that the uncharged quinolone molecules cross the OM through the lipid bilayer, while the negatively charged molecules are likely to pass through porin channels as magnesium chelates. Thus the relative contributions of the porin-mediated and lipid-mediated pathways are likely to depend on the protonation-deprotonation states of the drug, which will themselves be influenced by external pH. In addition, the charged species is proposed to accumulate in the periplasm due to the interior-negative Donnan potential across the OM [94]. This accumulation leads to high cytoplasmic levels as well, as the cytoplasm equilibrates very rapidly with the periplasm, even for drugs with oil/water partition coefficient less than 0.1. In porin-deficient mutants, quinolones still permeate through the outer membrane bilayer itself in their uncharged form, but do not accumulate in the periplasm because they are not sensitive to the Donnan potential, thus leading to decreased cytoplasmic concentrations and efficacy.
The uptake of tetracycline by E. coli cells was shown to be reduced in a mutant lacking OmpF [95], confirming the suggestion that tetracycline uses this pathway based on increased resistance in mutants with decreased ompF expression [96]. This accumulation, however, is not null in the absence of OmpF, and it positively correlates with pH, i.e. there is less influx of tetracycline at lower pH (pH 6.0) relative to neutral pH, or even 7.8 [95]. Tetracycline has a pKa of 7.7, and therefore exists mostly in a protonated form at a pH’s under the pKa. In this uncharged form, tetracycline is believed to enter cells by diffusion through the OM lipid barrier [94]. Thus, tetracycline, like fluoroquinolones, uses both a porin- and a lipid-mediated pathway, depending on its protonated status.
3.3. Porins and antibiotic resistance
As described above, porins provide a path through the OM to small hydrophilic antibiotics, such as β-lactams, as well as tetracycline, chloramphenicol and fluoroquinolones [11]. Any decrease in the ability or rate of entry of these compounds can lead to resistance. There is an abundance of reports of antibiotic resistance acquired through loss or functional change of porins in a large number of organisms, such as E. coli, P. aeruginosa, Neisseria gonorrhoeae, Enterobacter aerogenes and Klebsiella pneumoniae (see references [11, 28, 97–99] for reviews, and references therein). Although much of the mechanistic studies described above have focused on OmpF because of its well understood structural and functional properties relative to any other major porins, many of the reports of changes in porin expression often implicated both OmpF and OmpC. The role of minor porins (such as NmpC), or those expressed in specific conditions (such as PhoE), perhaps should not be underestimated, but there are far fewer reports on the involvement of these porins in antibiotic resistance (but see below). Still it appears that PhoE can serve as a conduit for entry of β-lactams (and be an even better one than OmpF and OmpC if the drug bears a negative charge) [75], as well as for chloramphenicol and tetracylcine [100].
It would be impractical in this review to cite all or even most of studies linking antibiotic resistance to general diffusion porins, but we can highlight some of the generally found common themes with specific examples. There are two major porin-based mechanisms for antibiotic resistance that have been reported in clinical isolates: 1) alterations of outer membrane profiles, including either loss/severe reduction of porins or replacement of one or two major porins by another; 2) altered function due to specific mutations reducing permeability.
Antibiotic resistance poses a daunting problem in hospital-acquired infections. Pagès and colleagues analyzed the porin content of 45 β-lactam resistant clinical isolates of E. aerogenes obtained from French hospitals [101]. Of those, 44% were shown to lack porin, as determined by immunodetection. The MIC’s of four antibiotics (cefepime, imipenem, cefotaxime and moxalactam) were drastically increased. Additionally, many strains displayed high constitutive or inducible β-lactamase activity, but some strains did not, and thus antibiotic resistance appears to originate essentially from the lack of porins in those strains. The increase in MIC’s for those porin-deficient strains was similar to those with robust β-lactamase activity, indicating that a reduction of porin-mediated permeability can be an efficient strategy for antibiotic resistance on its own.
Tetracylcine resistance can occur under antibiotic stress, by exposing sensitive E. coli cells to progressively increasing concentration of the antibiotic. The treatment, in fact, leads to a chromosome-mediated multiple antibiotic resistance (Mar phenotype), where the cells become insensitive to a variety of hydrophilic and lipophilic antibiotics [102, 103]. The response involves the coordinated change in the levels of multiple proteins including porins and drug efflux pumps, through mechanisms involving transcriptional and posttranscriptional regulation [104]. In particular, the upregulation of marA leads to increased levels of the small RNA micF, which inhibits translation of ompF RNA. Decreased OmpF levels are also postulated to originate from the periplasmic accumulation of other OM proteins, such as TolC and OmpX, which might titrate away the chaperones and assembly proteins required for membrane insertion of OM proteins [104]. Another example of upregulation of OmpX in coordination with a strong repression of general diffusion porins has also been documented for acquired resistance to a large number of antibiotics of a strain of Salmonella enterica Typhimurium after exposure to nalidixic acid [105]. In this case, repression also included other porins, besides OmpF, such as NmpC, LamB and Tsx.
