Summary
Using an orthogonal tRNA-synthetase pair, unnatural amino acids can be genetically encoded with high efficiency and fidelity; and over forty unnatural amino acids have been site-specifically incorporated into proteins in E. coli, yeast, or mammalian cells. Novel chemical or physical properties embodied in these amino acids enable new means for tailored manipulation of proteins. This review summarizes the methodology and recent progress in expanding this technology to eukaryotic cells. Applications of genetically encoded unnatural amino acids are highlighted with reports on labeling and modifying proteins, probing protein structure and function, identifying and regulating protein activity, and generating proteins with new properties. Genetic incorporation of unnatural amino acids provides a powerful method for investigating a wide variety of biological processes both in vitro and in vivo.
Introduction
The genetic code consists of 64 triplet codons specifying 20 canonical amino acids and 3 stop signals. This code is preserved in all three kingdoms of life with few reassignments (Knight et al., 2001). Although various amino acid derivatives have been identified in natural proteins, most of them are posttranslational modifications of the canonical amino acids (Uy and Wold, 1977). Two additional amino acids, selenocysteine (Bock et al., 1991) and pyrrolysine (Srinivasan et al., 2002), are also found to be incorporated into proteins co-translationally and thus regarded as natural expansion of the genetic code (Ambrogelly et al., 2006). With these amino acids and with the help of posttranslational modifications and cofactors, proteins are able to carry out various functions for sustaining life.
Given the important roles of proteins in biology, it is desirable to be able to manipulate proteins at will for understanding protein structure-function relationship, investigating protein-involved biological processes, and generating proteins and organisms with new properties. Therefore, tremendous efforts have been made on the incorporation of unnatural amino acids into proteins to introduce new functional groups apart from those found in the canonical amino acids, through chemical or biosynthetic means (Wang and Schultz, 2004). Chemical methods use chemistry to directly modify or prepare proteins. Early efforts focused on derivatizing reactive side chains of the protein, such as the thiol group of cysteine and the ε-amino group of lysine (Kaiser et al., 1985). The advance of stepwise solid-phase peptide synthesis makes possible the complete synthesis of peptides and small proteins (usually < 100 residues) containing any synthetically accessible unnatural amino acids (Kent, 1988). The protein size limitation is subsequently overcome by semisynthetic protein ligation methods, in which two or more peptide fragments are chemically ligated to make the full-length protein (Dawson et al., 1994, Muir, 2003). By contrast, biosynthetic methods harness the endogenous cellular machinery to translate the target protein, in which the unnatural amino acid is incorporated co-translationally. Bacterial strains auxotrophic for a particular amino acid have been used to globally replace the amino acid with a close structural analogue (Cohen and Cowie, 1957, Minks et al., 2000, Link et al., 2003). For the site-specific incorporation of unnatural amino acids, a suppressor tRNA is chemically acylated with an unnatural amino acid. When added into cell extracts or Xenopus oocytes that support protein translation, the acylated tRNA incorporates the attached unnatural amino acid in response to a stop or extended codon (Noren et al., 1989, Bain et al., 1989, Nowak et al., 1995). All of these methods have proved extremely valuable but each has limitations in either the lack of site selectivity, the in vitro nature of the method, or the low incorporation efficiency (Wang and Schultz, 2004).
The ability to genetically encode unnatural amino acids in the same manner as the canonical amino acids would enable specific changes to be precisely made in proteins directly in vivo, thus providing novel tools for understanding biology in molecular terms in the native settings. Here we review the methodology for genetically incorporating unnatural amino acids into proteins in live cells, and focus on recent advances in expanding the method to eukaryotic cells. Representative applications are discussed to illustrate the utility and scope of this technology in studying various biological questions.
Methodology
Similar to canonical amino acids, the genetic encoding of an unnatural amino acid requires a dedicated set of components including a tRNA, a codon, and an aminoacyl-tRNA synthetase (hereafter referred to as synthetase) (Figure 1) (Wang and Schultz, 2002). This tRNA/codon/synthetase set, termed the orthogonal set, must not crosstalk with endogenous tRNA/codon/synthetase sets and be functionally compatible with other components of the translation apparatus. Specifically, the orthogonal tRNA should not be recognized by any endogenous synthetase and it should decode the orthogonal codon, which is not assigned to any canonical amino acid. The orthogonal synthetase should not charge any endogenous tRNA but the orthogonal tRNA, and charge it with the unnatural amino acid only. The unnatural amino acid cannot be a substrate for any endogenous synthetase and be available in the cytoplasm of the host cell. Once expressed in cells, the orthogonal synthetase charges the orthogonal tRNA with the desired unnatural amino acid, and the acylated tRNA incorporates the attached unnatural amino acid in response to the unique codon into proteins by utilizing the endogenous translational machinery. As all elements are genetically encodable, this method has the potential to be applied to almost all genetically tractable organisms.
Figure 1.
A general method for genetically encoding unnatural amino acids in live cells.
Method Development Escherichia coli
E. coli was initially chosen as the host organism to develop this general method for genetically encoding unnatural amino acids in vivo, because its translational machinery had been extensively studied and genetic manipulation of E. coli is efficient and relatively straightforward. A unique codon is required to specify the unnatural amino acid, and the amber stop codon has been chosen for this purpose. There are three stop codons that do not code for amino acids but function as a signal for termination of translation. By reassigning a “blank” stop codon to an unnatural amino acid, any overlap and ambiguity with sense codons is avoided. The amber codon became the “blank” codon of choice in this approach because (i) the amber codon is the least used among the three stop codons in E. coli, and (ii) amber suppressor tRNAs have been identified or engineered in E. coli to readthrough the amber codon with canonical amino acids, and they do not significantly affect the growth rates of E. coli cells (Normanly et al., 1990). Therefore, specifying an unnatural amino acid with the amber codon was expected to minimize perturbations to the genetic code and to the health of the host.
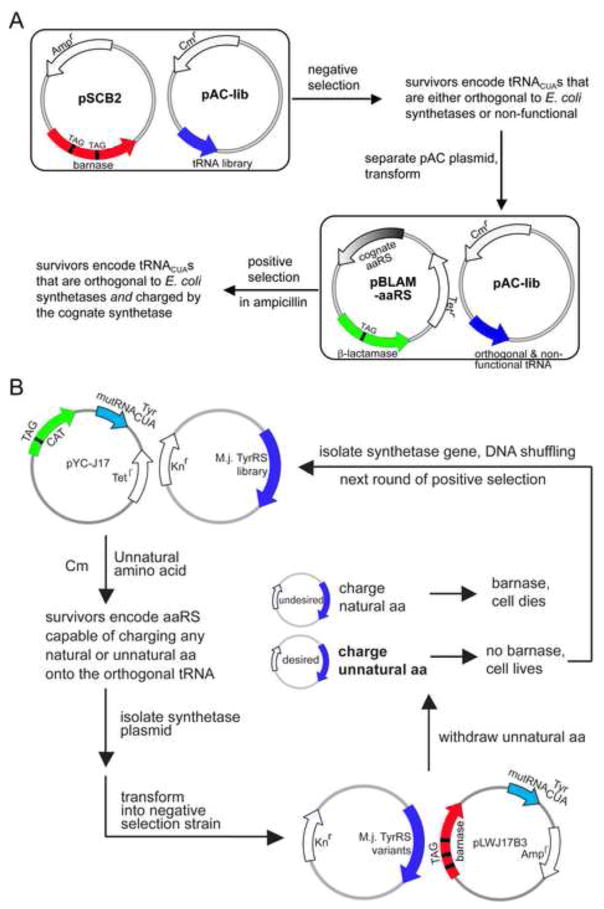
Endogenous tRNAs and synthetases recognize their cognate partners through delicate interactions between various regions (Ibba and Soll, 2000). As there is already a pool of tRNA/synthetase pairs inside cells, it is challenging to generate a new tRNA/synthetase pair orthogonal to existing ones and functional in translation. A successful strategy is to import a tRNA/synthetase pair from species in a different kingdom of life followed by fine tuning with selections. This strategy is based on early reports that in vitro cross-species aminoacylation is often low (Kwok and Wong, 1980). The first orthogonal tRNA/synthetase pair E. coli generated from archaeal bacteria was derived from the tRNATyr/tyrosyl-tRNA synthetase (TyrRS) from Methanococcus jannaschii (Wang et al., 2000). The anticodon of the tRNA is mutated to CUA for recognizing the amber codon UAG. This M. jannaschii tRNATyrCUA (MjtRNATyrCUA) and its cognate MjTyrRS function efficiently in translation in E. coli, but there is a small degree of aminoacylation of this MjtRNATyrCUA by endogenous E. coli synthetases. To make a completely orthogonal tRNA while maintaining its affinity for the cognate synthetase, a general strategy has been developed to evolve orthogonal tRNAs in E. coli (Figure 2A) (Wang and Schultz, 2001). A library of tRNA mutants is prepared by randomizing specific nucleotides, and the orthogonal tRNA is selected using a combination of negative and positive selections. The negative selection removes non-orthogonal tRNAs from the library, and the subsequent positive selection removes non-functional tRNAs but allows only orthogonal tRNAs with high affinity for the cognate synthetase to pass. This approach was applied to the MjtRNATyrCUA and the mutRNATyrCUA was identified that is orthogonal in E. coli and functions efficiently with MjTyrRS to translate the amber codon.
Figure 2.
