Abstract
The red blood cell membrane is specialized to exchange chloride and bicarbonate; usually the pH gradient, the chloride ratio, and the membrane potential are tightly coupled. We review the evidence that led to the ability to separately vary inside and outside pH in red cells. The effect of pH on Na pump activity and on the selectivity of the inside and the outside transport sites is reviewed. In red blood cells, at high pH, the outside site is not selective. An increase in protons leads to an increase in K+ affinity, thus making the site more selective. The pK for this site is different in rats and humans; because of the high conservation of residues in these two species, there are only a few possible residues that can account for this difference. On the inside, work from unsided preparations suggests that, at high pH, the transport site is highly selective for Na+. Once again, an increase in protons leads to an increase in K+ affinity, but now the result is a less selective site. During their maturation, reticulocytes lose many membrane proteins. The type and fractional loss is species dependent. For example, most reticulocytes lose most of their Na pumps, retaining about 100 pumps per cell, but animals from the order Carnivora lose all their pumps. We review some of the evidence that PKC phosphorylation of N-terminus serines is responsible for endocytosis in other cell types and species variation in this region.
INTRODUCTION
For over half a century, ion flux measurements across the red cell membrane have provided key information about how membrane transporters operate. Two of the best studied transporters are the anion exchanger and the Na pump. Interestingly, the anion exchanger is present at 1 million copies per red cell [1] whereas the Na pump is present at only about 100 copies per cell [2]. Not only flux measurements but also biochemical characterizations have been possible with these red cell proteins; even at low copy number it is possible to measure Na pump catalytic phosphorylation [2].
The rapid rate of Cl−/HCO3− exchange, for a long time, seemed to preclude the possibility of independently varying the inside and outside pH and the membrane potential. However, as this review will detail, red cellologists have developed techniques that exploit the red cell properties to make this possible. We will also discuss pH effects on the Na pump including our work on extracellular proton effects. The potential structural implications of the species differences in proton effects in red cells will also be examined in light of the recent report of the Na pump’s crystal structure in the Rb+ occluded conformation [3].
During maturation, the reticulocyte membrane keeps all of its anion exchanger but loses 98 to 100% of its Na pumps. The processes that shed the reticulocyte of Na pumps likely include endocytosis. We review some species differences with respect to the regulation of Na pump trafficking that may bear on reticulocyte maturation.
ANION EXCHANGER AND Na LEAK
In order to study the effect of extracellular pH on the Na pump in red cells, a key obstacle had to be overcome. The red cell membrane has a very high rate of Cl−/HCO3− exchange and the Cl− gradient sets the membrane potential. Thus for a long time, it seemed difficult, if not impossible, to independently vary the intracellular pH, the extracellular pH and the membrane potential. The ability to set pH on one side of the membrane independent of the pH on the other side and the membrane potential has been termed “pH clamp” [4–7]
EVIDENCE FOR LOW PROTON PERMEABIILITY
Jacobs and Parpart [8] studied the possibility of red blood cell proton transport; they used hemoglobin as their pH indicator and conducted their study at very low pH values. In spite of the fact that they used high proton concentrations, their data supported hydroxyl, but not proton, fluxes in red cells. Given the high proton concentrations studied, it is remarkable that the red cell membrane did not allow H+ to cross and this result certainly suggests the membrane is tight to protons. For our purposes, not only must the bilayer be tight to protons, but the proton flux mediated by transporters must be minimal as well.
Jennings [9] provided some of the first evidence that the background proton flux was low in red blood cells near neutral pH. Jennings set out to test a possible implication of the titratable model proposed by Gunn [10,11]. The titratable model very elegantly explained the different pH dependencies of the transport of chloride and sulfate by the red cell. In this model, as pH declined from 7 to more acidic values, the anion exchanger became titrated and this protonated form of the exchanger transported sulfate whereas the unprotonated form (at neutral pH) transported chloride. Jennings had two terrific insights. His first insight was that the proton might not only convert the exchanger from a chloride transporter to a sulfate transporter but that the proton might be cotransported along with the sulfate. The second insight was that this proton flux might be measurable-a remarkable thought since the bicarbonate flux is about 1000-times faster than the sulfate flux, even at bicarbonate concentrations of 1 mM. Jennings was able to reduce the bicarbonate concentration to vanishingly small amounts so that the proton movements on the anion exchanger would be larger for H-sulfate cotransport than for any residual Cl−/HCO3− exchange. He apparently had the intuition that any other proton pathway in the red cell would make a small contribution to background proton fluxes. Jennings very elegantly demonstrated H+ & SO4−2 cotransport on the red cell anion exchanger which remains a cornerstone to our understanding of anion exchanger mechanisms.
Two other studies provided the final basis for the pH clamp. It was found that extracellular DIDS irreversibly inhibited anion exchange in red cells and did not affect other transporters [1]. This apparent specificity is due to several reasons as DIDS is a reagent that reacts with unprotonated lysine residues. A key lysine that is extracellularly accessible on the anion exchanger has a particularly low pK, so that, at neutral pH, it is probably one of the few lysines available extracellularly to react with DIDS. When DIDS has access to the intracellular side of the membrane, it inhibits the Na pump and the plasma membrane calcium pump [12–16). Secondly, DIDS is an anion and so will preferentially go toward binding sites designed for anions. Since most anion sites (including ATP binding sites) have positive charges, lysine and arginine are likely to make up these sites; consequently, there is a 50% chance of DIDS reacting with an anion binding site in the absence of steric considerations. In contrast, lysines are present on many non-anion binding sites. Since DIDS does not inhibit other transporters in red cells when added from the outside it is tempting to think that these lysines are not critical for transport function. That is only a fair conclusion if the reaction is done at pH values above 9 where the lysines would be expected to be uncharged, otherwise it is more likely that DIDS simply did not react with extracellular lysines and thus no conclusion can be made about their functional importance. (For example, we have been unable to see any effects of extracellular DIDS pretreatment at pH 9 on the Na pump or the PMCA pump, suggesting that modification of extracellular lysines does not interfere with pump function. The recent Na pump crystal structure is also consistent with no critical extracellular lysine residues.)
The second study, by Knauf et al., [17], included an examination of the DIDS insensitive chloride, bicarbonate and hydroxyl transport. While DIDS inhibited 99% of chloride exchange, it only inhibited about 50% of chloride conductance. In the presence of DIDS, the hydroxyl flux was low. (Though the permeability constant is high reflecting the low hydroxyl concentration compared to chloride concentration.) Thus in the presence of DIDS, the membrane potential would still be determined by the chloride gradient, but if a pH gradient were established, the pathway(s) for proton/hydroxyl equilibration were slow enough that the pH, in the presence of reasonable amounts of buffer, could effectively be varied inside separately from outside and remain essentially constant for the flux measurements.
Two remarkable properties of the red blood cell anion exchanger are worth mentioning. First, the Cl−/HCO3− exchange rate is among the fastest known for membrane transporters.1 Second, the 1 million copies of the anion exchanger represents about 25% of the integral membrane protein of the red cell membrane.
