Abstract
IEX-1 (Immediate Early response gene X-1) is a stress-inducible gene. It suppresses production of reactive oxygen species (ROS) and protects cells from apoptosis induced by a wide range of stimuli, but the underlying mechanism is not known. This study reveals that IEX-1 targets the mitochondrial F1Fo-ATPase Inhibitor (IF1) for degradation, resulting in acceleration of ATP hydrolysis, concomitant with reduction in ROS production. A prominent role for IF1 degradation in the function of IEX-1 was corroborated by siRNA-mediated gene silencing of IF1 that recapitulated the effects of IEX-1 on ATP hydrolysis and ROS production. Moreover, progressive C-terminal truncation studies demonstrated that IEX-1 interacted with the C terminus of IF1 and the interaction might render IF1 prone to degradation by an as yet unidentified mitochondrial protease. In support of a physiological importance of IEX-1 in the modulation of IF1 expression, gene-targeted deletion of IEX-1 stabilized IF1 and reduced mitochondrial F1Fo-ATPase activity in vivo. The altered activity of the F1Fo enzyme may account for a metabolic switch from oxidative phosphorylation toward glycolysis in IEX-1 deficient cells. Thus, IEX-1 deficient cells were more susceptible to glucose deprivation than wild type counterparts and displayed increased glucose uptake and lactate production in hypoxic conditions. The cells were also relatively refractory to oligomycin-mediated inhibition of ATP production. The studies offer novel insights into the primary role of IEX-1 in regulating a balance between energy provision and ROS production.
Keywords: IEX-1, F1Fo-ATPase inhibitor, mitochondrial ROS homeostasis, protein degradation
Mitochondria are the site for the majority of cellular ATP synthesis, the main source for intracellular reactive oxygen species (ROS) production, and the key machinery for regulating cell death. These three biologic events are tied to the respiratory chain in the inner mitochondrial membrane. The electron transport chain consists of complex I, II, III, and IV, as well as an ATP synthase. The ATP synthase is comprised of the membrane-spanning Fo and the soluble F1 sectors, both of which are multiple protein complex.1,2 The F1Fo enzyme catalyzes the synthesis of ATP or oxidative phosphorylation utilizing the energy produced by the trans-membrane electrochemical proton gradient along the respiratory chain. A high membrane potential (Δψm) in the inner mitochondrial membrane as a consequence of cellular stress would slow the proton flux, giving rise to ROS formation.3 Depending on the amounts of ROS generated, an intermediate level of ROS can amplify oxidant-sensitive signaling to promote cell proliferation, whereas too much of ROS production causes oxidative stress or mitochondrial membrane disruption that can trigger either apoptosis or necrosis.4
Under conditions of oxygen deprivation, such as what occurs during ischemia or in the presence of an uncoupler of oxidative phosphorylation, the F1Fo-ATP synthase can switch from an ATP synthase to an ATPase, making it hydrolyze ATP produced in the cytosol by glycolysis.5 To preserve ATP, a natural inhibitor protein named F1Fo-ATPase Inhibitor (IF1) binds to the β subunit of the F1 complex through its inhibitory domain and inhibits ATP hydrolysis.6–8 Binding of IF1 to the F1 complex occurs only in a pH below neutrality and in de-energized inner mitochondrial membrane in response to ischemia.9 Under aerobic conditions, whether IF1 has a role or what role the IF1 plays in the control of the mitochondrial F1Fo-ATP synthase/ATPase activity remains poorly understood. Campanella et al.10 recently showed that IF1 overexpression increased the activity of F1Fo-ATP synthase by facilitation of dimerization of F1Fo-ATP synthase in aerobic cell culture. In contrast, decreased F1Fo-ATP synthase/ATPase activity and enhanced glycolysis were linked to increased IF1 expression in some tumors in early investigations.11,12 Lack of IF1 induces non-thyroidal hyper-metabolic syndrome in humans known as Luft’s disease, because of the altered morphology of mitochondrial cristae and uncoupling of mitochondrial respiration.13 These observations implicate that IF1 takes part in the regulation of mitochondrial F1Fo-ATP synthase/ATPase activity under both aerobic and anaerobic respiration, presumably through different mechanisms.
IEX-1 (immediate early responsive gene X-1) is known to protect cells from undergoing apoptosis induced by different stimuli, but to promote this process in response to deprivation of nutrition.14–16 Apart from the dual effects on apoptosis, IEX-1 induced by mechanical stress or aortic banding is associated with suppression in proliferation of vascular smooth muscle cells and repression of cardiomyocyte hypertrophy in several experimental models.17,18 The broad effects of IEX-1 hint that IEX-1 is involved in a widely conserved cell-signaling pathway. In accordance to this, we showed that overexpression of IEX-1 reduced mitochondrial ROS production both under basal conditions as well as immediately after exposure to apoptotic stimuli.19 To study the mechanism underlying IEX-1-mediated modulation of intracellular ROS production, a series of molecular and biochemical studies were carried out, revealing that IEX-1 targets IF1 for degradation in a manner dependent on its C terminus in this study. IEX-1-mediated modulation of IF1 degradation prevents an acute rise in mitochondrial membrane potential Δψm and ROS production triggered by apoptotic stimuli. In contrast, the absence of IEX-1 stabilized IF1, reduced F1Fo-ATPase activity, and stimulated a metabolic switch from oxidative phosphorylation toward glycolysis. These findings support a role for IF1 in the regulation of F1Fo-ATP synthase/ATPase activity in aerobic conditions and importance of IEX-1 in the control of IF1 degradation.
Results
IEX-1 interacts with IF1 and reduces its expression
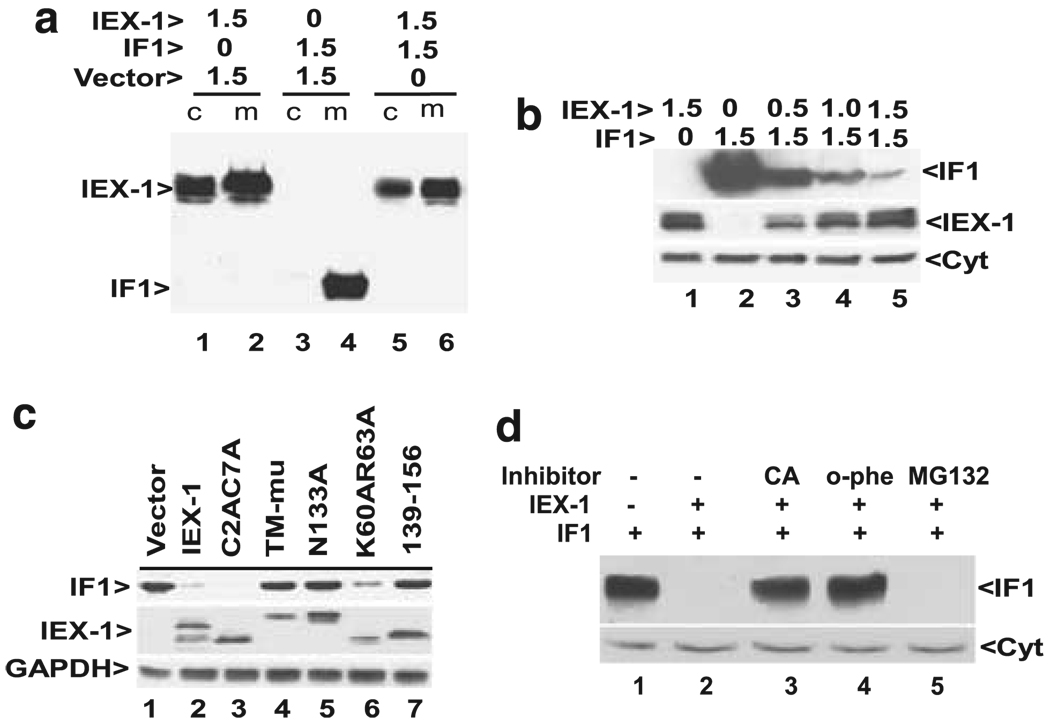
To investigate the mechanism underlying IEX-1-mediated suppression of intracellular ROS formation, we employed a yeast two-hybrid assay with the full length IEX-1 polypeptide as bait to identify IEX-1-interacting proteins. Having screened 3×106 transformant cells, we obtained five positive clones that encode either a partial or full length sequence of the mitochondrial F1F0-ATPase inhibitor IF16. The association of IEX-1 with IF1 is consistent with mitochondrial localization of IEX-1 and its role in regulating intracellular ROS homeostasis.19 Unexpectedly, we were unable to detect IF1 in either cytosol or mitochondrion-enriched fractions in subsequent confirmation studies, if IF1 was co-transfected with IEX-1 (Figure 1a, lanes 5 and 6). In the control where a plasmid harboring the IF1 cDNA was transfected along with a control vector, we observed a high level of IF1 expression exclusively in mitochondria (Figure 1a, lane 4), in agreement with a recent investigation.10 These results suggest that IEX-1 downregulates IF1 expression, rather than blocking its entry into mitochondria. Because IF1 was seen only in mitochondria, mitochondrion-enriched fraction was used in a subsequent investigation to assess IF1 expression unless otherwise indicated. To demonstrate a dose-dependent downregulation of IF1 by IEX-1, a fixed amount of IF1 was co-expressed with increasing amounts of IEX-1. We found that IF1 expression diminished proportionally with increasing amounts of IEX-1 (Figure 1b). Notably, in spite of a drastic decrease in IF1 expression after cotransfection with IEX-1, the co-transfection affected only slightly, if there was any, IEX-1 expression that was readily seen in both cytosol and mitochondrion-enriched fractions, regardless of whether IEX-1 was transfected alone or along with IF1 (Figure 1a, lanes 1 and 2 versus 5 and 6).
