Abstract
MHC-TGFcys33 ser transgenic mice have elevated levels of active Transforming Growth Factor (TGF)-beta1 in the myocardium. Previous studies have shown that these animals develop atrial, but not ventricular, fibrosis. Here we show that atrial fibrosis was accompanied with cardiomyocyte apoptosis. Although similar levels of cardiomyocyte apoptosis were present in the right and left atria of MHC-TGFcys33 ser hearts, the extent of fibrosis was more pronounced in the right atrium. Thus, additional factors influence the degree of atrial fibrosis in this model. Tritiated thymidine incorporation studies revealed cardiomyocyte cell cycle activity in left atrial cardiomyocytes, but not in right atrial cardiomyocytes. These observations suggested that cardiomyocyte cell cycle activation ameliorated the severity of atrial fibrosis. To directly test this hypothesis, MHC-TGFcys33 ser mice were crossed with MHC-cycD2 mice (which have constitutive cardiomyocyte cell cycle activity in the right atrium). Mice inheriting both transgenes exhibited right atrial cardiomyocyte cell cycle activity and a concomitant reduction in the severity of right atrial fibrosis, despite the presence of a similar level of cardiomyocyte apoptosis as was observed in mice inheriting the MHC-TGFcys33 ser transgene alone. These data support the notion that cardiomyocyte cell cycle induction can antagonize fibrosis in the myocardium.
Keywords: cardiomyocyte proliferation, cardiac myocyte apoptosis, heart regeneration
Progressive necrotic, apoptotic, and/or oncotic cardiomyocyte death underlies many forms of cardiovascular disease.1,2 Pathophysiologic cardiomyocyte loss is often accompanied with fibrosis, which is characterized by pronounced changes in the extracellular matrix including interstitial collagen deposition.3–5 Fibrosis increases myocardial stiffness and induces electrical heterogeneity, both of which contribute to the progressive loss of systolic and diastolic function, as well as susceptibility to cardiac arrhythmia and sudden death. Fibrosis is thought to be regulated in part by the activities of a number of hormone and cytokine systems, including TGF-beta family members, the renin-angiotensin-aldosterone system, acidic and basic FGF, endothelins, and TNF-alpha.1
Several approaches have been utilized to attempt to control the progression of myocardial fibrosis. These include pharmacologic modulation of the hormone/cytokine systems,6,7 as well as modulation of the activity of tissue metalloproteases which are able to degrade the extracellular matrix.8–10 Other strategies aimed at rendering cardiomyocytes more resistant to apoptotic and necrotic stimuli are also being pursued,11–13 with the notion that this would limit cardiomyocyte loss and in turn reduce fibrosis. In addition, replacement of lost cardiomyocytes might inhibit the progression of, and perhaps even promote the regression of, cardiac fibrosis. Potential approaches to promote cardiomyocyte replacement include cell cycle activation,14,15 direct replacement of lost cardiomyocytes via cellular transplantation,16 and recruitment and/or activation of cardiomyogenic stem cells.17,18
We have previously generated transgenic mice which over-express TGF-beta1 in the myocardium to directly examine the role of this cytokine in the genesis of myocardial fibrosis.19 The stringent post-translational regulation of TGF-beta1 necessitated the use of cDNAs encoding site-directed mutations that increased the level of active cytokine. Accordingly, mice were generated which carried a transgene comprising the cardiomyocyte-restricted alpha cardiac myosin heavy chain (MHC) promoter and a cDNA encoding a mutant TGF-beta1 molecule harboring a cysteine to serine substitution at amino acid residue #33. This mutation blocked covalent tethering of the TGF-beta1 latent complex to the extracellular matrix,20 resulting in an approximately 30-fold increase in the level of active TGF-beta1 in the adult transgenic hearts.19 Increased TGF-beta1 activity in these animals, which were designated MHC-TGFcys33 ser, did not induce ventricular fibrosis, as evidenced by histochemical and gene expression analyses. A delay in ventricular wound healing, which appeared to be secondary to cytokine-induced apoptosis of proliferating cardiac fibroblasts, was observed following myocardial injury in MHC-TGFcys33 ser transgenic hearts. These observations suggested that TGF-beta1 activity in itself was insufficient to promote fibrosis in the ventricles. Given the absence of overt fibrosis in the transgenic ventricles, it was surprising that age-dependent fibrosis was observed in the atria of these mice.19
In this study, the atrial phenotype in MHC-TGFcys33 ser transgenic mice was further characterized. The extent of fibrosis was observed to be more severe in the right atrium as compared to the left. Cardiomyocyte apoptosis was present at similar levels in the left and right atria, suggesting that factors in addition to cell death contributed to the differential atrial fibrosis. Surprisingly, tritiated thymidine incorporation studies revealed cardiomyocyte cell cycle activity in left atrium of MHC-TGFcys33 ser hearts, but not in the right atrium. These data suggested that the presence of cardiomyocyte cell cycle activity could ameliorate the severity of atrial fibrosis. To directly test this hypothesis, MHC-TGFcys33 ser mice were intercrossed with MHC-cycD2 mice,21 which exhibit constitutive right atrial cell cycle activity. Mice inheriting both transgenes exhibited right atrial cardiomyocyte cell cycle activity and a concomitant reduction in the severity of right atrial fibrosis, despite having a similar level of right atrial cardiomyocyte apoptosis as compared to mice inheriting the MHC-TGFcys33 ser transgene alone. Collectively, these data suggest that cardiomyocyte cell cycle activation can antagonize fibrosis, and support the notion that the controlled proliferation of cardiomyocytes can have a beneficial impact upon diseased hearts.
Materials and Methods
Transgenic mouse models
The generation and initial analyses of the MHC-TGFcys33 ser,19 MHC-cycD221 and MHC-nLAC22 transgenic mice have been described previously. Mice were maintained in a DBA/2J genetic background. All animal manipulations were performed according to NIH and Institutional Animal Care and Use Committee Guidelines.
