Abstract
Aims
Testing a relatively small genomic region with a few hundred SNPs provides limited information. Genome-wide association studies (GWAS) provide an opportunity to overcome the limitation of candidate gene association studies. Here, we report the results of a GWAS for the responses to an NSAID analgesic.
Materials & methods
European Americans (60 females and 52 males) undergoing oral surgery were genotyped with Affymetrix 500K SNP assay. Additional SNP genotyping was performed from the gene in linkage disequilibrium with the candidate SNP revealed by the GWAS.
Results
GWAS revealed a candidate SNP (rs2562456) associated with analgesic onset, which is in linkage disequilibrium with a gene encoding a zinc finger protein. Additional SNP genotyping of ZNF429 confirmed the association with analgesic onset in humans (p = 1.8 × 10−10, degrees of freedom = 103, F = 28.3). We also found candidate loci for the maximum post-operative pain rating (rs17122021, p = 6.9 × 10−7) and post-operative pain onset time (rs6693882, p = 2.1 × 10−6), however, correcting for multiple comparisons did not sustain these genetic associations.
Conclusion
GWAS for acute clinical pain followed by additional SNP genotyping of a neighboring gene suggests that genetic variations in or near the loci encoding DNA binding proteins play a role in the individual variations in responses to analgesic drugs.
Keywords: analgesic onset, GWAS, pain
Many serious diseases such as cancer, heart disease, AIDS and arthritis are often associated with unrelieved pain, despite major advances in the understanding of molecular mechanisms involved in pain and considerable pharmaceutical research and development in analgesia [1]. There are still few analgesic drug classes, all of which have safety problems, and as a consequence, pain related conditions frequently lack effective therapy and a wide variety of drugs are often used off-label for chronic pain in the absence of evidence of safety or efficacy for a pain indication [1]. Understanding the genetic basis of human variations in pain is critical for elucidating the molecular basis of pain sensitivity, variable responses to analgesic drugs and, eventually, to improved treatment of pain. The complexity of pain biology and the size of the human genome, combined with variable study designs, sample heterogeneity, small sample sizes, and phenotypic complexity, however, has resulted in inconsistent findings.
Individual variance in pain sensitivity and responses to analgesic drugs arise from a complex network of multiple gene polymorphisms and environmental factors. The contribution of each gene is likely to have a subtle effect on multiple mechanisms, making its signal difficult to detect. The relief of pain remains a largely unmet medical need with high prevalence of poorly controlled pain in various diseases and at the end of life. Understanding the role of genetics in pain and analgesic drug responses is critical to individualizing pain management, which can increase analgesic efficacy while reducing adverse drug reactions.
The discovery of genetic loci responsible for the individual variation in pain or responses to analgesics has proven challenging. Candidate gene association studies have identified a few genetic variations for pain but rarely have these associations been replicated by independent investigators [2–7]. Even if a polymorphism in a coding region does not result in an amino acid change, or if it is not in a coding sequence, it can still affect gene function by altering the stability, splicing or localization of the mRNA [8]. Noncoding RNAs are suggested to constitute a critical hidden layer of gene regulation in complex organisms [9]. Furthermore, current knowledge of human biology and the genome limits the list of candidate genes to investigate. Evaluating millions of SNPs across the whole genome can identify novel genes implicated in a complex phenotype like pain or responses to analgesic drugs. Genome-wide association studies (GWAS) can provide this information and overcome the limitation of candidate gene association studies as demonstrated for obesity [10], diabetes [11], rheumatoid arthritis [12], and behavioral traits [13].
Multiple copies of 1–50 kbp regions exist in the genome and are characterized as copynumber variation (CNV). This DNA variation involves segments that are smaller than those recognized microscopically, but larger than those that are readily detected by conventional sequence analysis. Hundreds of submicroscopic CNVs have been described in the human genome [14, 15]. Since many of the genetic differences between humans and other primates are a result of large duplications and deletions, it is reasonable to expect that differences in gene copy number (CN) could be a significant source of genetic variation between humans. Recent studies report that CNVs affect myocardial infarction and chemokine production [16–18]. Advanced technologies that enable us to perform whole-genome scan permits evaluation of both the qualitative aspect of genetic variation (SNP) and quantitative variation (CNV) and GWAS have been rapidly expanded in many complex traits.
