Abstract
Background
Previous comparative proteomic analysis on Plasmodium falciparum isolates of different adhesion properties suggested that protein phosphorylation varies between isolates with different cytoadherence properties. But the extent and dynamic changes in phosphorylation have not been systematically studied. As a baseline for these future studies, this paper examined changes in the phosphoproteome of parasitized red blood cells (pRBC).
Methods
Metabolic labelling with [35S] methionine on pRBC and 2D gel electrophoresis (2-DE) has previously been used to show the expression of parasite proteins and changes in protein iso-electric point (PI). 2-DE of different parasite strains was combined with immunoblotting using monoclonal antibodies specifically to phosphorylated serine/threonine and tyrosine, to obtain the phosphorylation profiles throughout the erythrocytic lifecycle. Affinity chromatography was used to purify/enrich phosphorylated proteins and these proteins from mature trophozoite stages which were identified using high-accuracy mass spectrometry and MASCOT search.
Results
2D-immunoblots showed that P. falciparum infection greatly increased phosphorylation of a set of proteins in pRBC, the dominant size classes for phosphorylated tyrosine proteins were 95, 60, 50 and 30 kDa and for phosphorylated serine/threonine were 120, 95, 60, 50, 43, 40 and 30 kDa. The most abundant molecules from 2D-gel mapping of phosphorylated proteins in ItG infected RBCs were identified by MALDI-TOF. A proteomic overview of phosphorylated proteins in pRBC was achieved by using complementary phosphorylated protein enrichment techniques combined with nano-flow LC/MS/MS analysis and MASCOT MS/MS ions search with phosphorylation as variable modifications. The definite phosphoproteins of pRBC are reported and discussed.
Conclusion
Protein phosphorylation is a major process in P. falciparum-parasitized erythrocytes. Preliminary screens identified 170 P. falciparum proteins and 77 human proteins as phosphorylated protein in pRBC, while only 48 human proteins were identified in the corresponding fractions from uninfected RBC. Refinement of the search to include significant ion scores indicating a specific phospho-peptide identified 21 P. falciparum proteins and 14 human proteins from pRBC, 13 host proteins were identified from normal RBC. The results achieved by complementary techniques consistently reflect a reliable proteomic overview of pRBC.
Background
Phosphorylation-dephosphorylation is the major control mechanism for many cellular functions, including the regulation of cell division, protein synthesis and transcription [1]. Phosphorylation determines many functions of proteins (e.g. enzymes, microtubules, histones and transcription factors) and regulates signal transduction to control cellular responses to a particular stimulation. Phosphorylation often occurs on multiple distinct sites on a given protein [2] and this multi-layer control magnifies the final signal and promotes sensitive regulation. In a study by Ptacek et al using proteome chip technology to determine the in vitro substrates recognized by the majority of yeast protein kinases, 4,192 phosphorylation events involving 1,325 different proteins were identified. Approximately one third of all eukaryotic proteins are phosphorylated [3,4]. This represents a broad spectrum of different biochemical functions and cellular roles. Since many yeast proteins and pathways are conserved, these results provide insights into the mechanisms and roles of protein phosphorylation in many eukaryotes.
Protein phosphorylation has been shown to be an important event in malaria infection. For example, Plasmodium falciparum infection leads to a dramatic increase in the phosphorylation level of erythrocyte protein 4.1 which forms a tight complex with the mature parasite-infected erythrocyte surface antigens [5-7]. The 11 N-terminal amino acids of membrane-associated band 3 are critical for tyrosine phosphorylation and deletion of these amino acids greatly reduced the ability of parasite invasion [8]. By using [g-32P] ATP and [32P] orthophosphate labelling combined with 2-DE analysis, Suetterlin et al have identified 59 P. falciparum specific phospho-proteins with molecular weights between 15 and 192 kDa in pRBC [9], including two HSP70 heat shock proteins, Pf-hsp and Pf-grp [10]. Plasmodium falciparum infection causes the plasma membrane of erythrocytes to become increasingly permeable to a variety of physiologically relevant solutes via the induction of new permeation pathways (NPPs) which is also thought to involve phosphorylation [11]. Approximately 100 kinases have been identified in the P. falciparum genome, represent 1.1–1.6% of the protein-coding genes of malaria parasite [12,13] and there is great interest in them as potential targets for drugs based on chemical inhibitors [14]. Examples of P. falciparum phosphorylation activity include a calcium-dependent protein kinase that prefers phosphorylation of proteins of the host erythrocytic membrane but very few parasite proteins [15] and P. falciparum phosphatases such as protein phosphatase 1 (PP1), which is responsible for regulation of the phosphorylation status of the membrane protein PfSBP1 [16]. As inhibitors of protein kinases and phosphatases can interfere with parasite growth [17], it is suggested that studies in this area may contribute to the development of novel therapies for malaria.
Severe malaria is believed to be caused by adhesion of P. falciparum pRBC in the small blood vessels of major organs [18]. This processes involved in the reaction of several parasite ligands with endothelium receptors, mainly, PfEMP1 [19-22], ICAM-1 [23] and CD36 [24,25]. Protein phosphorylation has also been shown to relate to cell adherence events. In epithelial cells, tyrosine phosphorylation of cadherins and their associated proteins has major effects on the stability of adhesion junctions [26]. Alteration of tyrosine phosphorylation status is associated with the expression of tyrosine kinase receptor K-sam, which may cause the cytoplasmic distribution of cadherin-catenin molecules and loose cell-cell adhesion in undifferentiated-type gastric cancers [27]. In malaria, CD36 ectodomain phosphorylation at Thr92 can regulate pRBC adhesion to CD36 and to human dermal microvascular endothelial cells (HDMECs) under flow conditions, and is regulated by a Src family kinase- and alkaline phosphatase dependent mechanism [28]. Lucchi et al demonstrated that pRBC cytoadherence to syncytiotrophoblast via chondroitin sulphate A (CSA) enhanced tyrosine phosphorylation of 93 kDa and 85 kDa proteins in these cells [29], but whether the phosphorylation was on protein of pRBC or BeWo cells has not been determined. Interestingly, a protein phosphatase inhibitor, levamisole reduced sequestration of P. falciparum trophozoites from malaria patients' peripheral blood, indicating a potential effect of dephosphorylation on Plasmodium pathogenesis [30]. It has been shown that parasite binding to endothelial cells activated MAPK signalling pathways and that the degree of phosphorylation was proportionate to the avidity of ICAM-1 binding [31]. Also related to the cytoadherence, phosphorylation of major erythrocyte proteins such as ankyrin and band 4.1 and 4.9 proteins can weaken the rigidity of the cytoskeleton thus reducing the binding efficiency of the infected erythrocyte [32].
It has been suggested that there is a correlation between parasite clones of different antigenic variants and cytoadherence, rosette formation and pathogenesis. These have led to studies on the diversity of these properties in P. falciparum isolates. One example is the line IT4/25/5 and its derivates [33]. ItG, A4 and C24 are representatives of parasite clones selected on C32 amelanotic melanoma cells (CD36) and HUVEC (ICAM-1), followed by sub-cloning. These three parasite clones have distinctive cytoadherence properties namely ItG binds strongly to ICAM-1 and to CD36, A4 binds strongly to CD36 and to ICAM-1, and C24 binds strongly to CD36 only. Another unrelated parasite line 3D7 is a low binding strain, which shows some binding to CD36 (although it can be selected for other binding specificities). It is clear that different parasite isolates have different binding phenotypes, but the molecules involved in adhesion and interaction with the host are still poorly understood. Moreover, very little is known about the phosphorylation status of the proteins involved in cytoadherence. Previous work on comparative proteomic analysis of parasite isolates with different adhesion properties found several changes in two-dimensional electrophoresis profiles in the protein isoelectric point (pI), consistent with protein phosphorylation as adding phosphoryl groups to a polypeptide chain supplies a negative charge to a protein [34], but the extent and dynamic changes of phosphorylation status have not been systematically studied. As a baseline for future studies, the changes in the phosphoproteome of different parasite lines at different stages of the asexual erythrocytic cycle were studied.
Methods
Plasmodium culture
Plasmodium falciparum isolates used in this study were: ItG [33,35] C24 [33], and 3D7 [36,37]. Parasites were cultured in vitro in group O+ human erythrocytes using previously described conditions [38]. Briefly, parasites were cultured in RPMI 1640 medium (supplemented with 37.5 mM HEPES, 7 mM D-glucose, 6 mM NaOH, 25 μg ml-1 gentamicin sulphate, 2 mM L-glutamine and 10% human serum) at a pH of 7.2 in a gas mixture of 96% nitrogen, 3% carbon dioxide and 1% oxygen. To minimize the effect of antigenic switching in culture, a batch of stabilates was prepared from post-selection cultures and used for no more than three weeks. To obtain specific lifecycle stages of P. falciparum, 5% sorbitol treatment or Plasmagel flotation was used. For this study, the trophozoites were obtained 24 to 32 hours after invasion and ring stages were obtained 6–10 hours after invasion.