Double-sieve selection with a mutant library for evolving orthogonal tRNAs or unnatural amino acid-specific synthetases. (A) Evolving orthogonal tRNAs in E. coli. The negative selection uses toxic barnase gene containing two TAG stop codons at permissive sites. tRNAs non-orthogonal to E. coli synthetases will be charged with amino acids and suppress the TAG codons to produce a functional barnase, which kills the cell. The positive selection uses the β–lactamase gene containing a TAG codon at a permissive site. tRNAs charged by the cognate synthetase and functional in translation will suppress the TAG codon and express active β–lactamase, which enables cells to survive in ampicillin. Cells containing non-functional tRNAs will be killed by ampicillin. (B) Evolving orthogonal synthetases to be specific for an unnatural amino acid in E. coli. The positive selection uses the chloramphenicol acetyl transferase (CAT) gene containing a permissive TAG stop codon in the presence of the unnatural amino acid. Mutant synthetases capable of charging the orthogonal with the unnatural or any natural amino acid will express full length CAT, enabling cells to survive in chloramphenicol (Cm). The negative selection uses the barnase gene containing permissive TAG codons. Synthetase mutants active toward natural amino acids will charge the to suppress TAG codons in the barnase gene, leading to cell death. In the absence of the unnatural amino acid, synthetase mutants specific for the unnatural amino acid will survive the negative selection. To improve activity and specificity, initial synthetase gene hits can be recombined using DNA shuffling and subject to next round of selection.
The next step is to change the substrate specificity of the orthogonal MjTyrRS from tyrosine to a desired unnatural amino acid, which is also achieved using two step selections (Figure 2B). A mutant synthetase library (> 109 members) is generated by randomizing five amino acid residues in the active site of the MjTyrRS and the selection of the residues to be randomized was enabled by the availability of B. steraothermophilus TyrRS crystal structure (Brick et al., 1989). The library is first positively selected in the presence of the unnatural amino acid, in which active synthetase mutants able to charge the orthogonal mutRNATyrCUA with the unnatural amino acid or any common amino acid are identified. The active synthetases are then subjected to a negative selection, in which synthetase mutants charging a common amino acid are eliminated. In the end, only synthetase mutants specific for the unnatural amino acid will survive both selections. Using this approach, a mutant synthetase specific for the unnatural amino acid, o-methyltyrosine (1, Figure 4), was identified from the MjTyrRS library. Together with the orthogonal mutRNATyrCUA evolved above, it enables the site-specific incorporation of o-methyltyrosine into proteins in response to the amber codon in E. coli. Western blot and mass spectrometric analyses of the expressed protein indicate that the translation fidelity for o-methyltyrosine is close to that of common amino acids (Wang et al., 2001).
Figure 4.
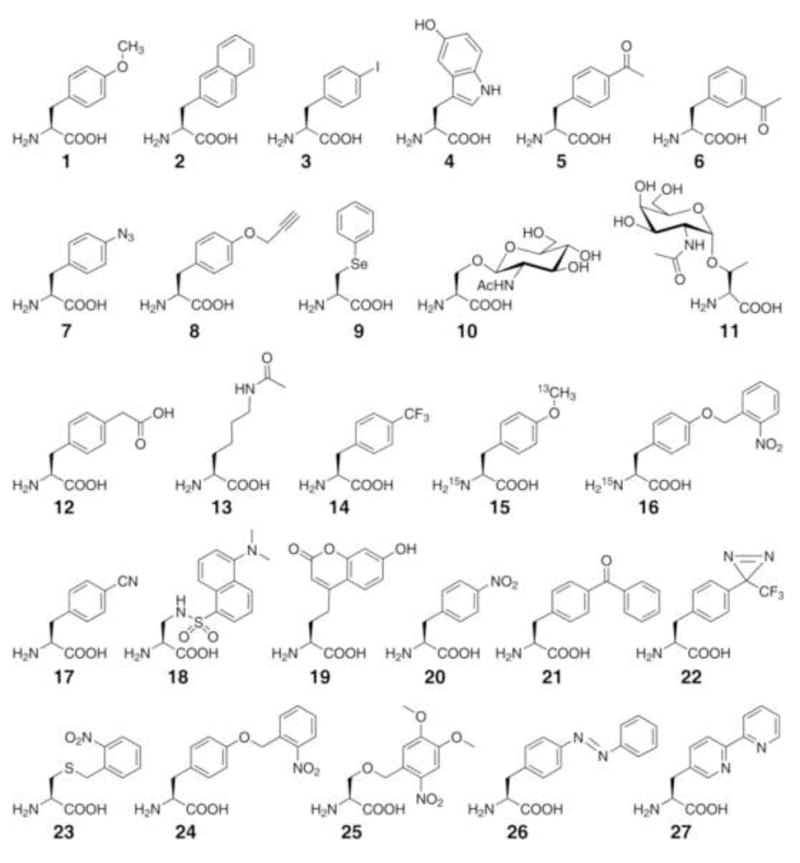
Structures for unnatural amino acids discussed in text.
This work represents the first expansion of the genetic code to include an unnatural amino acid using engineering approaches. Structurally similar to tyrosine and phenylalanine, o-methyltyrosine provides an excellent case to demonstrate the high translational fidelity that can be achieved using this approach. Meanwhile, it was important to test whether this method could be generally used to genetically encode various unnatural amino acids, especially those significantly deviating from common amino acids in structure. Subsequent experiments showed that L-3-(2-naphthyl)alanine (2), which represents a significant structural perturbation relative to tyrosine, can also be genetically incorporated into protein in E. coli (Wang et al., 2002). In the following years, the principle and methods developed herein have been used to genetically incorporate more than 40 different types of unnatural amino acids in E. coli (Wang et al., 2006). For most unnatural amino acids, suppression efficiencies range from 25% to 75% of the wild-type protein and translational fidelity is >99%. Yields of unnatural amino acid containing proteins are in the range of several milligrams to tens of milligrams per liter of cell culture. Additional orthogonal tRNA/synthetase pairs have since been generated (Anderson and Schultz, 2003), and four-base codons have also been used to encode unnatural amino acids in E. coli (Anderson et al., 2004). In addition, a biosynthetic pathway for p-aminophenylalanine was created in E. coli so that the strain is able to produce and genetically incorporate p-aminophenylalanine autonomously (Mehl et al., 2003).
Transition to Yeast
There are examples of eukaryotic organisms having natural changes in their genetic codes (Knight et al., 2001), suggesting that it is possible to artificially alter the genetic code in eukaryotes as well. The general strategy developed to genetically encode unnatural amino acids in E. coli should also be applicable to eukaryotic organisms. A number of tRNA/synthetase pairs derived from E. coli have been shown to be orthogonal in eukaryotic cells (Drabkin et al., 1996, Kowal et al., 2001), again due to the low cross kingdom aminoacylation. In particular, the E. coli tRNATyrCUA/TyrRS pair is orthogonal in yeast (Edwards and Schimmel, 1990, Kwok and Wong, 1980).
To evolve synthetases specific for unnatural amino acids in yeast, a general selection scheme analogous to that used in E. coli was set up, which also uses positive and negative selections with a mutant synthetase library. A synthetase library (108 in size) is similarly constructed by randomizing five active-site residues in the E. coli TyrRS that correspond to the five residues randomized in the MjTyrRS (Wang et al., 2001). Mutant synthetases are identified after several rounds of selection that, together with the E. coli tRNATyrCUA, incorporate a number of unnatural amino acids into proteins in yeast, albeit with rather low protein yields (about 0.05 mg/L) (Chin et al., 2003).
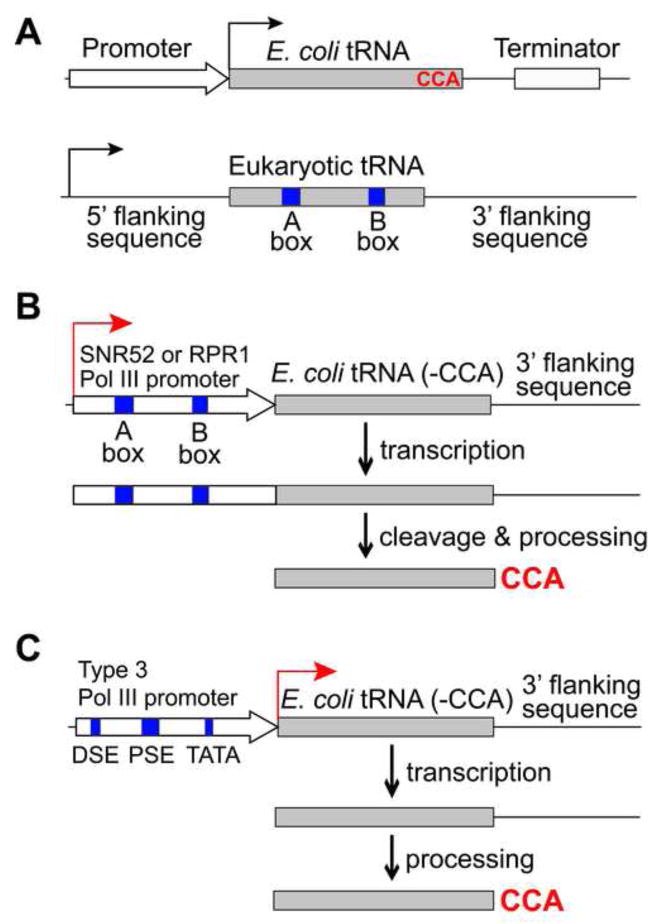
Low incorporation efficiency results in reduced yield of the full-length protein containing the unnatural amino acid and increase of truncated protein products terminated at the introduced amber codon, thus creating difficulties in employing genetically encoded unnatural amino acids strategy in yeast. To address this problem, new methods have been developed to efficiently express orthogonal prokaryotic tRNAs and to improve the target mRNA stability in yeast. Transcription and processing of tRNAs are different in prokaryotes and eukaryotes. A major distinction is that E. coli tRNAs are transcribed through promoters upstream of the tRNA gene, while eukaryotic tRNAs are transcribed through promoter elements within the tRNA known as the A- and B-box (Figure 3A) (Galli et al., 1981). The A- and B-box identity elements are conserved among eukaryotic tRNAs but are absent in many E. coli tRNAs. Creating the consensus A- and B-box sequences in E. coli tRNAs through mutation could cripple the tRNA (Sakamoto et al., 2002). The E. coli tRNATyrCUA has the B-box but no fully matched A-box. In the previously described system (Chin et al., 2003), only the tRNA structural gene was inserted in a plasmid, resulting in basal level of tRNA expression likely driven by a cryptic promoter elsewhere (Chen et al., 2007).
