Abstract
Comparative genomic analysis of important signaling pathways in C. briggase and C. elegans reveals both conserved features and also differences. To build a framework to address the significance of these features we determined the C. briggsae embryonic cell lineage, using the tools StarryNite and AceTree. We traced both cell divisions and cell positions for all cells through all but the last round of cell division and for selected cells through the final round. We found the lineage to be remarkably similar to that of C. elegans. Not only did the founder cells give rise to similar numbers of progeny, the relative cell division timing and positions were largely maintained. These lineage similarities appear to give rise to similar cell fates as judged both by the positions of lineally-equivalent cells and by the patterns of cell deaths in both species. However, some reproducible differences were seen, e.g., the P4 cell cycle length is more than 40% longer in C. briggsae than that in C. elegans (p < 0.01). The extensive conservation of embryonic development between such divergent species suggests that substantial evolutionary distance between these two species has not altered these early developmental cellular events, although the developmental defects of transpecies hybrids suggest that the details of the underlying molecular pathways have diverged sufficiently so as to not be interchangeable.
Keywords: C. briggsae, C. elegans, embryo, cell lineage, signaling pathway
Introduction
The deterministic embryonic development in C. elegans occurs through a complex interplay of lineally inherited factors and cell interactions to create rapidly a series of founder cells that in turn undergo a fixed series of divisions to produce the 558 cells of the embryo (Sulston et al., 1983; Lin et al., 1995; Hutter and Schnabel 1994). Contrary to early views, this developmental paradigm is not shared across the nematode phylum. Recent studies have uncovered the distantly related fresh water nematode Tobrilus diversipapillatus (clade II) that despite its similarity in mature body plan to other nematodes undergoes an initial series of proliferative divisions to create a layer of undifferentiated cells surrounding a large blastocoel. Development then proceeds with gastrulation initiated at the future mouth by the ingression of gut cell precursors (Schierenberg, 2005). The similarity of this development to the ‘classical’ type, which is widely distributed in the animal kingdom (Arendt, 2004), has led to the conjecture that C. elegans development represents a highly derived state, perhaps to accommodate its extremely rapid development. Variation in embryogenesis is also apparent in studies of other nematodes more closely related to C. elegans, a member of clade V. For example, founder cells in A. nanus (clade IV) can regulate their fate in a hierarchical manner after cell ablation (Wiegner and Schierenberg, 1999). In the more closely related Pellioditis marina (clade V) early divisions appear identical to those in C. elegans but by following the lineage until cells had taken up their final anatomical positions, investigators could detect differences in later divisions and cell fate (Houthoofd et al., 2003). Overall the lineage homology with C. elegans is high (about 95%), but fate homology was lower (about 75%). Partial lineage information for the more distantly related Halicephalobus sp. (clade IV) reveals a substantially different lineage (about 75%) and fate (about 57%) homologies. This broadening analysis of developmental patterns across the nematode phylum begins to provide insight into the evolutionary progression that gave rise to the C. elegans pattern.
C. briggsae is one of three nematode species most closely related to C. elegans and is increasingly the subject of detailed comparative analyses with C. elegans. Although the two species have similar larval and adult morphology, interspecific hybrids exhibit a variety of early embryonic defects (Baird and Yen, 2000) and their genomes are highly divergent. The two species shared a most recent common ancestor 60–110 million years ago using a molecular clock based on the arthopod-nematode split (Coghlan and Wolfe, 2002), and the average nucleotide divergence rate is estimated to be 1.8 synonymous substitutions per synonymous site (Stein et al., 2003). For comparison, the most recent common ancestor of mice and humans lived about 60 million years ago and the average substitution rate per site is 0.6 (Waterston et al., 2002). Although synteny is strongly conserved between C. elegans and C. briggsae, hundreds of chromosomal rearrangements are found between the two species and there has been substantial expansion and contraction of gene families (Hillier et al., 2007; Thomas, 2006). For example, the FTR (Fog Two Related) gene family has about 30 members in both species, but phylogenetic analysis suggests that each family is the result of species-specific expansions (Nayak et al., 2005). The C. elegans FTR family includes the gene fog-2, which is a recently derived gene critical for sperm production in hermaphrodites. This is consistent with the hypothesis that the self-fertilizing mode of reproduction shown by both species in fact was derived independently in each lineage (Kiontke et al., 2004).
