CONSPECTUS
Natural products, produced chiefly by microorganisms and plants, can be large and structurally complex molecules. These molecules are manufactured by cellular assembly lines, in which enzymes construct the molecules in a stepwise fashion. The means by which enzymes interact and work together in a modular fashion to create diverse structural features has been an active area of research; the work has provided insight into the fine details of biosynthesis.
A number of polycyclic aromatic natural products—including several noteworthy anticancer, antibacterial, antifungal, antiviral, antiparasitic, and other medicinally significant substances—are synthesized by polyketide synthases (PKSs) in soil-borne bacteria called actinomycetes. Concerted biosynthetic, enzymological, and structural biological investigations into these modular enzyme systems have yielded interesting mechanistic insights. A core module called the minimal PKS is responsible for synthesizing a highly reactive, protein-bound poly-β-ketothioester chain. In the absence of other enzymes, the minimal PKS also catalyzes chain initiation and release, yielding an assortment of polycyclic aromatic compounds. In the presence of an initiation PKS module, polyketide backbones bearing additional alkyl, alkenyl, or aryl primer units are synthesized, whereas a range of auxiliary PKS enzymes and tailoring enzymes convert the product of the minimal PKS into the final natural product. In this Account, we summarize the knowledge that has been gained regarding this family of PKSs through recent investigations into the biosynthetic pathways of two natural products, actinorhodin and R1128 (A–D).
We also discuss the practical relevance of these fundamental insights for the engineered biosynthesis of new polycyclic aromatic compounds. With a deeper understanding of the biosynthetic process in hand, we can assert control at various stages of molecular construction and thus introduce unnatural functional groups in the process. The metabolic engineer affords a number of new avenues for creating novel molecular structures that will likely have properties akin to their fully natural cousins.
1. INTRODUCTION
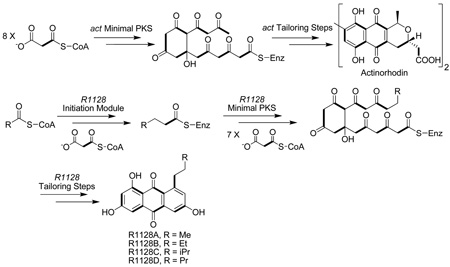
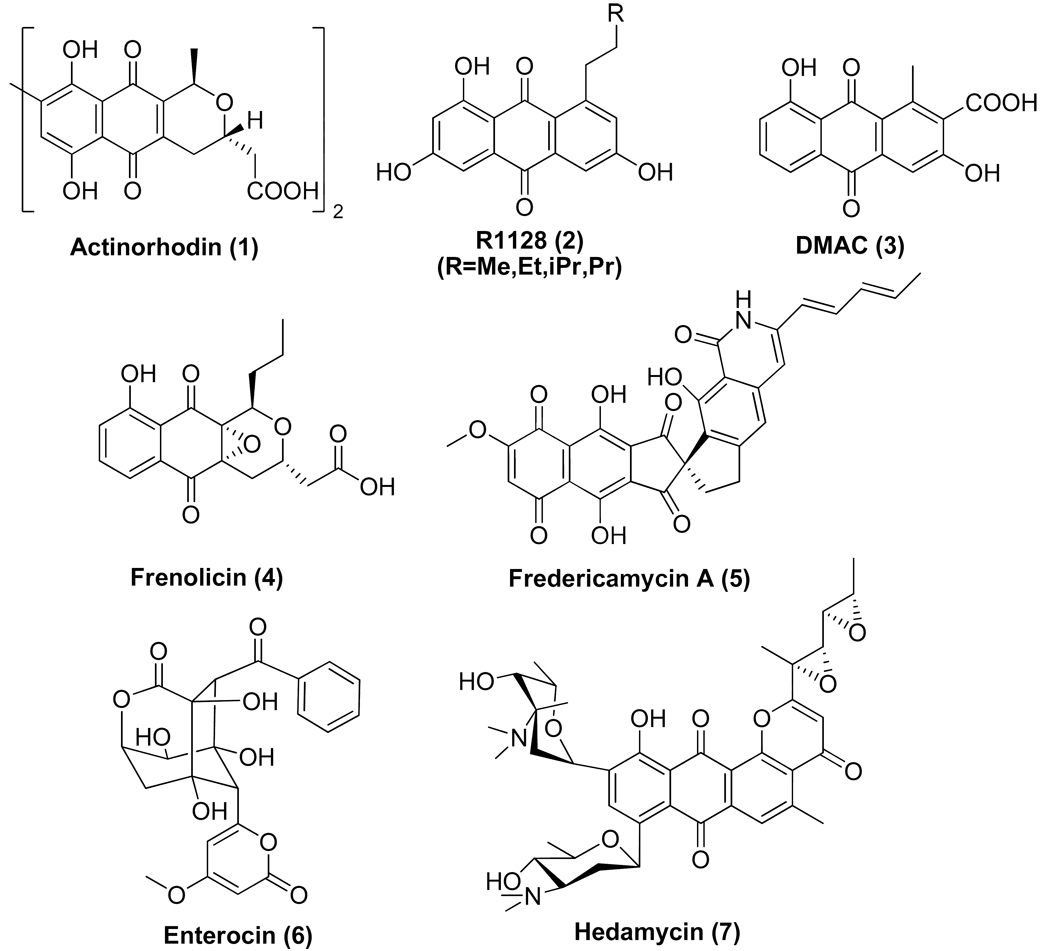
A number of polycyclic aromatic natural products are synthesized by polyketide synthases (PKSs) in soil-borne bacteria called actinomycetes. They exhibit anticancer (e.g. doxorubicin), antibacterial (e.g. oxytetracycline), antifungal (e.g. pradimicin), antiviral (e.g. A-74528), antiparasitic (e.g. frenolicin) and other related activities.1 These PKSs, also called type II PKSs because of their relationship to type II fatty acid synthases from bacteria and plants, are comprised of a set of highly conserved 5–50 kDa protein subunits. The chemistry, biology and biosynthesis of this family of polyketides have been the subjects of several comprehensive reviews in the past decade.2–6 This article provides a narrative of what has been learned within the past eight years regarding the biosynthetic logic of type II PKSs through structural, mechanistic and biosynthetic investigations into two prototypical synthases – the actinorhodin PKS and the R1128 PKS. The structures of actinorhodin (1) and R1128 (2), as well as those of related aromatic polyketides discussed in this article, are shown in Figure 1.