Figure 3.
General methods for efficient expression of prokaryotic tRNAs in eukaryotic cells using special Pol III promoters. (A) Gene elements for tRNA transcription in E. coli and eukaryotic cells. (B) Expression of prokaryotic tRNAs in yeast using an external SNR52 or RPR1 Pol III promoter, which contains the consensus A- and B-box sequences and is post-transcriptionally cleaved from the primary transcript. (C) A type-3 Pol III promoter, such as the H1 promoter, with a defined transcription initiation site is used to express prokaryotic tRNAs in mammalian cells.
One method for increasing tRNA expression is to use the 5′- and 3′-flanking sequences of an endogenous yeast suppressor tRNATyrCUA (SUP4) (Francis and RajBhandary, 1990, Drabkin et al., 1996, Kowal et al., 2001, Chen et al., 2007). The E. coli tRNATyrCUA without the -CCA trinucleotide is flanked by these sequences, and this cassette is repeated in tandem 3 to 6 times. A strong PGK1 RNA Polymerase II promoter is additionally placed upstream of the tandem arrangements (Chen et al., 2007). This combination gives an overall >50 fold increase in tRNA levels compared to the previous tRNA alone scheme. Together with an optimized expression of the synthetase, this approach improves the yield of unnatural amino acid containing proteins to 6–8 mg/L of yeast cells. A new method to efficiently express prokaryotic tRNAs in yeast involves an external promoter containing the consensus eukaryotic A- and B-box sequences (Figure 3B) (Wang and Wang, 2008). The promoter is placed upstream of the E. coli tRNA (without 3′-CCA) to drive the transcription of a primary RNA consisting of the promoter and the tRNA. The promoter is cleaved post-transcriptionally to yield the mature tRNA. Two yeast Pol III promoters, the RPR1 promoter and the SNR52 promoter, have been shown to efficiently drive the expression of the E. coli tRNATyrCUA in yeast. The expressed E. coli tRNATyrCUA is 6 to 9 fold more active in translation than the same tRNA transcribed using the SUP4 5′-flanking sequence. Interestingly, the increased activity is not a result of a higher tRNA transcription level, suggesting the importance of proper tRNA processing. The cleavage of the SNR52 or RPR1 promoter from the primary RNA transcript could directly generate the correct 5′ end of the tRNA. This external promoter method has also been used to express E. coli tRNALeuCUA in yeast, which also functions efficiently in translation, suggesting that it can be a general method for functionally expressing different prokaryotic tRNAs in yeast.
An important factor for unnatural amino acid incorporation in eukaryotes is the stability of the target mRNA. Eukaryotic cells have an mRNA surveillance mechanism, nonsense mediated mRNA decay (NMD), to identify mRNAs containing premature stop codons and target the mRNA for rapid degradation (Amrani et al., 2006). The amber stop codon is so far the most frequently used codon to encode unnatural amino acids, but NMD could shorten the target mRNA lifetime resulting in a lower protein yield. Inactivation of NMD would preserve the stability of the UAG-containing mRNA and thus enhance the incorporation efficiency. An NMD-deficient yeast strain was generated by knocking out the UPF1 gene, an essential component for NMD. The unnatural amino acid incorporation efficiency was indeed increased more than two-fold in the upf1Δ strain compared to the wild-type yeast (Wang and Wang, 2008). NMD in yeast shows a polar effect of nonsense codon positions (Cao and Parker, 2003). The steady-state mRNA level is reduced by NMD more significantly when the nonsense codon is closer to the 5′ end than to the 3′ end of an mRNA. Consistently, the increase of unnatural amino acid incorporation efficiency in the upf1Δ strain correlates with the position of the UAG codon: more than two-fold increase is measured when the amber codon is within the N-terminal two thirds of the gene while no significant increase is detected when it is within the C-terminal fourth of the coding region (Wang, Q., Wang, L., unpublished results). By using the external SNR52 promoter and the NMD-deficient upf1Δ strain, the overall yields of purified unnatural amino acid containing proteins have reached ~15 mg/L of yeast cells, approximately 300-fold higher than the previous system and comparable to the yield in E. coli.
Transition to Mammalian Cells
Genetic incorporation of unnatural amino acids into proteins in mammalian cells would face the same challenges as in yeast. Specifically, a major challenge is how to efficiently express prokaryotic orthogonal tRNAs and to ensure they are translationally competent in mammalian cells, due to the aforementioned difference in tRNA transcription and processing. In addition, another major challenge is how to evolve mutant orthogonal synthetases to be specific for an unnatural amino acid for use in mammalian cells. The latter arises because the transfection efficiency of mammalian cells is much lower than the transformation efficiency of E. coli or yeast, making it impractical to generate large synthetase mutant libraries inside mammalian cells. Moreover, survival-death selection in mammalian cells is not as efficient as in E. coli and yeast. Therefore, it is difficult to evolve a specific synthetase out of a large mutant synthetase library directly in mammalian cells.
The E. coli tRNATyrCUA -TyrRS pair is orthogonal in mammalian cells, but the E. coli tRNATyrCUA is not easily expressed in mammalian cells presumably due to the lack of a matched A-box. Fortunately, the B. stearothermophilus tRNATyrCUA happens to contain consensus A- and B-box sequences, is not charged by mammalian synthetases, and works with the heterologous E. coli TyrRS in suppressing the TAG codon. The B. stearothermophilus tRNATyrCUA (lacking 3′-CCA) was linked to the 5′-flanking sequence of the human tRNATyr, and nine tandem repeats of this gene arrangement was used for tRNA expression (Sakamoto et al., 2002). A small collection of designed active-site variants of the E. coli TyrRS was screened in vitro, and a mutant synthetase using 3-iodotyrosine (3) more effectively than Tyr is identified. This mutant synthetase is used with nine tandem B. stearothermophilus tRNATyrCUA to incorporate 3-iodotyrosine into proteins in Chinese hamster ovary (CHO) cells and HEK293 cells with approximately 95% fidelity (Sakamoto et al., 2002). In another attempt, mutations were introduced into the A-box region of the B. subtilis tryptophan opal suppressor tRNA (tRNATrpUCA) to create a consensus A-box (Zhang et al., 2004a). The 3′-CCA lacking mutant tRNA was expressed using the 5′- and 3′-flanking sequences from the Arabidopsis tRNATrp. Together with a rationally designed B. subtilis TrpRS mutant, this tRNA incorporates 5-hydroxytryptophan (4) into the foldon protein in HEK293 cells in response to the opal stop codon with ~97% fidelity.
Encouraging as these results are, it remained a challenge to incorporate various unnatural amino acids in mammalian cells with high efficiency and fidelity. The above approaches may not be generally applicable to other tRNA/synthetase pairs and various unnatural amino acids. As for tRNA expression, most bacterial tRNAs do not have the consensus eukaryotic A- and B-box sequences, and heterologous pairing of the tRNA with synthetase often results in low activity. Since the A- and B-box lie at nucleotides involved in tertiary interactions that support the L-shaped tRNA structure, mutations to create such consensus sequences, although they may not dramatically decrease the activity of the B. subtilis tRNATrpUCA, greatly impair the suppression ability of the E. coli TyrCUA (Sakamoto et al., 2002). In addition, using tandem repeats to boost tRNA expression level may eventually lead to lower expression, especially in stable cell lines, as repeating sequences are frequently associated with gene silencing (Hsieh and Fire, 2000).
A new method has been developed that involves the use of an external type-3 Pol III promoter to efficiently express functional prokaryotic tRNAs in mammalian cells (Figure 3C) (Wang et al., 2007). A type-3 Pol III promoter has its promoter elements exclusively upstream of the coding sequence, and does not require any intragenic elements (such as the A- and B-box) for transcription (Willis, 1993). Therefore, tRNAs with or without the consensus A- and B-box sequences are both expected to be transcribed by this type of promoter in mammalian cells. In addition, the transcription initiation site of some type-3 Pol III promoters, such as the HI promoter (Myslinski et al., 2001) and the U6 small-nuclear RNA promoter (Paule and White, 2000), is well defined, which could be used to generate the correct 5′ end of the tRNA without further post-transcriptional processing. Using a fluorescence-based translation assay, tRNA expression elements including the promoter, the 5′- and 3′-flanking sequence, and the 3′-CCA trinucleotide are linked to the E. coli tRNATyrCUA in various combinations and systematically tested for their ability to generate functional tRNAs in mammalian cells. The gene arrangement that yields tRNAs with the highest translational activity is the H1 promoter followed by the E. coli tRNATyrCUA (without 3′-CCA) and then by the 3′-flanking sequence of tRNAfMet. E. coli tRNATyrCUA expressed using this method is ~70 fold more active in translation than the one expressed with the 5′ and 3′-flanking sequences of human tRNAfMet. This method has also been used to express different E. coli tRNAs successfully, and it is effective in various mammalian cells such as HEK293, HeLa, and primary neurons.
For generating unnatural amino acid-specific synthetase mutants, it is difficult to predict a priori which active site residues need to be mutated. More often mutations at multiple sites are required to achieve high substrate specificity, suggesting that small collections of synthetase mutants will likely fall short. Mutants thus generated may still recognize common amino acids, as is the case with the synthetase used to incorporate 3--iodotyrosine.