Histology
Paraffin sections were prepared from Bouin's fixed samples using standard procedures.23 Cryosections (10 microns) were generated using standard histologic techniques.24 Sections were stained with picrosirius solution (0.1% Sirius Red in picric acid, Sigma Diagnostics, St. Louis MO) and counter-stained with Fast green (Fischer Scientific, Fair Lawn NJ). To quantitate atrial collagen content, images were captured with a digital camera and the red pixel content of the myocardium was measured using Adobe Photoshop 5.5 and Scion Images for Windows Beta 4.0.2 software as described.19
Apoptosis assays
For anti-activated caspase 3 immune assay, cryosections were processed according to the manufacturer’s specifications (Promega, Madison, WI). For ISEL analysis, cryosections were processed using the KLENOW-FragEL DNA Fragmentation Detection Kit (#QIA21, Oncogene Research, Boston MA). Signal was developed with biotinylated nucleotides and horseradish peroxidase (HRP) conjugated streptavidin, followed by incubation with diaminobenzidine. Beta-galactosidase activity was detected in the same sections via X-GAL staining as described.22 For TUNEL analysis, cryosections were processed using the TdT-FragEL DNA Fragmentation Detection kit according to the manufacturer's procedures (#QIA33, Oncogene Research, Boston MA). Signal was developed using biotinylated nucleotides and FITC-conjugated streptavidin (Roche Molecular Biochemicals, Indianapolis IN). Beta-galactosidase activity was detected in the same section via immune reactivity using a polyclonal anti-beta-galactosidase antibody (#55976, ICN, Aurora OH) and rhodamine-conjugated secondary antibody (#AQ301R, Chemicon, Temecuba CA).
Cardiomyocyte DNA synthesis assay
To monitor cardiomyocyte DNA synthesis, mice received a single injection of tritiated thymidine (200 µCi I.P. at 28 Ci/mM, Amersham, Arlington Heights IL) and were sacrificed 4 hours or two weeks later. Hearts were cryosectioned as described above and cardiomyocyte DNA synthesis was identified by co-localization of beta-galactosidase activity (dark blue signal following X-GAL staining) and silver grains as described.25
Statistical analysis
Results are expressed as mean ± S.E.M. The differences between right and left atria parameters were calculated with Student's paired t-test. The differences among MHC-TGFcys33 ser / (−), (−) / MHC-cycD2 and MHC-TGFcys33 ser/MHC-cycD2 mice were evaluated by ANOVA followed by Bonferroni post hoc test.
Results
Differential atrial fibrosis in MHC-TGFcys33 ser mice
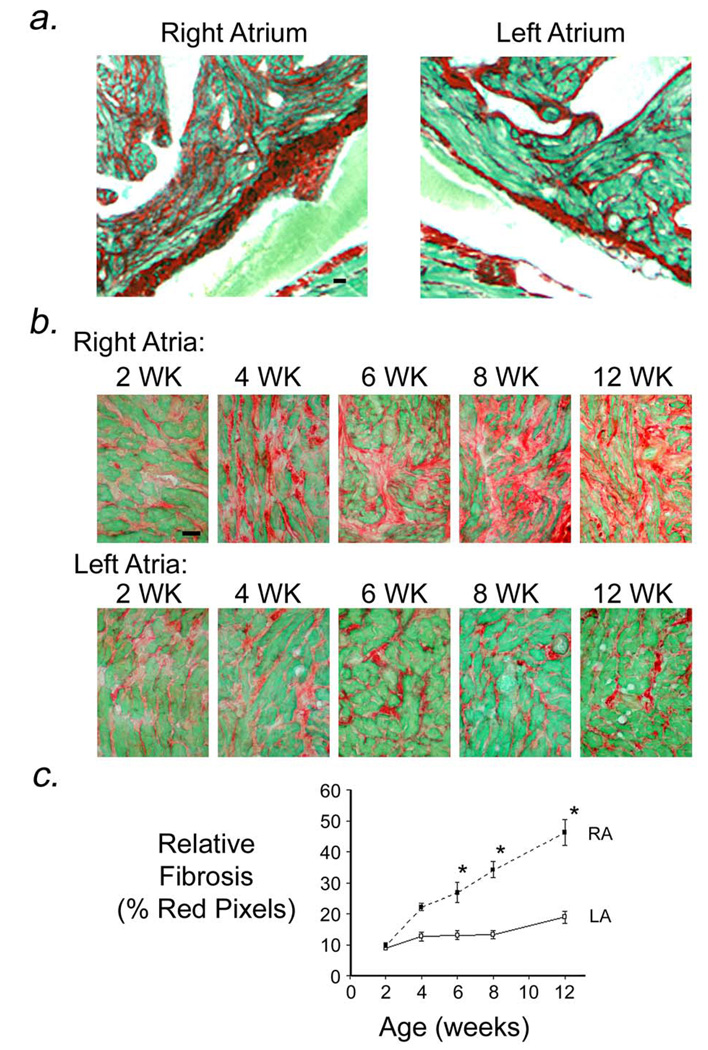
Previous studies demonstrated that MHC-TGFcys33 ser transgenic mice developed age-dependent atrial fibrosis.19 Histologic analysis of aged transgenic mice revealed that collagen deposition (red signal, Sirius Red stain) in the interstitial spaces between cardiomyocytes was markedly greater in the right atrium as compared to the left atrium (Figure 1a; heart is from an 18 month-old transgenic mouse). To further characterize the atrial fibrosis, MHC-TGFcys33 ser mice were crossed with MHC-nLAC mice (the presence of the MHC-nLAC reporter transgene facilitated the identification of cardiomyocyte nuclei in subsequent experiments, see below). Sections of hearts from 2, 4, 6, 8, and 12 week old MHC-TGFcys33 ser / MHC-nLAC double transgenic mice were stained with Sirius Red and Fast Green (Figure 1b). Interstitial collagen content was similar in left and right atria at 2 weeks of age. By 4 weeks of age collagen content appeared more pronounced in the right atrium, and this asymmetry in atrial collagen content was apparent at all subsequent ages examined. Quantitative image analysis confirmed similar collagen content in right and left transgenic atria at 2 weeks of age. Collagen content progressively increased with age in the right atria. In contrast, collagen content in the left atria remained relatively constant over the time points studied (Figure 1c).
Figure 1.
Atrial fibrosis in MHC-TGFcys33 ser transgenic hearts. (a) Sections from the right and left atria of an 18 month old MHC-TGFcys33 ser transgenic mouse, stained with Sirius Red and Fast Green. Bar = 20 microns. (b) Sections of right and left atria from individual MHC-TGFcys33 ser / MHC-nLAC transgenic hearts at 2, 4, 6, 8 and 12 weeks of age, stained with Sirius Red and Fast Green. Bar = 20 microns. (c) Quantitation of collagen content in the right (RA, filled squares) and left (LA, open squares) atria of MHC-TGFcys33 ser / MHC-nLAC mice at the indicated ages. Data is from 5 mice per time point. *: p<0.05, right atria vs. left atria.