The substitution of a narrower porin in lieu of the constitutively expressed large general-diffusion porins is another strategy for acquiring antibiotic resistance. For example, some clinical isolates from K. pneumoniae lack the large diffusion channels OmpK35 and OmpK36, but express a normally quiescent porin, OmpK37, which appears to form a smaller pore on the basis of sugar permeability [106]. This porin is akin to OmpN of E. coli and OmpS2 of S. typhi, two porin types which are normally strongly down-regulated in laboratory media conditions. The presence of OmpK37 combined with the absence of OmpK35 and OmpK36 lead to a drastic increase in the MIC’s of cefotaxime and cefoxitin, but not of carbapenems, indicating that these compounds might still be able to flux through OmpK37 as they do through P. aeruginosa OprD. This provides an explanation for the fact that K. pneumoniae infections resistant to most β-lactams can still be treated by carbapenems.
Altered function of porin leading to reduced permeation rate is another strategy found in antibiotic resistant bacteria. A hot spot for single or multiple mutations leading to such phenotype is the L3 loop, which delineates the constriction zone of general diffusion porins. A clinical isolate of E. aerogenes was found to have a glycine → aspartate substitution on the L3 loop of its major porin [107], which might lead to a distortion of the loop or further narrowing of the pore lumen, as in G119D of OmpF [83]. This mutant is characterized by a 3-fold decrease in porin conductance and a drastic reduction in cephalosporin sensitivity. It was found later on that this porin is Omp36, which is highly similar to E. coli OmpC [108]. This clinical isolate and two others from E. aerogenes, in fact, present multiple mutations in the porin gene, and are also highly resistant to cefepime, cefpirome and imipenem. Similar alterations in the amino acid composition of the N. gonorrhoeae porin Por have also be documented [109]. Here, a mutant with enhanced resistance to penicillin and tetracycline was found to have multiple mutations throughout the porin gene, and in particular in a region putatively homologous to the constricting L3 loop. Interestingly, six clinical isolates with similar resistance to penicillin also displayed single point mutations in the same region.
Finally, some bacterial species, such as P. aeruginosa, are intrinsically more resilient to antibiotic treatments, because of a low abundance of general diffusion porins, combined with numerous and highly efficient drug efflux mechanisms [11, 110]. As described above, OprF, the major porin of P. aeruginosa, is present in high abundance as a closed conformer, and exists as an open channel only at very low levels. Not surprisingly, acquired resistance to β-lactam antibiotics does not seem to involve loss or modification of OprF [111]. Resistance to carbapenems can be observed in mutants lacking the porin-specific OprD (see above), and in mutants with deletions in the L2 loop of OprD [88]. Carbapenem resistance via porin-delimitated pathways is not restricted to P. aeruginosa, as described above.
Concluding remarks
In conclusion, mechanisms affecting the barrier properties of the OM lipid bilayer itself or the expression and/or function of the general diffusion porin channels residing in the OM have an impact on the sensitivity of Gram-negative bacteria to many different types of antibiotics. Clearly any weakening of the LPS bilayer by targeting LPS synthesizing enzymes will sensitize bacteria to hydrophobic and some hydrophilic antibiotics, leading to the possibility of combinatorial drug therapy. A better understanding of the function of general diffusion porins, and in particular of the parameters that might lead to porin closure or inactivation, will allow a reassessment of the efficiency of penetration of the antibiotics using this pathway in different conditions. It is hoped that, as we further understand at the molecular level the structure and function of these OM macromolecules and of those that regulate them, scientists will be able to refine the current drug therapies or design new types of antibiotics that target these surface exposed entities.
