Abstract
The transcription factor, Sry-related High Mobility Group (HMG) box containing gene 9 (Sox9), plays a critical role in cartilage development by initiating chondrogenesis and preventing the subsequent maturation process called chondrocyte hypertrophy. This suppression mechanism by Sox9 on late-stage chondrogenesis partially results from the inhibition of Runt-related transcription factor 2 (Runx2), the main activator of hypertrophic chondrocyte differentiation. However, the precise mechanism by which Sox9 regulates late chondrogenesis is poorly understood.
In the present study, the transcriptional repressor vertebrate homolog of Drosophila bagpipe (Bapx1) was found to be a direct target of Sox9 for repression of Runx2 expression in chondrocytes. We identified a critical Sox9 responsive region in the Bapx1 promoter via a luciferase reporter assay. Analysis by chromatin immunoprecipitation and electrophoretic mobility shift assays indicated that Sox9 physically bound to this region of the Bapx1 promoter. Consistent with the notion that Bapx1 and Sox9 act as negative regulators of chondrocyte hypertrophy by regulating Runx2 expression, transient knockdown of Sox9 or Bapx1 expression by shRNA in chondrocytes increased Runx2 expression, as well as expression of the late chondrogenesis marker, Col10a1. Furthermore, while over-expression of Sox9 decreased Runx2 and Col10a1 expressions, simultaneous transient knockdown of Bapx1 diminished that Sox9 over-expressing effect.
Our findings reveal that the molecular pathway modulated by Bapx1 links two major regulators in chondrogenesis, Sox9 and Runx2, to coordinate skeletal formation.
Keywords: Sox9, Bapx1, Runx2, chondrocyte, chondrogenesis, shRNA
Introduction
Chondrogenesis is executed in multiple steps during endochondral ossification. Mesenchymal cells first proliferate, condense, become immature chondroblasts, transform into prehypertrophic chondrocytes, and differentiate into hypertrophic chondrocytes [1]. In immature chondroblasts (i.e., during early chondrogenesis), extracellular matrix (ECM) proteins such as type II collagen alpha 1 (Col2a1), type XI collagen alpha 2 (Col11a2), and aggrecan 1 (Agc1) are specifically expressed. On the other hand, in prehypertrophic and hypertrophic chondrocytes (i.e., during late chondrogenesis), expression of these proteins is decreased, while type X collagen alpha 1 (Col10a1), Indian hedgehog (Ihh), and osteopontin expression is increased [2]. In addition, Sox9 is dominantly expressed in early chondrogenesis, whereas Runx2 dominates as the critical transcription factor in late chondrogenesis [2, 3].
The transcription factor Sox9 is a critical regulator of chondrocyte-specific proteins, such as Col2a1, Col11a2, and Agc1, during cartilaginous development [4–6]. Analyses in genetically modified mice revealed that Sox9 promotes the early stage, but suppresses the late stage, of chondrogenesis [7–9]. On the contrary, the transcription factor Runx2 is a critical enhancer of chondrocyte maturation and osteoblast differentiation [10–14]. Although the relationship between Sox9 and Runx2 in chondrogenesis is poorly understood, Sox9 directly binds to Runx2 and inhibits its function [15].
Another transcription factor, Bapx1 (also known as Nkx3.2), plays a critical role during cartilage development. [16–18]. Bapx1 is a transcriptional repressor known to regulate Runx2 expression [19]. Interestingly, a recent study revealed that forced expression of Bapx1 throughout the cartilage template blocked chondrocyte hypertrophy, mirroring the Sox9 over-expression phenotype [20]. Furthermore, experiments using chicken somatic explants in culture have shown that Bapx1 and Sox9 are able to induce the expression of each other [21].
While Bapx1 is known to be a transcription factor crucial for chondrogenic differentiation [22], little is known about its transcriptional targets. Recently, Runx2 has been reported to be a target gene of Bapx1 [19], representing a critical transcription factor for osteoblast differentiation and also promoting hypertrophic chondrocyte differentiation in endochondral ossification [10–14, 23]. Furthermore, Sox9 promotes the early stage, but suppresses the late stage, of chondrogenesis [9]. These results suggest that Bapx1 potentially links the two major regulators of chondrogenesis, Sox9 and Runx2.
In the present study, Bapx1 expression was up-regulated by Sox9 in chondrocytes, and luciferase reporter assays revealed that Sox9 activated the Bapx1 promoter. Chromatin immunoprecipitation (ChIP) and electrophoretic mobility shift assays (EMSA) demonstrated that Sox9 physically bound to the Bapx1 promoter in chondrocytes. Additionally, our gain- and loss-of-function experiments showed that the expression of Runx2 and its target, Col10a1, was repressed by Sox9, coincident with increased Bapx1 expression. These data collectively demonstrate that Bapx1 is a direct Sox9 target in chondrocytes. Moreover, our findings strongly suggest that Sox9 represses Runx2 through Bapx1 regulation, resulting in suppression of hypertrophic chondrocyte differentiation.
Materials and Methods
Antibodies
Anti-Sox9 antibody (Chemicon) was used in western blots and electrophoretic mobility shift assays (EMSA). Anti-Bapx1 antibody (Abcam) was used in western blots. Anti-actin antibody (Sigma) was used as a control for quantitative determination of western blots. As secondary antibodies for western blots, horseradish peroxidase (HRP)-conjugated anti-rabbit immunoglobulin G (IgG) and anti-mouse IgG (Sigma) were used. Normal rabbit IgG (Santa Cruz Biotechnology Inc.) was used as a negative control in chromatin immunoprecipitation (ChIP) and EMSA experiments.
Cell culture
C3H10T1/2 cells were maintained in DMEM containing 10% FBS at 37°C in an atmosphere of 5% CO2. To induce chondrogenesis, 10 μl aliquots of a suspension containing 107 cells/ml were plated into wells of a 24-well plate containing culture medium. After 2 hours, 100 ng of human bone morphogenetic protein 2 (BMP-2) (PeproTech)/ml was added to the culture. Media was changed every 2 days.
Ribs from embryonic day 16.5 mice were used in the preparation of chondrocytes by digestion with collagenase. Murine chondrocytes were maintained in DMEM containing 10% FBS at 37°C in an atmosphere of 5% CO2.
Western blot
Cells were lysed with NP-40 lysis buffer (20 mM Tris-HCl (pH 7.6), 3 mM EDTA, 150mM NaCl, 0.5% NP-40, 50 mM NaF, 1 mM β-glycerophosphate, 5 mM Na4P2O7 □10 H2O, 1mM Na3VO4, 10 μg/ml PMSF) containing a Complete® protease inhibitor cocktail (Roche Diagnostics, Tokyo, Japan). Protein concentrations were measured using the DC Protein Assay kit (Bio-Rad Laboratories, CA, USA), and equal amounts of protein were subjected to SDS-polyacrylamide gel electrophoresis (PAGE). Separated proteins were electrically transferred to PVDF membrane, blocked with TBST (Tris-buffered saline containing 0.1% Tween-20) containing 5% skim milk, washed with TBST, and then incubated with primary antibody (appropriately diluted with blocking buffer) for either 1.5 hours at room temperature (RT) or overnight at 4°C. Subsequently, blots were washed with TBST, incubated with secondary antibody (diluted in blocking buffer) for 1 hour at RT, and washed with TBST. Proteins were detected using a chemiluminescence kit (Nacalai, Kyoto, Japan).