Cardiomyocyte apoptosis in MHC-TGFcys33 ser atria
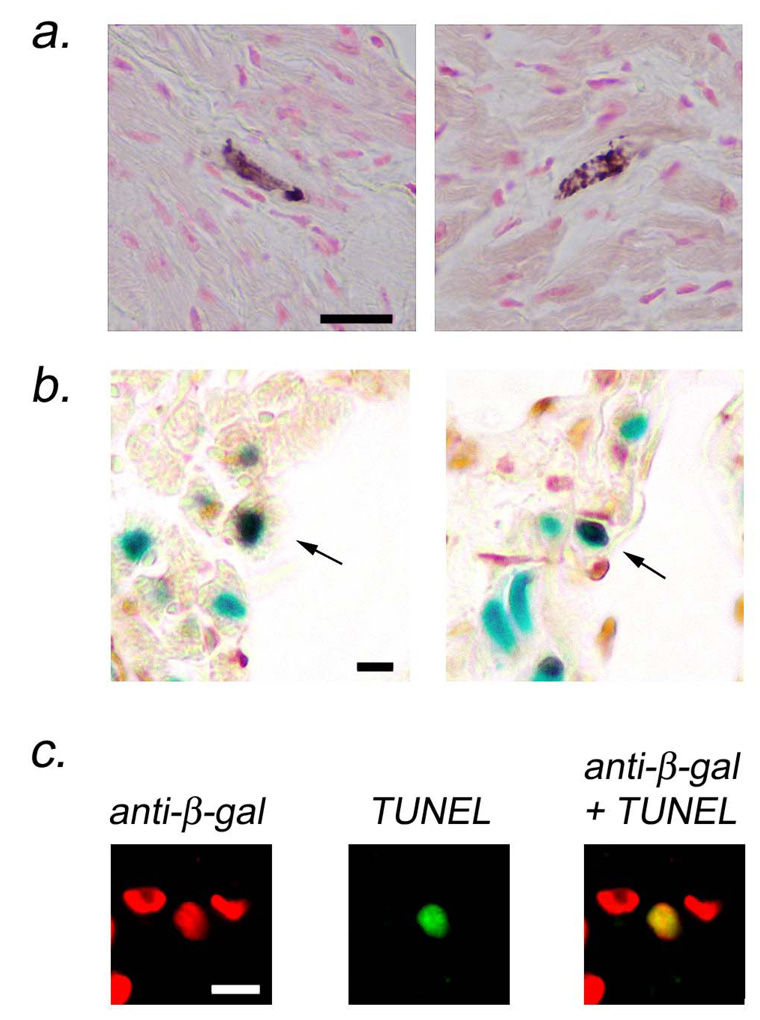
Activated caspase 3 immune assays were performed to determine if increased levels of cardiomyocyte apoptosis accompanied the increased fibrosis observed in the right atria of MHC-TGFcys33 ser mice. Since activated caspase 3 is distributed in the cytoplasm, cardiomyocytes at early stages of apoptosis were easily identified by morphologic criteria (i.e., rod-shaped cells, see Figure 2a). Similar levels of cardiomyocyte activated caspase 3 immune reactivity were observed in the right and left atria of the MHC-TGFcys33 ser hearts (0.45 ± 0.2 and 0.58 ± 0.18 activated caspase 3 positive cardiomyocytes per atrial section, respectively, n=8 mice; paired t-test, not significant). No apoptosis was detected when similar numbers of atrial cardiomyocytes were screened in age-matched non-transgenic littermates.
Figure 2.
Atrial cardiomyocyte apoptosis in MHC-TGFcys33 ser / MHC-nLAC double transgenic mice. (a) Activated caspase 3 immune assay for cardiomyocyte apoptosis. Cardiomyocytes at early stages of apoptosis were identified by the presence of activated caspase 3 immune reactivity (cytoplasmic brown signal in rod-shaped cells). Bar = 20 microns. (b) ISEL assay for cardiomyocyte apoptosis. Sections were stained with X-GAL to identify cardiomyocyte nuclei (blue signal) and for ISEL activity to identify apoptotic cells (HRP-conjugated reaction, dark brown signal). Apoptotic cardiomyocytes have brown and blue signals over the nucleus (arrows). Bar = 10 microns. (c) TUNEL assay for cardiomyocyte apoptosis. Sections were stained for beta-galactosidase immune reactivity to identify cardiomyocyte nuclei (rhodamine conjugated secondary antibody, red signal, left panel) and for TUNEL activity to identify apoptotic cells (FITC-conjugated secondary antibody, green signal, middle panel). Apoptotic cardiomyocytes are identified in the merged image (yellow signal, right panel). Bar = 10 microns.
ISEL and TUNEL analyses were performed to confirm these results. These analyses took advantage of the presence of the MHC-nLAC transgene, which targets expression of a nuclear localized beta-galactosidase reporter to cardiomyocytes.22,25 In the case of ISEL analyses, sections prepared from MHC-TGFcys33 ser / MHC-nLAC double transgenic mice were stained with X-GAL (blue signal over cardiomyocyte nuclei) and then processed for ISEL activity using biotinylated nucleotides and HRP-conjugated streptavidin (dark brown signal over apoptotic nuclei). ISEL-positive cardiomyocytes, which were identified by the superimposition of dark brown and blue signals, were readily seen (Figure 2b, arrows). In the case of TUNEL analysis, sections from MHC-TGFcys33 ser / MHC-nLAC double transgenic mice were processed simultaneously for beta-galactosidase immune reactivity (Figure 2c, left panel, red signal) and TUNEL activity (Figure 2c, middle panel, green signal). Merging the images confirmed that the TUNEL positive nucleus was from a cardiomyocyte (Figure 2c, right panel, yellow signal). In agreement with the activated caspase 3 immune reactivity assay, similar levels of cardiomyocyte ISEL activity and TUNEL signal were observed in the left and tright atria in MHC-TGFcys33 ser hearts. These data suggested that additional factors were likely to contribute to the differential levels of fibrosis seen in the left and right atria of the transgenic mice.