To circumvent the challenge of evolving unnatural amino acid-specific synthetases directly in mammalian cells, a transfer strategy is used. Since the E. coli tRNA TyrCUA/TyrRS pair is orthogonal in mammalian cells as well as in yeast, and the translational machinery of yeast is homologous to that of higher eukaryotes, it should be possible to evolve the specificity of this synthetase in yeast and transfer the optimized tRNA/synthetase pairs for use in mammalian cells. In one report, mutant synthetases that were evolved in yeast from the E. coli TyrRS specific for different unnatural amino acids (Deiters et al., 2003, Chin et al., 2003), together with the B. stearothermophilus tRNATyrCUA, have been successfully used in CHO cells and HEK293 cells for incorporating the corresponding unnatural amino acids (Liu et al., 2007). A high translational fidelity for the unnatural amino acid is confirmed with mass spectrometric analysis, and proteins could be produced up to 1 μg per 2 × 107 cells. In another report, mutant synthetases evolved in yeast from the E. coli TyrRS are used together with the cognate E. coli tRNATyrCUA expressed by the H1 promoter in mammalian cells (Wang et al., 2007). Because the H1 promoter also enables the functional expression of the E. coli tRNALeuCUA, mutant synthetases evolved in yeast from the E. coli LeuRS (Summerer et al., 2006) are also successfully transferred in mammalian cells to incorporate the fluorescent unnatural amino acid dansylalanine (Wang et al., 2007). Incorporation efficiencies are in the range of 13% to 41%, depending on the activity of the evolved synthetase. The incorporation fidelity is also high as measured by a sensitive fluorescence assay. Unnatural amino acids have been incorporated in HEK293 cells, HeLa cells, and even in primary neurons.
Applications
New chemical, physical, and biological properties can be introduced into proteins via the incorporation of unnatural amino acids, and thus unnatural amino acids have been exploited to study a wide range of biological problems involving proteins. In particular, the stability, specificity, and catalytic properties of multiple proteins have been extensively studied with unnatural amino acids incorporated with chemically acylated tRNAs, revealing fundamental principles for protein structure and function (Cornish et al., 1995). Microinjection of chemically acylated tRNAs into Xenopus oocytes enables the structure-function studies of integral membrane proteins with unnatural amino acids. Various ion channels and neurotransmitter receptors have been probed with unnatural amino acids using electrophysiological techniques, yielding novel insights on the function role of conserved amino acid residues and gating mechanism of this important family of proteins (Beene et al., 2003, Lummis, et al., 2005). Recent years have also seen elegant studies of signal transduction, selectivity of ion channels, and chromatin biology using unnatural amino acids that are introduced with native chemical ligation or expressed protein ligation (Hahn et al., 2007, Valiyaveetil et al., 2006, McGinty et al., 2008). There are already many excellent reviews on different aspects of this subject (Cornish et al., 1995, Hohsaka and Sisido, 2002, Beene et al., 2003, Link et al., 2003, Wang and Schultz, 2004, Schwarzer and Cole, 2005, Pellois and Muir, 2006), so here we will focus on applications of unnatural amino acids that are genetically encoded using the orthogonal tRNA/synthetase only. Representative examples are summarized to illustrate the usefulness of this technology in modifying and probing proteins, regulating protein functions, and generating new protein properties. We note that some of the unnatural amino acids discussed below have also been incorporated into proteins using other approaches, details of which can be found in the reviews cited above.
Labeling and Modifying Proteins
Reactive side chains of canonical amino acids such as cysteine and lysine have long been used for selective chemical modification of proteins, but the intrinsic selectivity is low unless the amino acid is absent or can be mutated in the target protein. A nonproteinogenic chemical group embodied by an unnatural amino acid can serve as a unique chemical handle for bio-orthogonal chemical reactions, through which a variety of reagents can be selectively appended to proteins in vitro. These reagents can be biophysical probes, tags, post-translational modifications, and groups modifying protein stability or activity. Currently, site-specific labeling of a target protein via unnatural amino acids directly inside live cells is still challenging.
The keto group is not present in any of the canonical amino acids, and reacts with hydrazides, alkoxyamines, and semicarbazides under aqueous, mild conditions to respectively produce hydrazone, oxime, and semicarbazone linkages that are stable under physiological conditions. The keto group has been genetically encoded in E. coli in the form of p-acetylphenylalanine (5) and specifically labeled in vitro with fluorescein hydrazide and biotin hydrazide with greater than 90% yield (Wang et al., 2003). Similarly, fluorescent dyes are selectively appended to the m-acetylphenylalanine (6) introduced into the membrane protein LamB in E. coli (Zhang et al., 2003). p-Acetylphenylalanine and p-benzoylphenylalanine (21) have also been incorporated into rhodopsin, a transmembrane G protein-coupled receptor, in mammalian cells (Ye et al., 2008). The purified mutant rhodopsin is labeled in vitro with fluorescein hydrazide.
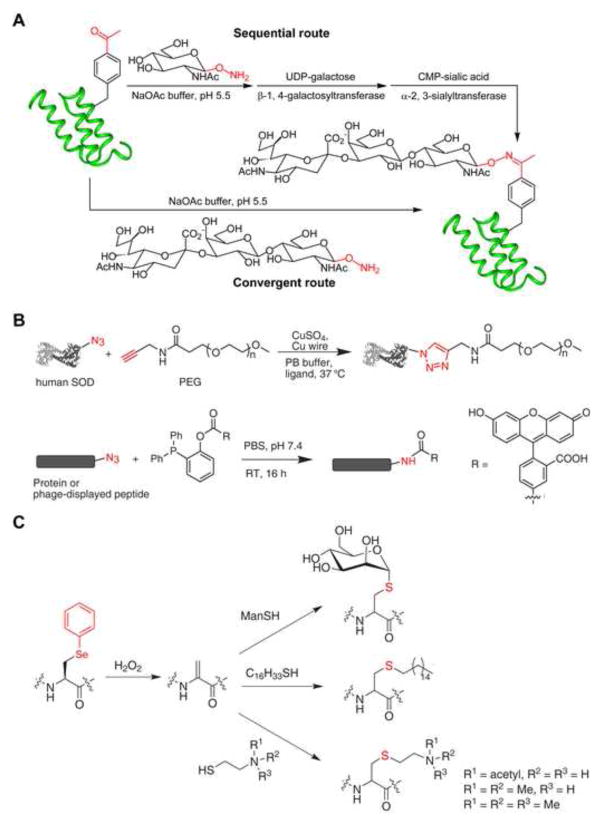
Natural glycoproteins are often present as a population of different glycoforms, which complicates glycan structure analysis and the study of glycosylation effects on protein structure and function. Generation of pure glycoprotein mimetics with defined glycan structures will be valuable for the systematic understanding of glycan function and the development of improved glycoprotein therapeutics. To prepare homogenous glycoprotein mimetics, sugars are synthesized in the aminooxy form and attached to p-acetylphenylalanine incorporated at defined sites in proteins (Figure 5A) (Liu et al., 2003). The attached sugar is subsequently elaborated by adding additional saccharides with glycosyltransferases to generate oligosaccharides with defined structures. Alternatively, an aminooxy-derivatized glycan can also be covalently coupled to the p-acetylphenylalanine containing protein in one step.
Figure 5.
Protein labeling and modification through the genetically incorporated chemical handles. (A) A general method for preparing glycoprotein mimetics with defined glycan structure using the keto group. (B) Protein modification with PEG through a copper-catalyzed cycloaddition reaction (top) and with fluorescein through Staudinger ligation (bottom). (C) Protein modification through phenylselenocysteine.
PEGylation, the attachment of polyethylene glycol (PEG) to proteins, increases protein stability and solubility. In comparison with using common amino acids, PEGylation using a unique chemical handle of an unnatural amino acid increases the homogeneity of the final protein product and provides flexibility in choosing PEGylation sites for optimizing protein activity. PEG has been attached to p-acetylphenylalanine introduced in the human growth hormone to afford a protein that retained wild-type activity but had a considerably improved half-life in serum (PG Schultz, personal communication). This approach can be extended to other therapeutic proteins to generate purer and more stable protein drugs.
Azide and acetylene are two additional nonproteinogenic chemical groups, which react with each other through a copper (I)-catalyzed [2+3] cycloaddition reaction, p-Azidophenylalanine (7) and p-propargyloxyphenylalanine (8) have been genetically incorporated into human superoxide dismutase-1 in E. coli and yeast (Deiters et al., 2003, Deiters et al., 2004). Purified mutant proteins are labeled in vitro with fluorescent dyes or PEG derivatized with the complementary functional group (Figure 5B). The requirement of Cu+ as catalyst may make it difficult to use this reaction inside live cells. The azide group can also react with phosphine derivatives through the Staudinger ligation. p-Azidophenylalanine has been incorporated into the Z-domain protein in E. coli or into peptides displayed on phage, and can be labeled with fluorescein-derived phosphines (Figure 5B) (Tsao et al., 2005).
Phenylselenide undergoes oxidative elimination in hydrogen peroxide followed by Michael addition with thiols (Figure 5C). Phenylselenocysteine (9) is genetically incorporated into GFP in E. coli (Wang et al., 2007). After treating with hydrogen peroxide, the resultant dehydroalanine is selectively labeled in vitro with thiol-containing mannopyranose to generate an analogue of glycosylated GFP, or with n-hexadecylmercaptan to generate a palmitoylated GFP with a non-hydrolyzable linkage. Similarly, phenylselenocysteine has been used to append analogues of methyl- or acetyl-lysine onto the purified Xenopus histone H3, important histone modifications that contribute to chromatin structure and accessibility (Guo et al., 2008). The H3 protein with an acetyl-lysine 9 mimic is deactylated by a histone deacetylation complex and phosphorylated by Aurora B kinase, suggesting that such chemically labeled histones are likely functional in nucleosomes to facilitate the analysis of chromatins.