RT-Qualitative and quantitative PCR
Total RNA was extracted with ISOGEN (Nippon Gene) according to the manufacturer’s protocol. Total RNA was reverse transcribed with Ready-To-Go You-Prime First-Strand Beads (GE Healthcare) with oligo-dT. To analyze gene expression, first strand cDNA was amplified via PCR, using gene-specific primer sets (Table 1) and PrimeStar™ HS DNA Polymerase (Takara), followed by agarose gel electrophoresis.
Table 1.
Primer sequences.
| a) Sequence for each primer set used in qualitative and quantitative RT-PCR | ||
|---|---|---|
| Gene Name | Forward primer sequence | Reverse primer sequence |
| Sox9 | 5′-TACGACTGGACGCTGGTGCC-3′ | 5′-CCGTTCTTCACCGACTTCCTCC-3′ |
|
| ||
| Col2al | 5′-TCATCGAGTACCGATCACAGA-3′ | 5′-GTTCGGGGGTTTTACAAGAAG-3′ |
|
| ||
| Bapxl | 5′-CCACTGCAGCCCTCCTACTA-3′ | 5′-GTGTCAAGTCCCAGCAGGAT-3′ |
|
| ||
| Runx2 | 5′-GGACGAGGCAAGAGTTTCAC-3′ | 5′-CCAGAGGCAGAAGTCAGAGG-3′ |
|
| ||
| Col10a1 | 5′-GTCCAAGAGGTGAACCTGGA-3′ | 5′-TCTGTGAGCTCCATGATTGC-3′ |
|
| ||
| Gapdh | 5′-CCTGGTCACCAGGGCTGC-3′ | 5′-CGCTCCTGGAAGATGGTGATG-3′ |
| b) Sequence for each primer set used in chromatin immunoprecipitation (ChIP)-PCR | ||
| Gene Name | Forward primer sequence | Reverse primer sequence |
|
| ||
| Col2al | 5′-GGGAGACCTCAGTCCTCCTT-3′ | 5′-GGAGGCTGTGCATTGTGG-3′ |
|
| ||
| Amh | 5′-GCTCAGGCCTCTGCAGTTAT-3′ | 5′-GGGTGGCCCTGCTTAT ATGT-3′ |
|
| ||
| Bapxl | 5′-GAGACGAATACCCAGCTCCA-3′ | 5′-GAGCCCAGAGTCGAAGAACA-3′ |
Quantitative gene expression analysis was performed via real-time PCR, using TaqMan Universal Master Mix reagents with TaqMan Probes or SYBR Green PCR master mix reagents (Applied Biosystems) with gene-specific primer sets (Table 1) on an ABI PRISM® 7900HT thermal cycler (Applied Biosystems). Col2a1, Runx2, Col10a1, and Gapdh mRNAs were measured using mouse TaqMan probes Mm00491889_m1, Mm00501578_m1, Mm00487041_m1, and Mm99999915_g1, respectively (Applied Biosystems). In each experiment, expression data was normalized to Gapdh expression.
Lentivirus production and transduction
Oligonucleotide sequences for short hairpin RNA (shRNA) targeting Sox9, Bapx1, or LacZ as a control were cloned into the pLenti4/BLOCK-iT™-DEST vector (Invitrogen), and Sox9 cDNA was cloned into the pLenti6 vector (Invitrogen). Each vector and ViraPower packaging vector (Invitrogen) were transfected to 293FT cells, and lentivirus was produced. Lentivirus was transduced into cells with 10μg/ml polybrene. The following oligonucleotide sequences were cloned into the pLenti4/BLOCK-iT™-DEST vector: LacZ (5’-AAATCGCTGATTTGTGTAGTCGGAGACGACTACACAAATCAGCGA-3’), Sox9 (5’-GCGACAACTTTACCAGTTTCACGAATGAAACTGGTAAAGTTGTCGC-3’), Sox9-2 (5’-GCGACGTCATCTCCAACATTGCGAACAATGTTGGAGATGACGTCGC-3’), and Bapx1 (5’-GAGATGTCAGCCAGCGTTTCACGAATGAAACGCTGGCTGACATCTC-3’)
Luciferase reporter assay and transfections
pGL4.12-Luc vector (Promega) including the indicated cloned genomic regions and thymidine kinase (TK) promoter was transfected into cultured cells. A Renilla luciferase reporter pRL-TK (Promega) was co-transfected as a control for evaluating transfection efficiency. Transfections were performed with Lipofectamine 2000 or LTX (Invitrogen) according to the manufacturer’s instructions. After 24 hours, cells were washed before measuring firefly and Renilla luciferase activities using the Dual-Glo™ Luciferase Assay System (Promega). In each experiment, firefly luciferase activity was normalized to Renilla luciferase activity.
Mutagenesis
Mutations were introduced using QuikChange® Site-Directed Mutagenesis (Stratagene) according to the manufacturer’s instruction.
Chromatin immunoprecipitation (ChIP)
Cells were cross-linked with 1% formaldehyde for 10 minutes at RT, followed by addition of glycine to a final concentration of 0.125 M. Next, cells were washed with PBS, collected, washed with cell lysis buffer (5mM PIPES pH 8.0, 85 mM KCl, 0.5% NP-40, and 10 μg of PMSF/ml) containing protease inhibitors, and resuspended in nuclear lysis buffer (50 mM Tris-HCl pH 8.1, 10 mM EDTA, 1% SDS, and 10 μg of PMSF/ml) containing protease inhibitors. Chromatin was sheared to approximately 200-1000 bp by sonication, prior to a 1/5 dilution in ChIP dilution buffer (16.7 mM Tris-HCl pH 8.1, 167 mM NaCl, 1.2 mM EDTA, 0.01% SDS, 1.1% Triton X-100, and 10 μg of PMSF/ml) containing protease inhibitors. Then, the chromatin solution was incubated with the indicated antibody and Dynabeads® M-280 sheep anti-rabbit IgG (Dynal Biotech) overnight at 4°C. Beads were washed three times with ChIP wash buffer (50 mM HEPES-KOH (pH 7.0), 0.5 M LiCl, 1 mM EDTA, 0.7% sodium deoxycholate, and 1% NP-40). Immunocomplexes were eluted from beads with ChIP elution buffer (50 mM Tris-HCl (pH 8.0), 10 mM EDTA, and 1% SDS) for 1 hour at 65°C. Eluates were then incubated overnight at 65°C to reverse cross-linking, prior to the addition of 0.5 mg of protease K/ml for 2 hours at 55°C. DNA was purified using the MinElute PCR purification kit (QIAGEN) and analyzed using the site-specific primer pair sets described in Table 1. Also, whole cell extract (WCE) was analyzed by PCR.
Electrophoretic mobility shift assay (EMSA)
DNA-protein binding was assayed with DNA probes that had been 32P-labeled by end-filling with Klenow fragment and Sox9 in the presence or absence of anti-Sox9 antibody or rabbit IgG. Anti-Sox9 antibody or rabbit IgG was pre-incubated with Sox9 for 30 minutes at 25°C before addition of radiolabeled probes. Reactions were performed in binding buffer [20mM HEPES (pH 7.9), 10% glycerol, 50 mM KCl, 0.05% NP-40, 0.5 mM EDTA, 0.5 mM DTT, and 1 mM PMSF] in the presence of 0.5 μg of poly (dG-dI), a nonspecific competitor, for 30 minutes at 25°C. Products of the binding reactions were separated by polyacrylamide gel electrophoresis (PAGE) on a 4% gel for 3 hours at 100 V.