Given that channels have ion flows at least 10–100 faster than transporters per protein per unit time, why didn’t the red cell evolve to have a bicarbonate/chloride channel and therefore require 10–100 times less bicarbonate/anion protein? In both a channel and a simple exchanger, the movements of chloride and bicarbonate are downhill; since the membrane potential of the red cell is −10 mV, there is only a slight difference between the electrochemical gradient which drives anions through channels, and the chemical gradient which drives anion movement on an electroneutral exchanger.
In most cells, having a large anion conductance would alter the membrane potential, but the red cell membrane potential is given by the chloride gradient, even though the chloride conductance of the red cell is 1000-times less than the chloride exchange on band 3. So membrane potential does not appear to be the reason.
Another distinctive property of the red cell is its very low permeability to cations, particularly Na. Our current electrophysiological instruments are only able to say that the most selective anion channel is at least 100:1 selective for anions over cations. The ratio of the chloride flux to the Na+ flux in red cells is roughly 1,000,000 to 1. So within our limits of detection, we can say that we don’t know of a chloride channel that is this selective. Thus, anion vs. cation selectivity may have driven the evolution of an anion exchanger over a channel.
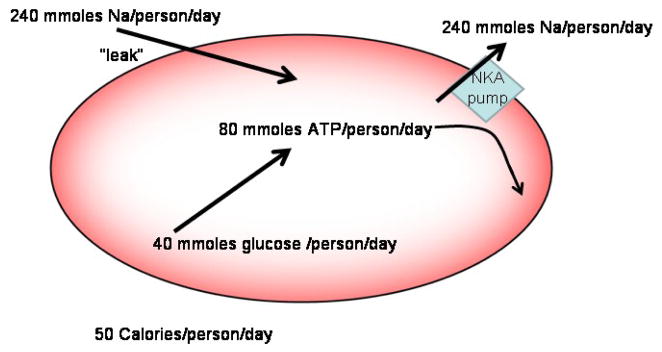
It has long been appreciated among the red cell transport community that having red cells that are tight to sodium probably is an important savings of energy, but we have not seen this quantified. That is, a substantial Na leak would require significant work by the Na pump to maintain the Na+ gradient (and cell volume). Since the Na pump is fueled by ATP, this implies burning of calories. Given that the Na pump flux in red cells is approximately 5 mmoles/liter packed cells per hour, that the human body contains about 2 liters of packed cells, and that red cells produce 2 net ATP molecules for every glucose molecule hydrolyzed, one can calculate that the red cell Na pump burns about 50 calories per day, which is about 2% of current daily suggested consumption. Clearly, if the Na+ leak were an order of magnitude larger (e.g. via an anion channel compared to an exchanger), the red cell Na pump energy consumption would be about 20% and represent a significant drain on energy resources.
It is interesting however, that in fact the major leak for Na+ is on the anion exchanger where Na+ masquerades as an anion by forming NaCO3− [18]; while this is a significant Na+ flux, it is minor in terms of the HCO3− flux. In contrast, the Na+-dependent bicarbonate transporters (NBCs) have a high rate of NaCO3− flux; NBCs and AEs are both members of the same large protein superfamily. Clearly, maintaining a low Na+ flux seems is energetically favorable for red cells.
NA PUMP SELECTIVITY AND PROTON EFFECTS ON NA PUMP
We will now review 4 studies examining the effect of pH on the Na pump. Three of these studies involved unsided preparations (shark rectal gland and dog kidneys) and one study used human and rat red blood cells and the pH clamp.
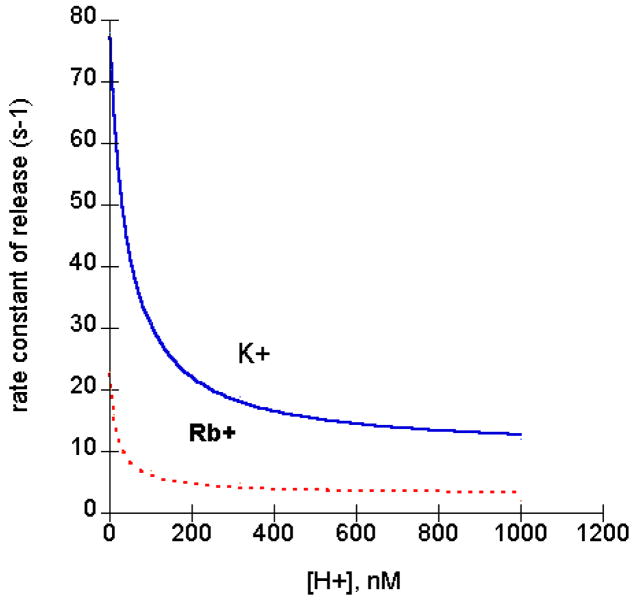
The Na,K ATPase activity pH profile is bell shaped, as is the case for many enzymes. What are the steps that are pH sensitive that account for this bell shape in the case of the Na pump? One of the steps is the rate of K+ release from the pump. The most thorough study on the pH dependence of K+ release was done by Forbush [19]. He developed a novel apparatus for fast measurements of K+ release. Essentially, he flowed buffer through a filter containing the 86Rb+ bound Na pump. For time constants (on the order of 100 sec−1) he needed to collect samples every 10 milliseconds-too fast for manual collection. He put 50 cuvettes on the perimeter of a record player turntable; at 33 rpm, in a single turn, each cuvette passes the filter outflow for about 10 msec. Using this apparatus Forbush found that protons inhibited K+ release[19]. We have replotted his data vs. proton concentration instead of pH and then fitted the data find the Ki for proton inhibition (see Fig. 2). The pK is 7.37 for K+ and 7.77 for Rb+. In addition, he noted that the rate of Rb+ release was about 2 to 5 times slower than for K+.
Fig. 2.