Metabolic labelling and fluorography
Parasites were synchronized with 5% sorbitol and left to grow to the trophozoite stage (28 hours after invasion). The parasitaemia was adjusted to 10% and cells washed with serum-free RPMI 1640 medium without methionine three times. In vitro metabolic labelling was carried out in RPMI medium lacking methionine, [35S] methionine was used at a concentration of 50 μCi ml-1. The culture was gassed and incubated at 37°C for four hours. The reaction was then stopped by centrifugation to remove radioisotopes. The labelled parasites were lysed as described previously and the Tris-insoluble pellet was subjected to 2D electrophoresis with pH 4–7 and 12.5% SDS PAGE. After electrophoresis, the gel was fixed and stained with Coomassie brilliant blue, immersed in Amplify (Amersham) for 30 min and dried for fluorography.
Two-dimensional electrophoresis (2-DE) and immunoblotting
Two-dimensional electrophoresis was carried out according to Nirmalan et al [39]. Briefly, synchronized parasite culture was incubated in 0.05% saponin for 10 min in PBS on ice to lyse erythrocytes. The lysate was collected by centrifugation at 10,000 × g for 5 min and washed three times with 10 mM Tris-HCl pH 7.4 with 1 × protease inhibitor cocktail (Roche). The whole lysate was then solubilized in 2-DE rehydration buffer [8 M urea, 2 M thiourea, 2% CHAPS, 65 mM dithiothreitol (DTT), and 0.5% ampholyte pH 4–7 or 3–10]. The sample was vortexed and sonicated on ice 10 times for 5 seconds followed by centrifugation at 15,000 × g for 10 min. The supernatant was subjected to 2-DE and the isoelectric focusing (IEF) was run on precast Amersham 11 cm pH 3–10 immobiline Drystrip gels using IPG phor IEF Unit (Amersham). The running programme consists of 10 h for 30 V, 40 min for 200 V, 1 h for 500 V, 4 h for 2,000 V and finally 8 h for 8,000 V. The voltage was increased gradually until a total of 80,000 vh was reached. The focused strips were equilibrated in 10 ml equilibration solution (50 mM Tris-HCl, pH 6.8, 6 M urea, 30% glycerol, 2% SDS) with reducing agent of 1% DTT for 10 min, and 10 ml equilibration solution with 4.5% iodoacetamide for another 10 min. The strips were then washed twice briefly with 1× SDS gel running buffer and loaded on 10 or 12.5% SDS-PAGE gels for second dimension separation. The gels were run at constant current 40 mA in a Laemmli's buffer system until the dye front reached the bottom of the gel [40]. For 2D immunoblots, 2D gels were transferred electrophoretically to Hybond ECL nitrocellulose (Amersham GE Healthcare UK). The membranes were blocked with blocking buffer (Sigma UK) at room temperature for 30 min before being probed with the anti-phosphorylated tyrosine (diluted 1/2,000 in blocking buffer, Cell Signalling) or anti-phosphorylated serine/theronine antibody (diluted 1/2,000 in blocking buffer, Qiagen) at 4°C, overnight. The membrane was washed six times in Tris/saline/Tween (TST: 0.01 M Tris pH 8.5/0.15 M NaCl/0.1% Tween 20) for 10 minutes each time. Goat anti-mouse IgG (H+L) horseradish peroxidase conjugate (Nordic 1/2,000) was used to localize antibody-antigen complexes. The blot was developed for chemi-luminescence signals using Lumigen Solution (Amersham GE Healthcare, UK) and the results viewed by fluorography.
Phosphatase treatment and purification of phosphorylated proteins
PhosProtein Purification Kit (Qiagen, Germany) was used to purify phosphorylated proteins from normal RBC and ItG infected pRBC according to the manufacturer's instructions. Briefly, 30 hours after invasion, pRBC were enriched by Plasmagel gel flotation from 250 ml culture (5 × 175 cm2 culture flasks with 10% parasitaemia), to give a packed pRBC pellet of 2.5 ml. Normal RBCs were cultured under the same conditions for at least two days. The pRBC or RBCs were washed with culture medium three times followed by the hypotonic lysis of integrated pRBC or RBC. The lysates were washed extensively with 50 mM Tris-HCl, pH 7.5, and cold saline to remove haemoglobin, all these washing buffers contained 1× proteanase inhibitor (complete mini, Roche, Germany). For phosphatase treatment experiments, samples were resolubilized in protein lysis buffer (2 M urea, 2% CHAPS, 25 mM Tris-base, pH 7.5), half of each sample was treated with 10 units (in 10 μl) Shrimp Alkaline Phosphatase (SAP) or equal volume of 1 × enzyme storage buffer as a control, and incubated for 10–15 min at 37°C. Subsequent protein purification followed the manufacturer's instructions. Phosphorylated proteins eluted from the column were precipitated with 80% acetone/20% TCA in a ratio of 1:1 at -20°C overnight. The proteins were recovered by centrifugation at 15,000 × g for 20 min at 4°C, washed with 90% ice-cold acetone twice and re-suspended in SDS sample buffer, boiled for 5 min prior to 1D SDS-PAGE electrophoresis.
Protein in gel digestion for MALDI and LC/MS/MS
Spot picking of phosphorylated proteins in 2D gels was guided by immunoblot images on duplicate gels. Enriched phosphorylated proteins were separated by 1D SDS-PAGE and excized into 34 slices. In gel digestion was performed as described previously [25], the excized protein was put into an Eppendorf Ultra Pure 1.5 ml centrifuge tube and cut into 1 mm3 cubes. The gel slices were dehydrated by the addition of 100 μl of 50% (v/v) acetonitrile/water and incubated at room temperature for 10 minutes. The dehydrant was then replaced by 100 μl of ammonium bicarbonate (50 mM) and incubated again at room temperature for 10 min. These last two steps were then repeated. The gel slices were dehydrated again with 100 μl of 50% acetonitrile for 10 min and dehydrated slices were incubated in 10 – 20 μl of sequence grade trypsin (Promega) (10 μg/ml) for 18 hours at 37°C. The supernatant was retained and the gel pellet treated with 20 μl of 70% acetonitrile (v/v in water) for 60 minutes at room temperature. The supernatant from this step was then removed and pooled with the previous supernatant. The combined supernatant was dried in a rotational vacuum concentrator (RVC2–18), resuspended in 15 μl water, dried again and resuspended in 15 μl of 0.1% formic acid (in water).
MALDI-TOF mass spectrometry
Samples from the in gel digestion were loaded in a sandwich manner with 1% cyano-4-hydroxycin-namic acid (Sigma) in 50% acetonitrile and 0.05% trifluoroacetic acid (TFA) onto a stainless steel target. High-resolution spectra were obtained using a Axima-CFR plus MALDI TOF instrument (Kratos Analytical, Manchester, UK) in reflectron mode. External calibration was performed using a mixed three point standard adjacent to the samples [25]. Acquisition and data processing are controlled by Launchpad software (Kratos Analytical, Manchester, UK).
Protein identification was performed by sending trypsin digested peptide masses to the P. falciparum and human databases of National Centre for Biotechnology Information (NCBI) using the MASCOT (Matrix Science) Peptide Mass Fingerprinting programme. Fixed carbamidomethyl modification and variable phosphorylation modifications were considered as parameters for the search programme. The mono-isotopic masses were used and the mass tolerance was set to 0.5 Da.
Nanoflow LC/MS/MS analysis and database searching
The tryptic peptides were solubilized in 0.5% formic acid and were separated by nanoflow high-performance liquid chromatography on a C18 reverse phase column (Dionex Ultimate 3000) and elution was performed with a continuous linear gradient of 50% acetonitrile for 30 min. The eluates were analysed by online LC-MS/MS by using an LTQ ion-trap mass spectrometer (Thermo Finnigan). The resulting MS/MS spectra were submitted to TurboSequest Bioworks version 3.1 and the individual spectra was merged into an mgf file before sending to MASCOT (v2.2) for searching. Searching was against NCBI P. falciparum and human databases and was performed using fixed carbamidomethyl and variable phosphorylation modifications. Peptide tolerance was set 0.8 Da, MS/MS tolerance was set 0.5 Da. Phosphorylated protein identities were considered significant if the protein score was over the 95% confidence limit and at least one phosphorylated site was unambiguously identified when a phosphorylated residue existed (matched) y- or b- ions in the peak lists of the fragment ions [providing evidence of observed neutral loss of H3PO4 from the precursor or identified intact phosphorylated residues of Serine (pS), Threonine (pT) and Tyrosine (pY)]. If the protein score reached a significant level but the ion score of phosphorylated peptide was under the 95% confidence limit, these were referred to as potential-phosphorylated proteins.