Results
Bapx1 is expressed in early chondrogenesis, and its expression is up-regulated by Sox9 in chondrocytes
The temporal sequence of Sox9 and Bapx1 expression in chondrogenesis was first examined by a well-established in vitro model, in which C3H10T1/2 cells are induced to undergo chondrogenesis by being plated as a high-density micromass culture in the presence of BMP-2 [24, 25]. During the chondrogenesis assay, expression of Sox9 and Bapx1 were examined by western blot. Sox9 protein increased for 5 days during chondrogenesis induction. However, in the next 4 days, the expression of Sox9 protein slightly decreased (Fig. 1a). Bapx1 expression mirrored that of Sox9; it increased for 5 days and decreased by day 9 (Fig. 1a). This parallel expression pattern of Sox9 and Bapx1 was also observed by RT-PCR analysis (Fig. 1b). Correspondingly, RT-PCR revealed that the Sox9 target gene, Col2a1, a marker gene of immature chondroblasts [4], increased until day 5 after chondrogenesis induction, but slightly decreased by day 9 (Fig. 1b). As marker genes of late stage chondogenesis, Runx2 and its target, Col10a1 [26], demonstrated significantly increased expression from days 5 to 9 (Fig. 1b). Thus, Bapx1 is expressed during early chondrogenesis, and its expression decreased in accordance with the incremental increase in expression of the mature chondrocyte markers Runx2 and Col10a1.
Fig. 1. Bapx1 is expressed during early chondrogenesis in C3H10T1/2 cells.
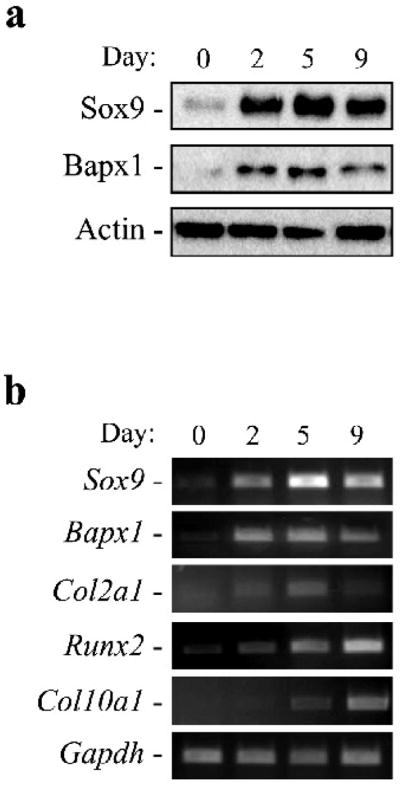
a. Western blot detection of Sox9 and Bapx1 proteins at the indicated time points. b. RT- PCR detection of mRNA of Sox9, Bapx1, Col2a1, Bapx1, Runx2, Col10a1, and Gapdh at several time points.
To test the potential relationship between Sox9 and Bapx1, we examined the effect of gain- or loss-of-function of Sox9 on Bapx1 expression. Sox9 expression was efficiently silenced by transducing cultured mouse chondrocytes with two different shRNA-expressing lentiviruses (Fig. 2a). Under these conditions, Col2a1 mRNA expression significantly decreased, confirming that Sox9 function is diminished in these chondrocytes (Fig. 2b).
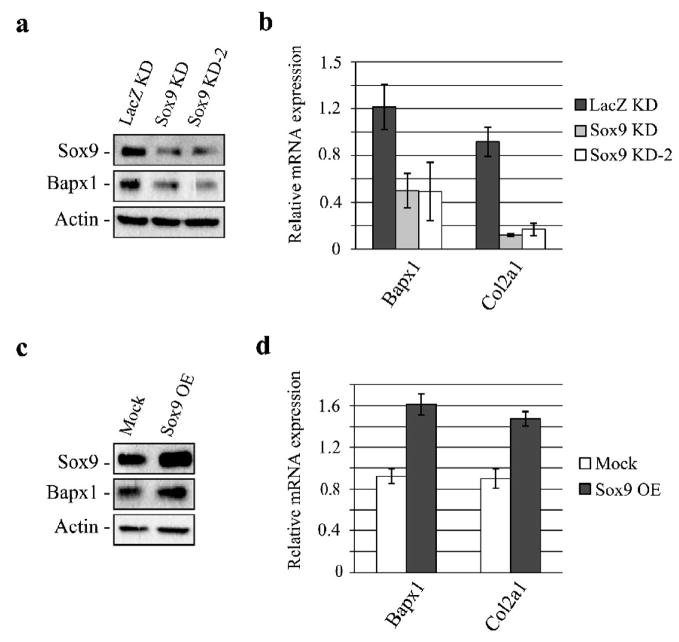
Fig. 2. Bapx1 expression is up-regulated by Sox9 in chondrocytes.
a. LacZ as a negative control or two Sox9-targeting shRNA expressing lentiviruses were transduced into cultured mouse chondrocytes and evaluated by western blot for knockdown (KD) efficiency and Bapx1 expression. b. Bapx1 and Col2a1 mRNA expression after Sox9 knockdown (KD) was examined by RT-quantitative PCR. Mean values (n=3) ± standard deviation are shown. c. Over-expression (OE) of Sox9 in cultured mouse chondrocytes and detection of Sox9 and Bapx1 proteins by western blot. d. Transcript levels of Bapx1 and Col2a1 in Sox9-over-expressing (OE) chondrocytes were assayed by RT-quantitative PCR. Mean values (n=3) ± standard deviation are shown.
Bapx1 expression in these cells was further assayed by western blot and RT-quantitative PCR. Bapx1 expression decreased in Sox9 knockdown chondrocytes (Fig. 2a and Fig. 2b). On the contrary, Sox9 over-expression through lentiviral transduction in cultured mouse chondrocytes (Fig. 2c) increased Bapx1 and Col2a1 expression (Fig. 2c, d). These results indicate that Sox9 is a critical activator of Bapx1 expression during early chondrogenesis.
Sox9 directly binds the Bapx1 promoter and enhances its activity
To assess whether Bapx1 is directly regulated by Sox9, Bapx1 promoter activity was examined by luciferase reporter assays. We cloned a promoter region of −4427 to +41 upstream of the Bapx1 transcriptional start site (TSS) and ligated it into a pGL4.12 vector containing a TK promoter and a luciferase reporter gene (Fig. 3a). We found that Sox9 expression increased Bapx1 promoter activity in a dose-dependent manner in C3H10T1/2 cells (Fig. 3b).