Figure 1.
Structures of selected actinomyces-derived aromatic polyketides [DMAC: 3,8-dihydroxy-1-methylanthraquinone-2-carboxylic acid].
2. PROTEIN SUBUNITS OF TYPE II PKSs
Methods for genetic manipulation of Gram–positive actinomycetes, pioneered by D. A. Hopwood and coworkers,7,8 provided the technical foundation for the cloning and sequencing of a number of aromatic polyketide biosynthetic gene clusters in the 1985–1995 timeframe. For example, the complete act gene cluster responsible for actinorhodin biosynthesis was cloned in 1984,9 and its PKS genes were fully sequenced by 1992.10 A ketosynthase (KS), chain length factor (CLF) and acyl carrier protein (ACP) are encoded within the act gene cluster; these subunits were designated the “minimal PKS’. At the time of cloning and initial characterization of these genes, the origin of the malonyl-CoA:ACP transacylase (MAT) activity of the PKS was unclear; later, it was shown that this enzyme is borrowed from the endogenous fatty acid synthase (vide infra).11,12 Additionally, the act gene cluster also encoded a ketoreductase (KR) responsible for reducing the C-9 ketone, a bifunctional aromatase/cyclase (ARO/CYC) that catalyzes dehydrative formation of the first aromatic ring, and a cyclase (CYC) that yields the second aromatic ring. Together, these subunits were necessary and sufficient to support the biosynthesis of the anthraquinone DMAC (3).13,14 By the mid-1990’s, at least ten more aromatic PKS gene clusters had been cloned from other actinomycetes, all of which encoded a KS, a CLF and an ACP gene together with homologs of some or all of the auxiliary genes. These findings provided the genetic foundation for subsequent mechanistic investigations into aromatic polyketide biosynthesis.
In contrast to the above PKSs, the biosynthetic gene cluster for the closely related R1128 anthraquinones (2) revealed not one but two KS and ACP genes in addition to the expected genes for downstream tailoring enzymes.15 This led to the speculation that R1128 biosynthesis required the action of two successive PKS modules, the first of which was dedicated to the construction of the alkyl appendage on the anthraquinone, and the second was the minimal PKS. An entirely analogous set of genes was also identified in the frenolicin (4) biosynthetic gene cluster,16,17 presumably to enable formation of the propyl moiety of this natural product. However, sequence analysis of the R1128 and frenolicin biosynthetic gene clusters raised several confounding questions. For example, a MAT-like protein was also encoded within both gene clusters, although its function was not clear. Also unclear was the source of ketoreductase (KR), dehydratase (DH) and enoylreductase (ER) activity required for the biosynthesis of the fully reduced alkyl moieties of these natural products. Last but not least, the need for a second ACP was unclear, given the prevailing consensus at the time that ACPs were functionally interchangeable. These and other mysteries were solved through the investigations summarized below.
3. ENZYMOLOGY OF TYPE II PKSs
The Minimal PKS
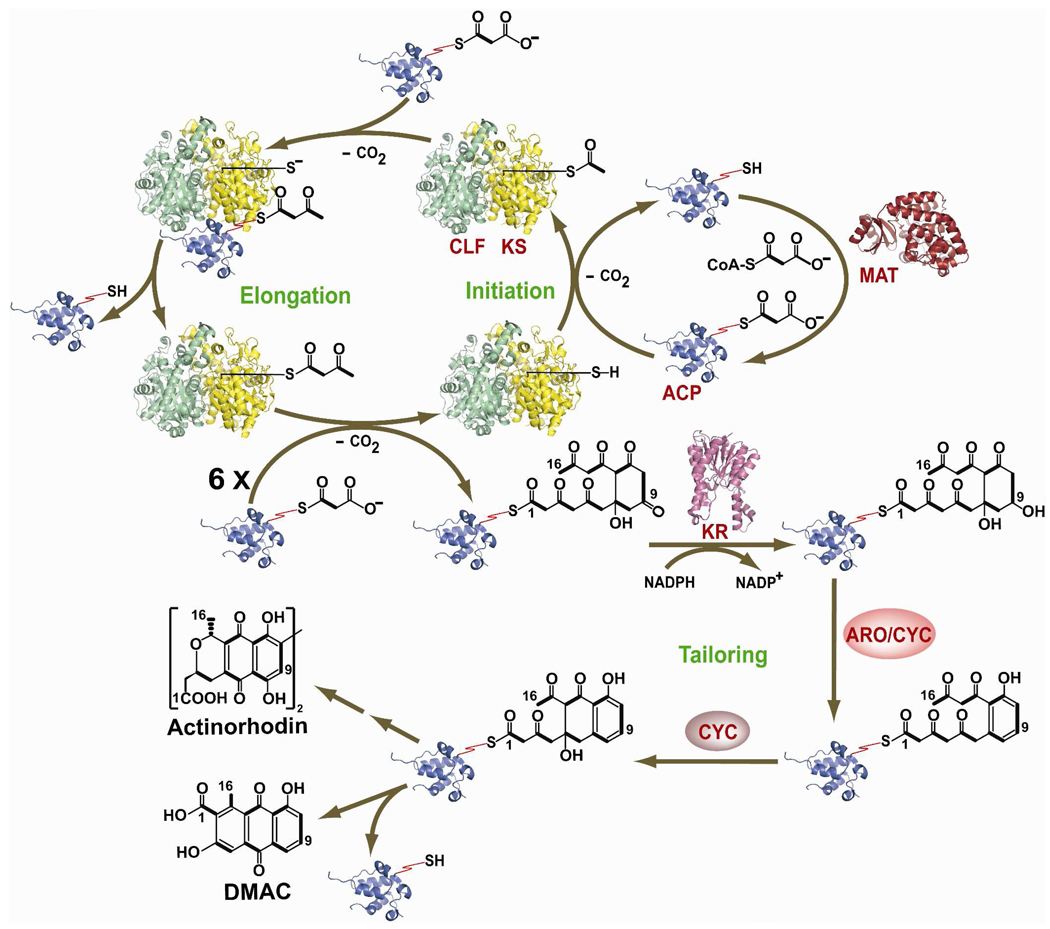
Cell-free synthesis of DMAC by a protein preparation from a recombinant strain of Streptomyces coelicolor paved the way for reconstituting and biochemically characterizing the act PKS (Figure 2).13 A key early observation was that the MAT from the S. coelicolor fatty acid synthase was necessary for PKS activity. This led to its inclusion as a component of the minimal PKS.11 As summarized in Figure 2, it is now well established that the C16 product of the act minimal PKS is primed by decarboxylation of malonyl-S-ACP, followed by an inter-thiol transfer of the resulting acetyl group onto the active site of the KS.18 This is followed by seven rounds of chain elongation, each of which involves decarboxylative C-C bond formation between a nucleophilic malonyl-S-ACP and the growing polyketide chain bound to the KS via an electrophilic thioester linkage. The full-length poly-β-ketoacyl chain is either released from the ACP via hydrolysis or cyclization, or is sequentially acted upon by the auxiliary PKS subunits like KR, ARO/CYC and CYC to yield DMAC (3). (Presumably quinone oxidation occurs spontaneously in the presence of molecular oxygen, although the potential contribution of a non-specific oxygenase has not been ruled out). Following its release from the ACP, tailoring enzymes such as methyl transferases, glycosyl transferases and oxygenases transform the polyketide product into a fully decorated natural product. In view of the extensive biochemical and structural characterization of the act PKS, it could be considered a prototype for type II PKSs.