Results
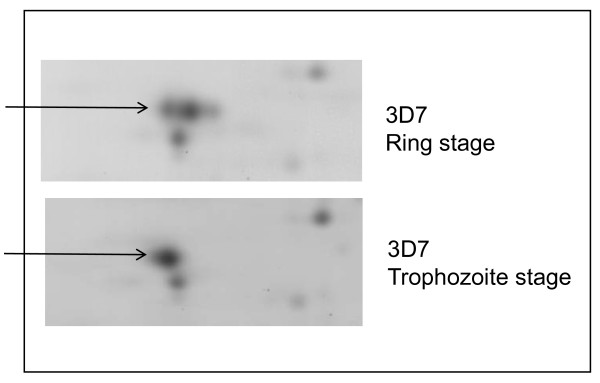
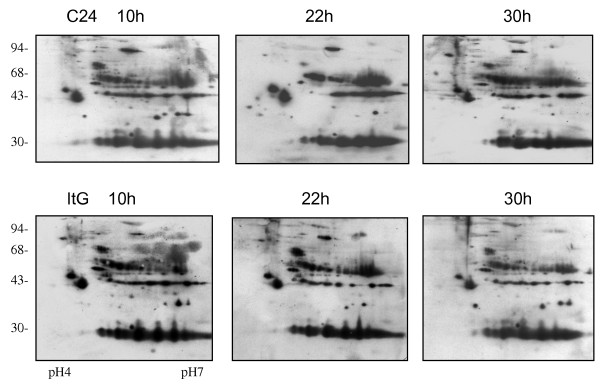
As reported previously [34], using a sensitive in vivo labeling technique together with 2D electrophoresis revealed a number of changes in P. falciparum protein profiles from the pRBC of different parasite lines (Additional file 1). These changes were not limited to proteins from parasite source, but were also found in host proteins in the erythrocyte, varying during erythrocytic development of the malaria parasite e.g., PI changes of human tropomycin between ring and trophozoite stage (Figure 1). To map the phosphorylated proteins of pRBC in 2D gels, specific antibodies to phosphorylated amino acids (tyrosine and serine/threonine) were used to probe total pRBC lysates from a developmental series. As shown in Figures 2 and Figure 3, P. falciparum infection increased protein phosphorylation along with the developmental time course. There were approximately 50 protein spots that reacted positively with anti-serine/threonine antibodies and 4–5 groups of spots that extended horizontally like a family of different isoforms. The results showed that the dominant size classes for phosphorylated tyrosine proteins were 95, 60, 50 and 30 kDa and for phosphorylated serine/threonine were 120, 95, 60, 50, 43, 40 and 30 kDa. The major difference between ItG and C24 was the delayed appearance of a set of phosphorylated tyrosine proteins with molecular weight from 70 to 95 kDa, such that they appeared in ItG at 22 hours post-invasion and appeared in C24 at 30 hours. There were many low abundant proteins with pI changes in both strains, especially in phosphorylated serine/threonine, but it was difficult to view them in stained gels. The phosphorylation antibodies also recognized protein spots of 30, 45 and 95 kDa on non-infected erythrocytes (see Additional file 2) indicating that phospho-proteins are present in normal RBC as well (see below). The results indicated that parasite infection produced major phosphorylation changes in RBC irrespective of the stage. The majority of the proteins that reacted with anti-phosphorylated serine/threonine antibodies were identified by using MALDI-TOF and MASCOT search with phosphorylation modifications. Figure 4 indicates the proteins identified with significant scores and details of these proteins are listed in Table 1.
Figure 1.
Detail from a 2D gel image of proteins from P. falciparum 3D7 line showing changes in pI between ring and trophozoite stages of pRBC. The Tris-insoluble 3D7 pRBC pellets of ring stage and trophozoite stage were extracted and separated on 2D electrophoresis on pH 4–7 IEF strips followed by 12.5% SDS-PAGE. The gels were stained with Coomassie blue. The indicated protein was identified as human tropomycin by MALDI-TOF mass spectrometry.
Figure 2.
Enhanced chemiluminescence images of immunoblots of phosphorylated serine/threonine proteins of different parasite lines at different time points on 2D gels. C24 and ItG parasites were synchronized by double sorbitol lysis and proteins extracted at the time points indicated after parasite invasion. The Tris-insoluble pRBC pellets were extracted and separated on 2D electrophoresis on pH 4–7 IEF strips followed by 12.5% SDS-PAGE. The gels were transferred on to nitrocellulose paper, and probed with antibodies to phosphorylated serine/threonine.
Figure 3.
Enhanced Chemiluminescence images of immunoblots of phosphorylated tyrosine proteins of different parasite lines at different time points on 2D gels. C24 and ItG parasites were synchronized by double sorbitol lysis and proteins extracted at the time points indicated after parasite invasion. The Tris-insoluble pRBC pellets were extracted and separated on 2D electrophoresis on pH 4–7 IEF strips followed by 12.5% SDS-PAGE. The gels were transferred on to nitrocellulose paper, and probed with antibodies to phosphorylated tyrosine.
Figure 4.
Phospho-proteins separated by 2D gel electrophoresis of ItG trophozoite pRBC and positively stained by anti-phosphorylated serine/threonine at 22 hours after invasion. The Tris-insoluble pRBC pellets were extracted and separated on 2-DE followed by immunoblotting. The duplicate gel stained with Coomassie blue was used to excise the corresponding protein spots and identify the proteins by MALDI-TOF or LC/MS/MS. All protein identities achieved significant protein scores in MASCOT search results.
Table 1.
Details of proteins identified from Figure 4
| Protein No. | Accession Number | Protein name | Mol. wt | PI | Peptide identified1 | Score |
| 1. | (AAP72014) | Apolipoprotein H | 37499 | 8.37 | 10 | 145 |
| 2. | NP_702487 | Glyceraldehyde-3-phosphate dehydrogenase | 37068 | 7.59 | 16 | 127 |
| 3. | P10988 | Actin-1 (Plasmodium) P10988 | 42044 | 5.27 | 12 | 123 |
| 4. | NP_001107609 | Erythrocyte membrane protein band 4.9 isoform | 43118 | 8.87 | 15* | 112 |
| 5. | XP_001352096 | Phosphoglycerate kinase | 45569 | 7.63 | 15 | 110 |
| 6. | AAV38387 | Adenylate kinase 1 | 21735 | 8.73 | 11 | 108 |
| 7. | CAB45236 | catalase | 59947 | 6.90 | 17 | 105 |
| 8. | XP_001347854 | GTP-binding nuclear protein ran/tc4 | 24974 | 7.72 | 23* | 98 |
| 9. | C3HU | complement C3 precursor | 188585 | 6.02 | 23* | 95 |
| 10. | XP_001352093 | s-adenosylmethionine synthetase, putative | 45272 | 6.28 | 19* | 91 |
| 11. | 1K1K_B | Carbonmonoxyhemoglobin C | 15970 | 7.98 | 12* | 88 |
| 12. | NP_705453 | Elonation factor 1 alpha | 49156 | 9.12 | 18, | 88 |
| 13. | BAB17688 | Heat Shock Protein hsp70homologue pfhsp70-3 | 71945 | 5.90 | 30* | 84 |
| 14. | AAD29608 | Kappa 1 immunoglobulin light chain | 26181 | 5.72 | 12* | 78 |
| 15. | XP_001350775 | Calcyclin binding protein, putative | 30547 | 8.36 | 10 | 73 |
| 16. | 1605217A | Ig gamma 1 | 25556 | 7.18 | 9 | 71 |
| 17. | XP_001347438 | Pf-hsp60 | 62911 | 6.71 | 24* | 66 |
| 18. | 1XQ9_A | Chain A phosphoglycerate mutase | 29891 | 8.31 | 23* | 57 |
Note: 1. * = phosphorylated peptides identified
To improve the sensitivity of detection, phosphorylated proteins purified/enriched by affinity chromatography techniques were separated by 1D SDS-PAGE and identified by nano-flow LC/MS/MS. Figure 5 shows the gel images of the proteins treated with either SAP or mock buffer and then subjected to a PhosProtein Purification column (Qiagen). The phosphorylated proteins were eluted after extensive washing, precipitated and separated in 1-DE. The arrows indicate bands that were excised for LC/MS/MS analysis. When the whole pRBC lysate was pre-treated with phosphatase, the level and extent of proteins seen on the gel (lane 2) and therefore available for affinity purification was greatly reduced (NB: equal amounts of proteins were loaded on the affinity column). 34 bands were excised from lane 1 and the proteins identified represent a shotgun phosphoproteome of ItG trophozoites. All the peptide ions generated from LC/MS/MS were searched against NCBI databases using MASCOT search with and without phosphorylation modifications.