Acknowledgments
Our own work on porins has been supported by NIH grant AI34905 and grant E-1597 from theWelch Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kadner RJ. Escherichia coli and Salmonella. In: Neidhart FC, editor. Cellular and Molecular Biology. ASM press; Washington, DC: 1996. pp. 58–87. [Google Scholar]
- 3.Kamio Y, Nikaido H. Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry. 1976;15:2561–2570. doi: 10.1021/bi00657a012. [DOI] [PubMed] [Google Scholar]
- 4.Nikaido H. Escherichia coli and Salmonella. In: Neidhart FC, editor. Cellular and Molecular Biology. ASM Press; Washington, DC: 1996. pp. 29–47. [Google Scholar]
- 5.Hirota Y, Suzuki H, Nishimura Y, Yasuda S. On the process of cellular division in Escherichia coli: a mutant of E. coli lacking a murein-lipoprotein. Proc Natl Acad Sci U S A. 1977;74:1417–1420. doi: 10.1073/pnas.74.4.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonntag I, Schwarz H, Hirota Y, Henning U. Cell envelope and shape of Escherichia coli: multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J Bacteriol. 1978;136:280–285. doi: 10.1128/jb.136.1.280-285.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugawara E, Nestorovich EM, Bezrukov SM, Nikaido H. Pseudomonas aeruginosa porin OprF exists in two different conformations. J Biol Chem. 2006;281:16220–16229. doi: 10.1074/jbc.M600680200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugawara E, Nikaido H. Pore-forming activity of OmpA protein of Escherichia coli. J Biol Chem. 1992;267:2507–2511. [PubMed] [Google Scholar]
- 9.Gerlach RG, Hensel M. Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens. Int J Med Microbiol. 2007;297:401–415. doi: 10.1016/j.ijmm.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 10.Kostakioti M, Newman CL, Thanassi DG, Stathopoulos C. Mechanisms of protein export across the bacterial outer membrane. J Bacteriol. 2005;187:4306–4314. doi: 10.1128/JB.187.13.4306-4314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruiz N, Kahne D, Silhavy TJ. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- 13.Raetz CR. Escherichia coli and Salmonella. In: Neidhart FC, editor. Cellular and Molecular Biology. ASM Press; Washington, DC: 1996. pp. 1035–1063. [Google Scholar]
- 14.Vaara M. Agents that increase the permeability of the outer membrane. Microbiol Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dame JB, Shapiro BM. Use of polymyxin B, levallorphan, and tetracaine to isolate novel envelope mutants of Escherichia coli. J Bacteriol. 1976;127:961–972. doi: 10.1128/jb.127.2.961-972.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mäkelä PH, Sarvas M, Calcagno S, Lounatmaa K. Isolation and genetic characterization of polymyxin-resistant mutants of Salmonella. FEMS Microbiol Lett. 1978;3:323–326. [Google Scholar]
- 17.Vaara M, Vaara T, Sarvas M. Decreased binding of polymyxin by polymyxin-resistant mutants of Salmonella typhimurium. J Bacteriol. 1979;139:664–667. doi: 10.1128/jb.139.2.664-667.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaara M, Vaara T, Jensen M, Helander I, Nurminen M, Rietschel ET, Makela PH. Characterization of the lipopolysaccharide from the polymyxin-resistant pmrA mutants of Salmonella typhimurium. FEBS Lett. 1981;129:145–149. doi: 10.1016/0014-5793(81)80777-6. [DOI] [PubMed] [Google Scholar]
- 19.Peterson AA, Fesik SW, McGroarty EJ. Decreased binding of antibiotics to lipopolysaccharides from polymyxin-resistant strains of Escherichia coli and Salmonella typhimurium. Antimicrob Agents Chemother. 1987;31:230–237. doi: 10.1128/aac.31.2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Z, Ribeiro AA, Lin S, Cotter RJ, Miller SI, Raetz CR. Lipid A modifications in polymyxin-resistant Salmonella typhimurium: PMRA-dependent 4-amino-4-deoxy-L-arabinose, and phosphoethanolamine incorporation. J Biol Chem. 2001;276:43111–43121. doi: 10.1074/jbc.M106960200. [DOI] [PubMed] [Google Scholar]