Figure 2.
Schematic representation of DMAC (3,8-dihydroxy-1-methylanthraquinone-2-carboxylic acid) biosynthesis by the act PKS. The cysteine thiol of the KS and the pantetheinyl thiol of the ACP are explicitly shown. Chain growth is initiated via decarboxylation of malonyl-ACP, followed by transfer of the resultant acetyl group onto the KS. Following seven rounds of chain elongation, the ACP-bound octaketide is released from the KS-CLF, whereafter it is modified by various auxiliary PKS subunits and tailoring enzymes into DMAC or actinorhodin. The atomic structures of all proteins shown, except for the act ARO/CYC and CYC, have been solved. The relevant PDB coordinate filenames are: 1TQY (act KS-CLF), 1OR5 (ACP), 1NM2 (MAT), 1X7H (act KR). [KS: Ketosynthase; CLF: Chain Length Factor; ACP: Acyl Carrier Protein; MAT: Malonyl-CoA:ACP Transacylase; KR: Ketoreductase; ARO: Aromatase; CYC: Cyclase, NADP: Nicotinamide Adenine Dinucleotide Phosphate]
Some aspects of the catalytic cycle shown in Figure 2 are noteworthy. First, the KS and CLF form a tight heterodimer with an elaborate pocket at their interface into which the growing polyketide chain is extruded.19 The “depth” of this pocket, dictated by specific residues in the CLF, determines the chain length of the polyketide product.20 Phylogenetic analysis suggests that chain length specificity has diversified repeatedly during evolution.21 Second, in contrast to the tight association of the KS and CLF, KS-ACP interactions are weak but specific.11,22 The weakness of these interactions leads to multiple KS-ACP exchanges during the formation of a single DMAC molecule.18 After the formation of each successive C-C bond, the growing chain is transferred back from the ACP to the KS before the proteins dissociate. When the full-length chain is formed, the ACP dissociates with the chain attached to its pantetheinyl arm. Cyclization of the first ring of the nascent polyketide chain occurs in the active site of KS-CLF. This regiospecific cyclization (C7–C12) may be spontaneous or catalyzed by the heterodimeric complex.
Naturally occurring KS-CLF heterodimers exhibit a fairly broad range of chain length preferences. For example, the act KS-CLF synthesizes an octaketide, the tcm KS-CLF synthesizes a decaketide, whereas the whiE KS-CLF synthesizes a dodecaketide. The pentadecaketide producing fdm KS-CLF from the fredericamycin (5) pathway produces the longest carbon skeleton reported so far.23 Some enzymes synthesize a mixture of products; for example, the fren KS-CLF from the frenolicin pathway makes both octaketides and nonaketides.24 Since ACPs serve as substrate carriers between the KS-CLF and upstream or downstream PKS subunits, one might hypothesize that it should be possible to genetically recombine KS-CLF heterodimers with heterologous initiation PKS modules or downstream PKS subunits, so long as the ACP carriers are compatible. Indeed, a number of empirical studies have borne out this prediction14,24–31 In the process, a range of novel polycyclic aromatic products have been generated, many of which lack close analogs among known natural products. Protein engineering may also be a promising avenue to engineer KS-CLF heterodimers with novel chain length specificity.20
The recognition between an ACP and a KS is essential for polyketide biosynthesis. Although the KSCLF and ACP subunits from many (but not all) type II PKSs are qualitatively interchangeable, the variable affinity can be kinetically distinguished and likely contributes to polyketide titers in at least some cases.32–35 An extreme example is actinorhodin and spore pigment biosynthesis in S. coelicolor. Even though both products are synthesized by closely related type II PKSs in the same host, their KSCLF and ACP subunits have orthogonal specificity.36 The solution structures of the act ACP, fren ACP and otc (oxytetracycline) ACP have been determined by NMR spectroscopy.37–39 Together with the X-ray structure of the act KS-CLF, this has provided insight into their relative docking orientation.19 However, re-engineering of this protein interface has not yet been successful, a testament to the difficulty associated with the general problem of engineering weak but specific protein-protein interactions. NMR spectroscopy has also shown that the ACP backbone adopts multiple conformations in dynamic equilibrium with each other.38 Presumably this is necessary in order to allow the ACP to dock with multiple enzyme partners during polyketide chain initiation, elongation, modification and release. Yet again, a solid understanding of the mechanistic principles for this extraordinary tolerance and specificity remains a major challenge towards a deeper understanding of assembly line biosynthesis of antibiotics.