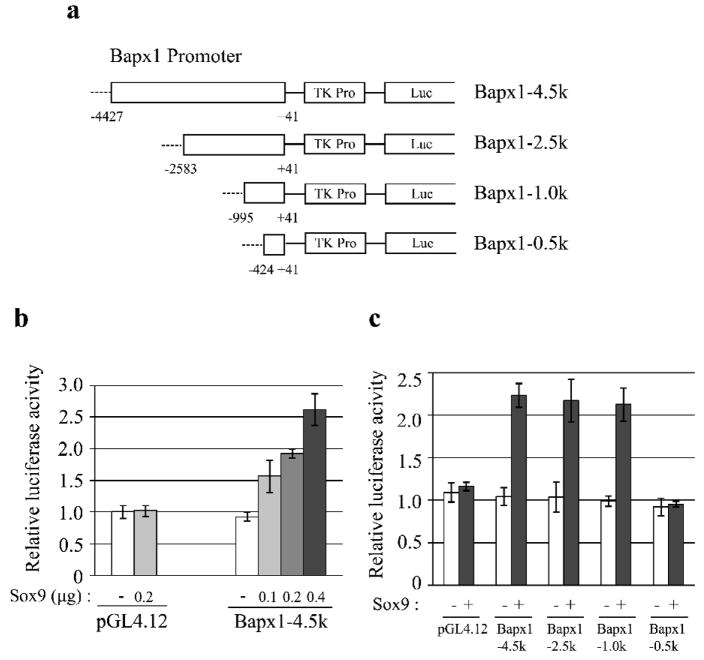
Fig. 3. Sox9-activated Bapx1 proximal promoter activity.
a. Luciferase reporter constructs containing the Bapx1 promoter are shown. Numbers indicate the position from the transcription start site (TSS). b. C3H10T1/2 cells in 24-well plates were co-transfected with 200 ng of the indicated luciferase reporter plasmid and the specified amounts of the Sox9-encoded plasmid. Mean values (n=3) of luciferase reporter activity ± standard deviation are shown. c. Mean values (n=3) of luciferase reporter activity ± standard deviation in C3H10T1/2 cells transfected with 200 ng of the indicated luciferase reporter plasmid and either 200 ng of Sox9-expressing or empty pcDNA3 plasmid are shown.
To identify the Sox9 responsive element, we further performed promoter analyses with serial deletions of the Bapx1 promoter (Fig. 3a). Deletions up to position −995 did not alter the incremental increase in luciferase activity due to Sox9 co-transfection; however, deletion to the position of −424 diminished this incremental activation (Fig. 3c). These results suggest that the Sox9 responsive element is located between position −995 and −424 upstream of the Bapx1 TSS.
In this Sox9 responsive region, we found multiple Sox9 binding motifs [(A/T)(A/T)CAA(T/A)G] (data not shown). Sox9 homodimers bind to enhancer regions of chondrocyte target genes containing inverted Sox9 binding sites separated by 3–4 bp that represent a Sox9 palindromic motif [27–30]. Based on this, we searched for a Sox9 palindromic motif in the Sox9 responsive element and found one at the position between −868 and −852 from the Bapx1 TSS (Fig. 4a). We further examined promoter activity with a fragment containing the putative Sox9 binding site corresponding to the region between −1152 and −727 (Fig. 4b). Sox9 increased the luciferase activity of this construct in a dose-dependent manner in C3H10T1/2 cells (Fig. 4c). Taken together, these results indicate that the Sox9 responsive element is located between −995 and −727 from the Bapx1 TSS.
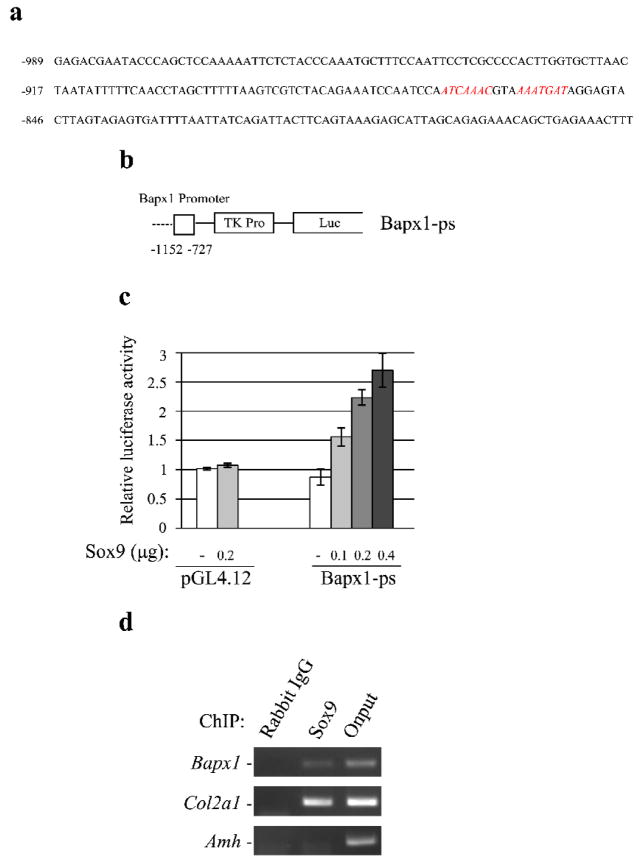
Fig. 4. Minimal Sox9 binding site in the Bapx1 promoter in chondrocytes.
a. Sequence of the Bapx1 promoter between −989 and −775 from the TSS. A potential Sox9 binding site is indicated in red. b. A luciferase reporter construct containing the Bapx1 minimum promoter is shown. Numbers indicate the position from the TSS. c. C3H10T1/2 cells were co-transfected with 200 ng of the indicated luciferase reporter plasmid and specified amounts of the plasmid-borne Sox9. Mean values (n= 3) of luciferase reporter activity ± standard deviation are shown. d. Chromatin-immunoprecipitated Sox9-binding non-enriched (rabbit IgG) DNA, enriched (Sox9) DNA, and 0.01% whole cell extract (WCE) DNA in chondrocytes was amplified by PCR using Sox9 site-specific primer sets for the Sox9 putative binding site, chondrocyte-specific target Col2a1, and Sertoli cell-specific target Amh.
To further determine whether Sox9 binds to the endogenous Bapx1 promoter, we performed ChIP using cultured mouse chondrocytes. Previous studies have detailed several Sox9 target genes and binding regions. For instance, Sox9 binds to the enhancer within the first intron of the chondrocyte-specific gene Col2a1 [31]. Consistent with this, we found that Sox9 bound to this region in cultured mouse chondrocytes by ChIP (Fig. 4d). We further found that Sox9 bound to the Bapx1 promoter containing putative Sox9 binding motifs in cultured mouse chondrocytes (Fig. 4d). Furthermore, our ChIP analysis did not detect Sox9 binding to an enhancer region of anti-Mullerian hormone (Amh), a well-known target in testis [32, 33] (Fig. 4d). Moreover, our RT-PCR analysis did not detect Amh transcripts in cultured mouse chondrocytes (data not shown). These results suggest that Bapx1 is a Sox9 target gene in chondrocytes.
Sox9 interacts with the Bapx1 promoter
We prepared oligonucleotide probes for the putative Sox9 binding regions (Fig. 5a) and performed EMSAs. Upon incubating the Bapx1-WT oligonucleotide probes with recombinant Sox9 protein, we detected two different bands, presumably corresponding to the binding of Sox9 as a monomer to one site (lower band) or as a dimer to both sites (upper band) (Fig. 5b). Addition of anti-Sox9 antibody to this reaction reduced the intensity of both bands, with an upward supershift observed, although incubation with anti-rabbit IgG antibody did not alter either the putative monomer or putative dimer bands (Fig. 5b).