- 21.Gunn JS, Miller SI. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J Bacteriol. 1996;178:6857–6864. doi: 10.1128/jb.178.23.6857-6864.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roland KL, Martin LE, Esther CR, Spitznagel JK. Spontaneous pmrA mutants of Salmonella typhimurium LT2 define a new two-component regulatory system with a possible role in virulence. J Bacteriol. 1993;175:4154–4164. doi: 10.1128/jb.175.13.4154-4164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, Miller SI. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 24.Prost LR, Sanowar S, Miller SI. Salmonella sensing of anti-microbial mechanisms to promote survival within macrophages. Immunol Rev. 2007;219:55–65. doi: 10.1111/j.1600-065X.2007.00557.x. [DOI] [PubMed] [Google Scholar]
- 25.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 26.Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, Miller SI. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- 27.Hancock RE, Farmer SW, Li ZS, Poole K. Interaction of aminoglycosides with the outer membranes and purified lipopolysaccharide and OmpF porin of Escherichia coli. Antimicrob Agents Chemother. 1991;35:1309–1314. doi: 10.1128/aac.35.7.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hancock RE, Bell A. Antibiotic uptake into gram-negative bacteria. Eur J Clin Microbiol Infect Dis. 1988;7:713–720. doi: 10.1007/BF01975036. [DOI] [PubMed] [Google Scholar]
- 29.Vaara M, Vaara T. Polycations sensitize enteric bacteria to antibiotics. Antimicrob Agents Chemother. 1983;24:107–113. doi: 10.1128/aac.24.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong C, Beis K, Nesper J, Brunkan-Lamontagne AL, Clarke BR, Whitfield C, Naismith JH. Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature. 2006;444:226–229. doi: 10.1038/nature05267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basle A, Rummel G, Storici P, Rosenbusch JP, Schirmer T. Crystal structure of osmoporin OmpC from E. coli at 2.0 A. J Mol Biol. 2006;362:933–942. doi: 10.1016/j.jmb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Cowan SW, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit RA, Jansonius JN, Rosenbusch JP. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 33.Buchanan SK, Smith BS, Venkatramani L, Xia D, Esser L, Palnitkar M, Chakraborty R, van der Helm D, Deisenhofer J. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat Struct Biol. 1999;6:56–63. doi: 10.1038/4931. [DOI] [PubMed] [Google Scholar]
- 34.Clantin B, Delattre AS, Rucktooa P, Saint N, Meli AC, Locht C, Jacob-Dubuisson F, Villeret V. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science. 2007;317:957–961. doi: 10.1126/science.1143860. [DOI] [PubMed] [Google Scholar]
- 35.Nikaido H, Rosenberg EY. Porin channels in Escherichia coli: studies with liposomes reconstituted from purified proteins. J Bacteriol. 1983;153:241–252. doi: 10.1128/jb.153.1.241-252.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakae T. Outer membrane of Salmonella typhimurium: Reconstitution of sucrose-permeable membrane vesicles. Biochemical And Biophysical Research Communications. 1975;64:1224–1230. doi: 10.1016/0006-291x(75)90823-2. [DOI] [PubMed] [Google Scholar]
- 37.Nikaido H, Rosenberg EY. Effect on solute size on diffusion rates through the transmembrane pores of the outer membrane of Escherichia coli. J Gen Physiol. 1981;77:121–135. doi: 10.1085/jgp.77.2.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benz R, Janko K, Boos W, Lauger P. Formation of large, ion-permeable membrane channels by the matrix protein (porin) of Escherichia coli. Biochim Biophys Acta. 1978;511:305–319. doi: 10.1016/0005-2736(78)90269-9. [DOI] [PubMed] [Google Scholar]
- 39.Benz R, Janko K, Lauger P. Ionic selectivity of pores formed by the matrix protein (porin) of Escherichia coli. Biochim Biophys Acta. 1979;551:238–247. doi: 10.1016/0005-2736(89)90002-3. [DOI] [PubMed] [Google Scholar]
- 40.Benz R, Schmid A, Hancock RE. Ion selectivity of gram-negative bacterial porins. J Bacteriol. 1985;162:722–727. doi: 10.1128/jb.162.2.722-727.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schindler H, Rosenbusch JP. Matrix protein from Escherichia coli outer membranes forms voltage-controlled channels in lipid bilayers. Proc Natl Acad Sci U S A. 1978;75:3751–3755. doi: 10.1073/pnas.75.8.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Im W, Roux B. Ions and counterions in a biological channel: a molecular dynamics simulation of OmpF porin from Escherichia coli in an explicit membrane with 1 M KCl aqueous salt solution. J Mol Biol. 2002;319:1177–1197. doi: 10.1016/S0022-2836(02)00380-7. [DOI] [PubMed] [Google Scholar]