Figure 5.
1D gel image of proteins eluted from a phospho-protein purification column. Lane 1, proteins with mock treatment, lane 2 proteins treated with SAP prior to purification/enrichment for phospho-proteins. The arrows indicate the cutting side and the numbers represent the bands excised and processed for LC/MS/MS analysis.
The proteins identified using searches including the phosphorylation modifications are listed in Additional file 3 (P. falciparum proteins from ItG-pRBC) and Additional file 4 (human erythrocyte proteins from ItG-pRBC). Additional file 5 contains the list of proteins identified from non-infected RBC. They include both confirmed and potential phosphorylated proteins. There were 170 P. falciparum proteins and 77 host proteins from purified ItG-pRBC phosphorylated protein pool, whereas in non-infected RBC, only 48 proteins were identified in the phosphorylated protein pool. To unambiguously identify phosphorylated peptides with phosphorylation sites, the phospho-peptides with significant ion score were further extracted to investigate their phosphorylation sites. These confirmed phosphorylated proteins are listed in Table 2. By using phosphorylation of serine/threonine and tyrosine as variable modifications, the selection criteria contains not only the total protein scores that reached a significant level, but also that protein hits included at least one significantly recognized phosphorylation site, giving further evidence in addition to their purification by affinity techniques for their phosphorylation status.
Table 2.
The proteins possess significant ion score for phosphorylated peptides with defined phosphorylation sites
| Plasmodium falciparum proteins from ItG-pRBCs: | ||||||
| No* | Accession no | Protein name | Phosphorylated peptide sequences | MW* | Site* | Score* |
| PF1 | PFB0100c | knob-associated His-rich protein | GASTTAGSTTGATTGANAVQSK | 69791 | T5, S8 | 103 |
| PF2 | PFE0870w | transcriptional regulator | FAYKSDEDDEGYNK | 133456 | S5 | 67 |
| PF3 | PF08_0137 | hypothetical protein PF08_0137 | KGSLGFDSFK | 147208 | S3 | 46 |
| PF4 | PF10_0159 | Glycophorin-binding protein (GBP-130) | SVVTEEQKVESDSEK | 90077 | S11 | 37 |
| PF6 | MAL7P1.38 | regulator of chromosome condensation protein | INEQSEGSNLPSEQNK | 79931 | S5 | 31 |
| PF7 | PFL0050c | hypothetical protein PFL0050c | AVPQNSGSNFDEFLDVK | 77525 | S8 | 32 |
| PF5 | PFI0265c | RhopH3 | GLEFYKSSLK | 104856 | S7 | 33 |
| PF8 | PFD1165w | protein kinase, conserved | INDPYDYLKSITNQEER | 75365 | S10 | 61 |
| PF9 | PFE1485w | hypothetical protein | ETNESIYIK | 225530 | T2, Y7 | 45 |
| PF10 | PF14_0648 | hypothetical protein | CDKEIYNLHIK | 238305 | Y6 | 36 |
| PF11 | PFE1600w | hypothetical protein | NYSTHENTYPTLK | 62720 | Y9 | 49 |
| PF12 | PF14_0434 | hypothetical protein | FSVFDSDDNSEEEIENK | 41909 | S6, S10 | 42 |
| IIEVHDNIPSPIIK | S10 | 75 | ||||
| DLNESPKNEPDIVYEEK | S5 | 59 | ||||
| PF13 | PF13_0214 | elongation factor 1-gamma | DDNNNNNNNDADNQHADLLSDDLAEK | 50491 | S20 | 48 |
| PF14 | MAL8P1.95 | hypothetical protein | NLENNEDDETNVGR | 37933 | T10 | 44 |
| PF15 | PF07_0008 | hypothetical protein | HYSLGGQPSSSGR | 27713 | S9 | 76 |
| NYSLGNLSTGTTSQGSTSSR | T11 | 76 | ||||
| YNTSNLASSSTTESSVSGLNTNEAHV | S10 | 78 | ||||
| NYSLGNLSTGTTSQGSTSSR | Y2 S16 | 58 | ||||
| YNTSNLASSSTTESSVSGLNTNEAHV | T12 | 92 | ||||
| PF16 | PFL1005c | chromodomain protein | NESPQWVEETNIR | 30999 | S3 | 55 |
| TGSDEEFEIGDILEIK | S3 | 98 | ||||
| PF17 | MAL7P1.174 | hypothetical protein | DGNDSSSEELDNTNVPTSKPK | 37984 | S5 | 63 |
| PF18 | PF13_0275 | hypothetical protein | YGSGSHDHEEEVVEA | 32877 | S3 | 35 |
| SSSNSHVESNTFQNK | S3 | 47 | ||||
| PF19 | XP_001351065.1 | RESA-like protein | LNTIGNLFEGEK | 36224 | T3 | 54 |
| PF20 | Q8IKH8_PLAF7 | ribosomal protein S3 | TGLTSILPDNISVLEPK | 24823 | S12 | 46 |
| PF21 | PFD0375w | hypothetical protein | EYMNNILYDQNK | 142869 | Y2 | 36 |
| Human proteins in ItG-pRBCs: | ||||||
| Hu1 | SPTB1 | spectrin beta isoform b | KEELGELFAQVPSMGEEGGDADLSIEK | 246894 | S13 | 89 |
| Hu2 | M28880 | Ankyrin | LGYISVTDVLK | 207138 | S5 | 62 |
| Hu3 | BAD92655 | Ankyrin 1 isoform 4 variant | ITHSPTVSQVTER | 208408 | S4 | 55 |
| Hu4 | CAA41149 | Erythrocyte alpha adducin | SPGSPVGEGTGSPPK | 81377 | S4 | 47 |
| AAVVTSPPPTTAPHK | S6 | 55 | ||||
| Hu5 | AAH56881 | ADD2 protein | TESVTSGPMSPEGSPSKSPSK | 78576 | S18 | 56 |
| SAGPQSQLLASVIAEKSR | S17 | 72 | ||||
| Hu6 | AAH04261 | Proline rich 7 protein | KIVTPFLSR | 28986 | T4 | 45 |
| Hu7 | MMHUE4 | Erythrocyte membrane protein band 4.1 | SLDGAAAVDSADR | 95750 | S1 | 65 |
| Hu8 | C3HU | Chain A, Complement Component C3 | SGIPIVTSPYQIHFTKTPK | 188585 | T17 | 21 |
| Hu9 | Q05764 | Beta-adducin | SPGSPVGEGTGSPPK | 81129 | S4 | 89 |
| Hu10 | CAH93400 | Erythrocyte membrane protein band 4.9 | SSSLPAYGR | 45600 | S3 | 38 |
| Hu11 | AAC50223 | Dematin 52 kDa subunit | RGAEEEEEEEDDDSGEEMK | 45726 | S14 | 50 |
| Hu12 | NP_001284 | Chloride ion current inducer protein I (Cln) | EPVADEEEEDSDDDVEPITEFR | 26084 | S11 | 55 |
| FEEESKEPVADEEEEDSDDDVEPITEFR | S17 | 60 | ||||
| Hu13 | CAA40340 | Glycophorin A | SPSDVKPLPSPDTDVPLSSVEIENPETSDQ | 14784 | S19, T27 | 43 |
| Hu14 | GFHUC | Glycophorin C | GTEFAESADAALQGDPALQDAGDSSR | S25 | 80 | |
| Human proteins from normal RBCs: | ||||||
| RBC1 | BAD92652 | Spectrin, beta, erythrocytic | DASVAEAWLIAQEPYLASGDFGHTVDSVEK | 268238 | S27 | 62 |
| LSSSWESLQPEPSHPY | S4 | 27 | ||||
| RBC2 | NP_000338 | Spectrin | TSPVSLWSR | 246468 | S2 | 26 |
| RBC3 | NP_976217 | Erythrocyte membrane protein band 4.1 | VSLLDDTVYECVVEK | 71955 | Y9 | 58 |
| SLDGAAAVDSADR | S1 | 81 | ||||
| RBC4 | AAL15446 | Erythrocyte membrane protein 4.1N | TETMTVSSLAIRK | 97405 | S7 S8 |
23 |
| RBC5 | NP_001969 | Erythrocyte membrane protein band 4.9 | RGAEEEEEEEDDDSGEEMK | 45514 | S14 | 70 |
| RBC6 | CAA34611 | Alt. ankyrin (variant 2.2) | ITHSPTVSQVTER | 189011 | S4 | 64 |
| RBC7 | NP_054909 | Adducin 1 (alpha) isoform c | AAVVTSPPPTTAPHK | 69985 | S6 | 58 |
| SPGSPVGEGTGSPPK | S4 | 74 | ||||
| RBC8 | AAH56881 | ADD2 protein | TESVTSGPMSPEGSPSKSPSK | 78576 | S18 | 54 |
| RBC9 | NP_002427 | Palmitoylated membrane protein 1 | TAELSPFIVFIAPTDQGTQTEALQQLQK | 52296 | T20 | 53 |
| RBC10 | 2GU8_A | Chain A, Discovery Of 2-Pyrimidyl-5-Amidothiophenes As Novel And Potent Inhibitors For Akt | TWDLCGTPEYLAPEIILSK | 39232 | T1 | 43 |
| RBC12 | 1BUW_B | Chain B, Crystal Structure Of S-Nitroso-Nitrosyl Hemoglobin A | GTFATLSELHADK | 15875 | S7 | 31 |
| RBC13 | 1DPF_A | Chain A, Crystal Structure Of A Mg-Free Form Of Rhoa Complexed With Gdp | DQFPAVYVPTVFENYVADIEVDGK | 20169 | Y7 | 23 |
No*: PF = Plasmodium protein; Hu = erythrocyte protein; MW* = protein molecular weight; site* = phosphorylated site identified by MASCOT; Score* = All phosphorylated peptide ion scores reached a significant level
GO slim software was used to classify these proteins into three categories: biological processes, cell components and molecular functions, producing three charts showing the percentage of proteins in specific categories (see Additional file 6). The height of each column indicates the numbers of proteins in each category, with different colours representing the proteins from the human host (blue) or the parasite (red).