Cardiomyocyte cell cycle activity in the left atria of MHC-TGFcys33 ser mice
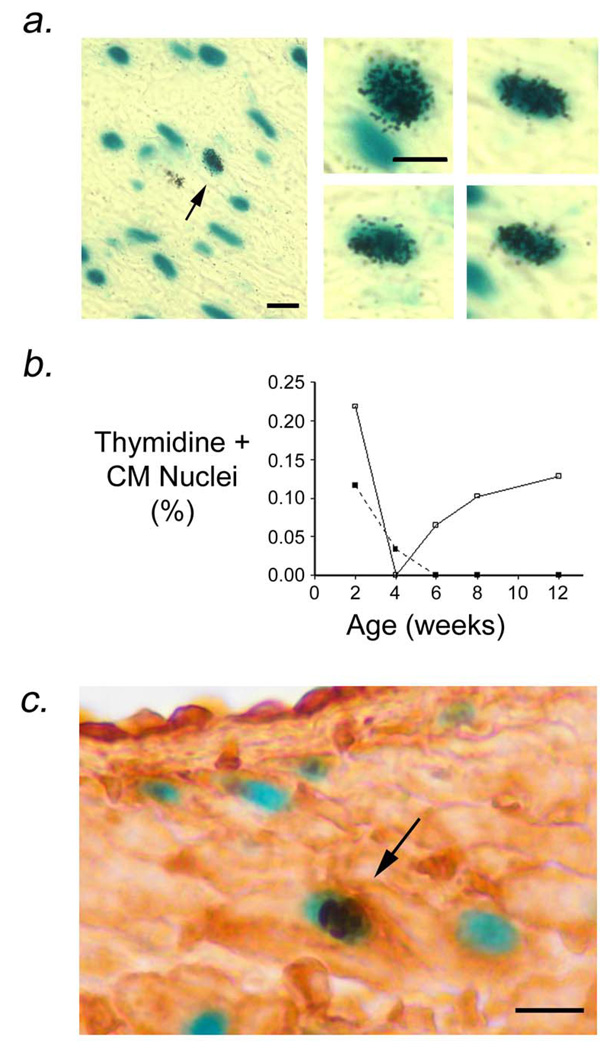
Tritiated thymidine incorporation assays were performed to monitor cell cycle activity in the atria of MHC-TGFcys33 ser mice. Once again, the experiments made use of the presence of the MHC-nLAC reporter transgene. MHC-TGFcys33 ser / MHC-nLAC double transgenic mice received a single injection of tritiated thymidine. The animals were sacrificed 4 hours later, and the hearts were sectioned, stained with X-GAL, and processed for autoradiography. Cardiomyocyte DNA synthesis was then quantitated by screening for the presence of silver grains over blue nuclei.22,25 Surprisingly, cardiomyocyte DNA synthesis was readily detected in the left atria of 12 week old MHC-TGFcys33 ser / MHC-nLAC double transgenic mice, but not in the right atria (Figure 3a, Table 1a).
Figure 3.
Cardiomyocyte DNA synthesis in left atria from MHC-TGFcys33 ser / MHC-nLAC double transgenic mice. (a) Low (left panel) and high (right panels) views of cardiomyocytes synthesizing DNA. Sections were stained with X-GAL to identify cardiomyocyte nuclei (blue signal) and processed for autoradiography to identify DNA synthesis (nuclear silver grains). Arrow in left panel indicates a cardiomyocyte nucleus which was synthesizing DNA. Bar = 10 microns. (b) Quantitation of cardiomyocyte DNA synthesis in right (filled squares) and left (open squares) atria of MHC-TGFcys33 ser at the indicated ages. Data is from 5 mice per time point. *: p<0.05, left atria vs. right atria. (c) Phosphorylated histone H3 immune reactivity (dark brown signal, horseradish peroxidase-conjugated secondary antibody) in cardiomyocyte nuclei (blue signal, X-GAL staining) of left atrial cardiomyocytes of MHC-TGFcys33 ser / MHC-nLAC double transgenic mice at 12 weeks of age. Bar = 10 microns.
Table 1.
Cardiomyocyte DNA synthesis levels in atria from MHC-TGFcys33 ser / MHC-nLAC double transgenic mice.
| Right Atrium | Left Atrium | |
|---|---|---|
| A. 12 week old mice: | ||
| % thymidine “+” nuclei | 0 | 0.10 ± 0.03 |
| Nuclei screened | 38,381 | 61,706 |
| Mice screened | 8 | 8 |
| B. Pulse/chase study: | ||
| % pulse thymidine “+” nuclei | 0 | 0.12 ± 0.05 |
| Nuclei screened | 11,846 | 20,217 |
| Mice screened | 3 | 3 |
| % chase thymidine “+” nuclei | 0 | 0.21 ± 0.05* |
| Nuclei screened | 14,642 | 22,661 |
| Mice screened | 3 | 3 |
| C. 26 week old mice: | ||
| % thymidine “+” nuclei | 0 | 0.11 ± 0.03 |
| Nuclei screened | 11,851 | 27,980 |
| Mice screened | 5 | 5 |
the difference in tritiated thymidine incorporation in the left atrial pulse and chase groups was significant when pooled data was analyzed with chi-square test (p<0.05)
To establish the time course of atrial cardiomyocyte DNA synthesis, thymidine incorporation analyses were performed on 2, 4, 6, 8, and 12 week old MHC-TGFcys33 ser / MHC-nLAC double transgenic mice. Cardiomyocyte DNA synthesis was detected in both the right and left atria of the MHC-TGFcys33 ser mice at two weeks of age (Figure 3b). Cardiomyocyte DNA synthesis was markedly down regulated in both atria by four weeks of age. These observations are in agreement with the marked decrease in cardiomyocyte cell cycle activity that occurs during neonatal life.26–28 At 6 weeks of age cardiomyocyte DNA synthesis was again apparent in the left atrium of MHC-TGFcys33 ser mice, and persisted at similar levels for the remainder of the experiment; in contrast, DNA synthesis did not resume in right atrial cardiomyocytes (Figure 3b).
Pulse chase experiments were performed to determine the stability of cardiomyocytes which synthesized DNA. Six 12 week old MHC-TGFcys33 ser / MHC-nLAC double transgenic animals were injected with tritiated thymidine. Three mice were sacrificed 4 hours later, and three were sacrificed 14 days later. The hearts were sectioned, processed for X-GAL staining and autoradiography, and the number of radiolabeled cardiomyocyte nuclei was determined for each time point (Table 1b). The increased tritiated thymidine labeling observed in the chase group indicated that left atrial cardiomyocytes which synthesized DNA were stable, and also suggested that a portion of these cells underwent karyokinesis and perhaps cytokinesis. This view was supported by the observation that phosphorylated histone H3 immune reactivity (a marker for mitosis)29 was detected in left atrial cardiomyocytes of MHC-TGFcys33 ser mice (Figure 3c, arrow). The frequency of cardiomyocyte phosphorylated histone H3 immune reactivity was approximately 1/15th of that observed for cardiomyocyte thymidine incorporation; similar ratios of cardiomyocyte DNA synthesis and mitosis have been reported in neonatal hearts30 and in the ventricle of adult transgenic mice with elevated cardiomyoycte cell cycle activity.21,31 DNA synthesis was also monitored in older MHC-TGFcys33 ser transgenic mice. Similar levels of left atrial cardiomyocyte DNA synthesis were observed in 12 and 26 week-old animals (Table 1c).