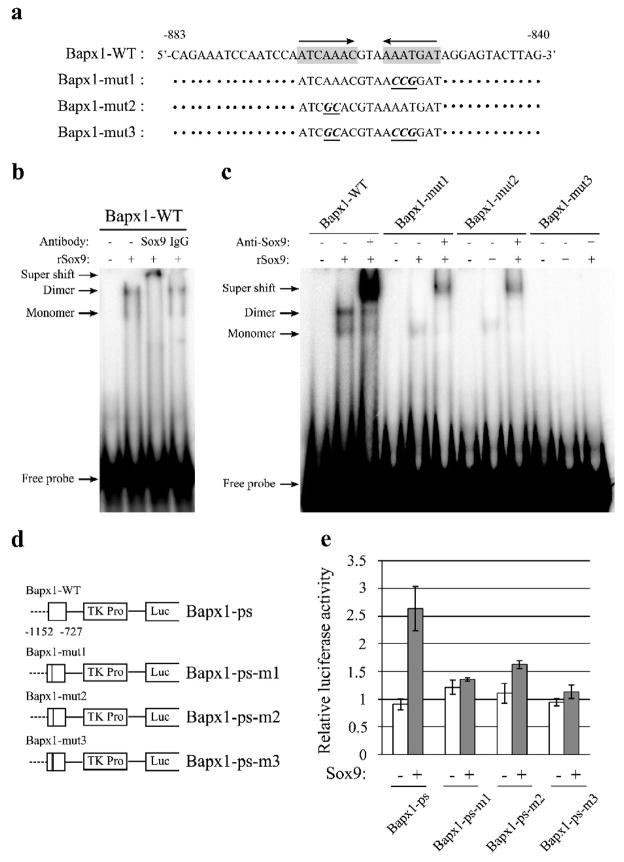
Fig. 5. Induction of Bapx1 promoter activity through the binding of a Sox9 homodimer.
a. Oligonucleotide sequences corresponding to the Bapx1 promoter are shown, with distance from the TSS indicated. Putative Sox9-binding consensus sequences are marked with a grey background; arrows denote site orientation. b. 32P-labeled oligonucleotide probes corresponding to the Bapx1 promoter complexed to Sox9 in the presence of anti-Sox9 antibody or control rabbit IgG were detected by electrophoretic mobility shift assays (EMSA). c. Binding of oligonucleotide probes with mutated putative Sox9 binding consensus sequences complexed to Sox9 protein (rSox9) was detected by electrophoretic mobility shift assays (EMSA). Mutations in one (mut1, 2) or both (mut3) Sox9-binding motifs were compared to wild-type (WT) in the presence or absence of anti-Sox9 antibody. d. Luciferase reporter constructs for Bapx1 Sox9-binding sites containing mutations are shown. e. C3H10T1/2 cells were transfected with the indicated reporter plasmids and co-transfected with Sox9-expressing or empty pcDNA3 plasmid. Mean values (n=3) of luciferase reporter activity ± standard deviation are shown.
Oligonucleotide probes with point mutations in one or both of the Sox9 motif sites were synthesized and EMSAs were performed. A single Sox9 motif point mutation resulted in a reduction of both bands, especially diminishing the upper band, whereas mutation of both sites completely abolished these bands (Fig. 5c). The supershifted band produced by incubation with anti-Sox9 antibody was reduced by a single mutation and abolished by dual mutations of the Sox9 motifs (Fig. 5c). These results indicated that Sox9 could bind as a monomer and/or dimer to the Bapx1 promoter site.
To further assess Sox9 binding and the contribution of this site to the Bapx1 promoter, a luciferase reporter assay was performed using a vector containing point mutations in one or both of the Sox9 binding sites (Fig. 5d). A single mutation in the Sox9 motif reduced, whereas dual mutations almost completely abolished, the incremental increase in luciferase activity by Sox9 in C3H10T1/2 cells (Fig. 5e). Taken together, these results indicate that Sox9 directly regulates Bapx1, and Sox9 homodimer binding is essential for this regulation.
Sox9 represses Runx2 by regulating Bapx1 in chondrocytes
Based on our finding that Bapx1 is a direct Sox9 target in chondrocytes, we speculated that Sox9 might repress Runx2 expression by enhancing Bapx1 expression, resulting in a suppression of terminal chondrocyte differentiation.
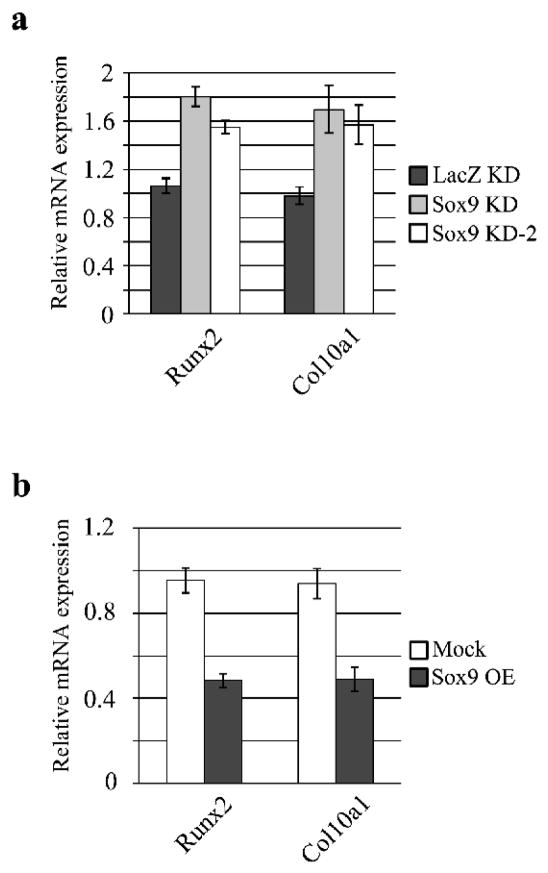
To test this hypothesis, Runx2 mRNA expression was assessed by quantitative PCR in Sox9 knockdown chondrocytes. Indeed, expression of Runx2 and its target, Col10a1, was increased after Sox9 knockdown in cultured mouse chondrocytes (Fig. 6a). Furthermore, over-expression of Sox9 inhibited the expression of Runx2 and Col10a1 mRNA (Fig. 6b).
Fig. 6. Sox9 represses Runx2 expression and function.
a. Runx2 and Col10a1 mRNA levels in Sox9 knockdown chondrocytes were assayed by RT-quantitative PCR. b. Runx2 and Col10a1 expression in Sox9 over-expressing chondrocytes were analyzed by quantitative RT-PCR.
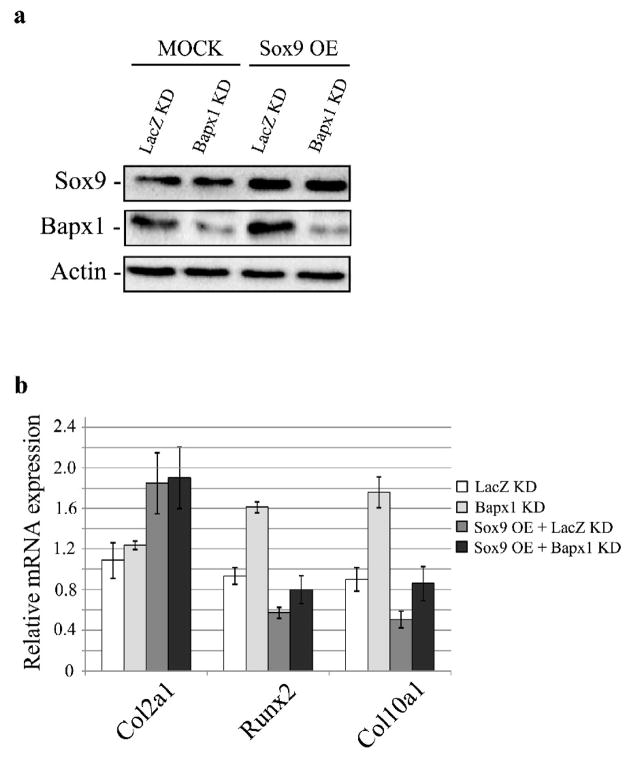
We next assessed the effect of Bapx1 knockdown on Runx2 and Col10a1 expression and found that while lentivirus-mediated knockdown of Bapx1 did not change Sox9 and Col2a1 expression, it increased Runx2 and Col10a1 expression (Figure 7a-b). We further found that Bapx1 knockdown diminished the suppressive effect of Runx2 and Col10a1 expression by over-expression of Sox9 (Figure 7a-b). These results suggest that Sox9 negatively regulates Runx2 by enhancing Bapx1 expression, leading to the inhibition of terminal chondrocyte differentiation.