Genes involved in early developmental pathways also show some differences between the two species. For example, whereas C. elegans has two Notch receptors, lin-12 and glp-1, Rudel and Kimble (2001 and 2002) found C. briggsae has at least two lin-12 related genes, although maintaining only a single glp-1 gene. RNAi experiments in C. briggsae suggested that the genes have related but distinct functions in germline development, embryogenesis and larval development in the two species.
Recently, we have developed methods that greatly facilitate the determination of the embryonic cell lineage in C. elegans (Bao et al., 2006; Boyle et al., 2006). Knowledge of the lineage, combined with the draft genome sequence for C. briggsae, could couple altered developmental events with molecular changes. With these motives in mind, we created a strain that enabled us to use our methods with adaptations to determine the cell lineage and cell migrations of C. briggsae. The results shed light on the recent evolution of the highly evolved development cycle of Rhabditid nematodes.
Materials and methods
Strain construction
In order to trace the C. briggsae embryonic cell lineage using StarryNite (Bao et al, 2006) and AceTree (Boyle et al., 2006), a strain that ubiquitously expresses nuclear localized green fluorescence protein (GFP) in soma after the 30-cell stage was constructed. A wild type C. briggsae strain AF16 was obtained from C. elegans Genetic Center (CGC) and maintained in the same way as that for C. elegans. A HIS-72::GFP fusion driven by his-72 native promoter (Bao et al., 2006) was introduced into AF16 by ballistic bombardment (Praitis et al., 2001) using GFP as a selection marker. About 105 worms synchronized in young adult stage were bombarded and split into 12 extra large (150 mm in diameters) peptone plates. After three days at room temperature, the food on the plates was exhausted and arrested larvae that ubiquitously express GFP were picked. A strain that ubiquitously expressed GFP after two successive generations was established. The resulting strain, designated RHW10040 was backcrossed into AF16 three times. All the C. briggsae strains were maintained at room temperature in the same way as that for C. elegans.
4D microscopy and lineaging
A Zeiss LSM 510 confocal microscope was used to collect image series as previously described (Murray et al., 2006) with modifications. A second channel was introduced by adding another track to collect both GFP and differential interference contrast (DIC) images simultaneously. A line step 4 and scanning speed 6 were used throughout the imaging process. Images were collected every minute at 31 focal planes up to 400 minutes from first cell division at 20°C. Only image stacks from the embryos that hatched normally were used for subsequent analysis.
The image stacks were processed and analyzed as described (Murray et al., 2006). AceTree (Boyle et al., 2006) was used to visualize the image stacks and edit the potential errors made by StarryNite. The RHW10040 embryos do not show GFP expression in somatic tissue until around 28 cell stage. The somatic cell lineage was manually traced to that stage using DIC image and followed up by StarryNite output. The strain also shows no germline expression, so the germline precursor P4 and its progeny, Z2 and Z3, were manually traced up to 350 cell stage. Given the similarity of cell division patterns between C. elegans and C. briggsae, a similar cell naming scheme was adopted (Bao et al., 2006) with one change. The naming rule for Cpp was changed to i0ya. It was also corrected in the AceTree (Boyle et al., 2006) for naming C. elegans cells. Cell identity after 350 cell stage in a few cases may not be consistent with what has been reported previously (Sulston et al., 1983) because StarryNite uses the orientation of the daughter nuclei to define orientation of cell division, rather than the orientation of the spindle pole (Sulston et al., 1983). Any differences between the two methods after the 350 cell stage have not yet been resolved. In addition, in certain lineages, we observed reproducible asymmetries in the timing of divisions of certain daughter cells in C. elegans (Bao et al., unpublished data) that are not apparent in the lineage diagrams of Sulston et al. (Sulston et al., 1983). For example, the Cxpa divisions are reproducibly delayed compared to those of Cxpp.
Identification of programmed cell death
The cells undergoing programmed cell death change their morphology and light refractivity in the same ways in C. briggsae as in C. elegans. Typically, a raised-button-like appearance can be observed in those cells undergoing apoptosis in living animals using DIC (Robertson and Thomson 1982). In addition, these cells are distinguishable in the GFP channel by their brighter, denser and smaller nuclei.