Besides using a chemical handle, changes can be directly incorporated into proteins in the form of unnatural amino acids, in particular, to mimic post-translational modifications. These modifications are common in eukaryotic proteins playing important roles in protein stability, localization, and function, but cannot be easily produced in bacterial cells. It is also difficult to prepare homogenously modified proteins from eukaryotic cells. Unnatural amino acids with the properties of post-translational modifications may allow the generation of homogenous proteins with the desired modifications to uncover their functional roles and possibly to control protein function.
Glycosylation is a frequent modification of eukaryotic proteins. Homogeneous glycoprotein mimetics with defined glycen structures can be prepared using the keto or phenylselenide-containing unnatural amino acids, but it leaves a small unnatural linkage between the protein and the first sugar. This linkage does not prevent glycosyltransferases from attaching additional sugars (Liu et al., 2003), but whether it affects other glycoprotein functions awaits more research. To prepare a native glycosylated protein, mutant synthetases have been evolved that site-specifically incorporate β-N-acetylglucosamine-O-serine (10) or N-acetylgalactosamine-α-Othreonine (11) into proteins in E. coli (Zhang et al., 2004b, Xu et al., 2004). Since there is no endogenous glycosyl modification in E. coli, genetic incorporation of glycosylated amino acids will help improve the production of proteins that require glycosylation for proper folding and function.
Phosphorylation of serine, threonine, and tyrosine by kinases and reversion by phosphatases are key switches in various signal transduction cascades. A phosphorylation mimic resistant to phosphatase can be used to identify the phosphorylation role of a specific residue, and to create stably activated proteins for functional assays. p-Carboxymethylphenylalanine (pCMF, 12), a nonhydrolyzable analog of phosphotyrosine, was found capable of mimicking the phosphorylated state of Tyr (Xie et al., 2007). Wild type human STAT1, after being phosphorylated at Tyr701, forms a homodimer and strongly binds a DNA duplex. The mutant STAT1 with Tyr701 substituted with pCMF also binds the same DNA duplex tightly, suggesting that pCMF could replace phosphotyrosine in the generation of constitutively active phosphoproteins.
Acetylation is another important reversible protein modification. For instance, acetylation of lysine residues in histone proteins controls the secondary structure of chromatin as well as gene expression levels. One method for generating acetyl- and methyl-lysine analogue tagged histones is described above using phenylselenocysteine. Nε-acetyllysine (13) is genetically incorporated into proteins in E. coli (Neumann et al., 2008). In the presence of a deacetylase inhibitor, manganese superoxide dismutase containing Nε-acetyllysine at residue Lys44 is produced; however, acetylation of Lys44 does not appear to affect the enzyme activity. In addition, Nε-acetyllysine and other lysine analogues are also incorporated into GRB2 in HEK293 cells (Mukai et al., 2008).
Probing Protein Structure and Function
The structure and function relationship of proteins can be studied with greater precision and accuracy when various biophysical probes are site-specifically incorporated into proteins. Genetically encoded biophysical probes further extend the potential of these studies into live cells, the native environment of proteins.
NMR spectra from large proteins or complexes are generally complicated to interpret. Specific NMR labels introduced at defined locations in a protein would greatly reduce the complexity and simplify signal assignment. Tri-fluoromethylphenylalanine (tfm-Phe, 14) was incorporated at the binding interface of two obligate dimers, nitroreductase and histidinol dehydrogenase, both of which contain active sites at the dimer interface (Jackson et al., 2007). Substrate or inhibitor induced conformational changes are evident by monitoring the 19F NMR chemical shifts. In another report, the binding of a small molecule ligand to the thioesterase domain of fatty acid synthase was studied by NMR using tfm-Phe, 13C and 15 C and N labeled o-methyltyrosine (15), and 15N labeled o-nitrobenzyltyrosine (16), at 11 different sites (Cellitti et al., 2008). Comparing the spectra of different mutants and the conformational changes upon addition of the small molecule, the binding site of this molecule is mapped on the protein. It is also worthy to note that photo-decaging of 16 to regenerate 15N labeled tyrosine provides a useful method of isotopic labeling of selected tyrosines without structural change.
p-Cyanophenylalanine (pCNPhe, 17) is a probe for infrared (IR) spectroscopy (Schultz et al., 2006). The stretching vibration of its nitrile group has strong absorption and a frequency (νCN) at ~2200 cm−1, which falls in the transparent window of protein IR spectra. pCNPhe was incorporated into myoglobin at His64, a site close to the iron center of the heme group, to examine ligand-bound states of the heme group. When the Fe(III) ligand is changed from water to cyanide in ferric myoglobin, νCN shifts from 2248 cm−1 to ~2236 cm−1. In the ferrous myoglobin, the linear Fe(II)CO complex shows a νCN absorption at 2239 cm−1 while the bent Fe(II)NO and Fe(II)O2 complexes at 2230 cm−1. These results demonstrate that the nitrile group is a sensitive probe for ligand binding and local electronic environment.
Fluorescent unnatural amino acids may complement the widely used fluorescent proteins in imaging protein expression, dynamics and function. Their smaller size can minimize potential perturbations to target proteins, and their fluorophores can be designed to report different micro environmental changes. Dansylalanine (18) contains the dansyl fluorophore, whose fluorescence intensity increases in hydrophobic surroundings. When incorporated on the surface of a β-barrel in human superoxide dismutase, dansylalanine shows little change in fluorescence after protein denaturation; when placed at an internal site of the β-barrel, its emission wavelength is red-shifted and intensity greatly decreased upon denaturation (Summerer et al., 2006). Therefore, fluorescence change can be used to infer when a residue is exposed to solvent during unfolding. L-(7-hydroxycoumarin-4-yl) ethylglycine (19) bears the coumarin fluorophore, which is sensitive to pH and polarity. It was genetically incorporated into myoglobin at two different helices, A or C (Wang et al., 2006). At 3M urea, coumarin placed in helix A and C shows a similar increase in fluorescence. However, at 2M urea fluorescence increases only when coumarin is incorporated in helix A, suggesting that helix C is not destabilized until urea concentrations above 3M. This result is consistent with previous NMR data, and suggests that coumarin fluorescence can also report local protein unfolding. On the other hand, p-nitrophenylalanine (20) quenches the fluorescence of nearby tryptophans (Tsao et al., 2006). When it is incorporated in a leucine zipper protein GCN4, the tryptophanyl fluorescence is quenched in a distance-dependent manner, making p-nitrophenylalanine a useful distance probe to monitor protein folding or conformational changes. Though still in its infancy, the use of genetically encoded fluorescent amino acids as real time optical reporters and biosensors in live cells is promising.
Introducing heavy atoms into protein crystals is a critical step for phase determination in X-ray crystallography. A new method to introduce heavy atoms into proteins is to genetically incorporate p-iodophenylalanine (3) into proteins in E. coli or yeast. p-Iodophenylalanine was incorporated into T4 lysozyme, and the mutant protein was crystallized (Xie et al., 2004). Diffraction data were collected with a laboratory CuKα X-ray source, and the structure was solved using single-wavelength anomalous dispersion phasing. A single iodinated amino acid among 164 residues results in a strong anomalous signal, about 3% of the total intensities, which compares favorably with the level achieved with selenomethionine using synchrotron beams. p-Iodophenylalanine causes little structural perturbation when substituted for Phe in the core of T4 lysozyme. This approach ensures that the heavy atom iodine is quantitatively introduced at a specific site of the target protein. The strong anomalous signal, the possibility of incorporation at multiple sites and in different cell types, and the use of an in-house X-ray source should facilitate solving protein structures in a high-throughput manner.
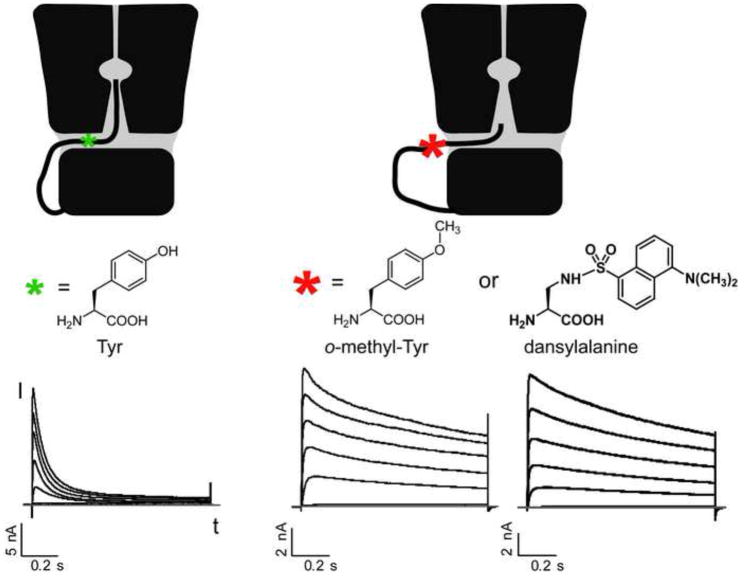
The side chain size of unnatural amino acids can be conveniently altered at atomic precision to provide desired changes for probing bulk effects of amino acids on protein structure and function. The fast inactivation mechanism of the voltage-gated K+ channel Kv1.4 was examined using unnatural amino acids in HEK293 cells (Wang et al., 2007). The classic ball-and-chain model for channel inactivation proposes that the N-terminal inactivation peptide forms a ball-like domain to occlude the channel exit for ions. In contrast, a new model hypothesizes that the inactivation peptide threads through a side portal and extends into the inner pore to block ion flow. To experimentally test this, Tyr19 in the inactivation peptide was initially mutated to Phe or Trp, which shows no difference in channel inactivation. However, mutation to o-methyltyrosine (1) results in a markedly slower inactivation, as does mutation to dansylalanine (18) (Figure 6). Modeling suggested that the diameter of the inactivation peptide, which is unchanged for Phe or Trp at this site but is larger for o-methyltyrosine and dansylalanine, is important for channel inactivation. This is likely due to the narrow width of the side portal in the channel, supporting the new model for channel inactivation.
Figure 6.