- 43.Alcaraz A, Nestorovich EM, Aguilella-Arzo M, Aguilella VM, Bezrukov SM. Salting out the ionic selectivity of a wide channel: the asymmetry of OmpF. Biophys J. 2004;87:943–957. doi: 10.1529/biophysj.104/043414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bredin J, Saint N, Mallea M, Molle EDG, Pages JM, Simonet V. Alteration of pore properties of Escherichia coli OmpF induced by mutation of key residues in anti-loop 3 region. Biochem J. 2002;363:521–528. doi: 10.1042/0264-6021:3630521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lauman B, Pagel M, Delcour AH. Altered Pore Properties and Kinetic Changes in Mutants of the Vibrio cholerae Porin OmpU. Molecular Membrane Biology in press. 2008 doi: 10.1080/09687680802454637. [DOI] [PubMed] [Google Scholar]
- 46.Liu N. PhD Thesis. University of Houston; 1999. Structure-Function Relationships of E. coli OmpC Porin - The Effects of Site-Directed Mutations on the Porin Channel Function. [Google Scholar]
- 47.Phale PS, Philippsen A, Widmer C, Phale VP, Rosenbusch JP, Schirmer T. Role of charged residues at the OmpF porin channel constriction probed by mutagenesis and simulation. Biochemistry. 2001;40:6319–6325. doi: 10.1021/bi010046k. [DOI] [PubMed] [Google Scholar]
- 48.Saint N, Lou KL, Widmer C, Luckey M, Schirmer T, Rosenbusch JP. Structural and functional characterization of OmpF porin mutants selected for larger pore size. II. Functional characterization. J Biol Chem. 1996;271:20676–20680. [PubMed] [Google Scholar]
- 49.Delcour AH. Solute uptake through general porins. Front Biosci. 2003;8:d1055–1071. doi: 10.2741/1132. [DOI] [PubMed] [Google Scholar]
- 50.Nikaido H. Porins and specific diffusion channels in bacterial outer membranes. J Biol Chem. 1994;269:3905–3908. [PubMed] [Google Scholar]
- 51.Arbing MA, Dahan D, Boismenu D, Mamer OA, Hanrahan JW, Coulton JW. Charged residues in surface-located loops influence voltage gating of porin from Haemophilus influenzae type b. J Membr Biol. 2000;178:185–193. doi: 10.1007/s002320010026. [DOI] [PubMed] [Google Scholar]
- 52.Baslé A, Delcour AH. In: Structure and Function of Bacterial and Eukaryotic Porins. Benz R, editor. Wiley-Interscience; 2004. pp. 79–98. [Google Scholar]
- 53.Bishop ND, Lea EJ, Mobasheri H, Spiro S. Altered voltage sensitivity of mutant OmpC porin channels. FEBS Lett. 1996;379:295–298. doi: 10.1016/0014-5793(95)01535-3. [DOI] [PubMed] [Google Scholar]
- 54.Delcour AH, Adler J, Kung C. A single amino acid substitution alters conductance and gating of OmpC porin of Escherichia coli. J Membr Biol. 1991;119:267–275. doi: 10.1007/BF01868731. [DOI] [PubMed] [Google Scholar]
- 55.Lakey JH, Lea EJ, Pattus F. OmpC mutants which allow growth on maltodextrins show increased channel size and greater voltage sensitivity. FEBS Lett. 1991;278:31–34. doi: 10.1016/0014-5793(91)80076-f. [DOI] [PubMed] [Google Scholar]
- 56.Phale PS, Schirmer T, Prilipov A, Lou KL, Hardmeyer A, Rosenbusch JP. Voltage gating of Escherichia coli porin channels: role of the constriction loop. Proc Natl Acad Sci U S A. 1997;94:6741–6745. doi: 10.1073/pnas.94.13.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simonet VC, Basle A, Klose KE, Delcour AH. The Vibrio cholerae porins OmpU and OmpT have distinct channel properties. J Biol Chem. 2003;278:17539–17545. doi: 10.1074/jbc.M301202200. [DOI] [PubMed] [Google Scholar]
- 58.Van Gelder P, Saint N, Phale P, Eppens EF, Prilipov A, van Boxtel R, Rosenbusch JP, Tommassen J. Voltage sensing in the PhoE and OmpF outer membrane porins of Escherichia coli: role of charged residues. J Mol Biol. 1997;269:468–472. doi: 10.1006/jmbi.1997.1063. [DOI] [PubMed] [Google Scholar]
- 59.Sen K, Hellman J, Nikaido H. Porin channels in intact cells of Escherichia coli are not affected by Donnan potentials across the outer membrane. J Biol Chem. 1988;263:1182–1187. [PubMed] [Google Scholar]