Cell cycle activation ameliorates fibrosis in the right atria of MHC-TGFcys33 ser mice
The data presented above raised the possibility that cardiomyocyte cell cycle activation ameliorated the extent of fibrosis in the left atria of MHC-TGFcys33 ser transgenic mice. To directly examine the consequences of cardiomyocyte cell cycle activation on atrial fibrosis, MHC-TGFcys33 ser mice were crossed with MHC-cycD2 mice. Previous studies have shown that expression of the MHC-cycD2 transgene was sufficient to drive cardiomyocyte DNA synthesis and proliferation in normal and diseased ventricles.21 MHC-TGFcys33 ser / (−), (−) / MHC-cycD2, and MHC-TGFcys33 ser / MHC-cycD2 transgenic mice were identified and sequestered. To facilitate quantitation of cardiomyocyte DNA synthesis, these mice also carried the MHC-nLAC reporter transgene; previous studies have shown that the presence of multiple transgenes utilizing the MHC promoter did not result in a reduction of expression levels.31,32 At 12 weeks of age, the mice received an injection of tritiated thymidine and the hearts were harvested and processed to monitor cardiomyocyte DNA synthesis, myocardial collagen content and cardiomyocyte apoptosis.
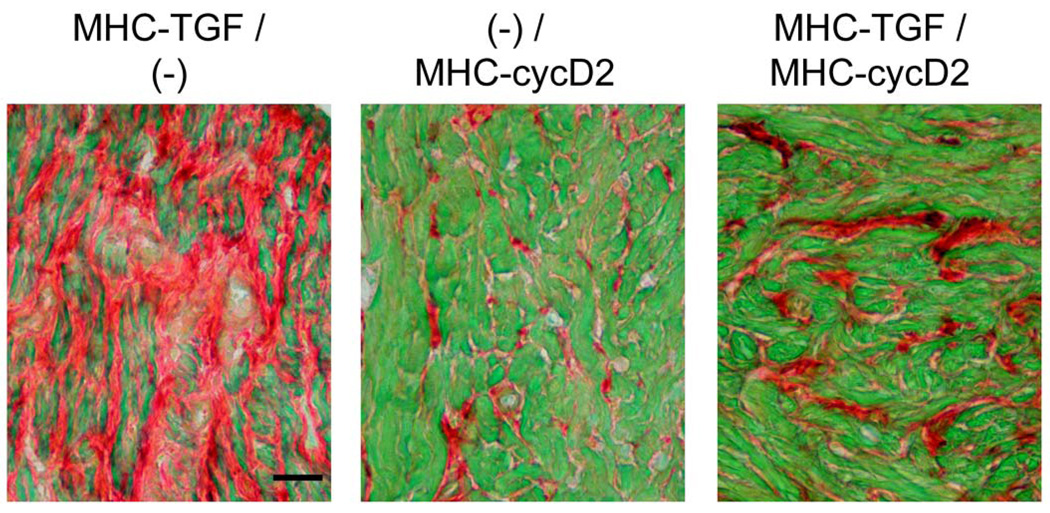
In agreement with the data presented above, no cardiomyocyte DNA synthesis was detected in the right atria of mice inheriting the MHC-TGFcys33 ser transgene alone (Table 2). In contrast, right atrial cardiomyocyte DNA synthesis was readily observed in mice inheriting the MHC-cycD2 transgene, indicating that targeted expression of cyclin D2 had similar effects on atrial and ventricular21 cardiomyocyte cell cycle induction. Right atrial cardiomyocyte DNA synthesis was also observed in mice inheriting both the MHC-cycD2 and MHC-TGFcys33 ser transgenes, and was accompanied with a marked reduction in right atrial collagen content (Figure 4, Table 2). Importantly, similar rates of cardiomyocyte apoptosis (as evidenced by activated caspase 3 immune reactivity) were observed in the right atria of mice inheriting the MHC-TGFcys33 ser transgene alone and in mice inheriting both the MHC-TGFcys33 ser and MHC-cycD2 transgenes. Analysis of adjacent sections revealed that the level of cardiomyocyte DNA synthesis (thymidine-positive, nLAC-positive nuclei per section) was very similar to the level of cardiomyocyte apoptosis (cell bodies with activated caspase-3 immune reactivity per section) in the right atria of MHC-TGFcys33 ser / MHC-cycD2 transgenic hearts (Table 2).
Table 2.
Cardiomyocyte apoptosis, cardiomyocyte DNA synthesis, and collagen content in right atria from MHC-TGFcys33 ser / (−), (−) / MHC-cycD2, and MHC-TGFcys33 ser / MHC-cycD2 transgenic mice.
| MHC-TGF / (−) |
(−) / MHC-cycD2 |
MHC-TGF / MHC-cycD2 |
|
|---|---|---|---|
| % thymidine “+” nuclei | 0 | 0.081 ± 0.018* | 0.055 ± 0.010* |
| Nuclei screened | 26,493 | 12,340 | 71,333 |
| Mice screened | 5 | 2 | 6 |
| Collagen content (%) | 42 ± 1.8 | 6.8 ± 1.8* | 18 ± 2.0*† |
| Mice screened | 5 | 2 | 6 |
| Activated caspase 3 “+” | 0.60 ± 0.31 | 0 | 0.83 ± 0.14 |
| cardiomyocytes / section | |||
| Mice screened | 5 | 2 | 6 |
| Total thymidine “+” | 0 | 1.3 ± 0.10* | 1.0 ± 0.31* |
| nuclei / section | |||
| Mice screened | 5 | 2 | 6 |
| Cardiomyocyte Number | 12.4 ± 1.0 | 21.8 ± 4.8# | 19.8 ± 0.7‡ |
| Mice screened | 5 | 2 | 6 |
All values are mean +/− SEM; statistical significance was determined by ANOVA followed by Bonferroni post hoc test.
p<0.05, vs. MHC-TGF / (−) mice
p<0.05, (−) / MHC-cycD2 vs MHC-TGF / MHC-cycD2
p<0.01, MHC-TGFcys33 ser / (−) vs. (−) / MHC-cycD2,
p<0.001, MHC-TGFcys33 ser / (−) vs. MHC-TGFcys33 ser / MHC-cycD2
Figure 4.
Reduced right atrial fibrosis in mice carrying both the MHC-TGFcys33 ser and the MHC-cycD2 transgenes. Right atria from MHC-TGFcys33 ser / (−) (left panel), (−) / MHC-cycD2 (middle panel) or MHC-TGFcys33 ser / MHC-cycD2 (right panel) mice, stained with Sirius Red and Fast Green. Bar = 20 microns.