Unnatural amino acids to probe the inactivation mechanism of ion channel Kv1.4. o-Methyltyrosine or dansylalanine extends the side chain length of tyrosine, which impedes the inactivation peptide from threading through the side portal of the ion channel and abolishes the fast inactivation, as shown in the current-time curve.
Identifying and Regulating Protein Activity
The identification of protein interactions and control of protein activities in vivo would ideally require methods that are noninvasive and easy to administer. Application of light is an attractive approach. Photo-responsive unnatural amino acids that can crosslink with nearby molecules, shed protecting groups, or change conformation have been developed and genetically incorporated into proteins. They enable the in vivo manipulation of proteins, which should have great potential in molecular and synthetic biology applications.
Crosslinking has been an important technique for uncovering protein interactions in cells. Site-specific photocrosslinking enabled by genetically encoded unnatural amino acids may help to pinpoint transient or weak interactions, to identify which region is involved, and to distinguish direct from indirect interactions. p-Azidophenylalanine (7), p-benzoylphenylalanine (pBpA, 21), and p-(3-trifluoromethyl-3H-diazirin-3-yl)-phenylalanine (TfmdPhe, 22) contain photocrosslinking side chains and have been genetically incorporated into proteins (Chin et al., 2002, Chin et al., 2002, Tippmann et al., 2007). pBpA is incorporated into the heat shock protein ClpB in E. coli at residue Tyr251, which is considered as the main substrate recognition residue (Schlieker et al., 2004). Biotinylated substrate peptides are shown to be crosslinked upon UV light exposure, but not if pBpA is incorporated elsewhere. pBpA is also incorporated into Ste2p, a GPCR that binds a short peptide pheromone α-factor, at several sites on the extracellular loops in yeast (Huang et al., 2008). At two sites biotinylated α-factor is indeed photocaptured and identified by Western blot. pBpA has also been incorporated into Grb2 (growth-factor-receptor-bound protein 2) in the ligand binding pocket of the SH2 domain in mammalian cells. When activated epidermal growth factor receptor (EGFR) and pBpA containing Grb2 are co-expressed and treated with UV light, a larger molecular weight band is detected suggesting the binding of EGFR to Grb2 (Hino et al., 2005). Genetically encoded photocrosslinkers should provide high specificity and be compatible with a wide range of biological processes, making this strategy attractive for probing protein complexes and interactions in vivo.
Photocaging of critical residues can be harnessed to regulate protein activity using light. A photoremovable protecting group is installed onto the suitable amino acid in the target protein, which masks the amino acid and renders the protein inactive. Photolysis releases the caging group and converts the amino acid to the wild-type active form, generating abrupt or localized changes. For example, mutation of the active-site Cys residue in the proapoptotic protease caspase 3 to o-nitrobenzylcysteine (23) in yeast leads to a catalytically inactive enzyme. UV illumination of the cell lysate converts ~40% of the caged caspase to the active enzyme (Wu et al., 2004). o-Nitrobenzyltyrosine (24) is incorporated into (β-galactosidase at Tyr503 in E. coli, which effectively reduced the activity of this enzyme to 5% of the wild-type form (Deiters et al., 2006). After a 30 minute exposure of bacterial cells to UV light, the enzyme regains activity to ~70% of wild-type levels. A photocaged serine, 4,5-dimethoxy-2-nitrobenzylserine (DMNB-Ser, 25), is incorporated at phosphoserine sites in the transcription factor Pho4 to control its phosphorylation in yeast (Lemke et al., 2007). DMNB-Ser blocks phosphorylation until photolysed with 405 nm light. Serine is then regenerated and subsequently phosphorylated, triggering the nuclear export of Pho4 (Figure 7A). Photocaging prevents post-translational modification of the selected residue until a desired moment, which is a more versatile method than mutating the residue to a null amino acid in examining signal transductions.
Figure 7.
Regulation of protein activity using photo-responsive amino acids. (A) Photocaged serine is used to control when the target serine is regenerated and phosphorylated. (B) The photo-isomerizable p-azophenyl-phenylalanine is used to control the binding of CAP to its promoter determined by gel mobility shift assay.
Photolysis of a caged residue is an irreversible process. Reversible modulation can be achieved with the photochromic azobenzene group, which undergoes a reversible cis-trans isomerization using light with different wavelengths (Figure 7B). The resultant change in geometry and/or dipole of the compound can be used to regulate protein activity. p-Azophenylphenylalanine (AzoPhe, 26) has been incorporated at Ile71 of the E. coli catabolite activator protein, a transcriptional activator (Bose et al., 2006). Its binding affinity for the promoter sequence decreases fourfold after irradiation at 334 nm, which converts the predominant trans AzoPhe to the cis-form. The isomerized cis AzoPhe is switched back to the trans-state by irradiation at >420 nm, after which the protein affinity for the promoter is completely recovered.
Generating New Protein Properties
Natural evolution affords numerous proteins for life needs, while directed protein evolution and rational engineering further increase the diversity of protein functions. Genetically encoding additional functional groups into proteins prompts the hope for generating totally novel and previously impossible protein properties, and some initial progresses are indeed encouraging.
Metal ions bound in proteins participate in a large number of catalytic and electron transfer reactions in cells. Rationally engineering a metalloprotein is challenging, as it is difficult to predict and control the amino acid shells surrounding the metal ion. Multidentate metal-chelating amino acids with coordinating atoms pre-orientated in the correct configuration may facilitate such effort. Bipyridylalanine (BpyAla, 27) has been shown to reversibly bind copper ions when genetically incorporated into T4 lysozyme (Xie et al., 2007). The Cu(II) binding property is exploited to oxidatively cleave the phosphosugar moiety of the nucleic acid backbone. BpyAla is incorporated into the catabolite activator protein (CAP) at Lys26, a site close to the protein-DNA interface (Lee and Schultz, 2008). In the presence of Cu(II) and 3-mercaptopropionic acid, the mutant CAP, originally a transcription factor, cleaves double-stranded DNA at its consensus sequence with high specificity. This method may be generally applied to other DNA binding proteins to map their preferred DNA sequences.
The distance probe p-nitrophenylalanine (20) has recently been rediscovered to stimulate potent immune responses for novel immunogenic applications. An animal usually will not mount a substantial immune response against a self protein. This immune self-tolerance has been shown to be broken by introducing p-nitophenylalanine into the antigen (Grünewald et al., 2008). Incorporation of this unnatural amino acid into tumor necrosis factor α (TNF-α) leads to a strong immune response directed at the unnatural mutant TNF-α in mice, even in the absence of adjuvant. The generated antibodies cross-react with native mouse TNF-α. The immune response is retained in mice after exposure to the mutant TNF-α, and protects mice against lipopolysaccharide-induced death. These striking observations have the potential for generating new therapies for cancer or protein-misfolding diseases.
Conclusions and Future Perspectives
Since the first unnatural amino acid was genetically encoded in live cells in 2001 (Wang et al., 2001), more than 40 unnatural amino acids have been incorporated into proteins in E. coli and about a dozen in yeast and in mammalian cells to date. It should be noted that this method currently cannot be used to incorporate unnatural amino acids that are incompatible with the ribosome (such as D-amino acids and β-amino acids) or toxic to cells (such as an analog that is structurally so similar to a common amino acid that the analog is charged by the cognate synthetase of the amino acid and misincorporated into the proteome) (Hartman et al., 2007). The novel functional groups embodied by these genetically encoded new amino acids will enable an increasing number of new experiments for biological research. The utility of many unnatural amino acids have been demonstrated in proof-of-principle experiments, and they are expected to be applied to various biological system for addressing challenging questions and uncovering unknowns in the coming years. Unnatural amino acids that are fluorescent, photo-responsive, bio-orthogonally reactive, or able to mimic various protein modifications would be particularly useful in addressing cell biology questions. Further improvement of the unnatural amino acid incorporation efficiency and detailed studies on the physiological effects of their incorporation would be important for effective application, especially in mammalian cells. The introduction of this technology into multicellular organisms would open the field to biological problems concerning how cells interact with one another in a variety of settings, allowing aspects of development, neural connectivity, and cellular signaling to be studied, to name only a few. The development of random unnatural amino acid mutagenesis may enable the evolution of protein properties that have never existed in nature. Lastly, genetic incorporation of unnatural amino acids has implications on various aspects of the origin and evolution of the genetic code itself, which remains to be explored.