Detection of the cell death relied on both the DIC and GFP images characteristic of dying cells. Death timings were assigned as close as possible to the last discernible time point of the dying cell. Two methods are used to identify a dying cell. First, given the conservation of cell lineage up to 350-cell stage, identities of cells known to die in C. elegans were used to examine the fate of its equivalents in C. briggsae. Lineage of these cells was followed up to the time point at which the characteristics of a dying cell were observed using both GFP and DIC images. Because of the density of nuclei after the last round of embryonic cell divisions, many of cells, especially some progeny of AB or MS, could not be traced reliably to the point they die. Second, starting from a DIC or GFP image of a late staged embryo, the cells with typical phenotypes of a dying cell were manually traced backward to the stages where reliable cell lineage was established. Thus, the identity of the dying cell was assigned. The whole process was repeated in at least two independent series where the identity of a dying cell agreed with one another.
Cell migrations
Embryonic cells undergo substantial migrations during embryogenesis. AceTree enables the ready observation of cell movements using a three-dimensional (3D) space-filling method (Boyle et al., 2006). The individual nuclei and cell divisions can be followed over time in the 3D movie. In addition, cell groups from a common ancestor can be differentially colored so that they can be compared for many features such as bilateral symmetry, migration directions and distances. For movie making, a single frame from each time point was saved as a JPEG file and resulting frames were used to produce movie using ImageReady according to supplier’s description.
Reliability
In order to achieve high confidence and reproducibility, a total of six embryonic series were traced to the 200 cell stage and lineage reproducibility was cross confirmed among all the series to verify the P4 division timing. Three of the six series were further traced to approximately 350 cell stage at which time point all of the cell identities can be cross confirmed between the three series. At this stage, most of the cells had completed the second to last round of embryonic divisions. Given the gradual degradation of GFP signal through the depth of embryo, one embryo mounted with opposite orientation in relation to the remaining two was included for maximization of ratio of signal/background which is critical for correct assignment of cell identity by StarryNite (Bao et al., 2006). Lineage tracing in C. briggsae is a de novo process without reference to that of C. elegans.
Ortholog assignment
Orthology is used to define genes separated from one another by speciation while paralogy describe those separated by gene duplication events (Fitch W. 1970). To identify members of the Notch and Wnt signaling pathways in C. briggsae and to establish their orthologous and paralogous relationships to the components of the pathways in C. elegans, we first identified C. elegans pathway components from the literature and WormBase (160). Using these we then examined InParanoid and TreeFam databases to find related genes based on the C. briggsae predicted gene set. For C. elegans genes where the InParanoid (O’Brien and Sonnhammer 2005) and TreeFam (Li et al., 2006) gave discordant results or no ortholog, we carried out tBLASTn searches against C. briggsae genome using default parameters (E value <10−10 and >40% identity over 50 amino acids). When tBLASTn revealed potential coding segments not part of an existing gene model, the region was used for ab initio gene prediction with FGENESH (Salamov and Solovyev, 2000). The resulting sequences were used for multiple alignments using ClustalW (Higgins and Sharp 1988) and construction of maximum likelihood (ML) tree using Phyml (Guindon and Gascuel, 2003) as described (Zhao et al., 2007). A C. briggsae gene is deemed as a C. elegans ortholog when the two form a unique cluster on a ML tree.
In several cases inspection of the available data led us to investigate possible gene prediction or sequencing/assembly errors. For lin-12, we found evidence for three copies in the whole genome shotgun assembly (CB25), all on supercontig cb25.fpc0201, with two lin-12-like genes (CBG06826 and CBG06829) present in inverted orientation between 175 kb and 200 kb and a third copy (CBG06951) at the end of the supercontig (700–705kb) but none of the predicted genes matched the sequence of the lin-12.1 cDNA or its related genomic segment (Rudel and Kimble, 2002). We also found a completed BAC sequence that spanned the region containing lin-12.1 (AC140918.1) that agreed with the lin-12.1 sequence. The BAC agreed with the cb25.fpc0201 sequence for most of its length, but differed in the region containing lin-12. A careful investigation of the cb25.fpc0201 supercontig and reassembly of the reads from the relevant regions revealed that there had been an assembly error and that the reads contributing to the third copy of the gene at the end of the contig could be readily assembled into the correctly assembled region. The corrected sequence thus agrees with the BAC sequence and has only two copies of the lin-12 gene. These have a head-to-head inverted orientation with ~11 kb separating the ATG start codons. The left copy corresponds to lin-12.1 and has the same orientation to the right flanking genes as lin-12 does in C. elegans, suggesting this is the true ortholog. The right copy is 95–100% identical to that but lacks the sequence corresponding to the carboxyl terminal end of lin-12.1, removing the PEST domain. Rudel and Kimble (2002) report recovering a second lin-12 cDNA, presumably corresponding to this copy and suggesting the gene is still expressed.