Discussion
The shotgun phosphoproteome of ItG-trophozoite pRBC revealed a total of 247 potential phosphorylated proteins with phosphorylated serine/threonine proteins (66.3%) more abundant than phosphorylated tyrosine proteins (33.7%). Most proteins identified by immune-blotting and 2D gel based MALDI-TOF techniques overlapped with proteins identified by affinity enrichment and LC/MS/MS analysis, supporting the specificity of techniques used. This is also indicated by the absence of abundant proteins in 2D gels (e.g. Pf-Enolase, Pf-Ornithine aminotransferase, human paraoxonase), indicated that the affinity chromatography technique selectively enriched a subset of proteins, ruling out general contamination of the samples, although this does not rule out a low-level of contamination.
Within the search algorithm variable modifications can be a powerful means of finding a match. MASCOT tests all possible arrangements of variable modifications to find the best match. For example, when Phosphorylation (Y) is selected, and a peptide contains 2 tyrosines, MASCOT will test for a match with the experimental data for that peptide containing 0, 1 or 2, tyrosine phosphorylation residues. Detection of phosphorylation is complicated because of site heterogeneity and the possibility of three fragmentation channels (intact fragments; neutral loss of HPO3 (80 Da); neutral loss of H3PO4 (98 Da). In MASCOT (v2.2), phosphorylated S, T and Y modification adds 80 Da, but whereas pY always stays intact in the spectrum, pS and pT can either stay intact or can lose 98 Da or occasionally 80 Da http://matrixscience/help/pt_mods_help.html. In MS/MS ions search, confidence that a protein has been identified correctly comes largely from multiple matches to peptides from the same protein. For a suspected phospho-peptide, the calculated monoisotopic mass should include the phosphorylated residue(s) with individual ion scores that reach a significant level (Table 2). The potential phospho-protein 'hits' (Additional file 3, 4 and 5) were also included where MASCOT has identified enriched proteins with phosphorylation modifications, but their ion score of individual phospho-peptides failed to reach a significant score level.
Immunoblotting of various erythrocytic stages suggested that serine/threonine phosphorylation takes place early in the erythrocytic cycle as the phosphoproteome at 10 hours after invasion was very similar to that at 30 hours after invasion (Figure 2). However, if comparing immunoblots of Figure 2 and 3 with normal RBC (Additional file 2), the reaction was dramatically increased indicating that malaria parasite infection induced pRBC phosphorylation. 2D immunoblotting of phosphorylated tyrosine was relatively weaker compared to that of serine/threonine, consistent with our LC/MS/MS data and the general observation that phosphorylated serine/threonine proteins are abundant and phosphorylated tyrosine is relatively rare, for example in yeast, in human cells [41] and in bacterium [42]. There was an indication of changes on tyrosine phospho-proteins around 80–95 kDa in the trophozoite stage (Figure 3), between ICAM-1 binding (ItG) and non-binding (C24) parasite lines. However we failed to confirm their phosphorylation status due to the limited amount of proteins recovered from protein spots of 2D gels. In the gel-based study using MALDI-TOF, only two human proteins were significantly identified around 80–95 kDa, therefore we extracted the data from Additional files 3 and 4 which were obtained from phospho-protein enrichment by affinity chromatography and 1DE separation. There were many proteins significantly identified with molecular weights from 75–100 kDa (Table 3). 14 proteins were identified as tyrosine phosphorylated proteins by using LC/MS/MS, however it is not possible to say whether they are those seen in the 2D Immunoblot in Figure 3 although the molecular weight and positions in the gel would support this. Further more specific studies will be required to differentiate the phosphorylation status in parasite strains with different binding properties and to test if internal phosphorylation contributes to cytoadherence.
Table 3.
Phosphoproteins from ItG trophozoites [75–100 kDa (see Fig. 5)]
| Accession number | Gene name | Mol Wt | PI | Score | Phosphorylation Details* |
| PF10_0159 | Glycophorin-binding protein 130 | 90077 | 5.08 | 779 | ST2 |
| PF08_0054 | Heat shock 70 kDa protein (HSP74) | 74754 | 5.51 | 444 | ST4 |
| BAG09363 | Rhoptry complex polypeptide RhopH3 | 104798 | 6.25 | 324 | ST3 |
| MAL7P1.38 | Regulator of chromosome condensation protein | 79931 | 6.47 | 323 | ST5, Y1 |
| PFL0055c | Protein with DNAJ domain | 108185 | 7.04 | 321 | ST1, Y1 |
| CAA28816 | PFRESAR2 NID (ring-infected erythrocyte surface antigen precursor) | 88816 | 4.92 | 181 | ST1, Y1 |
| Q8IFM1 | Protein kinase | 75365 | 9.08 | 153 | ST8, Y1 |
| PFF0250w | RNA binding protein, putative | 86204 | 6.47 | 146 | ST1 |
| Q9GTW3 | Glutamic acid-rich protein | 80987 | 4.91 | 135 | Y1 |
| Q25730 | Rhoptry associated protein-1 | 90423 | 6.69 | 121 | ST20, Y1 |
| Q8IJ23 | Polyadenylate-binding protein | 108656 | 8.35 | 110 | Y1 |
| PF11_0175 | Heat shock protein 101 | 103038 | 9.17 | 103 | ST1, Y1 |
| Q25869 | Heat-shock protein 86 | 86468 | 4.94 | 103 | ST3 |
| Q8I5H4 | Polyadenylate-binding protein, | 97455 | 8.96 | 94 | ST11 |
| Q8I3I6 | Beta adaptin protein | 102841 | 5.53 | 84 | ST2 |
| Q8I0V3 | Chaperonin cpn60, mitochondrial | 79898 | 4.93 | 76 | ST2 |
| E71608 | ATP-dept. acyl-CoA synthetase (TP) PFB0695c | 103277 | 8.83 | 63 | ST8 |
| PF10_0077 | Eukaryotic translation initiation factor 3 subunit | 84517 | 8.26 | 62 | ST1 |
| Q9NIH1 | P101/acidic basic repeat antigen | 86321 | 4.81 | 55 | ST1 |
| Q8I635 | Hypothetical protein | 77525 | 4.40 | 414 | ST3, Y1 |
| T18421 | Hypothetical protein C0140c | 89820 | 6.31 | 127 | ST7, Y1 |
| Q8IL87 | Hypothetical protein | 77412 | 9.07 | 90 | ST2, Y1 |
| Q8IL87 | Hypothetical protein.- | 77412 | 9.07 | 81 | ST4 |
| Q8I2G1 | Hypothetical protein PFI1735c.- | 82991 | 5.46 | 53 | ST3 |
| Host erythrocyte proteins | |||||
| Accession | Gene name | MW | PI | Score | Phospho-site |
| ADDB | Beta-adducin (Erythrocyte adducin subunit beta | 80854 | 5.51 | 1207 | ST7, Y1 |
| MMHUE4 | Erythrocyte membrane protein 4.1 | 95750 | 5.37 | 591 | ST3 |
| Q4VB86 | Hypothetical protein (EPB41 protein) | 83618 | 5.52 | 589 | ST3 |
| S18208 | Rabphilin-3A-interacting protein | 81260 | 5.67 | 456 | ST7 |
| AAD42222 | AF156225 NID: | 97567 | 5.22 | 382 | ST3 |
| ADDA | Alpha-adducin | 81304 | 5.60 | 153 | ST11, Y1 |
| CAC18967 | Sequence 3 from Patent WO0068693 | 85082 | 4.94 | 143 | ST1 Y1 |
| Q53FS6 | TNF receptor-associated protein 1 variant | 80326 | 8.05 | 72 | ST3 |
| I2C2 | Eukaryotic translation initiation factor 2C 2 | 97991 | 9.34 | 46 | ST1 |
* Phosphorylation details: The phosphorylated proteins were identified because they contained phosphorylated peptides. ST means serine/threonine, Y means tyrosine
The 30 kDa band seen with both anti-phospho tyrosine and serine/threonine antibodies shows some background, reacting with the secondary antibody only, but the intensity and extent of the reaction was greatly increased on the addition of the primary antibody. Moreover, it contains several known phospho-proteins, for example, calcyclin binding protein (gi23509063), adenylate kinase 2 (gi32394417), vesicle-associated membrane protein (gi23497447) and phosphoethanolamine N-methyltransferase (gi38018254). Some parasite erythrocyte membrane proteins such rifin (gi23613367) and stevor (gi23498318) have also seen when searching using MASCOT with phosphorylation modifications but they did not reach a significant level.