Materials & methods
Subjects
The study was approved by the Institutional Review Board of the National Institute of Dental and Craniofacial Research (NIH, MD, USA) and informed consent was obtained from all subjects. Among 221 patients of mixed ethnic population undergoing oral surgery, only European Americans (60 females and 52 males), with ages ranging from 17 to 35 years, were analyzed in this study to minimize population stratification and potential bias caused by age differences. It is usually not recommended that bony impacted third molars be removed when patients are out of this age range. Patients underwent standardized surgery by the same oral surgeon removing third molar teeth that included at least one bony impacted mandibular third molar. After receiving premedication with intravenous midazolam (4.9 ± 0.2 mg) and local anesthesia with 2% lidocaine (250.6 ± 43.0 mg) with epinephrine 1:100,000, a mucoperiosteal flap was raised and retracted, bone removed, and the teeth were sectioned as needed to facilitate extraction of the impacted lower third molars.
Genotyping
For GWAS, genomic DNA extracted from white blood cells with the Puregene™ DNA isolation kit (Gentra Systems Inc., MN, USA) was hybridized to SNP chips following manufacturer’s instruction (Affymetrix, CA, USA). Quality control procedures including call rates and the Hardy–Weinberg Equilibrium test were performed as suggested [19], and 255,785 SNPs that passed quality control filters were analyzed with dependent variables. Briefly, quality control procedures excluded 99,330 SNPs with call rates greater than 0.85, as well as 26,437 SNPs not in Hardy–Weinberg equilibrium (p < 0.001) in the control sample, yielding 374,975 SNPs available for analysis. The primary analytic modality involved computation of likelihood ratios (degrees of freedom [df] = 1) for the best possible genotypic split (e.g., recessive or dominant models) for each SNP, with the constraint that a minimum of ten subjects populate each split group (thereby excluding monomorphic and very rare SNPs), yielding 255,785 SNP splits. For GWAS statistical analyses, we used the recursive partitioning approach based on F-statistics, using Interactive Tree Analysis of Helix Tree 6.2.0 software (Golden Helix, MT, USA). The detailed description for the approach is available in the Appendix.
Next, we sought to extend the GWAS finding by conducting common SNP associations in the candidate gene region based on linkage disequilibrium (LD) of the HapMap data. The gene encoding zinc finger protein 429 (ZNF429) is in an LD with the SNP suggested from GWAS analysis. Genotyping was performed with Assays-by-Design SNP Genotyping Products (Applied Biosystems, CA, USA). A total of 5 ng of DNA was mixed in a well containing 2.5 µl of Taqman® universal master mix (Applied Biosystems) 0.25 µl of genotyping assay mix and 2.25 µl of DNAse free water. PCR was performed under the following conditions: 95°C, 10 min followed by 40 cycles of 92°C, 15 s and 60°C, 1 min in a Perkin–Elmer™ 9700 thermocycler (Perkin–Elmer Inc., MA, USA). Following PCR, fluorescence of each well was measured using the ABI Prism 7900 Sequence Detection System (Applied Biosystems). Genotype discrimination was performed using Taqman Sequence Detector version 2.0 software. Samples that failed to amplify were not included in the final analysis.
Clinical pain measurement
Clinically induced pain was recorded with a paper and pencil form of a 100 mm visual analog scale (VAS). After the extraction of the impacted third molars, pain was recorded every 20 min by VAS until subjects requested analgesic medication as the local anesthesia was eliminated and post-operative pain onset occurred. Ketorolac tromethamine (Toradol) was administered intravenously at the recommended dose (30 mg) and pain was recorded by VAS again at 15 min intervals for 180 min. When subjects requested analgesic medication as the local anesthesia dissipated (mean = 125 min; 95% confidence interval [CI]: 117–132 min) and post-operative pain (VAS, mean = 58.4 mm; 95% CI: 54.9–61.9) onset occurred. The maximum post-operative pain rating, post-operative pain onset time and the analgesic onset time after ketorolac administration were used as measures of clinical pain and the onset of nonsteroidal anti-inflammatory drug (NSAID) analgesia.