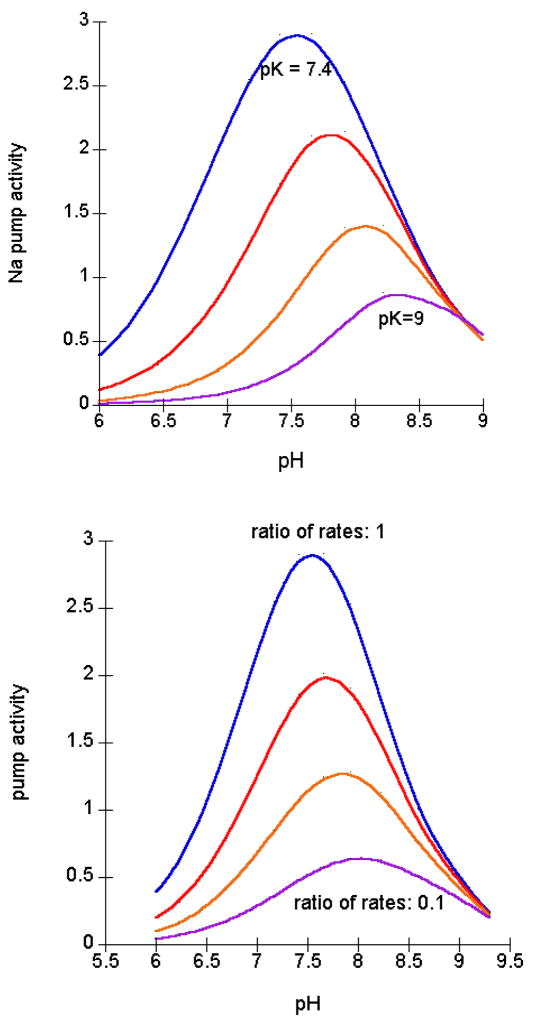
Forbush and Klodos [20] examined more closely which partial pump reactions were affected by pH to explain the bell shaped curve of Na,K ATPase activity. They found that protons increased the rate of the slowest Na+-dependent step. So the bell shaped curve for ATPase activity reflects 2 proton effects. If we start at pH 9, a Na+-dependent step is rate limiting. As protons are added, protons bind to a site with pK estimated by us from the Forbush and Klodos data to be ~ 8.5. This titration speeds up the Na+-dependent step and since it is rate limiting in this pH range, the overall ATP reaction speeds up. As the proton concentration is increased further, a second site, with pK ~7.4, is titrated and this slows down the deocclusion step, which eventually becomes rate limiting and this step then dominates the pH dependence of the ATPase reaction.
The location of the peak of the bell shaped pH dependence of ATPase varies in different conditions and species. The location of the peak reflects the balance of 2 ratios: the ratio of the affinity for the activating proton to the inhibiting proton, and the ratio of the rate of the slow Na+ step vs. the rate of K+-deocclusion. This is probably most easily seen by examining Forbush’s data[19]. When measuring just the rate of K+ and Rb+ release, we have the Ki for proton inhibition and the ratio of the rates for K+ and Rb+ release. During Na,K(Rb)-ATPase, the Na+-dependent steps are independent of Rb+ or K+ (since neither is bound). Fig. 3A shows the shift expected for changes in the Ki for the inhibitory proton with Rb and Fig. 3B, the shift expected for the changes in ratio of Na+-dependent steps to K+/Rb+ steps. Our fit of his data suggests that the combination with both changes fits the best.
Fig. 3.
Na ATPase
Skou [21] examined the pH dependence of Na-ATPase activity at low ATP (1 uM ATP). It should be noted that in the ATPase experiments, at each pH, the optimal Mg+2 was first determined so that the effects of pH on the formation of MgATP are eliminated. In Fig. 3 of [21] the pH optima is determined for Na-ATPase in the presence and absence of 2 mM K+. As described by others, at pH values less than about 7, addition of K+ actually inhibits the pump at this low ATP. (This is because K+ deocclusion is very very slow at low ATP). However, above pH 7, K+ actually stimulates the ATPase. As pH is decreased from pH 9 to pH 8, in the presence of K+, the enzyme is activated.
The bell shaped curve for Na-ATPase in the absence of K+ is a bit tougher to explain than for Na+K-ATPase. The Na+ steps are assumed to be common to both Na- and Na+K-ATPase. Yet the overall cycle rate for Na+K-ATPase is much faster, implying that the Na+-dependent steps are too fast to be rate limiting for Na-ATPase activity. The inhibition by protons (the downward slope at low pH) could reflect a similar effect to that seen in Na+K-ATPase. Presumably both cycles follow the sequence: transport site facing outside, transport site occluded, and transport site facing inside, or in terms of “gates”, outside gate open, both gates closed, inside gate open. When K+ is present, then the site is loaded with K+; in the absence of K+, the site could be empty, loaded with Na+, or loaded with H+ (H3O+). In any case, the conformational change from both gates closed to inside gate open must occur. When K+ is bound, we know from Forbush’s work that protons decrease the opening rate of the inside gate (deocclusion). It seems most convenient to assume that protons also decrease this rate when K+ is not present; obviously this rate is much, much slower in the absence of K+.
In Na,K-ATPase, the higher affinity effect of protons is to activate the slowest Na+ dependent step. Since this step is already much faster than the overall Na-ATPase cycle rate, this activation cannot account for the increase in rate from pH 9 to the peak for Na-ATPase. Protons could increase the rate of the closing of the outside gate in the absence of K+. Alternatively, protons could increase the rate of dephosphorylation from the K+-empty enzyme.
Internal Selectivity
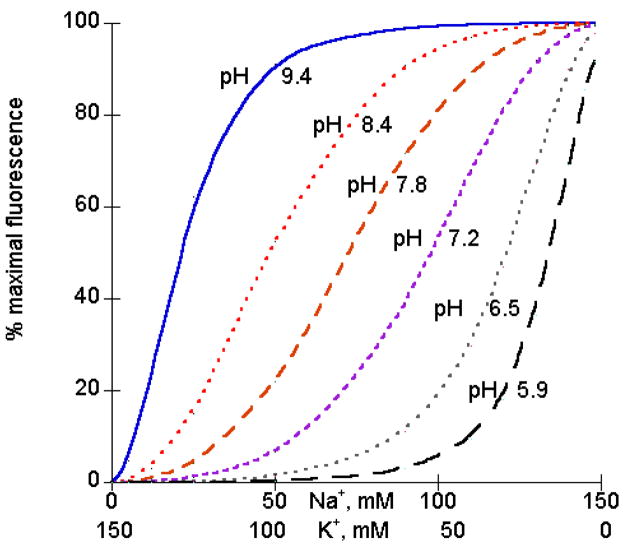
Skou and Esmann [22] examined equilibrium binding of Na+ and K+ to the internal sites of the Na pump. They made use of the fact that eosin binding to the pump changes its fluorescence and eosin binds better when Na is bound inside than when K+ is bound inside.2
As the proton concentration is increased from nanomolar to micromolar (from pH 9.4 to pH 5.9) there is a dramatic increase in the affinity for K+ vs. Na+. Skou and Esmann [22] concluded that protons increase the affinity for K+ or, equivalently, K+ binding increases the pK for protons. Since Na+ and K+ are binding inside, it is presumed that the proton is also binding inside (but see [23] for transmembrane proton effects). We have analyzed their data using a simple model with 2 Na+ ions or 2 K+ ions bound in the presence and absence of proton binding. As shown in Fig. 4, these equations provide a good fit to the data. We estimate that K+ binding increases the pK for protons by 3 pH units. This means that over a substantial pH range, when Na+ ions are bound, protons are not bound and when K+ ions are bound, protons are bound.
Fig. 4.
These curves closely match the data in Fig. 3 of [22], using pKa (for proton binding to the Na form) of 15 uM and pKb (for proton binding to the K form) of 30 nM.