Results
The embryonic cell lineages of C. elegans and C. briggsae are remarkably similar
To examine the embryonic cell lineage of C. briggsae we constructed a strain stably expressing a GFP::histone H3.3 fusion (see Materials and Methods ). Using automatic lineage tracing software package, StarryNite (Bao et al., 2006) and AceTree (Boyle et al, 2006) in combination with DIC images for the early divisions, we traced the lineages for three image series to about the 350-cell stage (approximately the time point that E gives rise to its 16th descendant) and for three additional series to approximately the 200-cell stage. Similar lineage trees were observed among all the image series. These series were compared to twenty previously described embryonic lineage seriess from C. elegans (Bao et al., 2006) by two methods. First, we manually compared the C. briggsae lineage trees to three embryonic lineage trees for C. elegans in an attempt to find obvious systematic changes. Second, we computationally compared the C. briggsae lineages against twenty C. elegans lineages for significant differences in cell cycle length and position.
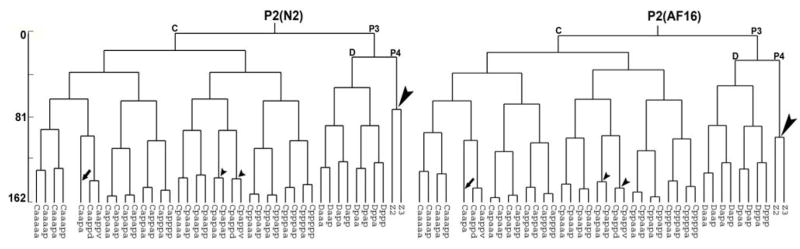
The C. briggsae cell lineage is very similar to that of C. elegans (Figure 1, Figure S1). Up to 350-cell stage, not only did the same founder cells give rise to an identical number of progeny, but also they divided with similar spindle orientation (for example, ABar) and with similar timing (Figure 1, Figure S1 and S2). Thus for every C. elegans cell there is a lineally equivalent C. briggsae cell with a comparable cell cycle length (accordingly, cells were named following the C. elegans convention (Sulston, et al., 1983)). For example, in C. briggsae, as in C. elegans, the E lineage divisions are slower compared to the MS lineage and the D lineage divisions are slower compared to C. Also the asymmetries of cell division timing seen between certain daughters of some C. elegans within MS, C and D founder lineages, were also observed in C. briggsae (Figure 1; Figure S1). For example, the cell cycle length of Caapa is 138.2±2.1 minutes whereas its sister Caapp lives for only 48.6±2.4 minutes in C. elegans; their cell cycle lengths in C. briggsae are 141.8±1.9 and 51.3±3.0 minutes respectively (Figure 1 and data not shown). Other asymmetries observed in both species include the longer cell cycle length of Capa versus Capp, Cppp versus Cppa, Daa versus Dap and Dpa versus Dpp (Figure 1). A close examination of the trees suggests that more subtle asymmetries are conserved as well (Figure 1, Figure S1).
Figure 1.
Conservation of asymmetries of cell division timing between C. elegans (left panel) and C. briggsae (right panel). As in C. elegans, P2 divides asymmetrically and gives rise to the founder cells, C and P3. P3 divides to produce founder cell D and germline precursor P4. In both species, Caapa divisions are significantly delayed (P<0.01) as indicated by arrows. Up to the fourth round of the C divisions and the third round of D divisions, the division timing is well conserved between the two species. One exception is the P4 division which is significantly delayed in C. briggsae compared to that of C. elegans (p<0.01, indicated by large arrowhead). The relative division timing for later rounds of cell division are more variable, presumably reflecting the accumulation of variation from preceding rounds of division. An example of natural variations between division timing of a sibling is indicated by small arrowheads. Time scale denotes minutes from division of P2 to division of E8 to give rise to E16 at 20 °C.
In order to compare the cell cycle length systematically, we compared the average cell cycle length for each cell between the six C. briggsae embryos and twenty C. elegans embryos. The cell cycle lengths were highly correlated to each other (r = 0.96) (Figure S3). The most significant outlier is P4. The average cell cycle length (from birth to the subsequent division) of P4 is 23 minutes longer (41.1%; n=6) in C. briggsae than that in C. elegans (p<0.01, Figure 1). This difference in timing results in a difference in the two species in the orientation and distance between the two P4 daughters (Z2 and Z3) in the 350-celled embryo (Figure 3). The earlier division of P4 in C. elegans is also associated with a shift of the Dp division orientation relative to that in C. briggsae (data not shown).