Downstream Enzymes
Many different types of enzymes participate in converting the nascent polyketide chain synthesized by the minimal PKS into a polycyclic aromatic natural product. Broadly speaking, they fall into two categories – those that recognize ACP-bound substrates (auxiliary PKS enzymes) and those that act upon freely diffusible substrates (tailoring enzymes). Whereas some of these downstream enzymes have broad substrate scope, others are highly specific. For example, the act KR is able to regiospecifically reduce the C-9 ketone of poly-β-ketoacyl-ACP substrates ranging from at least 16–24 carbons in length, and possibly longer.40,41 Structural studies suggest this observed blend of tolerance and specificity is due to the combined influence of an elaborate protein-protein interface between the ACP and KR as well as a distinct preference for the orientation in which the polyketide chain can dock next to the NADPH binding site of the KR.42–44 In contrast, the act ARO/CYC shows high specificity, and is neither able to accommodate longer chain substrates nor can it recognize fully unreduced poly-β-ketothioesters.24,27 Whereas a clearer understanding of the mechanism for this observation must await a high-resolution structure of this enzyme, the recent X-ray structure of a homolog from the tetracenomycin biosynthetic pathway provides some general guidelines for how this might occur.45 Similarly, the act CYC is also chain-length specific.27 Continued progress in rational engineering of new polycyclic aromatic compounds either requires harnessing a menu of naturally occurring auxiliary PKS and tailoring enzymes with a broad range of substrate scope, or the emergence of protein engineering principles by which the specificity of known enzymes such as the act ARO/CYC or act CYC can be predictably altered
The Initiation Module
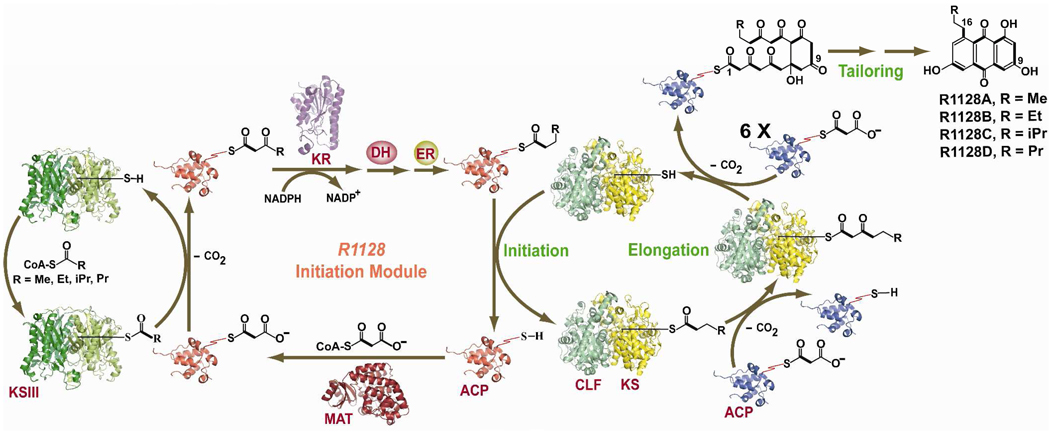
As introduced above, biosynthesis of the polyketide backbones of antibiotics such as R1128 and frenolicin is initiated by a PKS module that is distinct from the minimal PKS. These initiation modules are comprised of a dedicated KS (often referred to as a KSIII by analogy with bacterial fatty acid synthases) and ACP, which collaborate with a MAT, KR, DH and ER (all of which are borrowed from the host’s fatty acid synthase)46 to synthesize a diketide intermediate. Prior to our biosynthetic studies on R1128, studies on doxorubicin biosynthesis had revealed the requirement for an “extra” KS, which enables preferential incorporation of a propionyl-CoA derived primer unit instead of the more typical decarboxylative priming mechanism discussed above.47,48 However that chain initiation mechanism did not require the action of other enzymes, and a single ACP sufficed for doxorubicin biosynthesis. Our interest in R1128 was therefore motivated by a desire to understand how nature balances tolerance and specificity as she incorporates more elaborate primer units into the backbones of this class of polyketides.
The catalytic cycle of the initiation PKS module of the R1128 synthase is outlined in Figure 3. The KS is homodimeric, and does not require a partner CLF, presumably because chain length control is not relevant in this phase of R1128 biosynthesis. It has a marked preference for propionyl-CoA but can accommodate unbranched or branched acyl-CoA substrates up to four carbon atoms long,49 as verified by X-ray crystallography.50 It also has a strong specificity for its partner ACP and an absolute discrimination for the ACP from the minimal PKS module of the R1128 synthase.32 Remarkably, a single amino acid residue on the two ACPs account for most of this orthogonality. Presumably it is essential for the contemporaneous operation of distinct catalytic cycles by two dissociated PKS modules in the same cell. However, it raises the question of how the growing chain passes from one PKS module to the next. One theory is that the ACP from the initiation module can transfer an acyl chain to the KS of the minimal PKS, but the two proteins are unable to participate in a C-C bond forming reaction. If so, then this system may offer an attractive opportunity to dissect the general catalytic principles that differentiate these two types of reactions involving the same protein partners. Also unlike doxorubicin, the R1128 KS-CLF cannot be primed by malonyl-ACP decarboxylation when the initiation module is absent. Thus, an elaborate set of checkpoint mechanisms appear to be in operation as the carbon chain backbone of R1128 is fashioned.
Figure 3.
Schematic representation of the initiation PKS module of the R1128 synthase. The cysteine and pantetheinyl thiols of the two KSs and two ACPs, respectively are explicitly shown. Chain growth is initiated by the initiation module, followed by transfer to the elongation module. Those enzymes whose structures are known are shown as ribbon diagrams; their PDB coordinate file-names are: 1MZJ (ZhuH, KSIII), 2NM0 (β-Ketoacyl-Acyl Carrier Protein Reductase, KR); 1TQY (act KS-CLF), 1OR5 (ACP), 1NM2 (MAT). The ACP cartoons in red and blue refer to the ACP from initiation module and elongation module respectively. [KS: Ketosynthase; CLF: Chain Length Factor; ACP: Acyl Carrier Protein; MAT: Malonyl-CoA:ACP Transacylase; KR: Ketoreductase; DH: Dehydratase; ER: Enoyl Reductase; NADP: Nicotinamide Adenine Dinucleotide Phosphate]
Although it was initially proposed that the MAT homolog encoded with the R1128 gene cluster may participate in primer unit selection in the initiation module, it was subsequently shown that this enzyme is an ACP-specific thioesterase whose role is to purge the R1128 PKS of any acetyl groups that may accidently be transferred onto the pantetheinyl arm of the ACP,51 presumably because of the very high intracellular concentration of acetyl-CoA. Such thioesterases are infrequent participants in type II PKSs (although they also exist in the frenolicin and doxorubicin gene clusters), but are relatively commonplace in the gene clusters of multimodular PKSs and nonribosomal peptide synthetases, which draw from a much broader spectrum of building blocks to load onto carrier protein domains of their enzymatic assembly lines.52–54
It should be noted that the principles of chain initiation embodied in the R1128 synthase are representative but not identically conserved among type II PKSs that synthesize non-acetate primed antibiotics. For example, enterocin (6) biosynthesis is primed by a benzoyl primer unit, which is directly abstracted from a benzoyl-CoA precursor.55,56 In contrast, a much more elaborate chain initiation mechanism appears to have evolved for the biosynthesis of the hexadienyl unit that is elaborated into the anticancer product hedamycin (7). The hedamycin gene cluster57 encodes not just a KSIII-like protein but also two big, multidomain proteins harboring ketosynthases, acyl transferases, acyl carrier proteins, ketoreductases and dehydratases. Decoding hedamycin chain initiation mechanisms will undoubtedly add a new dimension to our appreciation of the versatility of type II PKSs.