Many phosphorylated proteins seen in this study using affinity enrichment and LC/MS/MS showed very high MASCOT scores and frequently appeared in the search results suggesting high abundance or multiple family members. Since the proteins were extracted from Tris-insoluble pellets of pRBC, this pool includes proteins of outer and inner membranes, cytoskeleton and nuclear proteins. One of these proteins from P. falciparum is from the family of cytoadherence linked asexual proteins (CLAG), with five members of this family identified in phosphorylated protein pool namely CLAG 2, 8, 9 and RhopH1/Clag3.1, RhopH1/Clag3.2. These proteins are the products of clag gene family and have been proposed as being involved in cellular interactions [43,44]. Other abundant phosphorylated proteins of interest were DNAJ protein (PFE1605w), which is a co-chaperone of HSP70 in E. coli where it forms an operon with DnaK (HSP70) to regulate its ATPase activity [45]. In P. falciparum DnaJ is also referred to as RESA (in Additional file 3) and is predicted to be exported outside the parasite as it contains a PEXEL motif [46]. ISWI protein homologue (PFF1185w) is a chromatin-remodelling protein involved in gene expression and the maintenance of higher order chromatin structure [47] and possesses variable binding functions to DNA and proteins. There were a number of heat shock phospho-proteins, including: HSP101 (PF11_0175), HSP86 (PF07_0029), HSP78 (PFI0875w), HSP70 (PFI0875w), HSP74 (PF08_0054), HSP60 (PF10_0153) and other proteins with chaperone activities such as 14-3-3 protein homologue (MAL8P1.69) and Chaperonin CPN60 (PFL1545c). Molecular chaperones are a large protein family with roles in unfolding proteins for translocation, assembly and degradation. It is particularly interesting that most of these proteins are not only phosphorylated, but also exported [48,49], making them candidates for communication with the external environment. Known phosphorylated proteins found in this study were protein kinases and phophatases. However, the numbers and types of these enzymes identified were limited, which may reflect the situation in vivo that only small amount of regulatory molecules are present suggesting that the study of protein kinases or phosphatases in P. falciparum may need to use specific affinity techniques to produce suitable yields [50].
A basic level of phosphorylation was observed in non-infected erythrocytes with phospho-proteins including spectrin, ankyrin, adducin and erythrocyte membrane proteins 4.1 and 4.9. This kind of phosphorylation may be caused by the age of erythrocytes and in vitro culture, or may be a self-adjustment by the erythrocyte to maintain its shape and survival. By subtracting normal erythrocyte data from data obtained from ItG-pRBC, it was possible to distinguish some erythrocyte proteins significantly modified by phosphorylation upon invasion. One interesting protein identified as phosphorylated host protein in ItG-pRBC is NP_001284, chloride ion current inducer protein I (Cln), also called chloride channel regulatory protein. Malaria parasites infection produces high permeability of erythrocyte membrane to a large variety of solutes [51]. The altered permeability is presumed to be due to the activation of endogenous dormant channels and chloride channels are very important to channel activity [52]. Our data on the phosphorylation of a chloride channel protein in response to parasite invasion supports the possibility that this may contribute to NPPs [11]. Another interesting host protein is complement component 3 (C3) which we found as a phospho-protein specifically after P. falciparum infection. C3 is a component of the plasma but increases its deposition on red cells and binding to haemoglobin in children with severe malarial anaemia [53]. C3 is a critical regulator of innate immunity, implicated in the regulation of T cell-mediated responses [54]. It enhances the opsonization of immune complexes by phosphorylation due to increased binding to IgG [55]. There have been reports on the changes of complement system during malaria infection [56,57] and the contribution of complement in protective immunity in malaria [58], but little information about the phosphorylation states of complement components and their roles during infection.
Conclusion
This study investigated the phosphorylation status of pRBC and dynamic changes of protein phosphorylation using Western-blots on 2D gels and/or phospho-protein enrichment coupled with high-accuracy mass spectrometry. The results have provided us with the basis of phosphorylation of RBC and pRBC for future research as well as identifying some interesting leads for further investigation of the role of protein phosphorylation in important parasite processes.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
YW: carried out the 2D-Immunoblot, MALDI, LC/MS/MS, data analysis and wrote the manuscript. MN: participated in the phosphatase treatment and phospho-protein enrichment. AQ: participated in protein identification using LC/MS/MS. DX: participated in LC/MS/MS analysis and data searching. JW: participated in the design of the study. AC: conceived of the study, and participated in its design, analysis of results analysis and writing the manuscript. All authors read and approved the final manuscript.
Supplementary Material
PI changes of pRBC from different parasite lines. Fluorography of proteins from trophozoite stage pRBC infected with 3D7 and ItG. At 20 hours after invasion, parasites were metabolically labelled with 50 μCi/ml [35S] methionine for 4 hours. Tris-insoluble pellets of pRBC were separated run on pH 4–7 IEF strips followed by 12% SDS-PAGE. Gels were stained with Coomassie blue, dried and exposed to X-ray film. Marked boxes show proteins with at least three fold changes between 3D7 and ItG. Enlarged images of corresponding boxes showing significant changes in the protein profiles. Arrows indicate the relative positions of the spots in different lines.
Immunoblot of normal RBC and secondary antibody control. Immunoblot of normal RBC separated by 2DE (using the same conditions as for Figures 2 &3) and probed with antibodies to phosphorylated serine/threonine (A) or tyrosine (B). Part C is an immunoblot of Tris-insoluble pellet of ItG-infected RBC probed with the secondary antibody only and developed by ECL.
Plasmodium falciparum phosphorylated proteins from ItG infected erythrocytes. Phosphorylated proteins purified/enriched by affinity chromatography techniques were separated by 1D SDS-PAGE and identified by nano-flow LC/MS/MS. Additional file 4 contains P. falciparum proteins from ItG-pRBC identified using searches including the phosphorylation modifications.
Host phosphorylated proteins from ItG infected erythrocytes. Phosphorylated proteins purified/enriched by affinity chromatography techniques were separated by 1D SDS-PAGE and identified by nano-flow LC/MS/MS. Additional file 5 contains human phosphorylated proteins from ItG-pRBC identified using searches including the phosphorylation modifications.
Phosphorylated proteins of non-infected erythrocytes purified from phospho-affinity column. Phosphorylated proteins purified/enriched by affinity chromatography techniques were separated by 1D SDS-PAGE and identified by nano-flow LC/MS/MS. Additional file 6 contains human phosphorylated proteins from normal RBC identified using searches including the phosphorylation modifications.
Gene ontology analysis of serine/threonine and tyrosine phosphorylated proteins. Gene ontology analysis of the serine/threonine and tyrosine phosphorylated proteins. A. Functional categories; B. Cellular components; C. Biological processes. The different colours indicate human (blue) or parasite (red) proteins.
Acknowledgments
Acknowledgements
The authors would like to thank Prof Andrew Tobin for his comments and suggestions on the manuscript and Dr. Duncan Robertson for help with LC/MS/MS analysis.
Contributor Information
Yang Wu, Email: ywu@liverpool.ac.uk.
Morag M Nelson, Email: morag.nelson@liverpool.ac.uk.
Andrew Quaile, Email: aquaile@liverpool.ac.uk.