Figure 3.
Comparison of cell positions and migration. Left panel: cell migration of founder cell progeny at 350 cell stage is well conserved between C. elegans (top) and C. briggsae (bottom). Progeny from each founder cell are color coded: ABa, red; ABp, blue; MS, cyan; E, green; C, magenta; D: pink; P4: yellow. Progeny of the same founder cell occupy a similar niche within the embryo. Right panel: bilaterally symmetric distribution of progeny involves the same set of founder cells in C. elegans (top) and C. briggsae (bottom). MSpaapp is the first cell to die (before 350 cell stage) in both species (marked grey). Note the orientation and distance between the two P4 progeny (yellow) are different between the two species. The progeny of hyp7 precursors (Cpaa and Caaa) appear less symmetric in C. elegans than that of C. briggsae presumably caused by the interdigitation event occurring at the stage. The positions of D progeny are well maintained between the two species.
The other notable outliers from the pairwise comparison were the four E descendents, Eal, Ear, Epl and Ear, whose cell cycle lengths are 16.7% longer in C. briggsae than in C. elegans (data not shown). However, the standard deviation is high for these cells for C. briggsae, suggesting such differences might represent more variation within one species rather than actual differences between the two species.
The same cells die in both species
Although the similarity of the lineage patterns between the two species is a strong indicator of a conserved developmental pathway, the 3-D image series allow the assessment of other indicators of cell fate. One such marker of cell fate is programmed cell death, which affects 113 of 671 embryonic cells (Sulston et al., 1983). The cell deaths affect only specific progeny and produce a distinctive morphology, both in DIC images, where the nuclei exhibit a raised button-like appearance and in GFP images, where the signal becomes condensed and intense.
We exploited this distinctive morphology to identify cell deaths in C. briggsae and then trace their lineages where possible. With the exception of MSpaapp, the cell deaths occurred after the 350 cell stage. To evaluate these later cell deaths, we manually traced sublineages through further divisions for cells that were sufficiently well-separated in our image series to confidently assign daughters and establish fates (see Materials and Methods). Of the sublineages in the MS lineage of C. elegans that produce14 cell deaths, we were able to follow the sublineages yielding 8 cell deaths in C. briggsae; all eight clearly involved equivalent cells (Figure 2, supplemental Table 1). Similarly in the AB and C lineages we could assign 18 additional cell deaths to specific lineages and 16 of these clearly involved lineally equivalent cells in the two species. Ambiguities in assigning the axis of cell division precluded unambiguous assignment of the two other deaths. Additional deaths were observed but not readily traced to specific lineages. However, of the cell deaths we could trace, our observations show that patterns of cell death are likely to be highly similar between the species.
Figure 2.
Cell death is well conserved between the two species. Shown are eight cell death events (crosses) identified in the MS lineage, all of which involved exactly the same set of cells in C. elegans (Sulston et al., 1983, data not shown). A bold bar denotes the death of its posterior daughter, MSpapapap, which was observed at a time beyond the cutoff used here (about 450 cell stage). Time scale denotes minutes at 20 °C from MS division to the arbitrary cutoff time point used for cell death identification.
Spatial positioning of founder cells and their descendants is well conserved between the two species
Cell positions during development provide an additional parameter to infer cell fate determinations. Using all six series, we compared the positions of lineally equivalent cells throughout their cell cycle lengths for all cells through the 200-cell stage. Along the x- (anterior posterior) axis, where orientation of each embryo is most reproducibly established, the average nuclear positions showed an extremely high correlation co-efficient (r = 0.99, Figure S4). Along the y- (left right) and z- (dorsal ventral) axis the correlation coefficients were slightly less (0.96 and 0.94 respectively); these slightly lower correlation coefficients may reflect at least in part the greater difficulty in reproducibly placing the embryos at the precise dorsal-venrtal or left-right orientations.