Acknowledgments
L.W. acknowledges the support from the Searle Scholar program, the Beckman Young Investigator Program, and the March of Dimes Foundation (Grant No. 5-FY08110).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambrogelly A, Palioura S, Söll D. Natural expansion of the genetic code. Nat Chem Biol. 2006;3:29–35. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- Amrani N, Sachs MS, Jacobson A. Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol. 2006;7:415–425. doi: 10.1038/nrm1942. [DOI] [PubMed] [Google Scholar]
- Anderson JC, Schultz PG. Adaptation of an orthogonal archaeal leucyl-tRNA and synthetase pair for four-base, amber, and opal suppression. Biochemistry. 2003;42:9598–9608. doi: 10.1021/bi034550w. [DOI] [PubMed] [Google Scholar]
- Anderson JC, Wu N, Santoro SW, Lakshman V, King DS, Schultz PG. An expanded genetic code with a functional quadruplet codon. Proc Natl Acad Sci U S A. 2004;101:7566–7571. doi: 10.1073/pnas.0401517101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain JD, Glabe CG, Dix TA, Chamberlin AR, Diala ES. Biosynthetic site-specific incorporation of a non-natural amino acid into a polypeptide. J Am Chem Soc. 1989;111:8013–8014. [Google Scholar]
- Beene DL, Dougherty DA, Lester HA. Unnatural amino acid mutagenesis in mapping ion channel function. Curr Opin Neurobiol. 2003;13:264–270. doi: 10.1016/s0959-4388(03)00068-0. [DOI] [PubMed] [Google Scholar]
- Bock A, Forchhammer K, Heider J, Leinfelder W, Sawers G, Veprek B, Zinoni F. Selenocysteine: the 21st amino acid. Mol Microbiol. 1991;5:515–520. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- Bose M, Groff D, Xie J, Brustad E, Schultz PG. The incorporation of a photoisomerizable amino acid into proteins in E. coli. J Am Chem Soc. 2006;128:388–389. doi: 10.1021/ja055467u. [DOI] [PubMed] [Google Scholar]
- Brick P, Bhat TN, Blow DM. Structure of tyrosyl-tRNA synthetase refined at 2.3 A resolution. Interaction of the enzyme with the tyrosyl adenylate intermediate. J Mol Biol. 1989;208:83–98. doi: 10.1016/0022-2836(89)90090-9. [DOI] [PubMed] [Google Scholar]
- Cao D, Parker R. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell. 2003;113:533–545. doi: 10.1016/s0092-8674(03)00353-2. [DOI] [PubMed] [Google Scholar]
- Cellitti SE, Jones DH, Lagpacan L, Hao X, Zhang Q, Hu H, Brittain SM, Brinker A, Caldwell J, Bursulaya B, et al. In vivo incorporation of unnatural amino acids to probe structure, dynamics, and ligand binding in a large protein by nuclear magnetic resonance spectroscopy. J Am Chem Soc. 2008;130:9268–9281. doi: 10.1021/ja801602q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Schultz PG, Brock A. An improved system for the generation and analysis of mutant proteins containing unnatural amino acids in Saccharomyces cerevisiae. J Mol Biol. 2007;371:112–122. doi: 10.1016/j.jmb.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Chin JW, Martin AB, King DS, Wang L, Schultz PG. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc Natl Acad Sci USA. 2002;99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JW, Santoro SW, Martin AB, King DS, Wang L, Schultz PG. Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J Am Chem Soc. 2002;124:9026–9027. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- Cohen GN, Cowie DB. Total replacement of methionine by selenomethionine in the proteins of Escherichia coli. C R Hebd Seances Acad Sci. 1957;244:680–683. [PubMed] [Google Scholar]
- Cornish VW, Mendel D, Schultz PG. Probing protein structure and function with an expanded genetic code. Angew Chem Int Ed Engl. 1995;34:621–633. [Google Scholar]
- Dawson PE, Muir TW, Clarklewis I, Kent SBH. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- Deiters A, Cropp TA, Mukherji M, Chin JW, Anderson JC, Schultz PG. Adding amino acids with novel reactivity to the genetic code of Saccharomyces cerevisiae. J Am Chem Soc. 2003;125:11782–11783. doi: 10.1021/ja0370037. [DOI] [PubMed] [Google Scholar]
- Deiters A, Cropp TA, Summerer D, Mukherji M, Schultz PG. Site-specific PEGylation of proteins containing unnatural amino acids. Bioorg Med Chem Lett. 2004;14:5743–5745. doi: 10.1016/j.bmcl.2004.09.059. [DOI] [PubMed] [Google Scholar]
- Deiters A, Groff D, Ryu Y, Xie J, Schultz PG. A genetically encoded photocaged tyrosine. Angew Chem Int Ed Engl. 2006;45:2728–2731. doi: 10.1002/anie.200600264. [DOI] [PubMed] [Google Scholar]
- Drabkin HJ, Park HJ, Rajbhandary UL. Amber suppression in mammalian cells dependent upon expression of an Escherichia coli aminoacyl-tRNA synthetase gene. Mol Cell Biol. 1996;16:907–913. doi: 10.1128/mcb.16.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards H, Schimmel P. A bacterial amber suppressor in Saccharomyces cerevisiae is selectively recognized by a bacterial aminoacyl-tRNA synthetase. Mol Cell Biol. 1990;10:1633–1641. doi: 10.1128/mcb.10.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis MA, RajBhandary UL. Expression and function of a human initiator tRNA gene in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:4486–4494. doi: 10.1128/mcb.10.9.4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli G, Hofstetter H, Birnstiel ML. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981;294:626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- Grünewald J, Tsao ML, Perera R, Dong L, Niessen F, Wen BG, Kubitz DM, Smider VV, Ruf W, Nasoff M, et al. Immunochemical termination of self-tolerance. Proc Natl Acad Sci USA. 2008;105:11276–11280. doi: 10.1073/pnas.0804157105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Wang J, Lee JS, Schultz PG. Site-specific incorporation of methyl- and acetyl-lysine analogues into recombinant proteins. Angew Chem Int Ed Engl. 2008;47:6399–6401. doi: 10.1002/anie.200802336. [DOI] [PubMed] [Google Scholar]
- Hahn ME, Pellois JP, Vila-Perello M, Muir TW. Tunable photoactivation of apost-translationally modified signaling protein and its unmodified counterpart in live cells. ChemBioChem. 2007;8:2100–2105. doi: 10.1002/cbic.200700404. [DOI] [PubMed] [Google Scholar]
- Hartman MC, Josephson K, Lin CW, Szostak JW. An expanded set of amino acid analogs for the ribosomal translation of unnatural peptides. PLoS ONE. 2007;2:e972. doi: 10.1371/journal.pone.0000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino N, Okazaki Y, Kobayashi T, Hayashi A, Sakamoto K, Yokoyama S. Protein photo-cross-linking in mammalian cells by site-specific incorporation of a photoreactive amino acid. Nat Methods. 2005;2:201–206. doi: 10.1038/nmeth739. [DOI] [PubMed] [Google Scholar]
- Hohsaka T, Sisido M. Incorporation of non-natural amino acids into proteins. Curr Opin Chem Biol. 2002;6:809–815. doi: 10.1016/s1367-5931(02)00376-9. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Fire A. Recognition and silencing of repeated DNA. Annu Rev Genet. 2000;34:187–204. doi: 10.1146/annurev.genet.34.1.187. [DOI] [PubMed] [Google Scholar]
- Huang LY, Umanah G, Hauser M, Son C, Arshava B, Naider F, Becker JM. Unnatural amino acid replacement in a yeast G protein-coupled receptor in its native environment. Biochemistry. 2008;47:5638–5648. doi: 10.1021/bi701866e. [DOI] [PubMed] [Google Scholar]
- Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- Jackson JC, Hammill JT, Mehl RA. Site-specific incorporation of a (19)F-amino acid into proteins as an NMR probe for characterizing protein structure and reactivity. J Am Chem Soc. 2007;129:1160–1166. doi: 10.1021/ja064661t. [DOI] [PubMed] [Google Scholar]
- Kaiser ET, Lawrence DS, Rokita SE. The chemical modification of enzymatic specificity. Annu Rev Biochem. 1985;54:565–595. doi: 10.1146/annurev.bi.54.070185.003025. [DOI] [PubMed] [Google Scholar]
- Kent SB. Chemical synthesis of peptides and proteins. Annu Rev Biochem. 1988;57:957–989. doi: 10.1146/annurev.bi.57.070188.004521. [DOI] [PubMed] [Google Scholar]
- Knight RD, Freeland SJ, Landweber LF. Rewiring the keyboard: evolvability of the genetic code. Nat Rev Genet. 2001;2:49–58. doi: 10.1038/35047500. [DOI] [PubMed] [Google Scholar]
- Kowal AK, Kohrer C, Rajbhandary UL. Twenty-first aminoacyl-tRNA synthetase-suppressor tRNA pairs for possible use in site-specific incorporation of amino acid analogues into proteins in eukaryotes and in eubacteria. Proc Natl Acad Sci U S A. 2001;98:2268–2273. doi: 10.1073/pnas.031488298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok Y, Wong JT. Evolutionary relationship between Halobacterium cutirubrum and eukaryotes determined by use of aminoacyl-tRNA synthetases as phylogenetic probes. Can J Biochem. 1980;58:213–218. doi: 10.1139/o80-029. [DOI] [PubMed] [Google Scholar]
- Lee HS, Schultz PG. Biosynthesis of a site-specific DNA cleaving protein. J Am Chem Soc. 2008;130:13194–13195. doi: 10.1021/ja804653f. [DOI] [PubMed] [Google Scholar]
- Lemke EA, Summerer D, Geierstanger BH, Brittain SM, Schultz PG. Control of protein phosphorylation with a genetically encoded photocaged amino acid. Nat Chem Biol. 2007;3:769–772. doi: 10.1038/nchembio.2007.44. [DOI] [PubMed] [Google Scholar]
- Link AJ, Mock ML, Tirrell DA. Non-canonical amino acids in protein engineering. Curr Opin Biotechnol. 2003;14:603–609. doi: 10.1016/j.copbio.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Liu H, Wang L, Brock A, Wong CH, Schultz PG. A method for the generation of glycoprotein mimetics. J Am Chem Soc. 2003;125:1702–1703. doi: 10.1021/ja029433n. [DOI] [PubMed] [Google Scholar]