Dong Xia, Email: dongxia@liverpool.ac.uk.
Jonathan M Wastling, Email: j.wastling@liverpool.ac.uk.
Alister Craig, Email: agcraig@liverpool.ac.uk.
References
- Cohen P. The origins of protein phosphorylation. Nat Cell Biol. 2002;4:E127–30. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- Cohen P. The regulation of protein function by multisite phosphorylation – a 25 year update. Trends Biochem Sci. 2000;25:596–601. doi: 10.1016/S0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- Ptacek J, Snyder M. Charging it up: global analysis of protein phosphorylation. Trends Genet. 2006;22:545–54. doi: 10.1016/j.tig.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, McCartney RR, Schmidt MC, Rachidi N, Lee SJ, Mah AS, Meng L, Stark MJ, Stern DF, De Virgilio C, Tyers M, Andrews B, Gerstein M, Schweitzer B, Predki PF, Snyder M. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- Jones GL, Edmundson HML. Protein phosphorylation during the asexual life cycle of the human malarial parasite Plasmodium falciparum. Biochim Biophys Acta. 1990;1053:118–124. doi: 10.1016/0167-4889(90)90002-U. [DOI] [PubMed] [Google Scholar]
- Murray MC, Perkins ME. Phosphorylation of erythrocyte membrane and cytoskeleton proteins in cells infected with Plasmodium falciparum. Mol Biochem Parasitol. 1989;34:229–236. doi: 10.1016/0166-6851(89)90051-0. [DOI] [PubMed] [Google Scholar]
- Magowan C, Liang J, Yeung J, Takakuwa Y, Coppel RL, Mohandas N. Plasmodium falciparum : Influence of malarial and host erythrocyte skeletal protein interactions on phosphorylation in infected erythrocytes. Exp Parasitol. 1998;89:40–49. doi: 10.1006/expr.1998.4261. [DOI] [PubMed] [Google Scholar]
- Perrotta S, Borriello A, Scaloni A, De Franceschi L, Brunati AM, Turrini F, Nigro V, del Giudice EM, Nobili B, Conte ML, Rossi F, Iolascon A, Donella-Deana A, Zappia V, Poggi V, Anong W, Low P, Mohandas N, Della Ragione F. The N-terminal 11 amino acids of human erythrocyte band 3 are critical for aldolase binding and protein phosphorylation: implications for band 3 function. Blood. 2005;106:4359–4366. doi: 10.1182/blood-2005-07-2806. [DOI] [PubMed] [Google Scholar]
- Suetterlin BW, Kappes B, Franklin RM. Localization and stage specific phosphorylation of Plasmodium falciparum phosphoproteins during the intraerythrocytic cycle. Mol Biochem Parasitol. 1991;46:113–122. doi: 10.1016/0166-6851(91)90205-K. [DOI] [PubMed] [Google Scholar]
- Kappes B, Suetterlin BW, Hofer-Warbinek R, Humar R, Franklin RM. Two major phosphoproteins of Plasmodium falciparum are heat shock proteins. Mol Biochem Parasitol. 1993;59:83–94. doi: 10.1016/0166-6851(93)90009-M. [DOI] [PubMed] [Google Scholar]
- Decherf G, Egee S, Staines HM, Ellory JC, Thomas SL. Anionic channels in malaria-infected human red blood cells. Blood Cells Mol Dis. 2004;3293:366–371. doi: 10.1016/j.bcmd.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Anamika , Srinivasan N, Krupa A. A genomic perspective of protein kinases in Plasmodium falciparum. Proteins. 2005;58:180–189. doi: 10.1002/prot.20278. [DOI] [PubMed] [Google Scholar]
- Ward P, Equinet L, Packer J, Doerig C. Protein kinases of the human malaria parasite Plasmodium falciparum: the kinome of a divergent eukaryote. BMC Genomics. 2004;5:79. doi: 10.1186/1471-2164-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerig C, Meijer L. Antimalarial drug discovery: targeting protein kinases. Expert Opin Ther Targets. 2007;11:279–290. doi: 10.1517/14728222.11.3.279. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Franklin RM, Kappes B. Plasmodium falciparum calcium-dependent protein kinase phosphorylates proteins of the host erythrocytic membrane. Mol Biochem Parasitol. 1994;66:329–343. doi: 10.1016/0166-6851(94)90159-7. [DOI] [PubMed] [Google Scholar]
- Blisnick T, Vincensini L, Fall G, Braun-Breton C. Protein phosphatase 1, a Plasmodium falciparum essential enzyme, is exported to the host cell and implicated in the release of infectious merozoites. Cell Microbiol. 2006;8:591–601. doi: 10.1111/j.1462-5822.2005.00650.x. [DOI] [PubMed] [Google Scholar]
- Ward GE, Fujioka H, Aikawa M, Miller LH. Staurosporine inhibits invasion of erythrocytes by malarial merozoites. Exp Parasitol. 1994;79:480–487. doi: 10.1006/expr.1994.1109. [DOI] [PubMed] [Google Scholar]
- Kyes S, Horrocks P, Newbold C. Antigenic variation at the infected red cell surface in malaria. Annu Rev Microbiol. 2001;55:673–707. doi: 10.1146/annurev.micro.55.1.673. [DOI] [PubMed] [Google Scholar]
- Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, Taraschi TF, Howard RJ. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, Ravetch JA, Wellems TE. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, Pinches R, Newbold CI, Miller LH. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendt AR, Simmons DL, Tansey J, Newbold CI, Marsh K. Intercellular adhesion molecule-1 is an endothelial cell adhesion receptor for Plasmodium falciparum. Nature. 1989;341:57–59. doi: 10.1038/341057a0. [DOI] [PubMed] [Google Scholar]
- Barnwell JW, Asch AS, Nachman RL, Yamaya M, Aikawa M, Ingravallo P. A human 88-kD membrane glycoprotein (CD36) functions in vitro as a receptor for a cytoadherence ligand on Plasmodium falciparum-infected erythrocytes. J Clin Invest. 1989;84:765–772. doi: 10.1172/JCI114234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oquendo P, Hundt E, Lawler J, Seed B. CD36 directly mediates cytoadherence of Plasmodium falciparum parasitized erythrocytes. Cell. 1989;58:95–101. doi: 10.1016/0092-8674(89)90406-6. [DOI] [PubMed] [Google Scholar]
- Daniel JM, Reynolds AB. Tyrosine phosphorylation and cadherin/catenin function. Bioessays. 1997;19:883–891. doi: 10.1002/bies.950191008. [DOI] [PubMed] [Google Scholar]
- Akimoto S, Ochiai A, Inomata M, Hirohashi S. Expression of cadherin-catenin cell adhesion molecules, phosphorylated tyrosine residues and growth factor receptor-tyrosine kinases in gastric cancers. Jpn J Cancer Res. 1998;89:829–836. doi: 10.1111/j.1349-7006.1998.tb00636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M, Hoang HL, Lee KM, Liu N, MacRae T, Montes L, Flatt CL, Yipp BG, Berger BJ, Looareesuwan S, Robbins SM. Ectophosphorylation of CD36 regulates cytoadherence of Plasmodium falciparum to microvascular endothelium under flow conditions. Infect Immun. 2005;73:8179–87. doi: 10.1128/IAI.73.12.8179-8187.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchi NW, Koopman R, Peterson DS, Moore JM. Plasmodium falciparum-infected red blood cells selected for binding to cultured syncytiotrophoblast bind to chondroitin sulfate A and induce tyrosine phosphorylation in the syncytiotrophoblast. Placenta. 2006;27:384–394. doi: 10.1016/j.placenta.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Dondorp AM, Silamut K, Charunwatthana P, Chuasuwanchai S, Ruangveerayut R, Krintratun S, White NJ, Ho M, Day NP. Levamisole inhibits sequestration of infected red blood cells in patients with falciparum malaria. J Infect Dis. 2007;196:460–6. doi: 10.1086/519287. [DOI] [PubMed] [Google Scholar]
- Jenkins N, Wu Y, Chakravorty S, Kai O, Marsh K, Craig AG. Plasmodium falciparum cytoadherence-related signalling in human endothelial cells. J Infect Dis. 2007;196:321–327. doi: 10.1086/518795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna EJ, Hitt AL. Cytoskeleton – plasma membrane interactions. Science. 1992;258:955–964. doi: 10.1126/science.1439807. [DOI] [PubMed] [Google Scholar]
- Roberts DJ, Craig AG, Berendt AR, Pinches R, Nash G, Marsh K, Newbold CI. Rapid switching to multiple antigenic and adhesive phenotypes in malaria. Nature. 1992;357:689–692. doi: 10.1038/357689a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Craig AG. Comparative proteomic analysis of metabolically labelled proteins from Plasmodium falciparum isolates with different adhesion properties. Malar J. 2006;5:67–80. doi: 10.1186/1475-2875-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ockenhouse CF, Tandon NN, Magowan C, Jamieson GA, Chulay JD. Identification of a platelet membrane glycoprotein as a falciparum malaria sequestration receptor. Science. 1989;243:1469–1471. doi: 10.1126/science.2467377. [DOI] [PubMed] [Google Scholar]