To extend this analysis and to illustrate the similarity of cell positions, we visually compared the positions of the founder cell progeny at the 350-cell stage in the two species (Figure 3). The cell groups and indeed the individual cells have migrated to remarkably similar positions. We also looked in detail at lineages that in C. elegans produce bilaterally symmetric cells. The lineally equivalent cells in C. briggsae also show similar bilateral symmetry. For example, ABplp and ABprp begin in asymmetric positions, with the former lying at the anterior left side and the later lying on posterior right side of the embryo respectively (See supplemental movie, Figure S5). Each gives rise to 64 progeny (mostly precursors of neuronal ganglia) that assume symmetric and equivalent positions after substantial migration in both species.
The major exceptions to the similarity of position of lineally equivalent cells were Z2 and Z3, the daughters of P4. These are significantly farther apart in C. elegans than C. briggsae ((p < 0.01, n=6), presumably reflecting the later division of P4 in C. briggsae. The delayed division also appears to affect the orientation of the spindle in the division of Dp, which is more dorsal/ventral in C. briggsae and anterior/posterior in C. elegans.
Discussion
Our results show that embryonic development in C. elegans and C. briggsae is remarkably similar as seen in their lineage patterns, in the cells undergoing programmed cell death and in position of cells through 350 cells. Although this similarity could result from convergent evolution, conservation of development since the most recent common ancestor seems more likely. This conservation over approximately 60–100 Myr, despite the substantial changes in the genome sequence (Stein et al., 2003), suggests that most changes in the pattern of cell division and migration in early development are deleterious and rapidly removed by purifying selection. Nonetheless, the defects observed in interspecific hybrids (Baird and Yen, 2000) indicate that the details of the underlying molecular networks have diverged. The conservation of individual cell cell cycle length and the relative timing of divisions may imply a substantial role for intercellular communication or at least coordination in early development.
The greatest difference we observed between the two lineages was in the timing of the P4 division. Later differences in germline development have been noted between the two species (Nayak et al., 2005; Hill et al., 2006) and as referred to above the hermaphroditic mode of reproduction was independently evolved in the two species (Kiontke et al., 2004) and involves different regulatory pathways. Whether the delay in P4 division in C. briggsae is part of this larger picture of rapid divergence of germline development will require further experiments.
The apparent plasticity of the Notch pathway genes seems paradoxical in the face of this conservation. The changes in the Notch genes contrast with those of the WNT pathway, where the conservation of 1:1 orthologs is obvious, but both pathways play essential roles in early cell fate decisions. Perhaps some of the changes in Notch ligands and targets involve later developmental steps, including gonad and vulva development (Chen and Greenwald, 2004), that are beyond the stages analyzed here. Gene duplications may have obscured the orthology relationships without changing the function of the genes involved in early development. Perhaps some of the observed differences simply reflect the transient effects of ongoing genomic duplications and rearrangements that are continuously disrupting the genome. Certainly, the second copy of the C. briggsae lin-12 gene (lin-12.2) seems likely to be on the path to becoming a pseudogene: it lacks the carboxyl-terminal domain and is more divergent from C. elegans lin-12. Similarly, one of the three copies of the arg-1/apx-1 ligands in C. briggsae, CBG10218, is probably a pseudogene, with a nonsense mutation in an exon encoding a critical DSL domain (Figure 4 and data not shown). Thus the observed differences in the Notch pathway genes may have little or no impact on early development.
Figure 4.
Comparison of Notch and Wnt signaling pathway components between C. elegans and C. briggsae. An individual maximum likelihood (ML) phylogenetic tree was built as described in Materials and Methods for Notch ligands, receptors and targets respectively. Similar trees were built for Wnt ligands, receptors and downstream effectors, β-catenin proteins. C. elegans genes are labeled in green and C. briggsae in blue. Numbers on the branches denote the percentage of replicates from 1000 bootstrapping (only >70% shown). Panel A: Notch pathway; panel B: Wnt pathway. A putative pseudogene is marked with star.
Presently our methods do not allow us to follow the lineage through its last round of cell division or to where the cells take up their eventual larval positions, so possibly some changes exist in these later lineages or in the eventual fate of the cells. However, should our present conclusions be extended, it would indicate that the C. elegans development pathway was largely established sometime after the split with Pellioditis marina (clade V) and before the most recent common ancestor with C. briggsae. It remains an open question as to when invariant lineage patterns were fixed in worm species such as Caenorhabditis species. Lineages of additional species, deeper into development should establish this time more firmly. The current set of nematode genome sequences consists almost entirely of species not significantly more divergent from C. elegans than C. briggsae. This work suggests the utility of sequencing additional genomes for nematodes with different levels of developmental variability. This would provide a platform for defining genomic changes associated with developmental changes.