Figure 1.
IEX-1-targeted degradation of IF1. (a) Reduction of IF1 expression by IEX-1. Equal amounts (1.5 µg) of IEX-1 construct and control vector or IF1 were co-transfected into CHO cells. Cytosol (c) and mitochondrion-enriched (m) fractions prepared from the resultant transfected cells were subjected into western blot analysis using both anti-IEX-1 Ab (the upper band) and anti-IF1 Ab (the lower band). One representative result of at least six experiments performed. (b) Decreased IF1 expression by IEX-1 in a dose-dependent fashion. A fixed amount of IF1 was transfected, along with increasing levels of IEX-1 for 48 h, followed by western blotting analysis of mitochondrion-enriched fractions using Ab specific for IF1, IEX-1 or cytochrome c (Cyt). Similar results were obtained from two separate experiments. (c) Effects of IEX-1 mutagenesis on IF1 expression. IF1 and IEX-1-Myc or its variants at a 1:1 ratio were cotransfected into 293T cells and analyzed by Ab directed at IF1, IEX-1 or GAPDH. One representative result of two experiments performed. (d) Inhibition of IF1 degradation by o-phenanthroline (o-phe). Equal amounts of IF1 and IEX-1 constructs were co-transfected into 293T cells for 48 h with an addition to the medium in the final 16 h of 10 µM carboxyatractyloside (CA), 1 µM o-phenanthroline (o-phe), or 10 µM MG-132. IF1 was analyzed as (b). Similar results were obtained from three independent studies
Our previous mutagenesis studies showed the requirement of distinct sequences of IEX-1 for its anti- and pro-apoptotic activities.19 To determine whether IEX-1-mediated downregulation of IF1 expression corrected with the anti- or pro-apoptotic activity of IEX-1, various mutants of IEX-1 were co-transfected with IF1 at a 1:1 ratio followed by the evaluation of IF1 expression. It was found that substitutions of three hydrophobic residues with hydrophilic ones in the transmembrane-like sequence (TM-mutant) failed to significantly reduce the IF1 expression (Figure 1c, lane 4). This also held the truth for IEX-1 with mutation at an N-linked glycosylation site (N133A, lane 5) or after truncation of the C terminus (139–156, lane 7). Remarkably, these same mutants were also unable to inhibit intracellular ROS production as previously shown.19 In contrast, mutations of two cysteines at positions 2 and 7 (C2AC7A) or nuclear localization sequence (K60AR63A), both of which retained the wild-type capacity of reducing ROS production, had little impact on IF1 expression (Figure 1c, lanes 3 and 6).19 Thus, IEX-1-mediated downregulation of IF1 is associated with its ability to suppress ROS production and thus its anti-apoptotic activity.
Downregulation of IF1 expression by IEX-1 through protein degradation
To address whether downregulation of IF1 by IEX-1 occurred at the level of transcription, IF1 mRNA was analyzed by RT-PCR using primers specific for the transfected IF1 in cells with or without co-transfection of IEX-1 and IF1. We found no alteration in the amount of IF1 mRNA expression, whether or not IEX-1 was co-transfected (data not shown), ruling out the possibility that IEX-1 regulates IF1 transcription directly. Importantly, IEX-1-mediated downregulation of IF1 was completely blocked by the divalent metal ion chelator o-phenanthroline that inhibits mitochondrial ATP-dependent proteases and prevents IF1 degradation in yeast (Figure 1d, lane 4).20 To corroborate the involvement of an ATP-dependent mitochondrial protease, a known inhibitor of the adenine nucleotide translocator (ANT), carboxyatractyloside, was also tested. Carboxyatractyloside stabilizes the nucleotide binding site of ANT on the cytoplasmic side of the inner mitochondrial membrane and blocks the exchange of matrix ATP and cytoplasmic ADP.21 It is expected that a blockade of ANT activity would specifically reduce ATP levels inside mitochondria, decreasing the activity of ATP-dependent mitochondrial proteases. When carboxyatractyloside was added to the culture, the inhibitor blocked IF1 degradation (Figure 1d, lane 3), in agreement with the participation of an ATP-dependent protease in IF1 degradation in mitochondria. IEX-1-targeted degradation of IF1 was not inhibited by proteasome inhibitors MG-132 (Figure 1d, lane 5), and aclacinomycin A or serine protease inhibitor, 3,4–dichloroisocoumarin (data not shown).
Functional and physical association of endogenous IEX-1 and IF1
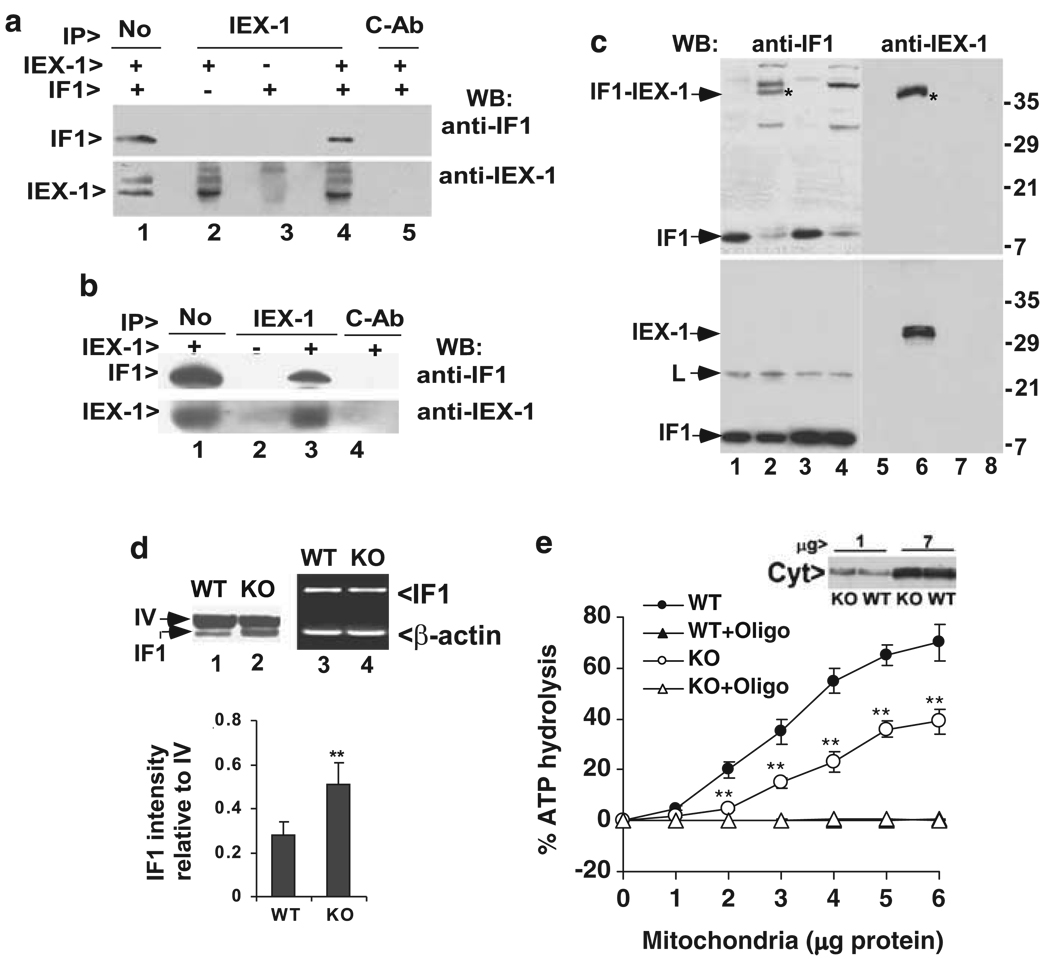
To address whether IEX-1 targeted IF1 for degradation by direct binding or by an indirect mechanism, immunoprecipitation (IP) using anti-IEX-1 antibody (Ab) followed by western blot with anti-IF1 Ab was carried out in cells transfected with both IEX-1 and IF1 and incubated in the presence of the protease inhibitor o-phenanthroline for the last 16 h. As can be seen in Figure 2a, IF1 was co-precipitated with IEX-1 by anti-IEX-1 Ab (lane 4), but not by control Ab (lane 5). The interaction did not take place when IF1 or IEX-1 construct was transfected alone (lanes 2 and 3). To address an interaction between endogenous IEX-1 and IF1, aortic tissues that express high levels of IEX-1 were dissected from wild-type (WT) and IEX-1 knockout (KO) mice, sliced into small pieces, and homogenized. Mitochondria were enriched from the resultant homogenates and subjected to immunoprecipitation. Once again, anti-IEX-1 Ab but not control Ab (lane 4) pulled down IF1 from aortic mitochondrial homogenates prepared from WT mice (lane 3), but not from IEX-1-deficient mice (lane 2) (Figure 2b). However, in a reverse study where anti-IF1 Ab was used in immunoprecipitation, it was unable to pull down IEX-1. Probably, binding of anti-IF1 Ab with IF1 interferes with IF1–IEX-1 interaction either directly or sterically. This possibility was argued for by progressive C-terminal truncation studies showing anti-IF1 Ab recognition of and IEX-1 interaction with IF1 at a similar region of IF1 (Figure 3).
Figure 2.