Our fits to the data of Forbush[19] and Skou and Esmann[22] suggest that protons decrease the rate of K+ deocclusion and increase the affinity for K+. This could reflect a single proton effect, as a decrease in off rate results in an increase in affinity. The pK values from our fits are very different-about 7.4 for Forbush and 9 for Skou and Esmann. Since these are apparent pKs based upon analysis of different types of data (equilibrium binding of cations vs. rates of deocclusion) it is possible that the actual pK values are the same. However, there are two other intriguing possibilities. One possibility is that the pK is species dependent, if so, then comparison of shark and pig Na pump sequences, along with the Na pump crystal structure [3] might provide some key insights. A second possibility concerns the effect of ATP on the pump. The Skou and Esmann data were obtained in the absence of ATP (and, presumably, in the absence of eosin binding to the kinetic low affinity ATP site). Forbush’s data were obtained in the presence of high ATP. ATP speeds up deocclusion rate considerably. One interpretation of the higher pK obtained by Skou and Esmann compared to Forbush is that part of the ATP effect is to shift the pK of the inhibitory proton.
SPECIFICALLY EXAMINING EXTRACELLULAR H EFFECTS IN RED CELLS
More recently, we decided to determine if protons were also involved in regulating the selectivity of extracellular Na+ and K+ binding [24]. We felt that measuring K+ fluxes in red blood cells would be an excellent system for these measurements. For convenience, 86Rb+ was used as a K+ congener for uptake and red cells were initially loaded with Na+ to saturate the inside sites and then treated with DIDS to reduce the proton permeability.
Two types of studies were done. In one case, we measured the [H+] activation of 86Rb+ uptake at fixed [Rb+] in the presence of different concentrations of inhibitory cations. Specifically, we compared Na+ and the Na+-like inhibitor guanidium [25] to the K+-like inhibitors, bretylium and TPA [26, 27]. Typically, in a standard inhibitor analysis, apparent Km and Vmax values would be calculated and the dependence of 1/Vmax and Km/Vmax on the [inhibitor] determined using a replot. The slope of Km/Vmax vs. [inhibitor] is proportional to the inhibitor Kd for binding to the empty transporter, whereas the slope of 1/Vmax vs. [inhibitor] is proportional to the inhibitor Kd for binding to the loaded transporter,
The problem with this analysis is that the calculation of Km/Vmax and the subsequent replot introduce errors that are not present in the original data. (e.g., the error for Km/Vmax is larger than the error for each separately.) Inspired by Knauf’s alternative analysis for ping pong equations [28], we rewrote the equation in the form:
where A = c + d × I and B = e + f × I
In this form, “d” includes the inhibitory constant for the unprotonated pump and “e” the inhibitory constant for the protonated pump. All the other parameters are independent of [inhibitor] and should be constants. In these studies, we compared external [H+] effects on ouabain-sensitive 86Rb+ uptake between human and rat red cells; for the human studies we deliberately used red cells from a single donor to minimize any between individual variability. With this analysis we directly determined the slope of A and B and so the errors in A and B are not propagated as in the Km/Vmax analysis.
When we plotted A vs. [inhibitor], the slopes for the different inhibitors were similar. Since the slope is proportional the inhibitory constant for binding to the unprotonated pump, it suggested that the unprotonated pump does not select between these different cationic inhibitors. In contrast, the plot of B vs. [inhibitor] revealed dramatic differences between slopes for these different inhibitors. The inhibitors fell into two classes: i. bretylium and TPA bound much better to the protonated pump, and much better than ii. guanidium and sodium. Consistent with the protonated pump preferring K+ or K+-like compounds over Na+ or Na+-like compounds [24].
In the second set of experiments, we measured the effect of [Rb+] on the [H+]-activation curves. This analysis is more complicated because the Km for a substrate, in the general case, is a complex expression including many rate constants; in contrast the Ki is the Kd for a dead end inhibitor. However, we learned from Phil Knauf the value of simplifying assumptions in order to examine what would occur in a simple case. Our general result was that Rb+-binding decreased the Km for protons; similarly, plotting the data as the effect of protons on [Rb+] activation shows that protons decreased the Km for Rb+. If we assume that Rb+ translocation is the rate-limiting step, which includes the deocclusion step, then our results suggest that protons cause an increase in Rb+ affinity. This was precisely the reverse of our expectation assuming that protons titrate a residue near the Rb+ binding site. Our results are remarkably similar to those of Skou’s: Protons increase the affinity for K+ and K+ increases the pK for protons. Though we feel Skou’s observations apply to an internal proton effect. Ours clearly applies to an external proton effect.
Our data are consistent with random binding of protons and Rb+(K+) (and K+-like inhibitors); after the first has bound, the affinity for the other increases about 10-fold. In contrast, the effect of protons on the affinity for Na+-like inhibitors is different. In humans, there is almost no change, whereas in rat, protons decrease the affinity for Na+. We cannot rule out the possibility that protons actually bind before K+ and help to induce the transport site. In this model, there is a nonspecific binding site for cations in the unprotonated pump form. When protons bind, a conformational change takes place which allows for higher K+ selectivity than the site in the absence of protons.
What potential extracelluar H+ sites exist on Na,K-ATPase?
Another interesting observation we found by comparing rat and human red cell Na,K-ATPase is that the pK for the unprotonated pump is about 1 pH unit different in the two cases, i.e. ~9.8 for rat and ~9.1 for human, see Table 1 [24]. A comparison of rat vs. human extracellular sequences reveals that there are very few differences that have potential titratable residues. One area is the M1M2 extracellular loop which contains two residues which provide ouabain-resistance to rat α1 (Fig. 4A). The other section is the M7M8 extracellular loop where some mild hydrophobic changes occur as well as a significant change from 873Val-Asp874 in human to 873Glu-Thr874 in rat (Fig. 4C). Alternatively, the titratable extracellular residue may actually be the same in both rat and human Na pumps, e.g. Glu309 (Fig. 4B). In this case, the different pK’s may arise via allosteric interactions from the changes in M1M2 or M7M8 (or both).