Supplementary Material
Acknowledgments
We thank James Thomas, Weiqing Li for helpful discussions and C. briggsae strains and Bhagwati P. Gupta C. briggsae mutant strains. We also thank other members of Waterston lab for insightful comments and suggestions. AF16 strain is provided by the Caenorhabditis Genetics Center (CGC) which is supported by the National Institute of Health National Center for Research Resources.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arendt D. Comparative aspects of gastrulation. In: Stern CD, editor. Gastrulation- from cells to embryo. New York: Cold Spring Harbor Laboratory Press; 2004. pp. 679–693. [Google Scholar]
- Austin J, Kimble J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell. 1987;51:589–599. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]
- Baird SE, Yen WC. Reproductive isolation in Caenorhabditis: terminal phenotypes of hybrid embryos. Evol Dev. 2000;2:9–15. doi: 10.1046/j.1525-142x.2000.00031.x. [DOI] [PubMed] [Google Scholar]
- Bao Z, Murray JI, Boyle T, Ooi SL, Sandel MJ, Waterston RH. Automated cell lineage tracing in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2006;103:2707–12. doi: 10.1073/pnas.0511111103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter M. Comparative genomics: two worms are better than one. Nature. 2003;426:395–396. doi: 10.1038/426395a. [DOI] [PubMed] [Google Scholar]
- Boyle TJ, Bao Z, Murray JI, Araya CL, Waterston RH. AceTree: a tool for visual analysis of Caenorhabditis elegans embryogenesis. BMC Bioinformatics. 2006;7:275. doi: 10.1186/1471-2105-7-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Greenwald I. The lateral signal for LIN-12/Notch in C. elegans vulval development comprises redundant secreted and transmembrane DSL proteins. Dev Cell. 2004;6:183–92. doi: 10.1016/s1534-5807(04)00021-8. [DOI] [PubMed] [Google Scholar]
- Coghlan A, Wolfe KH. Fourfold faster rate of genome rearrangement in nematodes than in Drosophila. Genome Res. 2002;12:857–867. doi: 10.1101/gr.172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WM. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- Good K, Ciosk R, Nance J, Neves A, Hill RJ, Priess JR. The T-box transcription factors TBX-37 and TBX-38 link GLP-1/Notch signaling to mesoderm induction in C. elegans embryos. Development. 2004;131:1967–78. doi: 10.1242/dev.01088. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–44. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Hill RC, de Carvalho CE, Salogiannis J, Schlager B, Pilgrim D, Haag ES. Genetic flexibility in the convergent evolution of hermaphroditism in Caenorhabditis nematodes. Dev Cell. 2006;10:531–8. doi: 10.1016/j.devcel.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Hillier LW, Miller RD, Baird SE, Chinwalla A, Fulton LA, Koboldt DC, Waterston RH. Comparison of C. elegans and C. briggsae Genome Sequences Reveals Extensive Conservation of Chromosome Organization and Synteny. PLoS Biol. 2007;5:e167. doi: 10.1371/journal.pbio.0050167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houthoofd W, Jacobsen K, Mertens C, Vangestel S, Coomans A, Borgonie G. Embryonic cell lineage of the marine nematode Pellioditis marina. Dev Biol. 2003;258:57–69. doi: 10.1016/s0012-1606(03)00101-5. [DOI] [PubMed] [Google Scholar]
- Hutter H, Schnabel R. glp-1 and inductions establishing embryonic axes in C. elegans. Development. 1994;120:2051–64. doi: 10.1242/dev.120.7.2051. [DOI] [PubMed] [Google Scholar]
- Kimble J, Simpson P. The LIN-12/Notch signaling pathway and its regulation. Annu Rev Cell Dev Biol. 1997;13:333–61. doi: 10.1146/annurev.cellbio.13.1.333. Review. [DOI] [PubMed] [Google Scholar]
- Kiontke K, Gavin NP, Raynes Y, Roehrig C, Piano F, Fitch DH. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc Natl Acad Sci USA. 2004;101:9003–8. doi: 10.1073/pnas.0403094101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Coghlan A, Ruan J, Coin LJ, Heriche JK, et al. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572–80. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Hill RJ, Priess JR. POP-1 and anterior-posterior fate decisions in C. elegans embryos. Cell. 1998;92:229–39. doi: 10.1016/s0092-8674(00)80917-4. [DOI] [PubMed] [Google Scholar]