Interaction of IF1 and IEX-1. (a) Association of IF1 with IEX-1 in mitochondria after overexpression. Mitochondria purified from CHO cells transfected with either IF1 alone or together with IEX-1 were lysed and immunoprecipitated with anti-IEX-1 Ab (lanes 2–4) or control IgG affinity-purified from pre-immune rabbit serum (c-Ab, lane 5), followed by immunoblotting with anti-IF1 Ab (top) or anti-IEX-1 Ab (bottom). Mitochondrial lysate equivalent to 10% of the starting materials for immunoprecipitation (IP) was shown in (lane 1). One representative result of three experiments performed. (b) Interaction of endogenous IEX-1 and IF1. Mitochondria were purified from aortic tissue of wild type (+) or IEX-1-deficient (−) mice and subjected to immunoprecipitation analysis as (a). One representative result of two experiments performed. (c) Formation of a protein complex of endogenous IF1 and IEX-1 with a cross-linker. Mitochondria purified from WT (lanes 1, 2, 5 and 6) or IEX-1 KO mice (lanes 3, 4, 7 and 8) were cross-linked with DSP prior to immunoprecipitation with anti-IF1 Ab. The immunoprecipitates were separated by non-reducing SDS-PAGE (the upper panel) or reducing SDS-PAGE (the lower panel) followed by immunoblotting with Ab specific for IF1 (lanes 1–4) or IEX-1 (lanes 5–8). Molecular mass is indicated on the right (kDa); an asterisk (*) indicates IF1-IEX-1 complex; and L is a mouse IgG light chain. One representative result of three experiments is shown. (d) IF1 expression in the absence of IEX-1. Mitochondria of aortic tissues dissected from indicated mice were purified as above and analyzed by immunoblotting with anti-IF1 Ab (lanes 1&2). One of the subunits of complex IV (IV) of the mitochondrial respiration chain is shown for protein loading control. IF1 intensity relative to the subunit of complex IV was analyzed by densitometry in Adobe Photoshop and expressed as means ± S.D. (n=5) in the lower panel. IF1 mRNA in aortic tissues from indicated mice was analyzed by RT-PCR (lanes 3 and 4). β-actin was run in parallel with IF1 in RT-PCR assay for controls. One representative result of two experiments performed. (e) Reduced ATPase activity in the absence of IEX-1. Mitochondria purified from aortic tissues of indicated mice were assayed for ATPase activity in the presence or absence of oligomycin (oligo) at 1 µg/mg mitochondrial protein. Mitochondrial quantity was determined by similar amount of cytochrome c (Cyt) presented in these two samples (inset) in which 1 or 7 µg of indicated mitochondria proteins were loaded in each lane. 32Pi release from [γ-32P]ATP was quantified on a scintillation counter and expressed as percentages of ATP hydrolysis relative to input [γ-32P]ATP as detailed in Materials and Methods. The data are the means ± S.D. of cumulative results of three independent experiments each in duplicate. **P<0.01, in the presence versus absence of IEX-1
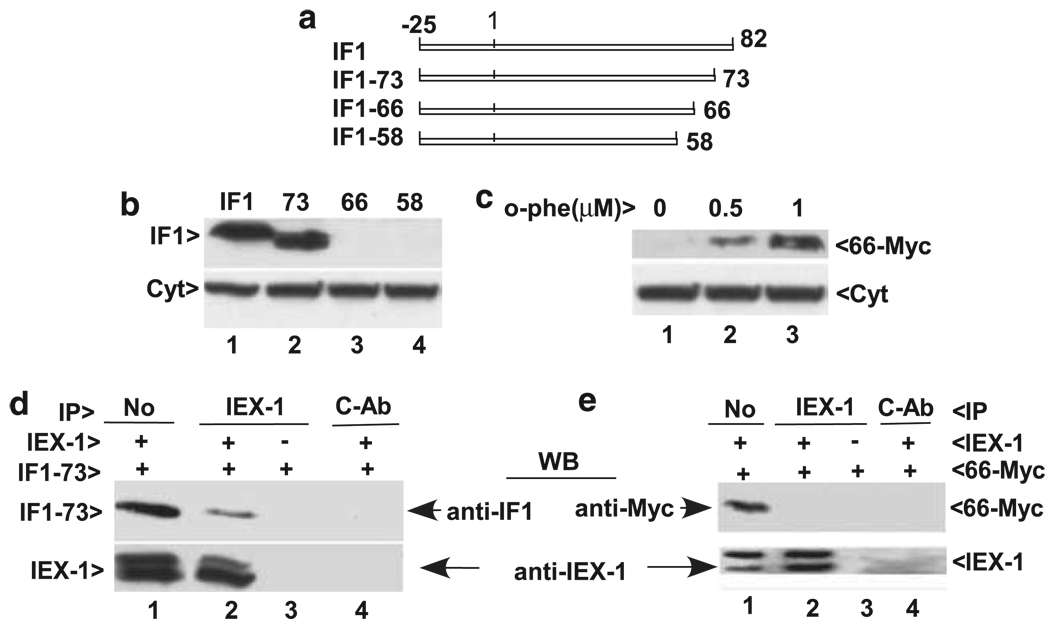
Figure 3.
The C terminus of IF1 is required for its interaction with IEX-1. (a) A schematic diagram of progressive C-terminal truncation of IF1. The numbers indicate amino residue position relative to mature protein and the mitochondrial signal peptide is located from –25 to 1. (b and c) The C terminus is required for IF1 stabilization. Cells transfected with various C-terminally deleted IF1 mutants were cultured in the presence of 1 µM o-phenanthroline overnight followed by immunoblotting with anti-IF1 Ab (the upper) or anti-cytochrome c (Cyt, the lower panel) in (b). Cells transfected by the IF1-66 mutant with Myc-tagged to its C terminus (66-Myc) were cultured in the presence or absence of varying concentrations of o-phenanthroline overnight followed by immunoblotting with anti-Myc Ab (the upper) or anti-cytochrome c (Cyt, the lower panel) in (c). (d and e) IEX-1 interacts with IF1-73 but not IF1-66. CHO cells were transfected with IEX-1, along with IF1-73 (d) or IF1-66-Myc (e) and cultured in the presence of 1 µM o-phenanthroline overnight. Immunoprecipitation (IP) was carried out with anti-IEX-1 Ab followed by immunoblotting with anti-IF1 (d) or anti-Myc Ab (e). Mitochondrial lysate equivalent to 10% of the starting materials for IP was loaded in lane 1 without immunoprecipitation (No). Similar results were obtained from three independent experiments
To circumvent Ab interference with IF1–IEX-1 interaction, protein crosslinking by DSP (dithiobis-succinimidyl-propionate) was carried out in mitochondria purified from IEX-1-deficient and WT mouse aortas before immunoprecipitation with anti-IF1 Ab. DSP is a cell permeable cross-linker that forms highly specific cross-linkages between lysines that are at 12 Å proximity. After cross-linking, IEX-1 could be co-precipitated with IF1 by anti-IF1 Ab. As can be seen in the upper panel of Figure 2c, a protein complex, marked in an asterisk, of approximately 40 kDa in size was recognized by both anti-IF1 Ab (lane 2) and anti-IEX-1 Ab (lane 6) in cross-linking samples prepared from WT mice. The protein complex was not present in non-cross-linked samples (Figure 2c, lanes 1 and 5) or in the absence of IEX-1, irrespective of cross-linking (lanes 3 and 4 and 7 and 8). IF1-IEX-1 complex was no longer seen in a reducing SDS-PAGE, and disappearance of IF1-IEX-1 complex was accompanied with increasing levels of non-cross-linked IF1 (Figure 2c, the lower left panel). We also observed IEX-1 with a correct molecular mass approximately 31 kDa under a reducing condition, in agreement with dissociation of IEX-1 from IF1–IEX-1 complex (Figure 2c, the lower right panel). In addition, when IF1 was evaluated in mitochondria isolated from aortic tissues, the level of IF1 expression was increased by almost one-fold in the absence compared with presence of IEX-1 as revealed by densitometric analysis (Figure 2d, lanes 1 and 2, and the lower panel). In sharp contrast to an increase in the level of IF1 protein, RT-PCR analysis gave rise to a similar level of IF1 mRNA in aortic tissues, whether or not IEX-1 was presented (Figure 2d, lanes 3 and 4). These results confirm stabilization of IF1 protein expression in vivo in the absence of IEX-1. Functionally, null mutation of IEX-1 significantly reduced mitochondrial ATPase activity that was measured by the ability of purified mitochondria to hydrolyze γ-32P-ATP in the absence versus the presence of oligomycin as detailed in Materials and Methods. A 20–30% decrease was observed in mitochondria purified from IEX-1 KO mice compared with those from WT mice (Figure 2e). The measured ATPase activity was specific for mitochondrial F1Fo-ATPase, as it was completely abolished by oligomycin, a specific inhibitor for the mitochondrial F1Fo-ATPase. Moreover, similar amounts of cytochrome c in the two samples excluded the possibility that the difference in ATPase activity in the absence or presence of IEX-1 was attributable to the mitochondrial quantity (Figure 2e, inset). Taken together, we conclude that downregulation of IF1 expression by IEX-1 in vivo is caused by its direct association with the inhibitor.