4. ENGINEERED BIOSYNTHESIS OF NOVEL POLYKETIDES USING TYPE II PKSs
From a technical standpoint, a key question encountered at the onset of any natural product biosynthetic engineering program is the choice of genetic tools, specifically the microbial host and plasmid(s) to be used. In the case of type II PKS products, a reliable and convenient host-vector system comprised of Streptomyces coelicolor CH999 and plasmid pRM5 have been widely adopted as the system of choice.14 Over the past two decades, a large number of polyketide pathways have been successfully implemented in this heterologous system.58
Conceptually, the central challenge for exploiting type II PKSs via biosynthetic engineering lies in understanding the scope of and limitations to the modularity of these multifunctional catalysts. Here it is worth elaborating upon the term “modularity”. In the context of multi-step catalysis, it could either refer to catalyst or substrate tolerance. In the case of type II PKSs, both have been shown to have considerable potential, albeit not without limitations.
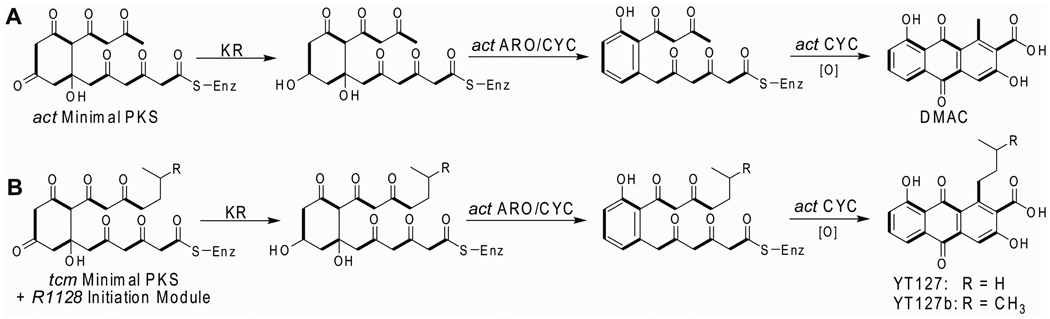
The initiation module is responsible for fabricating the functional group that primes aromatic polyketide biosynthesis, whereas the KS-CLF heterodimer controls the eventual chain length of the polyketide backbone. Simple steric considerations appear to dictate how primer and chain length selection is balanced, a phenomenon that can readily be rationalized by the nature of the substrate binding pocket in the KS-CLF.19 Bulkier and/or longer primer units yield products that have undergone correspondingly fewer elongation cycles by the minimal PKS, such that the overall chain length remains constant.29–31,59 Thus, if one wishes to selectively replace an acetyl primer-derived methyl substituent in an octaketide natural product with a C4 or C5 substituent, one must not only introduce an initiation module that synthesizes the desired acyl-ACP intermediate, but also replace the KS-CLF heterodimer of the octaketide PKS with a homolog with longer chain backbone (Figure 4: compare DMAC biosynthesis with YT127 biosynthesis). Of course, for all of this to occur as predicted, two additional requirements must be satisfied. First, the ACP must be able to transfer the appropriate intermediate between the two PKS modules. Based on a limited number of such experiments in the literature, there appears to be good compatibility between initiation and elongation modules of type II PKSs that synthesize antibiotics but not spore pigments.29,31 Second, downstream auxiliary PKS and tailoring enzymes must have tolerance toward the altered polyketide chain. In most instances studied so far, this appears to be the case. Therefore, this may be a general method for the regiospecific modification of any acetogenic polyketide natural product of a type II PKS. More ambitious exploitation of the interface between chain initiation and elongation in the context of aromatic polyketides will have to await mechanistic analyses of unusual natural systems such as those found in the hedamycin and fredericamycin biosynthetic pathways.
Figure 4.
Aromatic Polyketides produced by (A) act Minimal PKS and (B) R1128 Initiation Module and tcm Minimal PKS. The act minimal PKS synthesizes a C16 carbon chain backbone that is primed by acetyl group. The five and six-carbon primer unit in YT127 and YT127b respectively is derived from the initiation module of the R1128 PKS. In conjunction with this initiation module, the tcm minimal PKS therefore synthesizes an octaketide backbone rather than a decaketide backbone, so that the chain length of the resulting polyketide product remains constant.
Thus far, the ability to probe the tolerance and specificity of downstream enzymes that act upon nascent polyketide chains has been severely constrained by the scope of accessible substrates. Although the tailoring enzymes in a polyketide pathway (e.g. the C-3 ketoreductase involved in actinorhodin biosynthesis)60 recognize freely diffusible substrates, available crystal structures and biosynthetic data suggests that upstream enzymes such as the C-9 specific act KR or the act ARO/CYC catalyze reactions on ACP-bound substrates, and should therefore be considered as auxiliary components of the PKS. Analogs of these ACP-bound poly-β-ketone substrates are highly unstable in water and therefore difficult to study. Nonetheless, limited data suggests that, some of these enzymes (e.g. act KR and the urdGT2 glycosyltransferase from the urdamycin pathway)61 have broad specificity whereas others (e.g. act ARO/CYC) have narrower specificity. If the goal is to prepare a diverse library of compounds for screening applications, there is immense potential for identifying more members of the former category and exploiting their tolerance. However, if the goal is to prepare specific analogs of existing natural products of medicinal relevance, then the most specific tailoring enzyme(s) are the bottleneck(s), and one has little choice but to turn towards protein engineering as a way to expand their substrate scope in desired directions.