Table 1.
pK for extracellular protons binding to Na pump with transport site facing outside
| outside K+ bound | outside empty | outside Na+ bound | |
|---|---|---|---|
| human | 9.1 | 7.9 | 8.0 |
| rat | 9.8 | 8.7 | 7.8 |
Deducing molecular interactions between amino acid residues has made tremendous strides in the last decade with increased success in solving the high-resolution structures of many integral membrane proteins, including P-type ATPases. The sarco/endoplasmic reticulum Ca-ATPase (SERCA) has been crystallized in several conformations [29], whereas there is only a single crystal structure available for the Na,K-ATPase [3]. However, the Na,K-ATPase structure does include the two bound Rb+ ions which provides the first visualization of counter-ions in a P-type ATPase structure. The two Rb+ ions appear to be coordinated by side chains of residues in M4 (Glu327), M5 (Ser775, Asn776, Glu779), and M6 (Asp804), all of which had been previously implicated in cation binding via mutagenesis or chemical modification. The acidic residue Glu327 in the Na,K-ATPase is analogous to Glu309 in SERCA, which has been suggested to be a cytoplasmic gate for intracellular Ca2+ access to the cation ion binding site. Moreover the mobility of the “Glu309-gate” is influenced by movements in M1 which allosterically alter the position of Glu309 in M4 [30]. Mutagenesis studies of hydrophobic M1 residues in the Na,K-ATPase have been shown to significantly alter both Na+ and K+ affinities, consistent with a similar M1 regulation of Glu327 gating [31,32]. Of particular interest in this review is the observation that mutation of the conserved Leu99 in M1 (rat α1) caused a significant reduction in external K+ affinity [32], presumably via physical perturbations of M4 (and Glu327). Thus, if negative perturbations of M1 can decrease K+ext affinity, is it also possible that positive perturbations can increase K+ext affinity? If so, protonation of Arg113 or Asp124 (in rat) or both may stabilize M1 and increase the external affinity for K+, which would be consistent with our earlier observations that external H+ binding increases K+ext binding. Potassium binding is antagonistic to ouabain binding so it also fits with the low ouabain-sensitivity of rat α1.
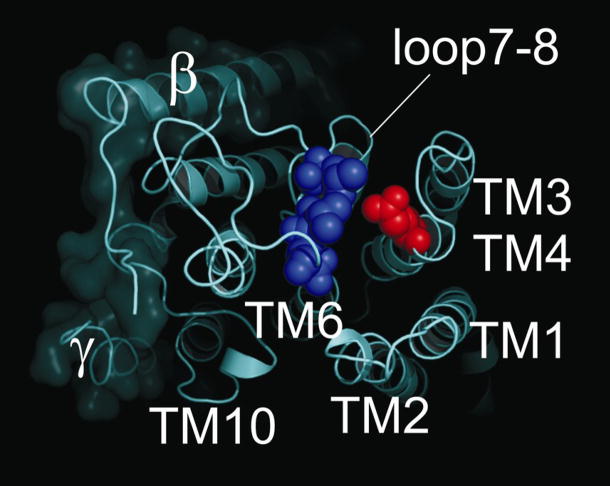
Although titration of Arg113 is an enticing target to explain our observations for the external pH effects in rat α1, it means that a different residue must account for external pH effects in human α1 (Gln113 in human; Fig. 4A). Another possibility is that the titratable residue is common among rat and human, but that the local environment surrounding the residue is different between the two species, accounting for the separate pK values. One intriguing residue is Glu309 in the M3M4 loop which is conserved among mammals. Capendeguy et al. [33] observed that mutating Glu309 to Cys increased Na,K-ATPase activity and the apparent affinity for K+ext in rat α1 expressed X. laevis oocytes. Moreover, subsequent modification of Glu309Cys with 2-[trimethylammonium]ethyl methanethiosulfonate (MTSET), which introduces a positive charge at this site, further enhanced the affinity for external K+. Recent work from the same laboratory demonstrated that the M3M4 loop is in close proximity with the M7M8 by demonstrating Cys-Cys disulfide bridge formation between mutants Tyr308Cys and Asp884Cys (numbering via Pig sequence [34]). Figure 5 shows the molecular structure of the pig enzyme looking down at the extracellular loops highlighting Glu309 in red and showing the close proximity to three residues in M7M8 which are substantially different between rat and human (i.e. Leu879, Val881, and Asn882). The changes in the rat sequence introduce threonine and glutamate into this region which certainly would alter the local environment around Glu309 compared to valine and asperagine. Another interesting observation is that the torpedo sequence has a glycine at the equivalent position of Glu309 and when it was mutated to a glutamate (i.e. Gly314Glu in Torpedo numbering), it caused a reduction Na,K-ATPase activity by about 50% [35]. The authors mentioned that the changes observed in extracellular K+ affinity were scattered within experimental error and thus could not be distinguished from wild-type values. Thus, in contrast to what one might predict, it appears that a negative charge in the M3M4 extracellular loop decreases the affinity for K+ext and masking this charge increases K+ binding and enzyme activity. Could the local environment (e.g. hydrophobic residues from M7M8) enhance the protonated state of Glu309, such that its pK is shifted 3–4 orders of magnitude and explain our previous pH effects? Experiments are currently underway to test this possibility.
Fig. 5.

Sequence alignment of extracellular loops M1M2, M3M4, and M7M8 from human, pig, and rat Na,K-ATPase α1. Transmembrane segments begin and end each stretch of sequence (labeled and bolded). The sequences between the indicated membrane spanning domains correspond to the respective extracellular loops. The significant amino acid differences between rat and human (and pig) are capitalized and underlined. In A, the Arg and Asp changes (i.e. R113 and D124 in this numbering) that confer ouabain-resistance are shown along with two additional substantial changes, Ala114Ser and Asn121Pro. In B, Glu309 is capitalized and underlined. In C, the SYGQ segment of M7M8 that physically interacts with the α-subunit is underlined. Also in C, Leu879, Val881, and Asn882 are capitalized and underlined. These are the residues that appear in blue in Fig. 6, which are in close proximity to Glu309 and are substantially different between rat and human.
NA PUMP TRAFFICKING
In the above section we described how comparing the kinetics of the Na pump in red cells of different species provides some insight on structure/function relationships. Another area where red cell Na pump behavior is different across species is the cell biological handling of Na pump during reticulocyte maturation. Reticulocytes of all species studied have a relatively high number of Na pumps per cell. In all species except for species in the order Carnivora, many Na pumps are removed and about 100 retained in the mature red cell. In contrast, in animals of the order Carnivora, all Na pumps are removed and this results in cells that are high in Na+ and low in K+ [36]. This result raises the question of how is the trafficking of the Na pump different so that in some species, some pumps are left and in other species, all pumps are removed.
RETICULOCYTE MATURATION
During reticulocyte maturation membrane proteins are lost. Perhaps the best characterized process of loss is of the transferrin receptors [37]. These receptors are first endocytosed, packaged in a multivescilar organelle and then released as exosomes by exocytosis. As mentioned, most, but not all, of the Na pumps are lost. In contrast, essentially all of the anion exchanger is retained.
The loss of Na pumps requires ATP [38]. Blostein and Grafova [39–41] considered two possible pathways for Na pump loss during reticulocyte maturation. One pathway was similar to the transferrin pathway, where the pumps are first endocytosed, then packaged in multivesicular structures and then released as exosomes. The exosomes produced by this process can have their membrane proteins facing inside out or right side out [42]. The second pathway was direct release of Na pump vesicles from the plasma membrane, a process now termed ectocytosis [37]. The latter pathway predicts that the vesicles would have their Na pumps right side out. Since the outside of the Na pump is resistant to trypsin, this model predicts that the vesicle with released Na pumps would not be proteolyzed by trypsin. In fact they found that vesicles with released Na pump could be degraded by trypsin, thus ruling out the second model.