If the presence of cyclin D2-induced cardiomyocyte DNA synthesis culminated in cell division, the date presented above would predict that there would be an increased number of right atrial cardiomyocytes in the MHC-TGFcys33 ser / MHC-cycD2 double transgenic mice as compared to MHC-TGFcys33 ser single transgenic animals. Accordingly, direct counts of cardiomyocyte cell bodies per unit area were performed. As expected, expression of cyclin D2 resulted in a statistically significant increase in the number of cardiomyocytes per unit area of right atrial tissue in the MHC-TGFcys33 ser / MHC-cycD2 mice as compared to MHC-TGFcys33 ser mice (Table 2).
Discussion
Transgene expression in the right atria of MHC-TGFcys33 ser mice resulted in cardiomyocyte apoptosis as evidenced by activated caspase 3, ISEL and TUNEL assays. Cardiomyocyte loss was accompanied by increased extracellular collagen deposition. Thus the progressive histopathology observed in the MHC-TGFcys33 ser transgenic mice was consistent with the process of reparative fibrosis in response to cardiomyocyte death.1 Although a similar level of cardiomyocyte apoptosis was apparent in MHC-TGFcys33 ser left and right atria at all ages examined, the extent of reparative fibrosis was markedly less in the left atrium. The presence of cell cycle activation in the left atrium suggested that cardiomyocyte proliferation could effectively ameliorate fibrosis.
Other observations argued against alternative mechanisms for the differential fibrosis in the MHC-TGFcys33 ser atria. For example, the levels of fibroblast DNA synthesis as measured by tritiated thymidine incorporation were very similar in right and left atria of the MHC-TGFcys33 ser transgenic mice (0.28 ± 0.039% and 0.36 ± 0.048%, respectively; n = 5 mice; paired t-test, not significant). The differential atrial collagen content was thus not attributable to differences in fibroblast cell cycle activity. In addition, the presence of similar levels of transgene-encoded mRNA in the right and left atria suggested that differential TGF-beta1 expression did not underlie the asymmetrical atrial fibrosis (see On Line Supplemental Data); unfortunately the lack of species-specific antibodies, and the presence of a marked induction of the endogenous TGF-beta1 gene,19 prohibited direct quantitation of transgene-encoded TGF-beta1 protein in mice carrying the MHC-TGFcys33 ser transgene alone or in combination with the MHC-cycD2 transgene. Collectively, these data suggested that cardiomyocyte cell cycle activity could ameliorate atrial fibrosis. This hypothesis was directly supported by the observation that cyclin D2-mediated cardiomyocyte proliferation markedly reduced the severity of fibrosis in the right atria of MHC-TGFcys33 ser / MHC-cycD2 double transgenic mice without impacting the level of cardiomyocyte apoptosis.
The mechanism by which expression of the MHC-TGFcys33 ser transgene induced atrial cardiomyocyte apoptosis, as well as left atrial cardiomyocyte cell cycle activity, was not clear. In vitro studies with cultured fetal or neonatal ventricular cardiomyocytes suggested that TGF-beta1 administration antagonized the mitogenic effects of other growth factors, but by itself has little effect on cardiomyocyte cell cycle activity.33,34 In light of this, the hypoplastic heart phenotype in mice expressing an activated TGF type 1 receptor (Alk5) under the regulation of the cardiac MHC promoter35 likely resulted from interplay between intrinsic (i.e. transgene expression) and extrinsic (i.e., cytokines) factors. In contrast, targeted expression of a constitutively active variant of the TGF-beta-activated kinase (TAK-1, a major effector of the TGF-beta1 signal transduction pathway) with the same MHC promoter did not induce a hypoplastic myocardial response.36 Rather these animals developed cardiac hypertrophy and exhibited cardiomyocyte apoptosis at an early age which progressed rapidly to overt heart failure and death. Moreover, this pro-apoptotic effect of TAK1 activation on ventricular myocardium in vivo contrasted with the apparent cardioprotective effect of TGF-beta1 administration during reperfusion injury of cultured adult cardiomyocytes.37
Thus, varying outcomes have been obtained following stimulation of the TGF-beta1 signal transduction pathway, depending upon the extent to which the pathway was activated as well as the degree to which pathway cross-talk occurred; this was particularly evident when a given effector molecule participated in multiple signaling pathways. The situation in MHC-TGFcys33 ser mice was further complicated as cardiomyocytes from different chambers of the heart exhibited strikingly different phenotypes. Indeed, with respect to cell cycle activity, TGF-beta1 expression induced cardiomyocyte DNA synthesis in left atrial cardiomyocytes, but not right atrial cardiomyocytes. Moreover, transgene expression did not influence the rate of ventricular cardiomyocyte DNA synthesis in normal adult hearts.19 In addition, experimental injury did not induce ventricular cardiomyocyte DNA synthesis in injured hearts.19 The presence of cardiac injury per se was probably not an overriding factor in the differential induction of atrial cardiomyocyte cell cycle activity in the MHC-TGFcys33 ser transgenic mice, as similar levels of apoptosis were present in left and right atria. Of interest, previous studies suggested a greater propensity for cell cycle induction in left atrial cardiomyocytes as compared other cardiomyocytes in the heart,30 a phenomenon that may be reflected in the results observed here.
Although direct evidence indicating that the MHC-TGFcys33 ser left atrial cardiomyocytes which synthesize DNA ultimately divided is lacking, the results of the pulse-chase experiment suggested that at least karyokinesis occured, a view supported by the presence of phosphorylated histone H3 immune. The failure to detect enlarged cardiomyocytes in histologic sections of the left atrium provided additional, albeit circumstantial, support of this viewpoint. Given that the MHC promoter used in this study was only expressed in differentiated cardiomyocytes, the observed results were most consistent with the notion that expression of the MHC-TGFcys33 ser transgene resulted in cell cycle activation in differentiated left atrial cardiomyocytes. Although the experiments as performed did not rule out the possibility that transgene-induced cardiac injury resulted in the activation of resident cardiomyogenic stem cell populations,17 the absence of cardiomyocyte cell cycle activity in the right atrium did not support this argument.