As mentioned above, modularity can also be manifested in the ability of the biosynthetic pathway to incorporate unnatural substrates. The natural substrates of most type II PKSs are malonyl-CoA and a handful of other simple acyl-CoAs such as acetyl-, propionyl-, and isobutyryl-CoA. Thus, exploiting this facet of type II PKS modularity requires the ability to engineer cellular metabolism to generate unnatural CoA thioesters (or close analogs). Thus far, relatively few studies have been conducted in this regard in the context of R112830 and enterocin62 biosynthesis. In the former case, unnatural acyl-CoA analogs have been generated in vivo via amino acid catabolic pathways, whereas in the latter case the broad tolerance of a CoA ligase has been exploited to convert benzoic acid analogs into the corresponding acyl-CoAs in vivo. This remains an exciting opportunity in natural product biosynthesis for the metabolic engineer.
In summary, type II PKSs are a fascinating family of multifunctional catalysts that synthesize a wide range of interesting and medicinally important natural products. In-depth studies on a few representative systems over the past decade have provided a strong foundation for considerably more powerful mechanistic investigations and engineering opportunities in the future. In the coming decade, we anticipate that bacterial aromatic polyketides will present an exciting opportunity for productive interplay between biological, biosynthetic and synthetic chemistry.
ACKNOWLEDGEMENTS
This work has been supported by a grant from the NIH (CA 77248). We thank our numerous colleagues who have contributed to our studies on the actinorhodin and R1128 synthases over the past 15 years.
Biographies
Abhirup Das received his MS degree from Indian Institute of Technology Kanpur, India in the year 2004. He is currently a PhD student in Chemistry. His research interest includes semi-synthesis and biosynthesis of medicinally useful aromatic polyketides.
Chaitan Khosla is a professor of Chemical Engineering, Chemistry and (by courtesy) Biochemistry at Stanford University. He received his Ph.D. in 1990 at the California Institute of Technology, and joined Stanford University in 1992 after completing his postdoctoral studies at the John Innes Centre, U.K. Since the beginning of his independent career, he and his students have investigated polyketide synthases in an effort to understand and exploit their modular architectures and mechanisms for practical biocatalysis. He is especially interested in the anti-infective properties of polyketide natural products.
REFERENCES
- 1.O'Hagan D. The Polyketide Metabolites. Ellis Horwood: Chichester; 1991. [Google Scholar]
- 2.Rawlings BJ. Biosynthesis of polyketides (other than actinomycete macrolides) Nat. Prod. Rep. 1999;16:425–484. doi: 10.1039/a900566h. [DOI] [PubMed] [Google Scholar]
- 3.Richardson M, Khosla C. Structure, Function, and Engineering of Bacterial Aromatic Polyketide Synthases. Comprehensive Natural Products Chemistry. 1999;1:473. [Google Scholar]
- 4.Shen B. Biosynthesis of Aromatic Polyketides. Topics in Current Chemistry. 2000;209:1. [Google Scholar]
- 5.Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat. Prod. Rep. 2007;24:162–190. doi: 10.1039/b507395m. [DOI] [PubMed] [Google Scholar]
- 6.Moore BS, Hertweck C. Biosynthesis and attachment of novel bacterial polyketide synthase starter units. Nat. Prod. Rep. 2002;19:70–99. doi: 10.1039/b003939j. [DOI] [PubMed] [Google Scholar]
- 7.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich: The John Innes Foundation; 2000. [Google Scholar]
- 8.Hopwood DA. Genetic Contributions to Understanding Polyketide Synthases. Chem. Rev. 1997;97:2465–2498. doi: 10.1021/cr960034i. [DOI] [PubMed] [Google Scholar]
- 9.Malpartida F, Hopwood DA. Molecular cloning of the whole biosynthetic pathway of a Streptomyces antibiotic and its expression in a heterologous host. Nature. 1984;309:462–464. doi: 10.1038/309462a0. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez-Moreno MA, Martinez E, Boto L, Hopwood DA, Malpartida F. Nucleotide sequence and deduced functions of a set of cotranscribed genes of Streptomyces coelicolor A3(2) including the polyketide synthase for the antibiotic actinorhodin. J. Biol. Chem. 1992;267:19278–19290. [PubMed] [Google Scholar]
- 11.Carreras CW, Khosla C. Purification and in vitro reconstitution of the essential protein components of an aromatic polyketide synthase. Biochemistry. 1998;37:2084–2088. doi: 10.1021/bi972919+. [DOI] [PubMed] [Google Scholar]
- 12.Summers RG, Ali A, Shen B, Wessel WA, Hutchinson CR. Malonyl-coenzyme A:acyl carrier protein acyltransferase of Streptomyces glaucescens: a possible link between fatty acid and polyketide biosynthesis. Biochemistry. 1995;34:9389–9402. doi: 10.1021/bi00029a015. [DOI] [PubMed] [Google Scholar]
- 13.Carreras CW, Pieper R, Khosla C. Efficient Synthesis of Aromatic Polyketides in Vitro by the Actinorhodin Polyketide Synthase. J. Am. Chem. Soc. 1996;118:5158–5159. [Google Scholar]
- 14.McDaniel R, Ebert-Khosla S, Hopwood DA, Khosla C. Engineered biosynthesis of novel polyketides. Science. 1993;262:1546–1550. doi: 10.1126/science.8248802. [DOI] [PubMed] [Google Scholar]
- 15.Marti T, Hu Z, Pohl NL, Shah AN, Khosla C. Cloning, nucleotide sequence, and heterologous expression of the biosynthetic gene cluster for R1128, a non-steroidal estrogen receptor antagonist. Insights into an unusual priming mechanism. J. Biol. Chem. 2000;275:33443–33448. doi: 10.1074/jbc.M006766200. [DOI] [PubMed] [Google Scholar]
- 16.GenBank Accession number: AF058302