- 60.Samartzidou H, Delcour AH. E.coli PhoE porin has an opposite voltage-dependence to the homologous OmpF. Embo J. 1998;17:93–100. doi: 10.1093/emboj/17.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nestorovich EM, Rostovtseva TK, Bezrukov SM. Residue ionization and ion transport through OmpF channels. Biophys J. 2003;85:3718–3729. doi: 10.1016/S0006-3495(03)74788-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baslé A, Qutub R, Mehrazin M, Wibbenmeyer J, Delcour AH. Deletions of single extracellular loops affect pH-sensitivity, but not voltage-dependence, of the E. coli porin OmpF. Protein Eng Des Sel. 2004;17:665–672. doi: 10.1093/protein/gzh078. [DOI] [PubMed] [Google Scholar]
- 63.Todt JC, Rocque WJ, McGroarty EJ. Effects of pH on bacterial porin function. Biochemistry. 1992;31:10471–10478. doi: 10.1021/bi00158a009. [DOI] [PubMed] [Google Scholar]
- 64.Duret G, Simonet V, Delcour AH. Modulation of Vibrio cholerae Porin Function by Acidic pH. Channels. 2007;1:70–79. doi: 10.4161/chan.3983. [DOI] [PubMed] [Google Scholar]
- 65.Müller DJ, Engel A. Voltage and pH-induced channel closure of porin OmpF visualized by atomic force microscopy. J Mol Biol. 1999;285:1347–1351. doi: 10.1006/jmbi.1998.2359. [DOI] [PubMed] [Google Scholar]
- 66.delaVega AL, Delcour AH. Cadaverine induces closing of E. coli porins. Embo J. 1995;14:6058–6065. doi: 10.1002/j.1460-2075.1995.tb00294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iyer R, Delcour AH. Complex inhibition of OmpF and OmpC bacterial porins by polyamines. J Biol Chem. 1997;272:18595–18601. doi: 10.1074/jbc.272.30.18595. [DOI] [PubMed] [Google Scholar]
- 68.Baslé A, Iyer R, Delcour AH. Subconductance states in OmpF gating. Biochim Biophys Acta. 2004;1664:100–107. doi: 10.1016/j.bbamem.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 69.Iyer R, Wu Z, Woster PM, Delcour AH. Molecular basis for the polyamine-ompF porin interactions: inhibitor and mutant studies. J Mol Biol. 2000;297:933–945. doi: 10.1006/jmbi.2000.3599. [DOI] [PubMed] [Google Scholar]
- 70.delaVega AL, Delcour AH. Polyamines decrease Escherichia coli outer membrane permeability. J Bacteriol. 1996;178:3715–3721. doi: 10.1128/jb.178.13.3715-3721.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Samartzidou H, Delcour AH. Distinct sensitivities of OmpF and PhoE porins to charged modulators. FEBS Lett. 1999;444:65–70. doi: 10.1016/s0014-5793(99)00030-7. [DOI] [PubMed] [Google Scholar]
- 72.Samartzidou H, Mehrazin M, Xu Z, Benedik MJ, Delcour AH. Cadaverine inhibition of porin plays a role in cell survival at acidic pH. J Bacteriol. 2003;185:13–19. doi: 10.1128/JB.185.1.13-19.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kobayashi Y, Takahashi I, Nakae T. Diffusion of beta-lactam antibiotics through liposome membranes containing purified porins. Antimicrob Agents Chemother. 1982;22:775–780. doi: 10.1128/aac.22.5.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zimmermann W, Rosselet A. Function of the outer membrane of Escherichia coli as a permeability barrier to beta-lactam antibiotics. Antimicrob Agents Chemother. 1977;12:368–372. doi: 10.1128/aac.12.3.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nikaido H, Rosenberg EY, Foulds J. Porin channels in Escherichia coli: studies with beta-lactams in intact cells. J Bacteriol. 1983;153:232–240. doi: 10.1128/jb.153.1.232-240.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nestorovich EM, Danelon C, Winterhalter M, Bezrukov SM. Designed to penetrate: time-resolved interaction of single antibiotic molecules with bacterial pores. Proc Natl Acad Sci U S A. 2002;99:9789–9794. doi: 10.1073/pnas.152206799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Danelon C, Nestorovich EM, Winterhalter M, Ceccarelli M, Bezrukov SM. Interaction of Zwitterionic Penicillins with the OmpF Channel Facilitates Their Translocation. Biophys J. 2006;90:1617–1627. doi: 10.1529/biophysj.105.075192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoshimura F, Nikaido H. Diffusion of beta-lactam antibiotics through the porin channels of Escherichia coli K-12. Antimicrob Agents Chemother. 1985;27:84–92. doi: 10.1128/aac.27.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vidal S, Bredin J, Pagès JM, Barbe J. Beta-lactam screening by specific residues of the OmpF eyelet. J Med Chem. 2005;48:1395–1400. doi: 10.1021/jm049652e. [DOI] [PubMed] [Google Scholar]