The first model requires that there is regulation of Na pump endocytosis and presumably that exocytosis also occurs. At the time of the Blostein and Grafova [41], there were little data suggesting that exocytosis or endocytosis of the Na pump was regulated (except for the regulation involved in generating polarity in epithelial and nerve cells). Blostein and Grafova [41] provided three lines of evidence in support of Na pump recycling in reticulocytes. 1. Normal reticulocytes lose Na pumps. If they are starved of ATP, a transient increase in the number of Na pumps is observed (see their Fig. 6B). 2. Chloroquine, which may interfere with endocytosis or endosomal pH, also, at short times, leads to an increase in surface Na pumps. 3. Ouabain also seems to slow the rate of loss of Na pumps. Taken in combination, these data support the notion that there is a pool of endosomal Na pumps that can traffic to the plasma membrane.
Fig. 6.
External view of the Na,K-ATPase structure[3]. Space-filling models illustrate Glutamate-309 in the M3M4 extracellular loop (red) and Leucine-879, Valine-881, and Asparagine-882 in the M7M8 extracellular loops (blue) of the pig renal enzyme. Transmembrane helices are labeled accordingly.
It is simplest to think that removal of Na pumps occurs by a process similar to transferrin receptor removal, that is, it starts with endocytosis of plasma membrane protein. For the Na pump there is the caveat that in animals not of the order Carnivora, about 100 Na pumps are left in the surface membrane in the mature red cell, whereas for transferrin receptors in the few species studied and for Na pumps in species from the order Carnivora, all the proteins are removed.
As far as we are aware, the signals for Na pump endocytosis in reticulocytes have not been studied further. However, Na pump endocytosis has been studied in other tissues and in these tissues PKC phosphorylation of serine residues plays a key role, but which serine residue(s) mediate the response varies between species. It would be interesting to determine if the same phosphorylation sites are involved in Na pump endocytosis in reticulocytes as has been described for Na pump endocytosis in kidney and in lung.
PKC and Na pump Trafficking
To date, most of the studies determining which serine residues are involved in PKC phosphorylation and regulation of trafficking has been done in renal cell models. The initial studies were done with rat Na pump, both because it was one of the first pumps cloned and also because its ouabain resistance made for easy selection of successfully transfected cells.
There are a number of papers that provide convincing data that support a role for Ser18 in the rat Na pump for being involved in PKC regulation of Na pump trafficking. To our mind, here are 4 key results:
Ser18 fits the criteria of a consensus PKC phosphorylation site which includes positive charges at nearby positions.
A mixture of PKC isoforms can phosphorylation the purified Na pump in vitro, and the stoichiometry approaches 1 phosphate per pump. [43].
Activation of PKC in OK or LLC-PK1 cells transfected with the rat Na pump under the appropriate conditions (particularly cell Na concentrations) alters Na pump trafficking [44]. Most of the Na pump trafficking studies were done using extracellular biotinylation with an impermeant reagent and then quantification.
Mutation of Ser18 to alanine reduces or eliminates the trafficking.
In conclusion, in rat Na pump Ser18 seems to be phosphorylated by PKC and this phosphorylation is a signal for trafficking of the Na pump. However, Ser18 is not conserved in other pumps. Ser11 has been considered as another possible site. However, the data are not as strong for this residue.
Ser11 does not fit the strict definition of a consensus PKC site. But there is a histidine residue at the +2 position. Beguin et al. [45] using molecular modeling found that when this residue was positively charged, it fit in the substrate site of PKC.
Some investigators have been able to detect PKC phosphorylation of purified pump from species other than rat. Feschenko and Sweadner [43] were able to phosphorylate rat Na pump with PKC, but they did not detect phosphorylation of pig or dog from [32P]ATP with PKC in contrast to their findings in rat. As they pointed out, they could not rule out that the pig or dog were already phosphorylated (in vivo, with “cold” Pi). Intriguingly, they found that the pig and dog Na pumps (in the presence of ouabain) promoted increased autophosphorylation of PKC, implying that PKC could bind to these pumps. In contrast to Feschenko and Sweadner’s results, Beguin et al. [45] were able to phosphorylate Na pump on the N-terminus with PKC in cane toad, duck, rabbit and sheep. Note that in all cases, the stoichiometry is much less than 1:1. This low stoichiometry raises great concerns, see [46]
It is true that in some cell types, (for example LLC-PK1 cells but not OK cells) activation of PKC leads to trafficking of pumps that lack Ser18 but have Ser11. In some cases, PKC activation leads to increased pump activity but apparently no change in trafficking, see, e.g., [47].
Mutation of Ser11 in these cases prevents trafficking [44,48]
Thus, the data are not as strong for supporting a direct PKC phosphorylation of Ser11.
One aspect that apparently has not received comment is that His13 is not conserved; in humans, other primates, and Xenopus, the residue is Gln and thus not positively charged. Interestingly, Beguin et al., [45] have shown that the endogenous Xenopus pump also responded to PMA as expected, so even though this pump lacks His13, it still responds similarly to other pumps. Note that Efendiev and Pedemonte [44] were testing “human” vs. rat pump, but they merely replaced the unique rat serine and did not also replace His-13 with Gln so their studies in LLC-PK1 cells do not address the issue. But Beguin et al., [45] studies in A6 cells (a Xenopus cell line) and Xenopus oocyte clearly indicate that the endogenous Xenopus pump responds to PMA even though it does not have His13 [45]. It seems to us that in the whole cell studies or cell homogenate studies, PKC might activate another kinase and it is this kinase that phosphorylates Ser11; this kinase is not present in the purified Na pump and PKC studies.
Motivated by our concern about the His13Gln change in humans and Xenopus, we have rechecked the human sequence and interestingly humans have a new Thr lacking in rat (and pig) in the N-terminus at residue 60 and it is a consensus PKC site. We feel that this residue may play the same role as Ser11 in other species. Amazingly, Xenopus has exactly the same two substitutions as human. Intriguingly, many species have both Thr60 and His18 so they might have 2 PKC sites. (Thr60 was noted by Beguin et al., [45] as a potential PKC site, but as far as we can tell, was not followed up on.)
It would be interesting to know whether reticulocyte maturation and changes in Na pump number are the same in species that lack His13. Note: dog has His13 so it cannot explain the difference seen in Carnivora.
Conclusions
In conclusion, we have shown that the recent crystal structure of the Na pump, combined with previous data on the pH dependence of the red cell Na pump in different species provide new information about potential important residues in the Na pump cycle. In addition, we reviewed the information about PKC regulation of Na pump trafficking in the context of reticulocyte maturation.
Fig. 1.
Contribution of Red Cell Na+ fluxes to daily energy utilization. The Na+ leak is about 0.25 moles per day per person. The Na pump pumps out 3 Na+ ions per ATP hydrolyzed. Human red cells produce about 2 ATP molecules per glucose.
Fig. 7.