The mechanistic underpinning of cardiomyocyte cell cycle activation in MHC-cycD2 transgenic mice was much more straightforward.38 D-type cyclins (cyclin D1, D2 and D3) are induced in response to growth factors and are obligate co-factors for the activity of cyclin dependent kinase. The active kinase complex phosphorylates members of the retinoblastoma protein family, resulting in the release of E2F transcription factors. E2F transcription factors induce expression of genes required for S-phase entry, thereby triggering cell cycle progression. Previous studies have shown that targeted expression of cyclins D1, D2 or D3 induced cell cycle activity in ventricular cardiomyocytes in uninjured hearts from transgenic mice.21,39 Interestingly, cardiomyocyte cell cycle activity persisted following myocardial infarction in mice which expressed cyclin D2 (but not cyclins D1 or D3), and was accompanied by a reduction in infarct size.21
In the current study, cyclin D2-induced right atrial cardiomyocyte cell cycle activity reduced the level of TGF-beta1 mediated fibrotic injury in mice inheriting both transgenes. Cardiomyocyte cell cycle activity was evidenced by tritiated thymidine incorporation. Although the absence of increased ploidy and multi-nucleation in MHC-cycD2 right atrial cardiomyocytes (see On Line Supplemental Data) suggested that cyclin D2-induced cardiomyocyte DNA synthesis was associated with cell division, the sensitivity of these analyses cannot preclude the possibility that the observed DNA synthesis contributed in part to multinucleation and/or endoreduplication. However, the observed increase in the number of cardiomyocytes per unit area right atrial tissue supports the notion that cell division occurred. Since transgene-mediated cyclin D2-induced cell cycle activation occurred independently of the transgene-mediated TGF-cys33 ser-induced fibrosis, the increase in cardiomyocyte cell number and concomitant reduction of atrial fibrosis in the right atria of MHC-TGF-cys33 ser / MHC-cycD2 double transgenic mice strongly supports the hypothesis that cell cycle activation can ameliorate structural damage to the heart.
The data presented here joins a growing number of studies in which cardiomyocyte cell cycle induction was observed in response to cardiac-restricted gene expression in transgenic mice.14,15 The results from the current study indicating that cell cycle induction ameliorated reparative fibrosis in the atrium complements and extends recent observations that cell cycle induction in the ventricle promotes favorable post-infarction remodeling31 and infarct regression.21 It is thus becoming apparent that cardiomyocyte cell cycle induction can promote myocardial repair in response to a variety of injuries. Successful therapeutic translation of these observations will likely require the development of small molecules that are able to mimic key aspects of transgene expression.
Acknowledgements
We thank D. Field for excellent technical assistance, and Drs. Mark Soonpaa, Michael Rubart, Pascal Lafontant, Wuqiang Zhu and Rethinasamy Prabhakar for comments on the manuscript. Supported by the NHLBI (LJF) and a Grant in Aid from the American Heart Association, Indiana Affiliate (HN).
References
- 1.Swynghedauw B. Molecular mechanisms of myocardial remodeling. Physiol Rev. 1999;79:215–262. doi: 10.1152/physrev.1999.79.1.215. [DOI] [PubMed] [Google Scholar]
- 2.Kostin S, Pool L, Elsasser A, Hein S, Drexler HC, Arnon E, Hayakawa Y, Zimmermann R, Bauer E, Klovekorn WP, Schaper J. Myocytes die by multiple mechanisms in failing human hearts. Circ Res. 2003;92:715–724. doi: 10.1161/01.RES.0000067471.95890.5C. [DOI] [PubMed] [Google Scholar]
- 3.Butt RP, Laurent GJ, Bishop JE. Collagen production and replication by cardiac fibroblasts is enhanced in response to diverse classes of growth factors. Eur J Cell Biol. 1995;68:330–335. [PubMed] [Google Scholar]
- 4.Boluyt MO, O'Neill L, Meredith AL, Bing OH, Brooks WW, Conrad CH, Crow MT, Lakatta EG. Alterations in cardiac gene expression during the transition from stable hypertrophy to heart failure. Marked upregulation of genes encoding extracellular matrix components. Circ Res. 1994;75:23–32. doi: 10.1161/01.res.75.1.23. [DOI] [PubMed] [Google Scholar]
- 5.Butt RP, Laurent GJ, Bishop JE. Mechanical load and polypeptide growth factors stimulate cardiac fibroblast activity. Ann N Y Acad Sci. 1995;752:387–393. doi: 10.1111/j.1749-6632.1995.tb17446.x. [DOI] [PubMed] [Google Scholar]
- 6.Aukrust P, Yndestad A, Damas JK, Gullestad L. Therapeutic potential of anticytokine therapy in congestive heart failure. Am J Cardiovasc Drugs. 2004;4:169–177. doi: 10.2165/00129784-200404030-00004. [DOI] [PubMed] [Google Scholar]
- 7.Remme WJ. Pharmacological modulation of cardiovascular remodeling: a guide to heart failure therapy. Cardiovasc Drugs Ther. 2003;17:349–360. doi: 10.1023/a:1027351808326. [DOI] [PubMed] [Google Scholar]
- 8.Siwik DA, Colucci WS. Regulation of matrix metalloproteinases by cytokines and reactive oxygen/nitrogen species in the myocardium. Heart Fail Rev. 2004;9:43–51. doi: 10.1023/B:HREV.0000011393.40674.13. [DOI] [PubMed] [Google Scholar]
- 9.Janicki JS, Brower GL, Gardner JD, Chancey AL, Stewart JA., Jr The dynamic interaction between matrix metalloproteinase activity and adverse myocardial remodeling. Heart Fail Rev. 2004;9:33–42. doi: 10.1023/B:HREV.0000011392.03037.7e. [DOI] [PubMed] [Google Scholar]
- 10.Heeneman S, Cleutjens JP, Faber BC, Creemers EE, van Suylen RJ, Lutgens E, Cleutjens KB, Daemen MJ. The dynamic extracellular matrix: intervention strategies during heart failure and atherosclerosis. J Pathol. 2003;200:516–525. doi: 10.1002/path.1395. [DOI] [PubMed] [Google Scholar]
- 11.Wencker D, Chandra M, Nguyen K, Miao W, Garantziotis S, Factor SM, Shirani J, Armstrong RC, Kitsis RN. A mechanistic role for cardiac myocyte apoptosis in heart failure. J Clin Invest. 2003;111:1497–1504. doi: 10.1172/JCI17664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crow MT, Mani K, Nam YJ, Kitsis RN. The mitochondrial death pathway and cardiac myocyte apoptosis. Circ Res. 2004;95:957–970. doi: 10.1161/01.RES.0000148632.35500.d9. [DOI] [PubMed] [Google Scholar]