- 80.Benson SA, Occi JL, Sampson BA. Mutations that alter the pore function of the OmpF porin of Escherichia coli K12. J Mol Biol. 1988;203:961–970. doi: 10.1016/0022-2836(88)90121-0. [DOI] [PubMed] [Google Scholar]
- 81.Misra R, Benson SA. Isolation and characterization of OmpC porin mutants with altered pore properties. J Bacteriol. 1988;170:528–533. doi: 10.1128/jb.170.2.528-533.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Misra R, Benson SA. Genetic identification of the pore domain of the OmpC porin of Escherichia coli K-12. J Bacteriol. 1988;170:3611–3617. doi: 10.1128/jb.170.8.3611-3617.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Simonet V, Malléa M, Pagès JM. Substitutions in the eyelet region disrupt cefepime diffusion through the Escherichia coli OmpF channel. Antimicrob Agents Chemother. 2000;44:311–315. doi: 10.1128/aac.44.2.311-315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jeanteur D, Schirmer T, Fourel D, Simonet V, Rummel G, Widmer C, Rosenbusch JP, Pattus F, Pages JM. Structural and functional alterations of a colicin-resistant mutant of OmpF porin from Escherichia coli. Proc Natl Acad Sci U S A. 1994;91:10675–10679. doi: 10.1073/pnas.91.22.10675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lou KL, Saint N, Prilipov A, Rummel G, Benson SA, Rosenbusch JP, Schirmer T. Structural and functional characterization of OmpF porin mutants selected for larger pore size. I. Crystallographic analysis. J Biol Chem. 1996;271:20669–20675. [PubMed] [Google Scholar]
- 86.Trias J, Dufresne J, Levesque RC, Nikaido H. Decreased outer membrane permeability in imipenem-resistant mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1989;33:1202–1206. doi: 10.1128/aac.33.8.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Trias J, Nikaido H. Outer membrane protein D2 catalyzes facilitated diffusion of carbapenems and penems through the outer membrane of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1990;34:52–57. doi: 10.1128/aac.34.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang H, Hancock RE. The role of specific surface loop regions in determining the function of the imipenem-specific pore protein OprD of Pseudomonas aeruginosa. J Bacteriol. 1996;178:3085–3090. doi: 10.1128/jb.178.11.3085-3090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Trias J, Nikaido H. Protein D2 channel of the Pseudomonas aeruginosa outer membrane has a binding site for basic amino acids and peptides. J Biol Chem. 1990;265:15680–15684. [PubMed] [Google Scholar]
- 90.Chapman JS, Georgopapadakou NH. Routes of quinolone permeation in Escherichia coli. Antimicrob Agents Chemother. 1988;32:438–442. doi: 10.1128/aac.32.4.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hirai K, Aoyama H, Irikura T, Iyobe S, Mitsuhashi S. Differences in susceptibility to quinolones of outer membrane mutants of Salmonella typhimurium and Escherichia coli. Antimicrob Agents Chemother. 1986;29:535–538. doi: 10.1128/aac.29.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cohen SP, Hooper DC, Wolfson JS, Souza KS, McMurry LM, Levy SB. Endogenous active efflux of norfloxacin in susceptible Escherichia coli. Antimicrob Agents Chemother. 1988;32:1187–1191. doi: 10.1128/aac.32.8.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chevalier J, Mallea M, Pages JM. Comparative aspects of the diffusion of norfloxacin, cefepime and spermine through the F porin channel of Enterobacter cloacae. Biochem J 348 Pt. 2000;1:223–227. [PMC free article] [PubMed] [Google Scholar]