Sequence alignment at the N-terminal of the Na pump. Sequence alignment of N-terminal of Na,K-ATPase α1. The amino acid differences are capitalized. The unique rat Ser is bolded and underlined. Histidine (blue H) is conserved in most species, but not human nor Xenopus (purple Q). At 60 a new potential PKC is formed in human compared to rat by Pro to Ser (red) mutation.
Acknowledgments
Work in our laboratories was supported by NIH GM 061583, NSF MCB-0347202, and NIH DK37512. We are both deeply grateful for the opportunity to have known Phil Knauf-he taught us a great deal about what good science and good life mean.
Footnotes
In this paper we use the term transporter for proteins such as exchangers, cotransporters, and pumps, that move only a few molecules per catalytic cycle and where the movement of the molecule requires conformational changes of the protein. This is different from the fundamental properties of ion channels. In channels, there are at least 2 conformations, an open and a closed conformation. After the channel changes from a closed to an open conformation, many hundreds to thousands of ions move through the open pore without requiring any significant protein conformational change.
In the 1980 paper, Skou and Esmann describe labeling the Na pump with eosin maleimide, but a later paper clarifies that they later determined that the signals were from the change of binding of unreacted eosin.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Knauf PA, Rothstein A. Chemical modification of membranes. I. Effects of sulfhydryl and amino reactive reagents on anion and cation permeability of the human red blood cell. J Gen Physiol. 1971;58:190–210. doi: 10.1085/jgp.58.2.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knauf PA, Proverbio F, Hoffman JF. Electrophoretic separation of different phophosproteins associated with Ca-ATPase and Na, K-ATPase in human red cell ghosts. J Gen Physiol. 1974;63:324–36. doi: 10.1085/jgp.63.3.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morth JP, Pedersen BP, Toustrup-Jensen MS, et al. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- 4.Milanick MA, Hoffman JF. Separate effects of internal and external pH on cation fluxes in human red blood cells as and external pH on cation fluxes in human red blood cells as studied by means of a pH clamp. J Gen Physiol. 1982;80:20a. [Google Scholar]
- 5.Dissing S, Hoffman JF. Ouabain-insensitive Na efflux from human red blood cells stimulated by outside H, Na, or Li ions. Journal of General Physiology. 1982;80:15a. [Google Scholar]
- 6.Dissing S, Hoffman JF. Anion-coupled Na efflux mediated by the human red blood cell Na/K pump. J Gen Physiol. 1990;96:167–93. doi: 10.1085/jgp.96.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milanick MA. Proton fluxes associated with the Ca pump in human red blood cells. Am J Physiol. 1990;258:C552–62. doi: 10.1152/ajpcell.1990.258.3.C552. [DOI] [PubMed] [Google Scholar]
- 8.Jacobs MH, Parpart AK. Is The Erythrocyte Permeable To Hydrogen Ions? Biol Bull. 1932;62:63–76. [Google Scholar]
- 9.Jennings ML. Proton fluxes associated with erythrocyte membrane anion exchange. J Membr Biol. 1976;28:187–205. doi: 10.1007/BF01869697. [DOI] [PubMed] [Google Scholar]
- 10.Gunn RB. A titratable carrier model for both mono- and di-valent anion transport in human red blood cells. In: Rorth M, Astrup P, editors. Oxygen Affinity of Hemoglobin and Red Cell Acid-Base Status. Munksgaard; Copenhagen: 1972. pp. 823–827. [Google Scholar]
- 11.Gunn RB. A titratable carrier for monovalent and divalent inorganic anions in red blood cells. In: Gerlach E, Moser K, Deutsch E, Wilmanns W, editors. Erythrocytes, Thrombocytes, Leucocytes, Recent Advances in Membrane and Metabolic Research. Georg Thieme Publishers; Stuttgart: 1973. pp. 77–79. [Google Scholar]
- 12.Fraile G, Romero PJ. Preferential inhibition of the human red cell Ca-ATPase by some disulfonic stilbene derivatives. Acta Cient Venez. 1987;38:448–52. [PubMed] [Google Scholar]
- 13.Pedemonte CH, Kaplan JH. Inhibition and derivatization of the renal Na,K-ATPase by dihydro-4,4′-diisothiocyanatostilbene-2,2′-disulfonate. Biochemistry. 1988;27:7966–73. doi: 10.1021/bi00420a056. [DOI] [PubMed] [Google Scholar]
- 14.Polvani C, Blostein R. Protons as substitutes for sodium and potassium in the sodium pump reaction. J Biol Chem. 1988;263:16757–63. [PubMed] [Google Scholar]
- 15.Vega FV, Cabero JL, Mårdh S. Inhibition of H,K-ATPase and Na,K-ATPase by DIDS, a disulphonic stilbene derivative. Acta Physiol Scand. 1988;134:543–7. doi: 10.1111/j.1748-1716.1998.tb08529.x. [DOI] [PubMed] [Google Scholar]
- 16.Gatto C, Lutsenko S, Kaplan JH. Chemical modification with dihydro-4,4′-diisothiocyanostilbene-2,2′-disulfonate reveals the distance between K480 and K501 in the ATP-binding domain of the Na,K-ATPase. Arch Biochem Biophys. 1997;340:90–100. doi: 10.1006/abbi.1997.9879. [DOI] [PubMed] [Google Scholar]
- 17.Knauf PA, Fuhrmann GF, Rothstein S, Rothstein A. The relationship between anion exchange and net anion flow across the human red blood cell membrane. J Gen Physiol. 1977;69:363–86. doi: 10.1085/jgp.69.3.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Callahan TJ, Goldstein DA. Anion inhibitor-sensitive unidirectional sodium movements in the human erythrocyte. J Gen Physiol. 1978;72:87–100. doi: 10.1085/jgp.72.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forbush B. Rapid release of 42K and 86Rb from an occluded state of the Na,K-pump in the presence of ATP or ADP. J Biol Chem. 1987;262:11104–15. [PubMed] [Google Scholar]
- 20.Forbush B, Klodos I. Rate-limiting steps in Na translocation by the Na/K pump. Soc Gen Physiol Ser. 1991;46:210–25. [PubMed] [Google Scholar]
- 21.Skou JC. Effects of ATP on the intermediary steps of the reaction of the (Na+ + K+)-ATPase. IV. Effect of ATP on K0.5 for Na+ and on hydrolysis at different pH and temperature. Biochim Biophys Acta. 1979;567:421–35. doi: 10.1016/0005-2744(79)90128-1. [DOI] [PubMed] [Google Scholar]
- 22.Skou JC, Esmann M. Effects of ATP and protons on the Na: K selectivity of the (Na+ + K+)-ATPase studied by ligand effects on intrinsic and extrinsic fluorescence. Biochim Biophys Acta. 1980;601:386–402. doi: 10.1016/0005-2736(80)90543-x. [DOI] [PubMed] [Google Scholar]