Fig. 7. Bapx1 knockdown diminishes the suppression of Runx2 expression and function by Sox9 over-expression in chondrocytes.
a. Over-expression of Sox9 and/or Bapx1 targeting shRNA in cultured mouse chondrocytes and detection of Sox9 and Bapx1 proteins by western blot. b. Col2a1, Runx2 and Col10a1 mRNA expression in chondrocytes with Sox9 over-expression (OE) and/or Bapx1 knockdown (KD) were assayed by quantitative RT-PCR. Mean values (n= 3) ± standard deviation are shown.
Discussion
Bapx1 is a direct Sox9 target gene
Various transcription factors are expressed and play essential roles during endochondral ossification. In immature chondrocytes, the transcription factors Sox9, Sox5, Sox6, and Bapx1, as well as several chondrocyte-specific ECM proteins, including Col2a1, Col11a2, and Agc1, are expressed. In contrast, these genes are down-regulated in differentiated mature chondrocytes; instead, Runx2 and the ECM protein, Col10a1, are expressed [2]. Previous studies have revealed the importance of each transcription factor in this process, but details including identification of target genes, downstream molecular mechanisms, and molecular crosstalk between these players were poorly understood.
Although past studies suggested that Sox9 increased Bapx1 expression [21], the mechanism behind this induction was unclear. In the present study, we identified Bapx1 as a direct Sox9 target gene in chondrocytes. Paired box gene 1 (Pax1), Pax9, and mesenchyme homeobox 1 (Meox1) proteins regulate Bapx1 expression by directly activating its promoter [34, 35]. Interestingly, binding sites for Pax1/Pax9 (position between −873 and −853), Meox1 (position between −828 and −823), and Sox9 (position between −868 and −852) are in close proximity within the Bapx1 promoter. These binding sites are concentrated between −880 and −820, with Pax1/Pax9 and Sox9 binding sites mostly overlapping. Based on these observations, it is possible that these factors cooperate to regulate Bapx1 expression.
However, Pax1/Pax9 and Meox1 expression is maintained in chondrogenic mesenchymal cells until they reach the pre-chondrocyte stage [36–40], whereas Sox9 expression is maintained throughout chondrocyte differentiation, except in hypertrophic chondrocytes [41–43]. Thus, Pax1/Pax9 and Meox1 expression occurs earlier than Sox9 expression, and these genes typically exhibit distinct expression patterns during chondrogenesis. Alternatively, Bapx1 is expressed in chondrogenic mesenchymal cells and subsequent chondroblast differentiation, so that its expression mirrors that of Sox9 [21, 44]. These observations suggest that Pax1/9 and Meox1 may regulate initial and/or early induction of Bapx1, and Sox9 contributes to maintain Bapx1 expression during subsequent chondrogenic differentiation. Nevertheless, whether these events actually happen during embryogenesis remains to be determined.
Runx2 repression is critical for hypertrophic suppression by Sox9
Bapx1 involvement in endochondral ossification was previously suggested by its function as a transcriptional repressor [22]. More recently, the direct targets of Bapx1 have been clarified. Runx2, a critical transcription factor for osteoblast differentiation that also regulates hypertrophic chondrocytes during endochondral ossification, was identified as a direct Bapx1 target [19]. Moreover, Sox9 promotes early chondrogenesis and suppresses hypertrophic chondrocyte differentiation [9], although the mechanisms remain to be elucidated. Additionally, retrovirus-mediated over-expression of Bapx1 in chick wings resulted in repressed hypertrophic differentiation [20]. These observations suggest a possibility that Sox9 suppresses hypertrophic differentiation by repressing Runx2 through regulating Bapx1 expression.
Our results showed that Sox9 knockdown resulted in incremental changes in Runx2 expression and its target gene, and inverse effects resulted from Sox9 over-expression in cultured chondrocytes. We further showed that Bapx1 knockdown diminished the suppressive effect of Runx2 expression and its target gene by over-expression of Sox9 in cultured chondrocytes. These findings strongly suggest that Sox9 suppresses Runx2 function indirectly, at least in part, by regulating Bapx1, leading to repression of hypertrophic differentiation.
A recent study indicated that Sox9 directly binds to Runx2 and blocks its function [15]. Our findings and reports by other investigators suggest that Runx2 repression is a critical event for Sox9 suppression of hypertrophic chondrocyte differentiation. During endochondral ossification, Sox9 and Runx2 expression are distinct events. Sox9 is expressed in immature chondroblasts, and Runx2 is expressed in mature chondrocytes. These findings may indicate that Sox9 sustains an immature state of chondroblasts by suppressing hypertrophic chondrocyte differentiation by repressing Runx2. In other words, Runx2 expression during chondrogenesis may function as a proliferation/differentiation switch.
However, a recent report revealed that S100A1 and S100B were direct Sox9 targets, and that they suppress hypertrophic chondrocyte differentiation [45]. Although a detailed mechanism was not worked out, it suggests a possibility for another mechanism of hypertrophic suppression by Sox9. Further analysis is required to clarify the precise mechanism for hypertrophic chondrocyte suppression by Sox9.
Bapx1 is a critical mediator of endochondral bone formation
In cartilaginous development, Sox9 enhances chondrogenesis by promoting the expression of chondrocyte-specific proteins, such as Col2a1, Col11a2, and Agc1 [4–6]. In addition, Sox9 plays a critical role in testicular development by regulating targets such as Amh and Ptgds in Sertoli cells [32, 33, 46]. However, the regulatory mechanism of tissue-specific target distribution is less well understood. Recent studies have shown that a missense mutation (A76E) in human SOX9 in an XY patient resulted in skeletal abnormalities without gender reversal by perturbing SOX9 dimerization, which results in dysregulation of chondrocyte-specific genes, but normal regulation of Sertoli cell-specific target [27, 28]. Furthermore, Sox9 homodimers may bind to enhancer regions of chondrocyte target genes containing inverted Sox9 binding motifs that represent a Sox9 palindromic motif [29, 30]. These reports suggest that dimer formation is critical for chondrocyte-specific gene regulation. Given that our results indicate that Bapx1 expression is regulated by Sox9 homodimer binding to the Bapx1 promoter, Bapx1 may be a critical regulator of endochondral bone formation. Indeed, Bapx1 homozygous mutant mice die perinatally and exhibit skeletal dysplasia, and transgenic mice over-expressing Bapx1 in limb buds exhibit preaxial polydactyly [47]. However, the abnormal skeletal phenotype of Bapx1 mutant mice was limited to the vertebral column and some craniofacial bones [16–18]. These findings may indicate that the necessity of Bapx1 for endochondral bone formation is limited to specific tissues, and other factors with similar functions play the role in other tissues.