- 17.Bibb MJ, Sherman DH, Omura S, Hopwood DA. Cloning, sequencing and deduced functions of a cluster of Streptomyces genes probably encoding biosynthesis of the polyketide antibiotic frenolicin. Gene. 1994;142:31–39. doi: 10.1016/0378-1119(94)90351-4. [DOI] [PubMed] [Google Scholar]
- 18.Dreier J, Khosla C. Mechanistic analysis of a type II polyketide synthase. Role of conserved residues in the beta-ketoacyl synthase-chain length factor heterodimer. Biochemistry. 2000;39:2088–2095. doi: 10.1021/bi992121l. [DOI] [PubMed] [Google Scholar]
- 19.Keatinge-Clay AT, Maltby DA, Medzihradszky KF, Khosla C, Stroud RM. An antibiotic factory caught in action. Nat. Struct. Mol. Biol. 2004;11:888–893. doi: 10.1038/nsmb808. [DOI] [PubMed] [Google Scholar]
- 20.Tang Y, Tsai SC, Khosla C. Polyketide chain length control by chain length factor. J. Am. Chem. Soc. 2003;125:12708–12709. doi: 10.1021/ja0378759. [DOI] [PubMed] [Google Scholar]
- 21.Ridley CP, Lee HY, Khosla C. Evolution of polyketide synthases in bacteria. Proc Natl Acad Sci U S A. 2008;105:4595–4600. doi: 10.1073/pnas.0710107105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dreier J, Shah AN, Khosla C. Kinetic analysis of the actinorhodin aromatic polyketide synthase. J. Biol. Chem. 1999;274:25108–25112. doi: 10.1074/jbc.274.35.25108. [DOI] [PubMed] [Google Scholar]
- 23.Wendt-Pienkowski E, Huang Y, Zhang J, Li B, Jiang H, Kwon H, Hutchinson CR, Shen B. Cloning, sequencing, analysis, and heterologous expression of the fredericamycin biosynthetic gene cluster from Streptomyces griseus. J. Am. Chem. Soc. 2005;127:16442–16452. doi: 10.1021/ja054376u. [DOI] [PubMed] [Google Scholar]
- 24.McDaniel R, Khosla SE, Hopwood DA, Khosla C. Engineered Biosynthesis of Novel Polyketides: Manipulation and Analysis of an Aromatic Polyketide Synthase with Unproven Catalytic Specificities. J. Am. Chem. Soc. 1993;115:11671–11675. [Google Scholar]
- 25.McDaniel R, Ebert-Khosla S, Fu H, Hopwood DA, Khosla C. Engineered biosynthesis of novel polyketides: influence of a downstream enzyme on the catalytic specificity of a minimal aromatic polyketide synthase. Proc. Natl. Acad. Sci. U. S. A. 1994;91:11542–11546. doi: 10.1073/pnas.91.24.11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDaniel R, Khosla SE, Hopwood DA, Khosla C. Engineered Biosynthesis of Novel Polyketides: actVII and actIV Genes Encode Aromatase and Cyclase Enzymes, respectively. J. Am. Chem. Soc. 1994;116:10855–10859. [Google Scholar]
- 27.McDaniel R, Ebert-Khosla S, Hopwood DA, Khosla C. Rational design of aromatic polyketide natural products by recombinant assembly of enzymatic subunits. Nature. 1995;375:549–554. doi: 10.1038/375549a0. [DOI] [PubMed] [Google Scholar]
- 28.Kramer PJ, Zawada RJX, McDaniel R, Hutchinson CR, Hopwood DA, Khosla C. Rational Design and Engineered Biosynthesis of a Novel 18-Carbon Aromatic Polyketide. J. Am. Chem. Soc. 1997;119:635–639. [Google Scholar]
- 29.Tang Y, Lee TS, Khosla C. Engineered biosynthesis of regioselectively modified aromatic polyketides using bimodular polyketide synthases. PLoS Biol. 2004;2:E31. doi: 10.1371/journal.pbio.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang Y, Lee TS, Lee HY, Khosla C. Exploring the Biosynthetic Potential of Bimodular Aromatic Polyketide Synthases. Tetrahedron. 2004;60:7659–7671. [Google Scholar]
- 31.Lee TS, Khosla C, Tang Y. Engineered biosynthesis of aklanonic acid analogues. J. Am. Chem. Soc. 2005;127:12254–12262. doi: 10.1021/ja051429z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang Y, Lee TS, Kobayashi S, Khosla C. Ketosynthases in the initiation and elongation modules of aromatic polyketide synthases have orthogonal acyl carrier protein specificity. Biochemistry. 2003;42:6588–6595. doi: 10.1021/bi0341962. [DOI] [PubMed] [Google Scholar]
- 33.Khosla C, Ebert-Khosla S, Hopwood DA. Targeted gene replacements in a Streptomyces polyketide synthase gene cluster: role for the acyl carrier protein. Mol. Microbiol. 1992;6:3237–3249. doi: 10.1111/j.1365-2958.1992.tb01778.x. [DOI] [PubMed] [Google Scholar]
- 34.Khosla C, McDaniel R, Ebert-Khosla S, Torres R, Sherman DH, Bibb MJ, Hopwood DA. Genetic construction and functional analysis of hybrid polyketide synthases containing heterologous acyl carrier proteins. J. Bacteriol. 1993;175:2197–2204. doi: 10.1128/jb.175.8.2197-2204.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherman DH, Kim ES, Bibb MJ, Hopwood DA. Functional replacement of genes for individual polyketide synthase components in Streptomyces coelicolor A3(2) by heterologous genes from a different polyketide pathway. J. Bacteriol. 1992;174:6184–6190. doi: 10.1128/jb.174.19.6184-6190.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee TS, Khosla C, Tang Y. Orthogonal protein interactions in spore pigment producing and antibiotic producing polyketide synthases. J. Antibiot. (Tokyo) 2005;58:663–666. doi: 10.1038/ja.2005.91. [DOI] [PubMed] [Google Scholar]
- 37.Crump MP, Crosby J, Dempsey CE, Parkinson JA, Murray M, Hopwood DA, Simpson TJ. Solution structure of the actinorhodin polyketide synthase acyl carrier protein from Streptomyces coelicolor A3(2) Biochemistry. 1997;36:6000–6008. doi: 10.1021/bi970006+. [DOI] [PubMed] [Google Scholar]
- 38.Li Q, Khosla C, Puglisi JD, Liu CW. Solution structure and backbone dynamics of the holo form of the frenolicin acyl carrier protein. Biochemistry. 2003;42:4648–4657. doi: 10.1021/bi0274120. [DOI] [PubMed] [Google Scholar]
- 39.Findlow SC, Winsor C, Simpson TJ, Crosby J, Crump MP. Solution structure and dynamics of oxytetracycline polyketide synthase acyl carrier protein from Streptomyces rimosus. Biochemistry. 2003;42:8423–8433. doi: 10.1021/bi0342259. [DOI] [PubMed] [Google Scholar]