Interaction of IEX-1 with the C terminus of IF1
Previous studies have shown that the C-terminal region of IF1 is not involved in its inhibitory action on the F1Fo-ATPase in yeast, but it is required for protecting the protein against degradation by mitochondrial proteases.20,22. Although the C-termini of two IF1 molecules interact with each other to form a dimer in uncoupled mitochondria as revealed by the protein crystal structural study of the soluble F1-IF1 complex,8 the C terminus of IF1 may be associated with several protective proteins in the inner mitochondrial membrane in coupled mitochondria.22–24 In support of this, removal of the C terminus of IF1 results in its degradation in yeast.20 IF1 degradation can be inhibited by culture of the cells in a medium supplemented with a mitochondrial protease inhibitor, o-phenanthroline.20 This inhibitor also blocked IF1 degradation in the presence of IEX-1 in mammalian cells (Figure 1d), suggesting a similar protease involved in IF1 degradation between yeast and mammalian cells. One possibility for IEX-1 targeting IF1 to degradation is that IEX-1 binds to the C terminus of IF1, resulting in its dissociation from protective proteins. To test this possibility, the C terminus of IF1 was progressively truncated through a PCR-based strategy (Figure 3a). Similar to what has been described in yeast,20 removal of nine amino residues (IF1-73) at the C terminus did not affect the stability of IF1 in the presence (Figure 3b, lane 2) or absence of the protease inhibitor o-phenanthroline (data not shown). Apparently, the last nine amino residues are dispensable in IF1 stability. However, truncation of 24 amino residues (IF1-58) led to its degradation even in the presence of the protease inhibitor o-phenanthroline (Figure 3b, lane 4), arguing for an indispensable role of the region spanning from 58 to 73 in IF1 stability in both mammalian and yeast cells.
Of note, IF1-66 was not seen by immunoblotting with anti-IF1 Ab in the presence of the o-phenanthroline inhibitor (Figure 3b, lane 3). But, when the mutant was tagged at the C terminus with Myc (IF1-66–Myc), anti-Myc Ab detected IF1-66 expression in the presence of o-phenanthroline in a dose-dependent manner, although the Ab was still unable to detect IF1-66-Myc in the absence of the inhibitor (Figure 3c), indicating inhibitor-dependent resistance to degradation, which was distinguished from IF1-73, a mutant that was refractory to degradation even in the absence of o-phenanthroline. The finding that IF1 Ab recognizes IF1-73 but not IF1-66 suggests that the region spanning from 66 to 73 contains an anti-IF1 Ab-recognizing epitope. Yet, loss of Ab recognizing sequence was not attributed to a failure of visualizing IF1-58 mutant, as IF1-58 mutant could not be seen even though it was tagged with Myc at its C terminus and cultured in the presence of o-phenanthroline (data not shown). As mentioned earlier, we were unable to pull down IEX-1 by anti-IF1 Ab without protein cross-linking. We suspected that anti-IF1 Ab might interfere with the interaction between IF1 and IEX-1, provided that the region from 66 to 73 was not only recognized by anti-IF1 Ab, but also required for IF1 interaction with IEX-1. To map this region for IF1–IEX-1 interaction, IEX-1 was co-transfected with IF1-73 or IF1-66-Myc in the presence of o-phenanthroline. Immunoprecipitation of resulting cell lysate with anti-IEX-1 Ab was followed by immunoblotting with anti-IF1 Ab (Figure 3d) or anti-Myc Ab (Figure 3e). No IF1-66-Myc was pulled down by anti-IEX-1 Ab (Figure 3e, lane 2). Under similar conditions, an association between IF1-73 and IEX-1 was evidenced (Figure 3d, lane 2). The data confirm that the region from 66 to 73 in IF1 is critical for interaction of IF1 with IEX-1.
IEX-1 affects mitochondrial activities in a manner similar to gene silencing of IF1
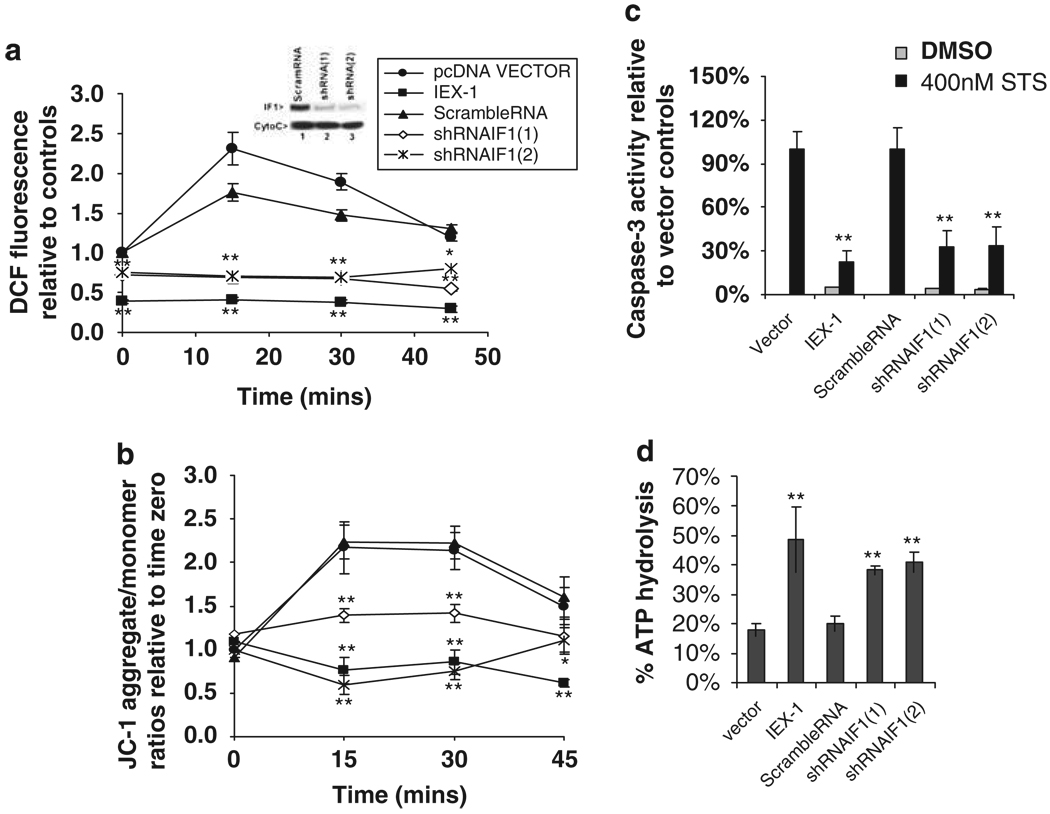
In theory, if IEX-1 targets IF1 for degradation, reduction of IF1 expression by gene silencing and IEX-1 overexpression both should have a similar impact on mitochondrial activities, such as ATP hydrolysis, mitochondrial membrane potential Δψm, and ROS production. Indeed, in cells infected with retrovirus carrying a cDNA encoding a short hairpin RNA specific for mouse IF1 (shRNAIF1) or transfected with an IEX-1 construct, ROS production, mitochondrial membrane potential Δψm, or caspase-3 activity all failed to increase following stimulation with staurosporin (STS) (Figure 4). Under similar conditions, we observed an acute increase in ROS production, mitochondrial membrane potential Δψm, and caspase-3 activity in cells expressing a scramble RNA or a control vector. The basal levels of ROS production also differed, with a significant decrease in cells expressing IEX-1, and to a lesser degree, in cells expressing shRNAIF1 (Figure 4a, time zero), presumably resulting from an increased ATPase activity in these cells (Figure 4d). In contrast, there was no significant difference at the basal level of mitochondrial membrane potential Δψm (Figure 4b, time zero). shRNAIF1(1) and shRNA(2) were targeting to different regions of IF1 mRNA and both downregulated IF1 by more than 85%, as verified by western blotting analysis (inset in Figure 4a). The ability of overexpressing IEX-1 or functional silence of IF1 to block an STS-induced increase in ROS production is in agreement with previous investigations showing that STS-induced apoptosis by acute increasing ROS production in mitochondria25 and overexpression of IEX-1 protected cells from STS-induced apoptosis.14,19 A shift of green fluorescence to orange/red fluorescence in cells bearing shRNAIF1 or overexpressing IEX-1 correlated with suppression of ROS production and caspase-3 activity in these cells following STS stimulation. The observations confirm that IEX-1 modulates mitochondrial membrane potential and ROS formation mainly through IF1 degradation in cells under stress. Moreover, a loss of IF1 inhibitory activity as manifested by increased ATP hydrolysis was demonstrated not only in cells bearing shRNAIF1, but also in cells expressing IEX-1, both of which resulted in a more than 50% increase in ATP hydrolysis as compared with that obtained in cells bearing a vector control or a scramble cDNA (Figure 4c). Similar effects of overexpression of IEX-1 and shRNAIF1 on ROS production were also observed in p65-deficient MEF cells (p65KO3T3) following 250 U/ml TNFα stimulation (data not shown), arguing that modulation of ROS production by IEX-1 through regulation of IF1 degradation occurred in cells stimulated with a physiological stimulus. The ability of IF1 gene silencing to recapitulate the effects of IEX-1 overexpression on ATP hydrolysis and ROS production argues strongly for a prominent role for IF1 degradation in IEX-1-mediated activity.
Figure 4.