- Delemarre BJ, Kaay HJ van der. Tropical malaria contracted the natural way in the Netherlands. Ned Tijdschr Geneeskd. 1979;123:1981–1982. [PubMed] [Google Scholar]
- Walliker D, Quakyi IA, Wellems TE, McCutchan TF, Szarfman A, London WT, Corcoran LM, Burkot TR, Carter R. Genetic analysis of the human malaria parasite Plasmodium falciparum. Science. 1987;236:1661–1666. doi: 10.1126/science.3299700. [DOI] [PubMed] [Google Scholar]
- Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- Nirmalan N, Sims PF, Hyde JE. Quantitative proteomics of the human malaria parasite Plasmodium falciparum and its application to studies of development and inhibition. Mol Microbiol. 2004;52:1187–99. doi: 10.1111/j.1365-2958.2004.04049.x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in-vivo and site-specific phosphorylation dynamics of signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Macek B, Mijakovic I, Olsen JV, Gnad F, Kumar C, Jensen PR, Mann M. The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol Cell Proteomics. 2007;6:697–707. doi: 10.1074/mcp.M600464-MCP200. [DOI] [PubMed] [Google Scholar]
- Smith JD, Craig AG. The surface of the Plasmodium falciparum- infected erythrocyte. Curr Issues Mol Biol. 2005;7:81–93. [PubMed] [Google Scholar]
- Holt DC, Gardiner DL, Thomas EA, Mayo M, Bourke PF, Sutherland CJ, Carter R, Myers G, Kemp DJ, Trenholme KR. The clag gene family of Plasmodium falciparum: Are there roles other than cytoadherence? Int J Parasitol. 1999;29:939–944. doi: 10.1016/S0020-7519(99)00046-6. [DOI] [PubMed] [Google Scholar]
- Saito H, Uchida H. Organization and expression of the dnaJ and dnaK genes of Escherichia coli K12. Mol Gen Genet. 1978;164:1–8. doi: 10.1007/BF00267592. [DOI] [PubMed] [Google Scholar]
- Watanabe J. Cloning and characterization of heat shock protein DnaJ homologues from Plasmodium falciparum and comparison with ring infected erythrocyte surface antigen. Mol Biochem Parasitol. 1997;88:253–258. doi: 10.1016/S0166-6851(97)00073-X. [DOI] [PubMed] [Google Scholar]
- Deuring R, Fanti L, Armstrong JA, Sarte M, Papoulas O, Prestel M, Daubresse G, Verardo M, Moseley SL, Berloco M, Tsukiyama T, Wu C, Pimpinelli S, Tamkun JW. The ISWI chromatin-remodeling protein is required for the maintenance of higher order chromatin structure. Mol Cell. 2000;5:355–356. doi: 10.1016/S1097-2765(00)80430-X. [DOI] [PubMed] [Google Scholar]
- Acharya P, Kumar R, Tatu U. Chaperoning a cellular upheaval in malaria: heat shock proteins in Plasmodium falciparum. Mol Biochem Parasitol. 2007;153:85–94. doi: 10.1016/j.molbiopara.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Botha M, Pesce ER, Blatch GL. The Hsp40 proteins of Plasmodium falciparum and other apicomplexa: regulating chaperone power in the parasite and the host. Int J Biochem Cell Biol. 2007;39:1781–1803. doi: 10.1016/j.biocel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Peck SC. Analysis of protein phosphorylation: methods and strategies for studying kinases and substrates. Plant J. 2006;45:512–522. doi: 10.1111/j.1365-313X.2005.02613.x. [DOI] [PubMed] [Google Scholar]
- Kirk K, Horner HA, Elford BC, Ellory JC, Newbold CI. Transport of diverse substrates into malaria-infected erythrocytes via a pathway showing functional characteristics of a chloride channel. J Biol Chem. 1994;269:3339–3347. [PubMed] [Google Scholar]
- Decherf G, Bouyer G, Egée S, Thomas SL. Chloride channels in normal and cystic fibrosis human erythrocyte membrane. Blood Cells Mol Dis. 2007;39:24–34. doi: 10.1016/j.bcmd.2007.02.014. [DOI] [PubMed] [Google Scholar]
- Odhiambo CO, Otieno W, Adhiambo C, Odera MM, Stoute JA. Increased deposition of C3b on red cells with low CR1 and CD55 in a malaria-endemic region of western Kenya: implications for the development of severe anemia. BMC Med. 2008;6:23. doi: 10.1186/1741-7015-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdei A, Fust G, Gergely J. The role of C3 in the immune response. Immunol Today. 1991;12:332–337. doi: 10.1016/0167-5699(91)90011-H. [DOI] [PubMed] [Google Scholar]
- Ekdahl KN, Nilsson B. Alterations in C3 activation and binding caused by phosphorylation by a casein kinase released from activated human platelets. J Immunol. 1999;162:7426–7433. [PubMed] [Google Scholar]
- Taylor PR, Seixas E, Walport MJ, Langhorne J, Botto M. Complement contributes to protective immunity against reinfection by Plasmodium chabaudi parasites. Infect Immun. 2001;69:3853–3859. doi: 10.1128/IAI.69.6.3853-3859.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goka BQ, Kwarko H, Kurtzhals JA, Gyan B, Ofori-Adjei E, Ohene SA, Hviid L, Akanmori BD, Neequaye J. Complement binding to erythrocytes is associated with macrophage activation and reduced haemoglobin in Plasmodium falciparum malaria. Trans R Soc Trop Med Hyg. 2001;95:545–549. doi: 10.1016/S0035-9203(01)90036-7. [DOI] [PubMed] [Google Scholar]
- Bergmann-Leitner ES, Scheiblhofer S, Weiss R, Duncan EH, Leitner WW, Chen D, Angov E, Khan F, Williams JL, Winter DB, Thalhamer J, Lyon JA, Tsokos GC. C3d binding to the circumsporozoite protein carboxy-terminus deviates immunity against malaria. Int Immunol. 2005;17:245–255. doi: 10.1093/intimm/dxh205. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PI changes of pRBC from different parasite lines. Fluorography of proteins from trophozoite stage pRBC infected with 3D7 and ItG. At 20 hours after invasion, parasites were metabolically labelled with 50 μCi/ml [35S] methionine for 4 hours. Tris-insoluble pellets of pRBC were separated run on pH 4–7 IEF strips followed by 12% SDS-PAGE. Gels were stained with Coomassie blue, dried and exposed to X-ray film. Marked boxes show proteins with at least three fold changes between 3D7 and ItG. Enlarged images of corresponding boxes showing significant changes in the protein profiles. Arrows indicate the relative positions of the spots in different lines.
Immunoblot of normal RBC and secondary antibody control. Immunoblot of normal RBC separated by 2DE (using the same conditions as for Figures 2 &3) and probed with antibodies to phosphorylated serine/threonine (A) or tyrosine (B). Part C is an immunoblot of Tris-insoluble pellet of ItG-infected RBC probed with the secondary antibody only and developed by ECL.
Plasmodium falciparum phosphorylated proteins from ItG infected erythrocytes. Phosphorylated proteins purified/enriched by affinity chromatography techniques were separated by 1D SDS-PAGE and identified by nano-flow LC/MS/MS. Additional file 4 contains P. falciparum proteins from ItG-pRBC identified using searches including the phosphorylation modifications.
Host phosphorylated proteins from ItG infected erythrocytes. Phosphorylated proteins purified/enriched by affinity chromatography techniques were separated by 1D SDS-PAGE and identified by nano-flow LC/MS/MS. Additional file 5 contains human phosphorylated proteins from ItG-pRBC identified using searches including the phosphorylation modifications.
Phosphorylated proteins of non-infected erythrocytes purified from phospho-affinity column. Phosphorylated proteins purified/enriched by affinity chromatography techniques were separated by 1D SDS-PAGE and identified by nano-flow LC/MS/MS. Additional file 6 contains human phosphorylated proteins from normal RBC identified using searches including the phosphorylation modifications.
Gene ontology analysis of serine/threonine and tyrosine phosphorylated proteins. Gene ontology analysis of the serine/threonine and tyrosine phosphorylated proteins. A. Functional categories; B. Cellular components; C. Biological processes. The different colours indicate human (blue) or parasite (red) proteins.