- Liu W, Brock A, Chen S, Chen S, Schultz P. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- Lummis SC, Beene DL, Lee LW, Lester HA, Broadhurst RW, Dougherty DA. Cis-trans isomerization at a proline opens the pore of a neurotransmitter-gated ion channel. Nature. 2005;438:248–52. doi: 10.1038/nature04130. [DOI] [PubMed] [Google Scholar]
- McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehl RA, Anderson JC, Santoro SW, Wang L, Martin AB, King DS, Horn DM, Schultz PG. Generation of a bacterium with a 21 amino acid genetic code. J Am Chem Soc. 2003;125:935–939. doi: 10.1021/ja0284153. [DOI] [PubMed] [Google Scholar]
- Minks C, Alefelder S, Moroder L, Huber R, Budisa N. Towards new protein engineering: in vivo building and folding of protein shuttles for drug delivery and targeting by the selective pressure incorporation (SPI) method. Tetrahedron. 2000;56:9431–9442. [Google Scholar]
- Muir TW. Semisynthesis of proteins by expressed protein ligation. Annu Rev Biochem. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- Mukai T, Kobayashi T, Hino N, Yanagisawa T, Sakamoto K, Yokoyama S. Adding 1-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem Biophys Res Commun. 2008;371:818–822. doi: 10.1016/j.bbrc.2008.04.164. [DOI] [PubMed] [Google Scholar]
- Myslinski E, Ame JC, Krol A, Carbon P. An unusually compact external promoter for RNA polymerase III transcription of the human H1RNA gene. Nucleic Acids Res. 2001;29:2502–2509. doi: 10.1093/nar/29.12.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H, Peak-Chew SY, Chin JW. Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat Chem Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- Noren CJ, Anthony-Cahill SJ, Griffith MC, Schultz PG. A general method for site-specific incorporation of unnatural amino acids into proteins. Science. 1989;244:182–188. doi: 10.1126/science.2649980. [DOI] [PubMed] [Google Scholar]
- Normanly J, Kleina LG, Masson JM, Abelson J, Miller JH. Construction of Escherichia coli amber suppressor tRNA genes. III Determination of tRNA specificity. J Mol Biol. 1990;213:719–726. doi: 10.1016/S0022-2836(05)80258-X. [DOI] [PubMed] [Google Scholar]
- Nowak MW, Kearney PC, Sampson JR, Saks ME, Labarca CG, Silverman SK, Zhong WG, Thorson J, Abelson JN, Davidson N, et al. Nicotinic receptor binding site probed with unnatural amino acid incorporation in intact cells. Science. 1995;268:439–442. doi: 10.1126/science.7716551. [DOI] [PubMed] [Google Scholar]
- Paule MR, White RJ. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellois JP, Muir TW. Semisynthetic proteins in mechanistic studies: using chemistry to go where nature can’t. Curr Opin Chem Biol. 2006;10:487–491. doi: 10.1016/j.cbpa.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Hayashi A, Sakamoto A, Kiga D, Nakayama H, Soma A, Kobayashi T, Kitabatake M, Takio K, Saito K, et al. Site-specific incorporation of an unnatural amino acid into proteins in mammalian cells. Nucleic Acids Res. 2002;30:4692–4699. doi: 10.1093/nar/gkf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlieker C, Weibezahn J, Patzelt H, Tessarz P, Strub C, Zeth K, Erbse A, Schneider-Mergener J, Chin JW, Schultz PG, et al. Substrate recognition by the AAA+ chaperone ClpB. Nat Struct Mol Biol. 2004;11:607–615. doi: 10.1038/nsmb787. [DOI] [PubMed] [Google Scholar]
- Schultz KC, Supekova L, Ryu Y, Xie J, Perera R, Schultz PG. A genetically encoded infrared probe. J Am Chem Soc. 2006;128:13984–13985. doi: 10.1021/ja0636690. [DOI] [PubMed] [Google Scholar]
- Schwarzer D, Cole PA. Protein semisynthesis and expressed protein ligation: chasing a protein’s tail. Curr Opin Chem Biol. 2005;9:561–569. doi: 10.1016/j.cbpa.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- Summerer D, Chen S, Wu N, Deiters A, Chin JW, Schultz PG. A genetically encoded fluorescent amino acid. Proc Natl Acad Sci USA. 2006;103:9785–9789. doi: 10.1073/pnas.0603965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tippmann EM, Liu W, Summerer D, Mack AV, Schultz PG. A genetically encoded diazirine photocrosslinker in Escherichia coli. ChemBioChem. 2007;8:2210–2214. doi: 10.1002/cbic.200700460. [DOI] [PubMed] [Google Scholar]
- Tsao ML, Tian F, Schultz PG. Selective Staudinger modification of proteins containing p-azidophenylalanine. ChemBioChem. 2005;6:2147–2149. doi: 10.1002/cbic.200500314. [DOI] [PubMed] [Google Scholar]
- Tsao ML, Summerer D, Ryu Y, Schultz PG. The genetic incorporation of a distance probe into proteins in Escherichia coli. J Am Chem Soc. 2006;128:4572–4573. doi: 10.1021/ja058262u. [DOI] [PubMed] [Google Scholar]
- Uy R, Wold F. Posttranslational covalent modification of proteins. Science. 1977;198:890–896. doi: 10.1126/science.337487. [DOI] [PubMed] [Google Scholar]
- Valiyaveetil FI, Leonetti M, Muir TW, Mackinnon R. Ion selectivity in a semisynthetic K+ channel locked in the conductive conformation. Science. 2006;314:1004–1007. doi: 10.1126/science.1133415. [DOI] [PubMed] [Google Scholar]
- Wang J, Xie J, Schultz PG. A genetically encoded fluorescent amino acid. J Am Chem Soc. 2006;128:8738–8739. doi: 10.1021/ja062666k. [DOI] [PubMed] [Google Scholar]
- Wang J, Schiller SM, Schultz PG. A biosynthetic route to dehydroalanine-containing proteins. Angew Chem Int Ed Engl. 2007;46:6849–6851. doi: 10.1002/anie.200702305. [DOI] [PubMed] [Google Scholar]
- Wang L, Magliery TJ, Liu DR, Schultz PG. A new functional suppressor tRNA/aminoacyl-tRNA synthetase pair for the in vivo incorporation of unnatural amino acids into proteins. J Am Chem Soc. 2000;122:5010–5011. [Google Scholar]
- Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. [Google Scholar]
- Wang L, Schultz PG. A general approach for the generation of orthogonal tRNAs. Chem Biol. 2001;8:883–890. doi: 10.1016/s1074-5521(01)00063-1. [DOI] [PubMed] [Google Scholar]
- Wang L, Brock A, Schultz PG. Adding L-3-(2-Naphthyl)alanine to the genetic code of E. coli. J Am Chem Soc. 2002;124:1836–1837. doi: 10.1021/ja012307j. [DOI] [PubMed] [Google Scholar]
- Wang L, Schultz PG. Expanding the genetic code. Chem Commun. 2002:1–11. doi: 10.1039/b108185n. [DOI] [PubMed] [Google Scholar]
- Wang L, Zhang Z, Brock A, Schultz PG. Addition of the keto functional group to the genetic code of Escherichia coli. Proc Natl Acad Sci U S A. 2003;100:56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Schultz PG. Expanding the genetic code. Angew Chem Int Ed Engl. 2004;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- Wang L, Xie J, Schultz PG. Expanding the genetic code. Annu Rev Biophys Biomol Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- Wang Q, Wang L. New methods enabling efficient incorporation of unnatural amino acids in yeast. J Am Chem Soc. 2008;130:6066–6067. doi: 10.1021/ja800894n. [DOI] [PubMed] [Google Scholar]
- Wang W, Takimoto J, Louie GV, Baiga TJ, Noel JP, Lee KF, Slesinger PA, Wang L. Genetically encoding unnatural amino acids for cellular and neuronal studies. Nat Neurosci. 2007;10:1063–1072. doi: 10.1038/nn1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis IM. RNA polymerase III. Genes, factors and transcriptional specificity. Eur J Biochem. 1993;212:1–11. doi: 10.1111/j.1432-1033.1993.tb17626.x. [DOI] [PubMed] [Google Scholar]
- Wu N, Deiters A, Cropp TA, King D, Schultz PG. A genetically encoded photocaged amino acid. J Am Chem Soc. 2004;126:14306–14307. doi: 10.1021/ja040175z. [DOI] [PubMed] [Google Scholar]
- Xie J, Wang L, Wu N, Brock A, Spraggon G, Schultz PG. The site-specific incorporation of p-iodo-L-phenylalanine into proteins for structure determination. Nat Biotechnol. 2004;22:1297–1301. doi: 10.1038/nbt1013. [DOI] [PubMed] [Google Scholar]
- Xie J, Liu W, Schultz PG. A genetically encoded bidentate, metal-binding amino acid. Angew Chem Int Ed Engl. 2007;46:9239–9242. doi: 10.1002/anie.200703397. [DOI] [PubMed] [Google Scholar]
- Xie J, Supekova L, Schultz PG. A genetically encoded metabolically stable analogue of phosphotyrosine in Escherichia coli. ACS Chem Biol. 2007;2:474–478. doi: 10.1021/cb700083w. [DOI] [PubMed] [Google Scholar]
- Xu R, Hanson SR, Zhang Z, Yang YY, Schultz PG, Wong CH. Site-specific incorporation of the mucin-type N-acetylgalactosamine-alpha-O-threonine into protein in Escherichia coli. J Am Chem Soc. 2004;126:15654–15655. doi: 10.1021/ja044711z. [DOI] [PubMed] [Google Scholar]
- Ye S, Köhrer C, Huber T, Kazmi M, Sachdev P, Yan EC, Bhagat A, Rajbhandary UL, Sakmar TP. Site-specific incorporation of keto amino acids into functional G protein-coupled receptors using unnatural amino acid mutagenesis. J Biol Chem. 2008;283:1525–1533. doi: 10.1074/jbc.M707355200. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Smith BA, Wang L, Brock A, Cho C, Schultz PG. Anew strategy for the site-specific modification of proteins in vivo. Biochemistry. 2003;42:6735–6746. doi: 10.1021/bi0300231. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Alfonta L, Tian F, Bursulaya B, Uryu S, King DS, Schultz PG. Selective incorporation of 5-hydroxytryptophan into proteins in mammalian cells. Proc Natl Acad Sci USA. 2004a;707:8882–8887. doi: 10.1073/pnas.0307029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Gildersleeve J, Yang YY, Xu R, Loo JA, Uryu S, Wong CH, Schultz PG. A new strategy for the synthesis of glycoproteins. Science. 2004b;303:371–373. doi: 10.1126/science.1089509. [DOI] [PubMed] [Google Scholar]