Similar effects of IEX-1 overexpression and functional silencing of IF1 on mitochondrial functions. CHO cells were transfected with an IEX-1 construct or a control vector or infected with shRNAIF1-containing retrovirus or scramble RNA-containing control virus. IF1 expression in the cells was analyzed by immunoblotting using anti-IF1 Ab (inset in a). The cells were stimulated with 400 nM staurosporin (STS) for indicated times to assess intracellular ROS production (a) and mitochondrial membrane potential (b). Intracellular ROS levels were evaluated by measuring oxidation of DCF substrate. DFC fluorescence intensity was normalized to vector control at time zero. The data are cumulative results of two independent experiments with each in duplicate. Mitochondrial membrane potential was determined by mitochondrial membrane potential-dependent accumulation of fluorescence probe JC-1 with a Spectra MAX GEMINI microplate fluorometer using 485/530 nm and 535/590 nm as excitation/emission wavelength. The data are mean alterations ± S.D. of the red/green fluorescence ratios relative to vector controls at time zero and represent one of three independent experiments with each performed in triplicate. (c) Caspase-3 activity. CHO cells bearing indicated constructs were stimulated with STS for 6 h. Caspase-3 activation was measured by a caspase-3-specific fluorometric substrate either immediately (0 min) or 10 min after addition of the substrate by fluoroMax-3 spectrometer using 380/460 mm as excitation/emission. Caspase3 activity is expressed as percentage increases (means ± S.D.) in fluorescence intensity at 10 min relative to zero time point and normalized to vector controls (100%) for comparison. One representative result of three independent experiments performed is shown. (d). ATP hydrolysis. Mitochondria were purified from cells carrying an indicated construct made in a without STS treatment. ATPase activity of purified mitochondria equivalent to 5 µg protein was assayed in each sample as Figure 2e. The data are the means ± S.D. of cumulative results of three independent experiments each in duplicate. Statistic significance: *P<0.05 or **P<0.01, respectively relatively to vector controls
Increased glycolysis in the absence of IEX-1
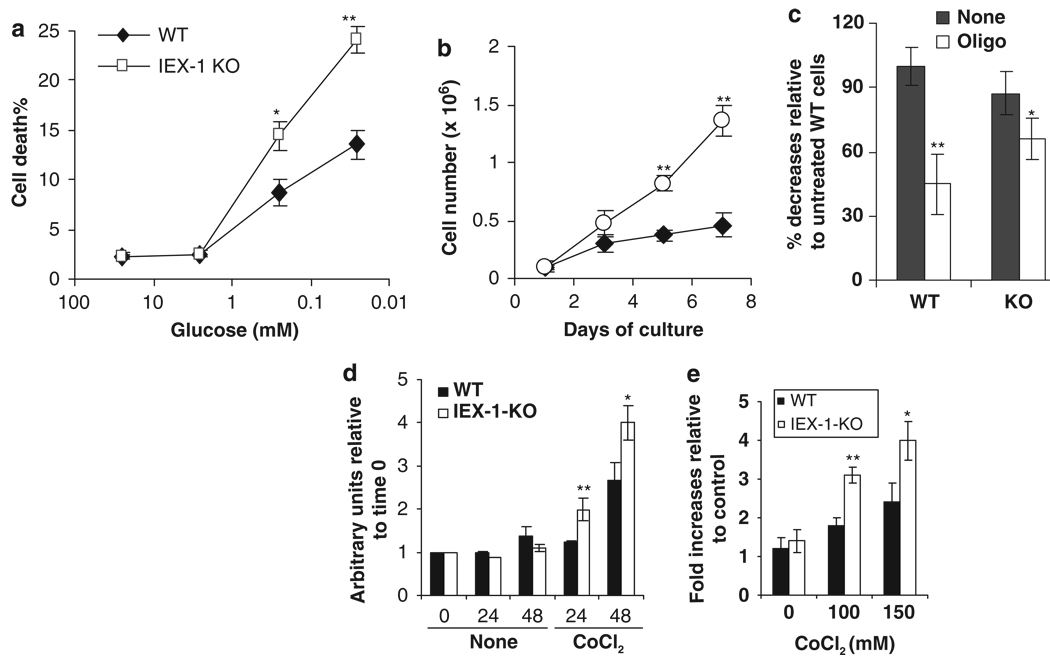
IF1 is highly expressed in cell lines or tumor cells when compared with primary tissues.10–12 Increasing expression of IF1 has been linked to decreased F1Fo-ATPase activity and a rise in the level of glycolysis in some tumors.11,12 To determine whether or not increased IF1 expression in IEX-1-deficient cells was associated with elevation in glycolysis, embryonic fibroblasts (MEFs) prepared from IEX-1 KO mice and WT mice were cultured in a medium containing decreasing concentrations of glucose. As can be seen in Figure 5a, IEX-1-deficient cells were more susceptible to glucose deprivation than WT cells, manifested by a significant increase in cell death in the absence compared with the presence of IEX-1 at glucose concentrations ≤0.25mM (P<0.001). The increased glucose dependence of cells lacking IEX-1 suggests relatively high levels of glycolysis in the cells. Consistent with this, IEX-1-deficient cells were grown much better than WT cells over a 1-week period of culturing in complete medium (Figure 5b), due to promotion of cell growth by glycolysis.26 Moreover, oligomycin that specifically blocks F1Fo-ATP synthase/ATPase activity reduced total intracellular ATP levels by more than 50% in WT cells, but only 20% in IEX-1-deficient cells after 1 h treatment (P<0.01, Figure 5c). The results suggest that contribution of the mitochondrial oxidative phosphorylation to the global cellular ATP is diminished in the absence of IEX-1. A smaller decline in the total ATP level in IEX-1 KO cells is unlikely due to a blockade of ATP hydrolysis with increased IF1 expression in the cells, because oligomycin inhibits F1Fo-ATPase activity strongly (Figure 2e). A relatively low level of total ATP in control IEX-1 KO cells compared with WT cells is consistent with diminished mitochondrial oxidative phosphorylation and increased glycolysis, as glycolysis is relatively insufficient in ATP production compared with oxidative phosphorylation. Moreover, while culture media from IEX-1-deficient MEFs and WT controls were assayed for lactate production, lack of IEX-1 enhanced lactate production significantly in the presence of CoCl2, a hypoxia-inducing compound (Figure 5d), concomitant with elevated glucose uptake by the cells (Figure 5e). The data argue that null mutation of IEX-1 makes cells prone to a metabolic switch from oxidative phosphorylation toward glycolysis.
Figure 5.
A metabolic shift in the absence of IEX-1. MEFs prepared from IEX-1 KO and WT control embryos were cultured at decreasing concentrations of glucose and viable and dead cells were counted 48 h later by trypan blue exclusion (a). Or, the cells were cultured in complete medium and viable cells were counted at indicated days (b). Intracellular ATP contents were measured in the macrophages cultured in the presence (oligo) or absence (None) of 0.5 µM oligomycin for 1 h (c). The data are expressed as mean percentages ± S.D. relative to control WT cells that are arbitrarily set as 100%. Lactate accumulation in the media was measured at indicated times in the presence or absence of 150 µM CoCl2 (d) and glucose uptake in the cells were analyzed after 48 h culture in the presence of varying concentrations of CoCl2 (e). Total number of viable cells or protein concentration from viable cells is used for normalization of lactate accumulation and glucose uptake, respectively. The data represent the means ± S.D. of triplicate wells of three (a, c, d) and two (b and e) independent experiments. Statistic significance: *P<0.05 or **P<0.01, respectively in the presence versus absence of IEX-1
Discussion
Our studies demonstrate a critical role for IEX-1 in the control of IF1 degradation in a cell under stress. Overexpression of IEX-1 diminished the level of IF1 expression and prevented stress-induced ROS production in favor of cell survival. In contrast, null mutation of IEX-1 resulted in the elevation of IF1 expression and reduction of mitochondrial F1Fo-ATPase activity. The IEX-1 deficiency-induced enhancement of IF1 expression may affect oxidative phosphorylation in mitochondria, as implicated by a metabolic switch from oxidative phosphorylation toward glycolysis in IEX-1-deficient cells. Although questions remain regarding the mechanism underlying IF1-mediated inhibition of F1Fo-ATPase activity in respirating mitochondria, a high level of IF1 expression is likely to reduce F1Fo-ATPase activity. The reduction, in conjunction with increased mitochondrial membrane potential induced by cellular stress would enhance ROS production due to proton retardation along the respiration chain in coupled mitochondria.3 The role of IEX-1 in the control of IF1 stability and mitochondrial ROS homeostasis in cells under stress is consistent with its varying, sometimes conflicting effects on cell survival and growth described in different cell types,27 because ROS are well known to play a dual role as both deleterious and beneficial species. The study identifies a novel mechanism by which IEX-1 takes part in a balance between bioenergic metabolism and ROS production in mitochondria in response to an external signal through controlling IF1 degradation, although other mechanisms may also exist.
IF1 is well known to block the activity of F1Fo-ATPase during oxygen deprivation by its binding with the β subunit of F1 complex.8 However, this model of IF1’s action hardly explains where IF1 is located and what a role IF1 plays in respirating mitochondria, in which the pH is relatively low on the cytoplasmic side of the inner mitochondrial membrane and relatively high inside the matrix of mitochondria.28 Utilizing in situ cross-linking of endogenous IF1, Lopex-Mediavialla et al.23 found IF1 almost exclusively in the inner mitochondrial membrane in respirating mitochondria, although the association was disputed by others.29 Binding of IF1 to the inner mitochondrial membrane doesn’t rely on pH or mitochondrial membrane potential, distinguished from its binding to the F1 complex in uncoupled mitochondria.22,23,30 Disruption of IF1 binding to the inner mitochondrial membrane by a specific Ab-enhanced F1Fo-ATPase activity,30 arguing that IF1 in the inner mitochondrial membrane in respirating mitochondria acts as an inhibitor for F1Fo-ATPase as well. Consistent with an inhibitory role for IF1 in F1Fo-ATPase activity in respirating mitochondria, a relatively low activity of F1Fo-ATPase and high level of glycolysis are associated with increasing levels of IF1 expression in some tumors.11,12 Moreover, increased F1Fo-ATPase activity in respirating mitochondria was also demonstrated in IF1-deficient patients who developed non-thyroidal hypermetabolic syndrome named Luft’s disease.13 Similarly, increased IF1 expression reduces the F1Fo-ATPase activity in the absence of IEX-1 in the current study. Recently, Campanella et al.10 showed that IF1 overexpression increased the formation of dimeric ATP synthase complexes and F1Fo-ATP synthase activity while suppressing its ATPase activity under physiological conditions, in agreement with dimerization of F1Fo-ATP synthase reducing the activity of F1Fo-ATPase as shown earlier.31 These observations raise an intriguing possibility that IF1 may suppress F1Fo-ATPase activity in respirating mitochondria by promoting formation of dimeric F1Fo, although further investigations are needed to conclude this.