- 94.Nikaido H, Thanassi DG. Penetration of lipophilic agents with multiple protonation sites into bacterial cells: tetracyclines and fluoroquinolones as examples. Antimicrob Agents Chemother. 1993;37:1393–1399. doi: 10.1128/aac.37.7.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thanassi DG, Suh GS, Nikaido H. Role of outer membrane barrier in efflux-mediated tetracycline resistance of Escherichia coli. J Bacteriol. 1995;177:998–1007. doi: 10.1128/jb.177.4.998-1007.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cohen SP, McMurry LM, Levy SB. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J Bacteriol. 1988;170:5416–5422. doi: 10.1128/jb.170.12.5416-5422.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Achouak W, Heulin T, Pages JM. Multiple facets of bacterial porins. FEMS Microbiol Lett. 2001;199:1–7. doi: 10.1111/j.1574-6968.2001.tb10642.x. [DOI] [PubMed] [Google Scholar]
- 98.Poole K. Outer membranes and efflux: the path to multidrug resistance in Gram-negative bacteria. Curr Pharm Biotechnol. 2002;3:77–98. doi: 10.2174/1389201023378454. [DOI] [PubMed] [Google Scholar]
- 99.Poole K. Resistance to beta-lactam antibiotics. Cell Mol Life Sci. 2004;61:2200–2223. doi: 10.1007/s00018-004-4060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mortimer PG, Piddock LJ. The accumulation of five antibacterial agents in porin-deficient mutants of Escherichia coli. J Antimicrob Chemother. 1993;32:195–213. doi: 10.1093/jac/32.2.195. [DOI] [PubMed] [Google Scholar]
- 101.Charrel RN, Pages JM, De Micco P, Mallea M. Prevalence of outer membrane porin alteration in beta-lactam-antibiotic-resistant Enterobacter aerogenes. Antimicrob Agents Chemother. 1996;40:2854–2858. doi: 10.1128/aac.40.12.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.George AM, Levy SB. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J Bacteriol. 1983;155:531–540. doi: 10.1128/jb.155.2.531-540.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.George AM, Levy SB. Gene in the major cotransduction gap of the Escherichia coli K-12 linkage map required for the expression of chromosomal resistance to tetracycline and other antibiotics. J Bacteriol. 1983;155:541–548. doi: 10.1128/jb.155.2.541-548.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Viveiros M, Dupont M, Rodrigues L, Couto I, Davin-Regli A, Martins M, Pages JM, Amaral L. Antibiotic stress, genetic response and altered permeability of E. coli. PLoS ONE. 2007;2:e365. doi: 10.1371/journal.pone.0000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dowd SE, Killinger-Mann K, Brashears M, Fralick J. Evaluation of gene expression in a single antibiotic exposure-derived isolate of Salmonella enterica typhimurium 14028 possessing resistance to multiple antibiotics. Foodborne Pathog Dis. 2008;5:205–221. doi: 10.1089/fpd.2007.0062. [DOI] [PubMed] [Google Scholar]
- 106.Domenech-Sanchez A, Hernandez-Alles S, Martinez-Martinez L, Benedi VJ, Alberti S. Identification and characterization of a new porin gene of Klebsiella pneumoniae: its role in beta-lactam antibiotic resistance. J Bacteriol. 1999;181:2726–2732. doi: 10.1128/jb.181.9.2726-2732.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.De E, Basle A, Jaquinod M, Saint N, Mallea M, Molle G, Pages JM. A new mechanism of antibiotic resistance in Enterobacteriaceae induced by a structural modification of the major porin. Mol Microbiol. 2001;41:189–198. doi: 10.1046/j.1365-2958.2001.02501.x. [DOI] [PubMed] [Google Scholar]
- 108.Thiolas A, Bornet C, Davin-Regli A, Pages JM, Bollet C. Resistance to imipenem, cefepime, and cefpirome associated with mutation in Omp36 osmoporin of Enterobacter aerogenes. Biochem Biophys Res Commun. 2004;317:851–856. doi: 10.1016/j.bbrc.2004.03.130. [DOI] [PubMed] [Google Scholar]
- 109.Gill MJ, Simjee S, Al-Hattawi K, Robertson BD, Easmon CS, Ison CA. Gonococcal resistance to beta-lactams and tetracycline involves mutation in loop 3 of the porin encoded at the penB locus. Antimicrob Agents Chemother. 1998;42:2799–2803. doi: 10.1128/aac.42.11.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Livermore DM. Of Pseudomonas, porins, pumps and carbapenems. J Antimicrob Chemother. 2001;47:247–250. doi: 10.1093/jac/47.3.247. [DOI] [PubMed] [Google Scholar]
- 111.Bratu S, Landman D, Gupta J, Quale J. Role of AmpD, OprF and penicillin-binding proteins in beta-lactam resistance in clinical isolates of Pseudomonas aeruginosa. J Med Microbiol. 2007;56:809–814. doi: 10.1099/jmm.0.47019-0. [DOI] [PubMed] [Google Scholar]