- 23.Milanick MA, Gunn RB. Proton inhibition of chloride exchange: asynchrony of band 3 proton and anion transport sites? Am J Physiol. 1986;250:C955–69. doi: 10.1152/ajpcell.1986.250.6.C955. [DOI] [PubMed] [Google Scholar]
- 24.Milanick MA, Arnett KL. Extracellular protons regulate the extracellular cation selectivity of the sodium pump. J Gen Physiol. 2002;120:497–508. doi: 10.1085/jgp.20028573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gatto C, Helms JB, Prasse MC, Huang SY, Zou X, Arnett KL, Milanick MA. Testing Homology Models of P-type ATPases via Kinetics: similarities and differences between cytoplasmic cation access in Na,K-ATPase and PMCA. Biochemistry. 2006;45:13331–13345. doi: 10.1021/bi060667j. [DOI] [PubMed] [Google Scholar]
- 26.Helms JB, Arnett KL, Gatto C, Milanick MA. Bretylium, an organic quaternary amine, inhibits the Na,K-ATPase by binding to the extracellular K-site. Blood cells, Mol, & Dis. 2004;32:394–400. doi: 10.1016/j.bcmd.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 27.Gatto C, Helms JB, Prasse MC, Arnett KL, Milanick MA. Tetrapropylammonium, an extracellular cation site probe in Na,K-ATPase, reveals how ATP and Pi alter access to the transport site. Am J Physiol – Cell Physiol. 2005;289:C302–C311. doi: 10.1152/ajpcell.00043.2005. [DOI] [PubMed] [Google Scholar]
- 28.Restrepo D, Cronise BL, Snyder RB, Knauf PA. A novel method to differentiate between ping-pong and simultaneous exchange kinetics and its application to the anion exchanger of the HL60 cell. J Gen Physiol. 1992;100:825–46. doi: 10.1085/jgp.100.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toyoshima C. Structural aspects of ion pumping by Ca2+-ATPase of sarcoplasmic reticulum. Arch Biochem Biophys. 2008;476:3–11. doi: 10.1016/j.abb.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 30.Obara K, Miyashita N, Xu CI, et al. Structural role of countertransport revealed in Ca2+ pump crystal structure in the absence of Ca2+ Proc Nat Acad Sci. 2005;102:14489–14496. doi: 10.1073/pnas.0506222102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Einholm AP, Toustrup-Jensen M, Andersen JP, Vilsen B. Mutation of Gly-94 in transmembrane segment M1 of Na,K-ATPase interferes with Na and K binding in E2P conformation. Proc Nat Acad Sci. 2005;102:11254–11259. doi: 10.1073/pnas.0501201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Einholm AP, Andersen JP, Vilsen B. Importance of Leu99 in Transmembrane Segmen M1 of the Na,K-ATPase in the binding and occlusion of K. J Biol Chem. 2007;282:23854–23866. doi: 10.1074/jbc.M702259200. [DOI] [PubMed] [Google Scholar]
- 33.Capendeguy O, Chodanowski P, Michielin O, Horisberger JD. Access of Extracellular Cations to their Binding Sites in Na,K-ATPase: Role of the Second Extracellular Loop of the a Subunit. J Gen Physiol. 2006;127:341–352. doi: 10.1085/jgp.200509418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Capendeguy O, Iwaszkiewicz J, Michielin O, Horisberger JD. The fourth extracellular loop of the α-subunit of Na,K-ATPase: Functional evidence for close proximity with the second extracellular loop. J Biol Chem. 2008;283:27850–27858. doi: 10.1074/jbc.M802194200. [DOI] [PubMed] [Google Scholar]
- 35.Eguchi H, Morii M, Takahashi Y, et al. Functional consequences of various leucine mutations in the M3/M4 loop of the Na,K-ATPase alpha-Subunit. J Membr Biol. 2008;221:133–40. doi: 10.1007/s00232-007-9091-3. [DOI] [PubMed] [Google Scholar]
- 36.Maede Y, Inaba M. (Na,K)-ATPase and Ouabain binding in reticulocytes from dogs with high K and low K erythrocytes and their changes during maturation. J Biol Chem. 1985;260:3337–43. [PubMed] [Google Scholar]
- 37.Johnstone RM. Revisiting the road to the discovery of exosomes. Blood Cells Mol Dis. 2005;34:214–9. doi: 10.1016/j.bcmd.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 38.Weigensberg AM, Blostein R. Energy depletion retards the loss of membrane transport during reticulocyte maturation. Proc Natl Acad Sci U S A. 1983;80:4978–82. doi: 10.1073/pnas.80.16.4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blostein R, Grafova E. Characteristics of membrane transport losses during reticulocyte maturation. Biochem Cell Biol. 1987;65:869–75. doi: 10.1139/o87-113. [DOI] [PubMed] [Google Scholar]
- 40.Blostein R, Grafova E. Factors affecting transport changes associated with reticulocyte maturation. Biomed Biochim Acta. 1987;46:S172–6. [PubMed] [Google Scholar]
- 41.Blostein R, Grafova E. Decrease in Na(+)-K(+)-ATPase associated with maturation of sheep reticulocytes. Am J Physiol. 1990;259:C241–50. doi: 10.1152/ajpcell.1990.259.2.C241. [DOI] [PubMed] [Google Scholar]
- 42.Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–79. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 43.Feschenko MS, Sweadner KJ. Conformation-dependent phosphorylation of Na,K-ATPase by protein kinase A and protein kinase C. J Biol Chem. 1994;269:30436–44. [PubMed] [Google Scholar]
- 44.Efendiev R, Pedemonte CH. Contrary to rat-type, human-type Na,K-ATPase is phosphorylated at the same amino acid by hormones that produce opposite effects on enzyme activity. J Am Soc Nephrol. 2006;17:31–8. doi: 10.1681/ASN.2005070681. [DOI] [PubMed] [Google Scholar]
- 45.Béguin P, Peitsch MC, Geering K. alpha 1 but not alpha 2 or alpha 3 isoforms of Na,K-ATPase are efficiently phosphorylated in a novel protein kinase C motif. Biochemistry. 1996;35:14098–108. doi: 10.1021/bi960516o. [DOI] [PubMed] [Google Scholar]
- 46.Mahmmoud YA, Cornelius F. Protein kinase C phosphorylation of purified Na,K-ATPase: C-terminal phosphorylation sites at the alpha- and gamma-subunits close to the inner face of the plasma membrane. Biophys J. 2002;82:1907–19. doi: 10.1016/S0006-3495(02)75540-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Féraille E, Béguin P, Carranza ML, et al. Is phosphorylation of the alpha1 subunit at Ser-16 involved in the control of Na,K-ATPase activity by phorbol ester-activated protein kinase C? Mol Biol Cell. 2000;11:39–50. doi: 10.1091/mbc.11.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khundmiri SJ, Bertorello AM, Delamere NA, Lederer ED. Clathrin-mediated endocytosis of Na+,K+-ATPase in response to parathyroid hormone requires ERK-dependent phosphorylation of Ser-11 within the alpha1-subunit. J Biol Chem. 2004;279:17418–27. doi: 10.1074/jbc.M311715200. [DOI] [PubMed] [Google Scholar]