Given the importance of F1Fo-ATP synthase activity in a variety of cellular functions, the activity of IF1 must be delicately regulated in respirating mitochondria, but we know little about the regulation to date. The level of IF1 expression appears to be varying considerably in different tissues and cell types, and the varying levels of IF1 are directly associated with its activity.10 Thus, one of the key mechanisms for modulating IF1 activity is to control its expression level. This study suggests that IEX-1 is involved in the control of the level of IF1 expression and thus F1Fo-ATPase activity. However, IEX-1 itself does not possess protease activity and cannot bind to ATP that is required for a chaperon protein. How IEX-1 targets IF1 for degradation is unknown at present. One of the possibilities would be that interaction of IEX-1 with the C terminus of IF1 displaces it from protective proteins, making it prone to degradation by an as yet unidentified protease.32 IF1 has been shown to interact with three proteins through its C terminus in the inner membrane in yeast.22 The observation that removal of the C terminus leads to IF1 degradation suggests that proteins in association with the C terminus of IF1 may protect it from degradation.20 Conceivably, dissociation of IF1 with its protective proteins by interaction of IEX-1 with IF1 at its C terminus may be the key to IEX-1-mediated regulation of IF1 degradation. IEX-1 may represent one of the mechanisms in the control of IF1 degradation in phosphorylating mitochondria, by which it links an external signaling directly to the F1Fo-ATPase activity.
Mitochondrial malfunction is associated with a variety of disorders with clinical outcomes largely dependent on the magnitude and tissue distribution of the impairment. IEX-1 is abundant in vascular tissues, which are metabolically active and contain a large number of mitochondria.33 An importance of IEX-1 in the regulation of mitochondrial F1Fo-ATPase through the control of IF1 degradation explains the development of hypertension and cardiac hyperplasia in IEX-1-deficient mice.34 A progressive decline in the activity of mitochondrial oxidative respiration leading to diminished capacity for ATP production but elevated ROS production is one of critical factors for hypertension development in both humans and animal models. This may hold the truth in IEX-1-deficient mice, as IEX-1 deficiency-induced hypertension was effectively prevented by treatment with anti-oxidant N-acetylcysteine (article in preparation). Likewise, a defect in mitochondrial function causing heart diseases and hypertension has been described in mice with respiratory chain-deficiency, gene-targeted deletion of the uncoupling protein 2 or selective deletion of the cardiac/skeletal muscle-specific adenine nucleotide translocator isoform1, or overexpression of UCP1 in the artery wall.35–38
Mitochondria have evolved multiple defense mechanisms to protect their functional integrity under various physiological and pathological conditions. These include anti-oxidants for scavenging ROS and preventing oxidation of mitochondrial membrane lipids, proteins and DNA, the Bcl-2 family for controlling mitochondrial membrane permeability, UCPs for limiting the rate of free radical generation, and so on. Among all these mechanisms, IEX-1-mediated control of IF1 degradation may represent one of the earliest measurements of protection against cellular stress.
Materials and Methods
Animals
IEX-1 KO and WT control mice on the mixed 129Sv/C57BL/6 background were generated by gene targeting (article in preparation).34 Both female and male mice were used and housed in conventional cages at the animal facilities of Massachusetts General Hospital in compliance with institutional guidelines.
Yeast two-hybrid assay
To investigate the mechanism underlying IEX-1-regulated ROS homeostasis, a GAL4-based yeast two-hybrid assay was performed to identify proteins that interacted with IEX-1 using the Matchmaker two-hybrid system 3 according to the manufacturer’s instructions (Clontech, Palo Alto, CA). Briefly, IEX-1 cDNA was in frame cloned into the DNA-binding domain of GAL4 in the shuttle vector pGBKT7 as bait. Yeast strain AH109 transformed with pGBKT7-IEX-1 was used to screen, by yeast mating, pretransformed HeLa matchmaker cDNA fusion library cloned into the GAL4 activation domain vector pGADT7 in yeast Y187. We also transformed pGBKT7-IEX-1 along with pGADT7 empty vector or pGBKT7-lam vector alone into AH109 as controls. More than 106 transformant cells were screened and the positive yeast clones were validated by co-transforming pGBKT7-IEX-1 and positive pGADT7 library plasmids each into AH109 yeast cells. The confirmed clones were sequenced and sequence homology was searched with the BLAST algorithm through the National Center for Biotechnology Information web site.
Co-immunoprecipitation assay
CHO cells were transfected with IEX-1, IF1 plasmid either alone or together at a 1:2 ratio and incubated for 48 h in complete medium with the addition of 1 µM O-phenanthroline, an inhibitor for mitochondrial proteases, in the final 16 h of incubation. The cells were harvested, washed, and lysed with a lysis buffer (Tris 50mM pH 7.5, 137mM NaCl, 0.5% NP-40, 10% glycerol, 1mM EDTA, 1mM orthovanadate, 20mM β-glycerophosphate, 20mM NaF, 1mM sodium pyrophosphate and 1% of a protease inhibitor cocktail).14 In some experiments, mitochondria were enriched by differential centrifugations as previously described19 and lysed with the lysis buffer. The resulting cell or mitochondrial lysate was pre-cleared with agarose beads for 1 h at 4 °C with gentle shaking followed by immunoprecipitation by a polyclonal anti-IEX-1 Ab and protein G-conjugated PLUS-agarose (Invitrogen) at 4 °C overnight. The polyclonal Ab was raised in rabbit against IEX-1 peptide sequence 51–75 and affinity-purified by the peptide-conjugated agarose beads as described.33 The pellets were washed extensively with lysis buffer and the binding proteins were eluted by SDS-PAGE protein loading buffer and subjected to immunoblotting analysis with Ab against IF1 (Invitrogen) or IEX-1. Ab-specific proteins were visualized with SuperSignal West Femto Maximum sensitivity for a substrate horseradish peroxidase (Pierce).
To evaluate interaction of endogenous IEX-1 with IF1 in aortic tissues, the entire length of aorta from the iliac bifurcation to the aortic arch was dissected free and cleaned off clotted blood, debris and fibroadipose tissue from 3-month-old WT and IEX-1 KO mice. The aorta was sliced into small pieces, and dounced–homogenized in a homogenizing buffer (H buffer; 210mM mannitol, 70mM sucrose, 1mM EDTA, 5mM HEPES and 1% protease inhibitor cocktail, pH 7.4). Mitochondrion-enriched fraction was subjected to immunoprecipitation as above. In cross-linking studies, mitochondria were purified by a continuous sucrose gradient from 0.8 to 1.5M in the homogenizing buffer, washed and suspended at 5mg protein per ml in 200 µl crosslinking buffer (pH 8.0, 250mM Sucrose, 2mM MgATP, 25mM HEPES, 0.4% n-dodecyl-β-D-maltoside, 1% of a protease inhibitor cocktail, and 1mM phenylmethylsulfonyl fluoride) and cross-linked with 2mM DSP (dithiobissuccinimidyl-propionate) on ice for 2 h. The reaction was stopped by the addition of two volumes of 3×lysis buffer. The cross-linked samples were sonicated and centrifuged at 16 000×g at 4 °C for 20 min at a microcentrifuge to remove insoluble material. The resultant supernatant was subjected to immunoprecipitation by anti-IF1 Ab. Ab-specific protein complexes were analyzed by reducing or non-reducing SDS-PAGE, followed by immunoblotting with anti-IEX-1 or anti-IF1 Ab as above.
RT-PCR
Total RNA was isolated from aortas of WT and IEX-1 KO mice or cells transfected with indicated plasmids using RNeasy Mini Kit according to the manufacturer’s instructions (Qiagen). Contaminated DNA was removed by digestion with DNase I. The resultant RNA was reverse transcribed using ThermoScript reverse transcriptase and random hexamer primers (Invitrogen). IF1 and β-actin genes were amplified by TaqDNA polymerase with the following primers: IF1 forward, 5′-GAGGCTTCGGTTCGGATCAGTCC-3′ and reverse 5′-CCGCCACTGTGCTGGATATCTGC-3′; and β-actin forward, 5′-CTTCTTTG CAGCTCCTTCGTTG-3′ and reverse, 5′-TCGTCCCAGTTGGTAACAATGC-3′. RNA samples that had not been reverse transcribed were run in parallel as negative controls. The PCR products were separated in 1% agarose and visualized by the Kodak GelLogic 200 imaging system (Kodak Inc.).
Retrovirus-transduced short hairpin RNA (shRNA)
Short hairpin 21-nucleotide base targeting sequences of IF1 were selected by RNAi designer (Invitrogen) and subcloned into pRS shRNA vector to obtain shRNAIF1(1), 5′-GCTGAAGAGGAACGATATTTC-3′; and shRNAIF1(2), 5′-GAAATTGAGCGC CATAAGCAGAA GATCAA-3′. The resultant constructs were transfected into human embryonic kidney 293 T cells (HEK 293 T), along with an equal amount of a retrovirus packaging plasmid pCL-Ampho. After 48 h culture, the supernatants containing replicate-deficient retrovirus were harvested and filtered through a 0.45 µm low protein-binding filter (Whatman, Clifton, NJ, USA). CHO cells were infected with shRNAIF1-containing virus or control virus in the presence of 8 µg/ml polybrene for 2–3 h and then incubated in a fresh medium for another 48 h before IF1 protein expression and function analyses.