- 40.Fu H, Khosla SE, Hopwood DA, Khosla C. Engineered Biosynthesis of Novel Polyketides: Dissection of the Catalytic Specificity of the act Ketoreductase. J. Am. Chem. Soc. 1994;116:4166–4170. [Google Scholar]
- 41.Tin-Wein Y, Shen Y, McDaniel R, Floss HG, Khosla C, Hopwood DA, Moore BS. Engineered Biosynthesis of Novel Polyketides from Streptomyces Spore Pigment Polyketide Synthases. J. Am. Chem. Soc. 1998;120:7749–7759. [Google Scholar]
- 42.Hadfield AT, Limpkin C, Teartasin W, Simpson TJ, Crosby J, Crump MP. The crystal structure of the actIII actinorhodin polyketide reductase: proposed mechanism for ACP and polyketide binding. Structure. 2004;12:1865–1875. doi: 10.1016/j.str.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Korman TP, Hill JA, Vu TN, Tsai SC. Structural analysis of actinorhodin polyketide ketoreductase: cofactor binding and substrate specificity. Biochemistry. 2004;43:14529–14538. doi: 10.1021/bi048133a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korman TP, Tan YH, Wong J, Luo R, Tsai SC. Inhibition kinetics and emodin cocrystal structure of a type II polyketide ketoreductase. Biochemistry. 2008;47:1837–1847. doi: 10.1021/bi7016427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ames BD, Korman TP, Zhang W, Smith P, Vu T, Tang Y, Tsai SC. Crystal structure and functional analysis of tetracenomycin ARO/CYC: implications for cyclization specificity of aromatic polyketides. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5349–5354. doi: 10.1073/pnas.0709223105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang Y, Lee HY, Kim CY, Mathews I, Khosla C. Structural and functional studies on SCO1815: a beta-ketoacyl-acyl carrier protein reductase from Streptomyces coelicolor A3(2) Biochemistry. 2006;45:14085–41093. doi: 10.1021/bi061187v. [DOI] [PubMed] [Google Scholar]
- 47.Bao W, Sheldon PJ, Hutchinson CR. Purification and properties of the Streptomyces peucetius DpsC beta-ketoacyl:acyl carrier protein synthase III that specifies the propionate-starter unit for type II polyketide biosynthesis. Biochemistry. 1999;38:9752–9757. doi: 10.1021/bi990751h. [DOI] [PubMed] [Google Scholar]
- 48.Bao W, Sheldon PJ, Wendt-Pienkowski E, Hutchinson CR. The Streptomyces peucetius dpsC gene determines the choice of starter unit in biosynthesis of the daunorubicin polyketide. J. Bacteriol. 1999;181:4690–4695. doi: 10.1128/jb.181.15.4690-4695.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meadows ES, Khosla C. In vitro reconstitution and analysis of the chain initiating enzymes of the R1128 polyketide synthase. Biochemistry. 2001;40:14855–14861. doi: 10.1021/bi0113723. [DOI] [PubMed] [Google Scholar]
- 50.Pan H, Tsai S, Meadows ES, Miercke LJ, Keatinge-Clay AT, O'Connell J, Khosla C, Stroud RM. Crystal structure of the priming beta-ketosynthase from the R1128 polyketide biosynthetic pathway. Structure. 2002;10:1559–1568. doi: 10.1016/s0969-2126(02)00889-4. [DOI] [PubMed] [Google Scholar]
- 51.Tang Y, Koppisch AT, Khosla C. The acyltransferase homologue from the initiation module of the R1128 polyketide synthase is an acyl-ACP thioesterase that edits acetyl primer units. Biochemistry. 2004;43:9546–9555. doi: 10.1021/bi049157k. [DOI] [PubMed] [Google Scholar]
- 52.Sattely ES, Fischbach MA, Walsh CT. Total biosynthesis: in vitro reconstitution of polyketide and nonribosomal peptide pathways. Nat. Prod. Rep. 2008;25:757–793. doi: 10.1039/b801747f. [DOI] [PubMed] [Google Scholar]
- 53.Walsh CT. The chemical versatility of natural-product assembly lines. Acc. Chem. Res. 2008;41:4–10. doi: 10.1021/ar7000414. [DOI] [PubMed] [Google Scholar]
- 54.Staunton J, Weissman KJ. Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 55.Xiang L, Moore BS. Characterization of benzoyl coenzyme A biosynthesis genes in the enterocin-producing bacterium "Streptomyces maritimus". J. Bacteriol. 2003;185:399–404. doi: 10.1128/JB.185.2.399-404.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Izumikawa M, Cheng Q, Moore BS. Priming type II polyketide synthases via a type II nonribosomal peptide synthetase mechanism. J. Am. Chem. Soc. 2006;128:1428–1429. doi: 10.1021/ja0559707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bililign T, Hyun CG, Williams JS, Czisny AM, Thorson JS. The hedamycin locus implicates a novel aromatic PKS priming mechanism. Chem. Biol. 2004;11:959–969. doi: 10.1016/j.chembiol.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 58.Ridley CR, Khosla C. Encyclopedia of Microbiology. 2009 in press. [Google Scholar]
- 59.Nicholson TP, Winfield C, Westcott J, Crosby J, Simpson TJ, Cox RJ. First in vitro directed biosynthesis of new compounds by a minimal type II polyketide synthase: evidence for the mechanism of chain length determination. Chem. Commun. (Camb) 2003:686–687. doi: 10.1039/b300847a. [DOI] [PubMed] [Google Scholar]
- 60.Ichinose K, Surti C, Taguchi T, Malpartida F, Booker-Milburn KI, Stephenson GR, Ebizuka Y, Hopwood DA. Proof that the ACTVI genetic region of Streptomyces coelicolor A3(2) is involved in stereospecific pyran ring formation in the biosynthesis of actinorhodin. Bioorg. Med. Chem. Lett. 1999;9:395–400. doi: 10.1016/s0960-894x(99)00011-6. [DOI] [PubMed] [Google Scholar]
- 61.Mittler M, Bechthold A, Schulz GE. Structure and action of the C-C bond-forming glycosyltransferase UrdGT2 involved in the biosynthesis of the antibiotic urdamycin. J. Mol. Biol. 2007;372:67–76. doi: 10.1016/j.jmb.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 62.Kalaitzis JA, Izumikawa M, Xiang L, Hertweck C, Moore BS. Mutasynthesis of enterocin and wailupemycin analogues. J. Am. Chem. Soc. 2003;125:9290–9291. doi: 10.1021/ja035973o. [DOI] [PubMed] [Google Scholar]