- Lin R, Thompson S, Priess JR. pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell. 1995;83:599–609. doi: 10.1016/0092-8674(95)90100-0. [DOI] [PubMed] [Google Scholar]
- Mello CC, Draper BW, Priess JR. The maternal genes apx-1 and glp-1 and establishment of dorsal-ventral polarity in the early C. elegans embryo. Cell. 1994;77:95–106. doi: 10.1016/0092-8674(94)90238-0. [DOI] [PubMed] [Google Scholar]
- Mickey KM, Mello CC, Montgomery MK, Fire A, Priess JR. An inductive interaction in 4-cell stage C. elegans embryos involves APX-1 expression in the signaling cell. Development. 1996;12:1791–1798. doi: 10.1242/dev.122.6.1791. [DOI] [PubMed] [Google Scholar]
- Moskowitz IP, Rothman JH. lin-12 and glp-1 are required zygotically for early embryonic cellular interactions and are regulated by maternal GLP-1 signaling in Caenorhabditis elegans. Development. 1996;122:4105–4117. doi: 10.1242/dev.122.12.4105. [DOI] [PubMed] [Google Scholar]
- Murray JI, Bao Z, Boyle TJ, Waterston RH. The lineaging of fluorescently-labeled Caenorhabditis elegans embryos with StarryNite and AceTree. Nat Protoc. 2006;1:1468–76. doi: 10.1038/nprot.2006.222. [DOI] [PubMed] [Google Scholar]
- Nayak S, Goree J, Schedl T. fog-2 and the evolution of self-fertile hermaphroditism in Caenorhabditis. PLoS Biol. 2005;3:e6. doi: 10.1371/journal.pbio.0030006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves A, Priess JR. The REF-1 family of bHLH transcription factors pattern C. elegans embryos through Notch-dependent and Notch-independent pathways. Dev Cell. 2005;8:867–879. doi: 10.1016/j.devcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- O’Brien RM, Sonnhammer ELL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Research. 2005;33:476–485. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M, Polakis P. Wnt signaling in oncogenesis and embryogenesis--a look outside the nucleus. Science. 2000;287:1606–9. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–26. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess JR, Schnabel H, Schnabel R. The glp-1 locus and cellular interactions in early C. elegans embryos. Cell. 1987;51:601–11. doi: 10.1016/0092-8674(87)90129-2. [DOI] [PubMed] [Google Scholar]
- Robertson AG, Thomson JN. Morphology of programmed cell death in the ventral nerve cord of Caenorhabiditis elegans larvae. J Embryol Exp Morphol. 1982;67:89–100. [Google Scholar]
- Rocheleau CE, Downs WD, Lin R, Wittmann C, Bei Y, et al. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell. 1997;90:707–16. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- Rudel D, Kimble J. Conservation of glp-1 regulation and function in nematodes. Genetics. 2001;157:639–54. doi: 10.1093/genetics/157.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel D, Kimble J. Evolution of discrete Notch-like receptors from a distant gene duplication in Caenorhabditis. Evol Dev. 2002;4:319–33. doi: 10.1046/j.1525-142x.2002.02027.x. [DOI] [PubMed] [Google Scholar]
- Salamov AA, Solovyev VV. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–22. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierenberg E. Unusual cleavage and gastrulation in a freshwater nematode: developmental and phylogenetic implications. Dev Genes Evol. 2005;215:103–8. doi: 10.1007/s00427-004-0454-9. [DOI] [PubMed] [Google Scholar]
- Skiba F, Schierenberg E. Cell lineages, developmental timing, and spatial pattern formation in embryos of free-living soil nematodes. Dev Biol. 1992;151:597–610. doi: 10.1016/0012-1606(92)90197-o. [DOI] [PubMed] [Google Scholar]
- Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, et al. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:E45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Thomas JH. Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res. 2006;16:1017–30. doi: 10.1101/gr.5089806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wiegner O, Schierenberg E. Regulative development in a nematode embryo: a hierarchy of cell fate transformations. Dev Biol. 1999;215:1–12. doi: 10.1006/dbio.1999.9423. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Thomas JH, Chen N, Sheps JA, Baillie DL. Comparative Genomics and Adaptive Selection of the ATP-Binding-Cassette Gene Family in Caenorhabditis Species. Genetics. 2007;175:1407–18. doi: 10.1534/genetics.106.066720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.