Inhibition of IF1 degradation by various protease inhibitors
CHO cells were co-transfected with equal amounts of IEX-1 and IF1 plasmids for 36 h, after which different protease inhibitors were added and incubated overnight. Following inhibitors were used: proteasome inhibitors including aclacinomycin A (50 nM) and MG-132 (10 µM), a serine protease inhibitor 3,4–Dichloroisocoumarin (20 µM), an inhibitor for the adenine nucleotide translocator (ANT) named carboxyatractyloside (10 µM, Sigma), and a known mitochondrial protease inhibitor, O-phenanthroline (1 µM, Sigma). Cells were harvested and mitochondrion-enriched fractions were prepared and lysed as described. IF1 expression levels in mitochondrion-enriched fractions were evaluated by immunoblotting with anti-IF1 Ab.
Determination of ATPase activity
Mitochondria were purified from CHO cells transfected with different plasmids or aortic tissues as described. Mitochondria equivalent to indicated amounts of proteins were suspended in 20 µl of cold ATPase reaction buffer (25mM HEPES pH 7.4, with 25mM KCL, 3mM MgCl2, 5mM potassium cyanide, 1mM ATP, and with or without oligomycin at 1 µg/mg mitochondrial proteins).39 The reaction was started at 30 °C after addition of 40 µM [γ-32P]ATP, allowed to proceed for 5 min, and stopped with 1ml quenching solution (1 N sulfuric acid, 0.5% ammonium molybdate). The release of 32Pi from [γ-32P]ATP formed a phosphomolybdate complex that was extracted by addition of 1.5 ml isobutanol.40 After centrifugation, 32Pi from [γ-32P]ATP in the upper phase was counted on a MicroBeta 1450 scintillation counter (Wallac, Finland). Background controls were the reaction without crude mitochondria, and three reactions for each construct were measured in parallel. Percentages of ATP hydrolysis were determined in a formula: γ-32Pi counts in experimental sample – background/input [γ-32P]ATP counts – background × 100%.
Detection of intracellular ROS
Intracellular ROS were assayed by measuring intracellular oxidation of 2′, 7′-dichlorofluorescein (DCF) as described.19 The substrate is 5-(and-6)-chloromethyl -2′, 7′ –dichlorofluorescein diacetate, acetyl ester (CM-H2DCFDA) (Invitrogen). It is nonfluorescent until removal of the acetate groups by intracellular oxidation and thus serves as a cell-permeate indicator for ROS. To measure intracellular ROS, the substrate was added directly to cell culture at a final concentration of 5 µM and incubated for 15 min at 37 °C followed by two washes with PBS. DCF fluorescence intensity was measured randomly at six different fields per well with Spectra MAX GEMINI EM microplate fluorometer using 488/525 nm as excitation/emission wavelength that gave an average fluorescence intensity. Background controls were cells transfected with the same construct without substrate loading and two wells for each construct were measured in parallel.
Measurement of caspase-3 activity
Relative caspase-3 activities that are widely used to quantify apoptosis were measured in a cell lysate using a caspase-3 fluorometric substrate as described earlier.19 The caspase-3 fluorometric substrate, acetyl-Asp-Glu-Val-Asp-7-amido-4-methylcoumarin was hydrolyzed by active caspase-3 specifically, releasing fluorescent 7-amido-4-methylcoumarin that was measured at 0 and 10 min, with the fluoroMax-3 spectrometer using 380/460 nm as excitation/emission wavelength. A blank was measured in parallel without the substrate and was subtracted from all the measurements.
Analysis of mitochondrial membrane potential (Δψm)
Mitochondrial membrane potential changes were analyzed following stimulation with staurosporin using the fluorescent potentiometric dye JC-1 (5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethyl-benzimidazolyl-carbocyanine iodide) per the manufacturer’s instruction (Molecular probes). JC-1 is a cationic dye that exhibits potential-dependent accumulation in the mitochondrial membrane, indicated by a fluorescence emission shift from green (525 nm) to red (590 nm). To measure Δψm, the probe was added directly into the cell culture at a final concentration of 10 µg/ml and incubated for 15 min at 37°C followed by two washes with PBS. JC-1 fluorescence was measured randomly at six different fields per well with a Spectra MAX GEMINI EM microplate fluorometer (Molecular Devices). Under this concentration, JC-1 aggregate/monomer ratios declined to the lower level (~3) within 5 minus after 1 µM carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) was added to the cells, which induces maximal depolarization. In contrast, addition of 2.5 µg/ml oligomycin increased the ratio maximally (~16). Aggregate/monomer ratios were normalized to untreated conditions (100%) for each sample tested in triplicate. Background controls were cells transfected with the same construct without JC-1 addition.
Glucose uptake
Primary MEFs were generated from timed pregnant (E13.5) IEX-1-KO and WT mice, cultured in DMEM supplemented with 10% fetal bovine serum, and used at passages 2–3. Nearly confluent MEFs were incubated for 10 min at 37°C with 5% CO2 in DMEM medium containing 0.1mM 2-deoxyglucose and 10 nM 2-[3H]Deoxyglucose (2-DG) (PerkinElmer Life Sciences), after which cells were washed with ice-cold PBS, lysed by the addition of 2ml of 10mM NaOH containing 0.1% Triton X-100, and assayed for 2-[3H]Deoxyglucose incorporation by a 1450 MicroBeta scintillation counter (Wallac, Turku, Finland). The protein content was determined by a Bio-Rad protein assay kit (Bio-Rad Laboratory Inc., CA, USA) for normalization.
Lactate production
Lactate levels in the culture medium were determined using a lactate assay kit as per the manufacturer’s instruction (Biomedical Research Service, State University of New York at Buffalo, USA). The lactate assay kit is based on the reduction of the tetrazolium salt 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-phenyltetrazolium chloride in an NADH-coupled enzymatic reaction to formazan, which is water-soluble and exhibits an absorption maximum at 492 nm. Total viable cell number was used for normalization.
Measurement of intracellular ATP level
IEX-1 KO and WT macrophages were cultured in fresh, complete medium in the presence or absence of 0.5 µM oligomycin for 1 h, after which cellular ATP contents were measured by an intracellular ATP kit (BioThema AB) as per the manufacturer’s instruction. The luminescence was monitored by a MicroBeta JET luminometer (PerkinElmer). ATP content in WT cells in the absence of oligomycin was arbitrarily set as 100%.
Statistical analysis
One-way ANOVA was used to analyze the significance of experimental and relevant controls, except for Figure 2d where the Student’s two-tailed t-test was used.
Acknowledgements
We thank members in Dr Wu’s group for their stimulating discussion and Drs Kenneth D Bloch and Robert E Edwards for critical reading and reviewing of the paper. This work is supported by the National Institutes of Health Grants AI050822 and AI070785, a Research Scholar Grant RSG-01-178-01-MGO from the American Cancer Society, a Senior Research Award from the Crohn’s and Colitis Foundation of America, and a Grant-in-Aid award from the American Heart Association (to MXW).