In this study, we focused on Bapx1 function as a transcriptional repressor of Runx2, given that Bapx1 targets, except for Runx2, were virtually unknown. Analysis of Bapx1-deficient mice by other researchers has revealed that Bapx1 is also necessary for somite proliferation and differentiation into chondrocytes [16–18]. Moreover, an autoregulatory loop between Bapx1 and Sox9 was demonstrated in a chick somite culture system, although the mechanism of Sox9 up-regulation by Bapx1 was not clarified [21]. In the present study, our Bapx1 knockdown chondrocytes did not exhibit altered expression of Sox9 and Col2a1. The possibility exists that during embryogenesis, depending on the specific stage of development and tissues involved, Bapx1 up-regulates Sox9, resulting in the promotion of proliferation and early chondrogenesis. Further analyses are needed to determine precise Bapx1 functions during chondrogenesis.
In the present study, we revealed that Bapx1 is a direct Sox9 target gene. Our findings strongly suggest that Sox9 repressed Runx2 through Bapx1 regulation, resulting in suppressed hypertrophic chondrocyte differentiation, thereby promoting normal endochondral ossification.
Acknowledgments
We thank members of the Department of Regenerative Medicine for discussions and technical support. This study was supported by grants from JST SORST, Genome Network Project (MEXT), Grants-in Aid for Scientific Research (MEXT), Research on Child Health and Development, Research on Publicly Essential Drugs and Medical Devices (The Japan Health Sciences Foundation), and NIH (AR50631) (H. A.).
Abbreviations
- Bapx1
vertebrate homolog of Drosophila bagpipe
- Col10a1
type X collagen alpha 1
- Col2a1
type II collagen alpha 1
- Gapdh
glyceraldehyde-3-phosphate dehydrogenase
- KD
knockdown
- Runx2
runt-related transcription factor 2
- Sox9
Sry-related High Mobility Group box containing gene 9
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Provot S, Schipani E. Molecular mechanisms of endochondral bone development. Biochem Biophys Res Commun. 2005;328:658–665. doi: 10.1016/j.bbrc.2004.11.068. [DOI] [PubMed] [Google Scholar]
- 2.Lefebvre V, Smits P. Transcriptional control of chondrocyte fate and differentiation. Birth Defects Res C Embryo Today. 2005;75:200–212. doi: 10.1002/bdrc.20048. [DOI] [PubMed] [Google Scholar]
- 3.Goldring MB, Tsuchimochi K, Ijiri K. The control of chondrogenesis. Journal of cellular biochemistry. 2006;97:33–44. doi: 10.1002/jcb.20652. [DOI] [PubMed] [Google Scholar]
- 4.Lefebvre V, Huang W, Harley VR, Goodfellow PN, de Crombrugghe B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Molecular and cellular biology. 1997;17:2336–2346. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bridgewater LC, Lefebvre V, de Crombrugghe B. Chondrocyte-specific enhancer elements in the Col11a2 gene resemble the Col2a1 tissue-specific enhancer. The Journal of biological chemistry. 1998;273:14998–15006. doi: 10.1074/jbc.273.24.14998. [DOI] [PubMed] [Google Scholar]
- 6.Sekiya I, Tsuji K, Koopman P, Watanabe H, Yamada Y, Shinomiya K, Nifuji A, Noda M. SOX9 enhances aggrecan gene promoter/enhancer activity and is up-regulated by retinoic acid in a cartilage-derived cell line, TC6. The Journal of biological chemistry. 2000;275:10738–10744. doi: 10.1074/jbc.275.15.10738. [DOI] [PubMed] [Google Scholar]
- 7.Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nature genetics. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 8.Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes & development. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akiyama H, Lyons JP, Mori-Akiyama Y, Yang X, Zhang R, Zhang Z, Deng JM, Taketo MM, Nakamura T, Behringer RR, McCrea PD, de Crombrugghe B. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes & development. 2004;18:1072–1087. doi: 10.1101/gad.1171104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 11.Inada M, Yasui T, Nomura S, Miyake S, Deguchi K, Himeno M, Sato M, Yamagiwa H, Kimura T, Yasui N, Ochi T, Endo N, Kitamura Y, Kishimoto T, Komori T. Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Dev Dyn. 1999;214:279–290. doi: 10.1002/(SICI)1097-0177(199904)214:4<279::AID-AJA1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 12.Kim IS, Otto F, Zabel B, Mundlos S. Regulation of chondrocyte differentiation by Cbfa1. Mech Dev. 1999;80:159–170. doi: 10.1016/s0925-4773(98)00210-x. [DOI] [PubMed] [Google Scholar]
- 13.Takeda S, Bonnamy JP, Owen MJ, Ducy P, Karsenty G. Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Genes & development. 2001;15:467–481. doi: 10.1101/gad.845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ueta C, Iwamoto M, Kanatani N, Yoshida C, Liu Y, Enomoto-Iwamoto M, Ohmori T, Enomoto H, Nakata K, Takada K, Kurisu K, Komori T. Skeletal malformations caused by overexpression of Cbfa1 or its dominant negative form in chondrocytes. J Cell Biol. 2001;153:87–100. doi: 10.1083/jcb.153.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou G, Zheng Q, Engin F, Munivez E, Chen Y, Sebald E, Krakow D, Lee B. Dominance of SOX9 function over RUNX2 during skeletogenesis. Proc Natl Acad Sci U S A. 2006;103:19004–19009. doi: 10.1073/pnas.0605170103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lettice LA, Purdie LA, Carlson GJ, Kilanowski F, Dorin J, Hill RE. The mouse bagpipe gene controls development of axial skeleton, skull, and spleen. Proc Natl Acad Sci U S A. 1999;96:9695–9700. doi: 10.1073/pnas.96.17.9695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tribioli C, Lufkin T. The murine Bapx1 homeobox gene plays a critical role in embryonic development of the axial skeleton and spleen. Development. 1999;126:5699–5711. doi: 10.1242/dev.126.24.5699. [DOI] [PubMed] [Google Scholar]
- 18.Akazawa H, Komuro I, Sugitani Y, Yazaki Y, Nagai R, Noda T. Targeted disruption of the homeobox transcription factor Bapx1 results in lethal skeletal dysplasia with asplenia and gastroduodenal malformation. Genes Cells. 2000;5:499–513. doi: 10.1046/j.1365-2443.2000.00339.x. [DOI] [PubMed] [Google Scholar]
- 19.Lengner CJ, Hassan MQ, Serra RW, Lepper C, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Nkx3.2-mediated repression of Runx2 promotes chondrogenic differentiation. The Journal of biological chemistry. 2005;280:15872–15879. doi: 10.1074/jbc.M411144200. [DOI] [PubMed] [Google Scholar]
- 20.Provot S, Kempf H, Murtaugh LC, Chung UI, Kim DW, Chyung J, Kronenberg HM, Lassar AB. Nkx3.2/Bapx1 acts as a negative regulator of chondrocyte maturation. Development. 2006;133:651–662. doi: 10.1242/dev.02258. [DOI] [PubMed] [Google Scholar]