- 13.Bock-Marquette I, Saxena A, White MD, Dimaio JM, Srivastava D. Thymosin beta4 activates integrin-linked kinase and promotes cardiac cell migration, survival and cardiac repair. Nature. 2004;432:466–472. doi: 10.1038/nature03000. [DOI] [PubMed] [Google Scholar]
- 14.Pasumarthi KB, Field LJ. Cardiomyocyte cell cycle regulation. Circ Res. 2002;90:1044–1054. doi: 10.1161/01.res.0000020201.44772.67. [DOI] [PubMed] [Google Scholar]
- 15.Dowell JD, Field LJ, Pasumarthi KB. Cell cycle regulation to repair the infarcted myocardium. Heart Fail Rev. 2003;8:293–303. doi: 10.1023/a:1024738104722. [DOI] [PubMed] [Google Scholar]
- 16.Dowell JD, Rubart M, Pasumarthi KB, Soonpaa MH, Field LJ. Myocyte and myogenic stem cell transplantation in the heart. Cardiovasc Res. 2003;58:336–350. doi: 10.1016/s0008-6363(03)00254-2. [DOI] [PubMed] [Google Scholar]
- 17.Dimmeler S, Zeiher AM, Schneider MD. Unchain my heart: the scientific foundations of cardiac repair. J Clin Invest. 2005;115:572–583. doi: 10.1172/JCI24283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassink RJ, Dowell JD, Brutel de la Riviere A, Doevendans PA, Field LJ. Stem cell therapy for ischemic heart disease. Trends Mol Med. 2003;9:436–441. doi: 10.1016/j.molmed.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Nakajima H, Nakajima HO, Salcher O, Dittie AS, Dembowsky K, Jing S, Field LJ. Atrial but not ventricular fibrosis in mice expressing a mutant transforming growth factor-beta(1) transgene in the heart. Circ Res. 2000;86:571–579. doi: 10.1161/01.res.86.5.571. [DOI] [PubMed] [Google Scholar]
- 20.Brunner AM, Marquardt H, Malacko AR, Lioubin MN, Purchio AF. Site-directed mutagenesis of cysteine residues in the pro region of the transforming growth factor beta 1 precursor. Expression and characterization of mutant proteins. J Biol Chem. 1989;264:13660–13664. [PubMed] [Google Scholar]
- 21.Pasumarthi KB, Nakajima H, Nakajima HO, Soonpaa MH, Field LJ. Targeted expression of cyclin D2 results in cardiomyocyte DNA synthesis and infarct regression in transgenic mice. Circ Res. 2005;96:110–118. doi: 10.1161/01.RES.0000152326.91223.4F. [DOI] [PubMed] [Google Scholar]
- 22.Soonpaa MH, Koh GY, Klug MG, Field LJ. Formation of nascent intercalated disks between grafted fetal cardiomyocytes and host myocardium. Science. 1994;264:98–101. doi: 10.1126/science.8140423. [DOI] [PubMed] [Google Scholar]
- 23.Luna LG, Gridley MF. Armed Forces Institute of Pathology (U.S.) Manual of histologic staining methods of the Armed Forces Institute of Pathology. 3rd ed. New York: Blakiston Division McGraw-Hill; 1968. [Google Scholar]
- 24.Bullock GR, Petrusz P. Techniques in immunocytochemistry. London; New York: Academic Press; 1982. [Google Scholar]
- 25.Soonpaa MH, Field LJ. Assessment of cardiomyocyte DNA synthesis in normal and injured adult mouse hearts. Am J Physiol. 1997;272:H220–H226. doi: 10.1152/ajpheart.1997.272.1.H220. [DOI] [PubMed] [Google Scholar]
- 26.Soonpaa MH, Kim KK, Pajak L, Franklin M, Field LJ. Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol. 1996;271:H2183–H2189. doi: 10.1152/ajpheart.1996.271.5.H2183. [DOI] [PubMed] [Google Scholar]
- 27.Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol. 1996;28:1737–1746. doi: 10.1006/jmcc.1996.0163. [DOI] [PubMed] [Google Scholar]
- 28.Clubb FJ, Jr, Bishop SP. Formation of binucleated myocardial cells in the neonatal rat. An index for growth hypertrophy. Lab Invest. 1984;50:571–577. [PubMed] [Google Scholar]
- 29.Wei Y, Mizzen CA, Cook RG, Gorovsky MA, Allis CD. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proc Natl Acad Sci U S A. 1998;95:7480–7484. doi: 10.1073/pnas.95.13.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rumiantsev PP. Growth and hyperplasia of cardiac muscle cells. London, U.K.; New York, N.Y., U.S.A.: Harwood Academic Publishers; 1991. [Google Scholar]
- 31.Nakajima H, Nakajima HO, Tsai SC, Field LJ. Expression of mutant p193 and p53 permits cardiomyocyte cell cycle reentry after myocardial infarction in transgenic mice. Circ Res. 2004;94:1606–1614. doi: 10.1161/01.RES.0000132279.99249.f4. [DOI] [PubMed] [Google Scholar]
- 32.Pasumarthi KB, Tsai SC, Field LJ. Coexpression of mutant p53 and p193 renders embryonic stem cell-derived cardiomyocytes responsive to the growth-promoting activities of adenoviral E1A. Circ Res. 2001;88:1004–1011. doi: 10.1161/hh1001.090878. [DOI] [PubMed] [Google Scholar]
- 33.Kardami E. Stimulation and inhibition of cardiac myocyte proliferation in vitro. Mol Cell Biochem. 1990;92:129–135. doi: 10.1007/BF00218130. [DOI] [PubMed] [Google Scholar]
- 34.Florini JR, Ewton DZ. Actions of anabolic hormones and growth factors on cultured neonatal heart cells. Growth Regul. 1995;5:28–35. [PubMed] [Google Scholar]
- 35.Charng MJ, Frenkel PA, Lin Q, Yamada M, Schwartz RJ, Olson EN, Overbeek P, Schneider MD. A constitutive mutation of ALK5 disrupts cardiac looping and morphogenesis in mice. Dev Biol. 1998;199:72–79. doi: 10.1006/dbio.1998.8905. [DOI] [PubMed] [Google Scholar]
- 36.Zhang D, Gaussin V, Taffet GE, Belaguli NS, Yamada M, Schwartz RJ, Michael LH, Overbeek PA, Schneider MD. TAK1 is activated in the myocardium after pressure overload and is sufficient to provoke heart failure in transgenic mice. Nat Med. 2000;6:556–563. doi: 10.1038/75037. [DOI] [PubMed] [Google Scholar]
- 37.Yang BC, Zander DS, Mehta JL. Hypoxia-reoxygenation-induced apoptosis in cultured adult rat myocytes and the protective effect of platelets and transforming growth factor-beta(1) J Pharmacol Exp Ther. 1999;291:733–738. [PubMed] [Google Scholar]
- 38.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 39.Soonpaa MH, Koh GY, Pajak L, Jing S, Wang H, Franklin MT, Kim KK, Field LJ. Cyclin D1 overexpression promotes cardiomyocyte DNA synthesis and multinucleation in transgenic mice. J Clin Invest. 1997;99:2644–2654. doi: 10.1172/JCI119453. [DOI] [PMC free article] [PubMed] [Google Scholar]