Abbreviations
- Ab
antibody
- ANT
adenine nucleotide translocator
- DCF
dichlorofluorescein
- DSP
dithiobis-succinimidyl-propionate
- IEX-1
immediate early response gene X-1
- IF1
F1Fo-ATPase inhibitor
- KO
knockout
- ROS
reactive oxygen species
- shRNA
short hairpin RNA
- STS
staurosporin
- UCP
uncoupling protein
- WT
wild type
References
- 1.Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 2.Boyer PD. The ATP synthase—a splendid molecular machine. Annu Rev Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 3.Kadenbach B. Intrinsic and extrinsic uncoupling of oxidative phosphorylation. Biochim Biophys Acta. 2003;1604:77–94. doi: 10.1016/s0005-2728(03)00027-6. [DOI] [PubMed] [Google Scholar]
- 4.Jezek P, Hlavata L. Mitochondria in homeostasis of reactive oxygen species in cell, tissues, and organism. Int J Biochem Cell Biol. 2005;37:2478–2503. doi: 10.1016/j.biocel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Schwerzmann K, Pedersen PL. Regulation of the mitochondrial ATP synthase/ATPase complex. Arch Biochem Biophys. 1986;250:1–18. doi: 10.1016/0003-9861(86)90695-8. [DOI] [PubMed] [Google Scholar]
- 6.Pullman ME, Monroy GC. A naturally occurring inhibitor of mitochondrial adenosine triphosphatase. J Biol Chem. 1963;238:3762–3769. [PubMed] [Google Scholar]
- 7.Green DW, Grover GJ. The IF(1) inhibitor protein of the mitochondrial F(1)F(0)-ATPase. Biochim Biophys Acta. 2000;1458:343–355. doi: 10.1016/s0005-2728(00)00085-2. [DOI] [PubMed] [Google Scholar]
- 8.Cabezon E, Montgomery MG, Leslie AG, Walker JE. The structure of bovine F1-ATPase in complex with its regulatory protein IF1. Nat Struct Biol. 2003;10:744–750. doi: 10.1038/nsb966. [DOI] [PubMed] [Google Scholar]
- 9.Di Pancrazio F, Mavelli I, Isola M, Losano G, Pagliaro P, Harris DA, et al. In vitro and in vivo studies of F(0)F(1)ATP synthase regulation by inhibitor protein IF(1) in goat heart. Biochim Biophys Acta. 2004;1659:52–62. doi: 10.1016/j.bbabio.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Campanella M, Casswell E, Chong S, Farah Z, Wieckowski MR, Abramov AY, et al. Regulation of mitochondrial structure and function by the F1Fo-ATPase inhibitor protein, IF1. Cell Metab. 2008;8:13–25. doi: 10.1016/j.cmet.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Chernyak BV, Dukhovich VF, Khodjaev EY. The effect of the natural protein inhibitor on H+ATPase hepatoma 22a mitochondria. FEBS Lett. 1987;215:300–304. doi: 10.1016/0014-5793(87)80166-7. [DOI] [PubMed] [Google Scholar]
- 12.Luciakova K, Kuzela S. Increased content of natural ATPase inhibitor in tumor mitochondria. FEBS Lett. 1984;177:85–88. doi: 10.1016/0014-5793(84)80986-2. [DOI] [PubMed] [Google Scholar]
- 13.Sjostrand FS. Molecular pathology of Luft disease and structure and function of mitochondria. J Submicrosc Cytol Pathol. 1999;31:41–50. [PubMed] [Google Scholar]
- 14.Garcia J, Ye Y, Arranz V, Letourneux C, Pezeron G, Porteu F. IEX-1: a new ERK substrate involved in both ERK survival activity and ERK activation. EMBO J. 2002;21:5151–5163. doi: 10.1093/emboj/cdf488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Schlossman SF, Edwards RA, Ou CN, Gu J, Wu MX, et al. Impaired apoptosis, extended duration of immune responses, and a lupus-like autoimmune disease in IEX-1-transgenic mice. Proc Natl Acad Sci USA. 2002;99:878–883. doi: 10.1073/pnas.022326699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schilling D, Pittelkow MR, Kumar R. IEX-1, an immediate early gene, increases the rate of apoptosis in keratinocytes. Oncogene. 2001;20:7992–7997. doi: 10.1038/sj.onc.1204965. [DOI] [PubMed] [Google Scholar]
- 17.Schulze PC, de Keulenaer GW, Kassik KA, Takahashi T, Chen Z, Simon DI, et al. Biomechanically induced gene iex-1 inhibits vascular smooth muscle cell proliferation and neointima formation. Circ Res. 2003;93:1210–1217. doi: 10.1161/01.RES.0000103635.38096.2F. [DOI] [PubMed] [Google Scholar]
- 18.Lehoux S, Tedgui A. All strain, no gain: stretch keeps proliferation at bay via the NF-kappaB response gene iex-1. Circ Res. 2003;93:1139–1141. doi: 10.1161/01.RES.0000108693.79326.A8. [DOI] [PubMed] [Google Scholar]
- 19.Shen L, Guo J, Santos-Berrios C, Wu MX. Distinct domains for anti- and pro-apoptotic activities of IEX-1. J Biol Chem. 2006;281:15304–15311. doi: 10.1074/jbc.M600054200. [DOI] [PubMed] [Google Scholar]
- 20.Ichikawa N, Fukuda M, Hashimoto T, Tagawa K. The carboxyl-terminal region of the yeast ATPase inhibitor is indispensable for the stability of the protein in mitochondria. J Biochem (Tokyo) 1998;124:886–891. doi: 10.1093/oxfordjournals.jbchem.a022203. [DOI] [PubMed] [Google Scholar]
- 21.Klingenberg M. The ADP-ATP translocation in mitochondria, a membrane potential controlled transport. J Membr Biol. 1980;56:97–105. doi: 10.1007/BF01875961. [DOI] [PubMed] [Google Scholar]
- 22.Ichikawa N, Nakabayashi K, Hashimoto T. A yeast mitochondrial ATPase inhibitor interacts with three proteins that are easy to dissociate from the mitochondrial inner membrane. J Biochem (Tokyo) 2002;132:649–654. doi: 10.1093/oxfordjournals.jbchem.a003269. [DOI] [PubMed] [Google Scholar]
- 23.Lopez-Mediavilla C, Vigny H, Godinot C. Docking the mitochondrial inhibitor protein IF1 to a membrane receptor different from the F1-ATPase beta subunit. Eur J Biochem. 1993;215:487–496. doi: 10.1111/j.1432-1033.1993.tb18058.x. [DOI] [PubMed] [Google Scholar]
- 24.Minauro-Sanmiguel F, Bravo C, Garcia JJ. Cross-linking of the endogenous inhibitor protein (IF1) with rotor (gamma, epsilon) and stator (alpha) subunits of the mitochondrial ATP synthase. J Bioenerg Biomembr. 2002;34:433–443. doi: 10.1023/a:1022514008462. [DOI] [PubMed] [Google Scholar]
- 25.Kruman I, Guo Q, Mattson MP. Calcium and reactive oxygen species mediate staurosporine-induced mitochondrial dysfunction and apoptosis in PC12 cells. J Neurosci Res. 1998;51:293–308. doi: 10.1002/(SICI)1097-4547(19980201)51:3<293::AID-JNR3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 26.Deberardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Wu MX. Roles of the stress-induced gene IEX-1 in regulation of cell death and oncogenesis. Apoptosis. 2003;8:11–18. doi: 10.1023/a:1021688600370. [DOI] [PubMed] [Google Scholar]
- 28.Matsuyama S, Llopis J, Deveraux QL, Tsien RY, Reed JC. Changes in intramitochondrial and cytosolic pH: early events that modulate caspase activation during apoptosis. Nat Cell Biol. 2000;2:318–325. doi: 10.1038/35014006. [DOI] [PubMed] [Google Scholar]
- 29.Husain I, Jackson PJ, Harris DA. Interaction between F1-ATPase and its naturally occurring inhibitor protein. Studies using a specific anti-inhibitor antibody Biochim Biophys Acta. 1985;806:64–74. doi: 10.1016/0005-2728(85)90082-9. [DOI] [PubMed] [Google Scholar]
- 30.Zanotti F, Raho G, Gaballo A, Papa S. Inhibitory and anchoring domains in the ATPase inhibitor protein IF1 of bovine heart mitochondrial ATP synthase. J Bioenerg Biomembr. 2004;36:447–457. doi: 10.1023/B:JOBB.0000047327.68173.9b. [DOI] [PubMed] [Google Scholar]
- 31.Garcia JJ, Morales-Rios E, Cortes-Hernandez P, Rodríguez-Zavala JS. The inhibitor protein (IF1) promotes dimerization of the mitochondrial F1F0-ATP synthase. Biochemistry. 2006;45:12695–12703. doi: 10.1021/bi060339j. [DOI] [PubMed] [Google Scholar]
- 32.Arlt H, Steglich G, Perryman R, Guiard B, Neupert W, Langer T. The formation of respiratory chain complexes in mitochondria is under the proteolytic control of the m-AAA protease. EMBO J. 1998;17:4837–4847. doi: 10.1093/emboj/17.16.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feldmann KA, Pittelkow MR, Roche PC, Kumar R, Grande JP. Expression of an immediate early gene, IEX-1, in human tissues. Histochem Cell Biol. 2001;115:489–497. doi: 10.1007/s004180100284. [DOI] [PubMed] [Google Scholar]
- 34.Sommer SL, Berndt TJ, Frank E, Patel JB, Redfield MM, Dong X, et al. Elevated Blood Pressure and Cardiac Hypertrophy Following Ablation of the gly96/IEX-1 Gene. J Appl Physiol. 2005;100:707–716. doi: 10.1152/japplphysiol.00306.2005. [DOI] [PubMed] [Google Scholar]
- 35.Graham BH, Waymire KG, Cottrell B, Trounce IA, MacGregor GR, Wallace DC. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nat Genet. 1997;16:226–234. doi: 10.1038/ng0797-226. [DOI] [PubMed] [Google Scholar]
- 36.Hansson A, Hance N, Dufour E, Rantanen A, Hultenby K, Clayton DA, et al. A switch in metabolism precedes increased mitochondrial biogenesis in respiratory chain-deficient mouse hearts. Proc Natl Acad Sci USA. 2004;101:3136–3141. doi: 10.1073/pnas.0308710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blanc J, Alves-Guerra MC, Esposito B, Rousset S, Gourdy P, Ricquier D, et al. Protective role of uncoupling protein 2 in atherosclerosis. Circulation. 2003;107:388–390. doi: 10.1161/01.cir.0000051722.66074.60. [DOI] [PubMed] [Google Scholar]
- 38.Bernal-Mizrachi C, Gates AC, Weng S, Imamura T, Knutsen RH, DeSantis P, et al. Vascular respiratory uncoupling increases blood pressure and atherosclerosis. Nature. 2005;435:502–506. doi: 10.1038/nature03527. [DOI] [PubMed] [Google Scholar]
- 39.Aggeler R, Coons J, Taylor SW, Ghosh SS, Garcia JJ, Capaldi RA, et al. A functionally active human F1F0 ATPase can be purified by immunocapture from heart tissue and fibroblast cell lines. Subunit structure and activity studies. J Biol Chem. 2002;277:33906–33912. doi: 10.1074/jbc.M204538200. [DOI] [PubMed] [Google Scholar]
- 40.Feschenko MS, Sweadner KJ. Phosphorylation of Na,K-ATPase by protein kinase C at Ser18 occurs in intact cells but does not result in direct inhibition of ATP hydrolysis. J Biol Chem. 1997;272:17726–17733. doi: 10.1074/jbc.272.28.17726. [DOI] [PubMed] [Google Scholar]