- 21.Zeng L, Kempf H, Murtaugh LC, Sato ME, Lassar AB. Shh establishes an Nkx3.2/Sox9 autoregulatory loop that is maintained by BMP signals to induce somitic chondrogenesis. Genes & development. 2002;16:1990–2005. doi: 10.1101/gad.1008002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murtaugh LC, Zeng L, Chyung JH, Lassar AB. The chick transcriptional repressor Nkx3.2 acts downstream of Shh to promote BMP-dependent axial chondrogenesis. Dev Cell. 2001;1:411–422. doi: 10.1016/s1534-5807(01)00039-9. [DOI] [PubMed] [Google Scholar]
- 23.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 24.Denker AE, Haas AR, Nicoll SB, Tuan RS. Chondrogenic differentiation of murine C3H10T1/2 multipotential mesenchymal cells: I. Stimulation by bone morphogenetic protein-2 in high-density micromass cultures. Differentiation; research in biological diversity. 1999;64:67–76. doi: 10.1046/j.1432-0436.1999.6420067.x. [DOI] [PubMed] [Google Scholar]
- 25.Haas AR, Tuan RS. Murine C3H10T1/2 multipotential cells as an in vitro model of mesenchymal chondrogenesis. Methods in molecular biology (Clifton, NJ. 2000;137:383–389. doi: 10.1385/1-59259-066-7:383. [DOI] [PubMed] [Google Scholar]
- 26.Zheng Q, Zhou G, Morello R, Chen Y, Garcia-Rojas X, Lee B. Type X collagen gene regulation by Runx2 contributes directly to its hypertrophic chondrocyte-specific expression in vivo. J Cell Biol. 2003;162:833–842. doi: 10.1083/jcb.200211089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernard P, Tang P, Liu S, Dewing P, Harley VR, Vilain E. Dimerization of SOX9 is required for chondrogenesis, but not for sex determination. Human molecular genetics. 2003;12:1755–1765. doi: 10.1093/hmg/ddg182. [DOI] [PubMed] [Google Scholar]
- 28.Sock E, Pagon RA, Keymolen K, Lissens W, Wegner M, Scherer G. Loss of DNA-dependent dimerization of the transcription factor SOX9 as a cause for campomelic dysplasia. Human molecular genetics. 2003;12:1439–1447. doi: 10.1093/hmg/ddg158. [DOI] [PubMed] [Google Scholar]
- 29.Bridgewater LC, Walker MD, Miller GC, Ellison TA, Holsinger LD, Potter JL, Jackson TL, Chen RK, Winkel VL, Zhang Z, McKinney S, de Crombrugghe B. Adjacent DNA sequences modulate Sox9 transcriptional activation at paired Sox sites in three chondrocyte-specific enhancer elements. Nucleic acids research. 2003;31:1541–1553. doi: 10.1093/nar/gkg230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Genzer MA, Bridgewater LC. A Col9a1 enhancer element activated by two interdependent SOX9 dimers. Nucleic acids research. 2007;35:1178–1186. doi: 10.1093/nar/gkm014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krebsbach PH, Nakata K, Bernier SM, Hatano O, Miyashita T, Rhodes CS, Yamada Y. Identification of a minimum enhancer sequence for the type II collagen gene reveals several core sequence motifs in common with the link protein gene. The Journal of biological chemistry. 1996;271:4298–4303. doi: 10.1074/jbc.271.8.4298. [DOI] [PubMed] [Google Scholar]
- 32.De Santa Barbara P, Bonneaud N, Boizet B, Desclozeaux M, Moniot B, Sudbeck P, Scherer G, Poulat F, Berta P. Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Mullerian hormone gene. Molecular and cellular biology. 1998;18:6653–6665. doi: 10.1128/mcb.18.11.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arango NA, Lovell-Badge R, Behringer RR. Targeted mutagenesis of the endogenous mouse Mis gene promoter: in vivo definition of genetic pathways of vertebrate sexual development. Cell. 1999;99:409–419. doi: 10.1016/s0092-8674(00)81527-5. [DOI] [PubMed] [Google Scholar]
- 34.Rodrigo I, Hill RE, Balling R, Munsterberg A, Imai K. Pax1 and Pax9 activate Bapx1 to induce chondrogenic differentiation in the sclerotome. Development. 2003;130:473–482. doi: 10.1242/dev.00240. [DOI] [PubMed] [Google Scholar]
- 35.Rodrigo I, Bovolenta P, Mankoo BS, Imai K. Meox homeodomain proteins are required for Bapx1 expression in the sclerotome and activate its transcription by direct binding to its promoter. Molecular and cellular biology. 2004;24:2757–2766. doi: 10.1128/MCB.24.7.2757-2766.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deutsch U, Dressler GR, Gruss P. Pax 1, a member of a paired box homologous murine gene family, is expressed in segmented structures during development. Cell. 1988;53:617–625. doi: 10.1016/0092-8674(88)90577-6. [DOI] [PubMed] [Google Scholar]
- 37.Peters H, Neubuser A, Kratochwil K, Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes & development. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peters H, Wilm B, Sakai N, Imai K, Maas R, Balling R. Pax1 and Pax9 synergistically regulate vertebral column development. Development. 1999;126:5399–5408. doi: 10.1242/dev.126.23.5399. [DOI] [PubMed] [Google Scholar]
- 39.Candia AF, Hu J, Crosby J, Lalley PA, Noden D, Nadeau JH, Wright CV. Mox-1 and Mox-2 define a novel homeobox gene subfamily and are differentially expressed during early mesodermal patterning in mouse embryos. Development. 1992;116:1123–1136. doi: 10.1242/dev.116.4.1123. [DOI] [PubMed] [Google Scholar]
- 40.Candia AF, Wright CV. Differential localization of Mox-1 and Mox-2 proteins indicates distinct roles during development. Int J Dev Biol. 1996;40:1179–1184. [PubMed] [Google Scholar]
- 41.Wright E, Hargrave MR, Christiansen J, Cooper L, Kun J, Evans T, Gangadharan U, Greenfield A, Koopman P. The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nature genetics. 1995;9:15–20. doi: 10.1038/ng0195-15. [DOI] [PubMed] [Google Scholar]
- 42.Ng LJ, Wheatley S, Muscat GE, Conway-Campbell J, Bowles J, Wright E, Bell DM, Tam PP, Cheah KS, Koopman P. SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev Biol. 1997;183:108–121. doi: 10.1006/dbio.1996.8487. [DOI] [PubMed] [Google Scholar]
- 43.Zhao Q, Eberspaecher H, Lefebvre V, De Crombrugghe B. Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev Dyn. 1997;209:377–386. doi: 10.1002/(SICI)1097-0177(199708)209:4<377::AID-AJA5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 44.Tribioli C, Frasch M, Lufkin T. Bapx1: an evolutionary conserved homologue of the Drosophila bagpipe homeobox gene is expressed in splanchnic mesoderm and the embryonic skeleton. Mech Dev. 1997;65:145–162. doi: 10.1016/s0925-4773(97)00067-1. [DOI] [PubMed] [Google Scholar]
- 45.Saito T, Ikeda T, Nakamura K, Chung UI, Kawaguchi H. S100A1 and S100B, transcriptional targets of SOX trio, inhibit terminal differentiation of chondrocytes. EMBO reports. 2007;8:504–509. doi: 10.1038/sj.embor.7400934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilhelm D, Hiramatsu R, Mizusaki H, Widjaja L, Combes AN, Kanai Y, Koopman P. SOX9 regulates prostaglandin D synthase gene transcription in vivo to ensure testis development. The Journal of biological chemistry. 2007;282:10553–10560. doi: 10.1074/jbc.M609578200. [DOI] [PubMed] [Google Scholar]
- 47.Tribioli C, Lufkin T. Bapx1 homeobox gene gain-of-function mice show preaxial polydactyly and activated Shh signaling in the developing limb. Dev Dyn. 2006;235:2483–2492. doi: 10.1002/dvdy.20867. [DOI] [PubMed] [Google Scholar]