Data analysis
A total of 11 SNPs from the ZNF429 in an LD with the most statistically significant SNP in Helix Tree results were analyzed. To resolve phase unknown genotypes and estimate population frequencies in unrelated individuals we employed PHASE method, a probability based Bayesian algorithm. To quantify LD (allelic association between marker and genes), widely used D′ and r2 were calculated. A value of zero implies independence, whereas 1.0 means complete co-transfer. Haploblocks based on CI method were generated by Haploview version 3.11 with identical genomic region of HapMap.
For the CNV analysis, Partek GS version 6.2 (Partek, MO, USA) was used to compare CNs between slow analgesic onset (≥ 30 min, n = 7) and rapid analgesic onset (≤ 5 min, n = 33) patients. According to the manufacturer’s recommendation for generating baselines for CN analysis (n > 25), rapid analgesic onset group was defined as 5 min or less. Slow analgesic onset was defined conversely as 5% of the patients with the slowest reported onset of analgesia.
Results
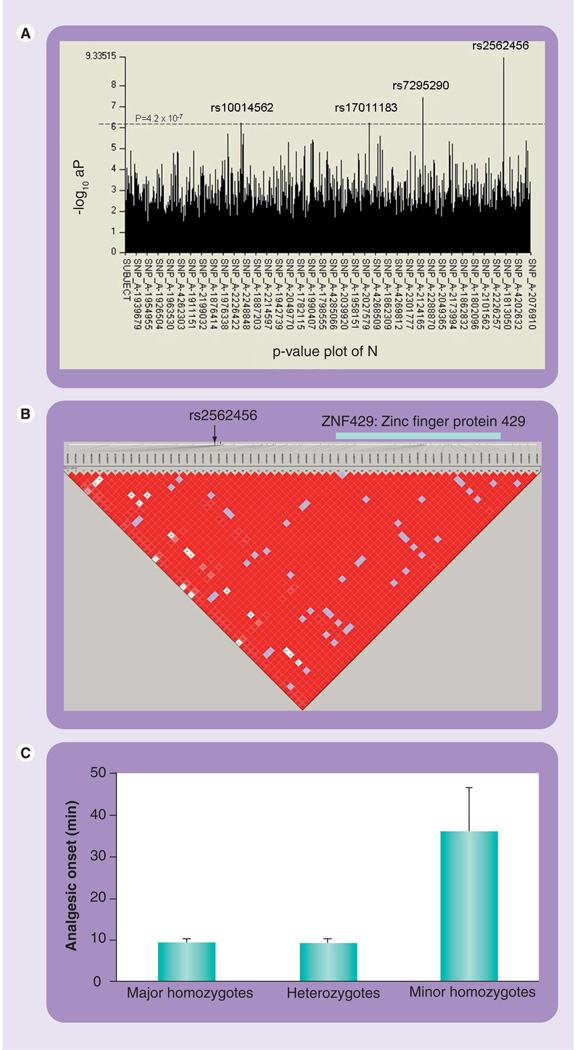
Genome-wide association study analysis revealed a significant association (p < 3.3 × 10−8, p-value threshold was conservatively calculated for 500,000 SNPs × 3 tests from Bonferroni multiple test correction) with analgesic onset after ketorolac administration with Helix Tree analysis (FIGURE 1A). This revealed that a SNP (rs2562456, p = 2.3 × 10−10) of an unknown gene LOC400680, which is not annotated in current National Center for Biotechnology Information (NCBI) assembly, is significantly associated with analgesic onset at the false-discovery rate (FDR) of 1.18 × 10−4. Haploview analysis combining with the HapMap identified that this SNP is in an LD with SNPs in ZNF429 (FIGURE 1B). Additional 11 SNPs of ZNF429 region (TABLE 1, NT_011295.10) including four tag SNPs suggested by HapMap data were genotyped. It revealed that the D′ among these 11 SNPs are 1.0 (TABLE 2) and four SNPs are significantly associated with analgesic onset (rs2650825, rs1879234, rs2562408, rs2562466). Homozygous minor allele subjects of these SNPs show approximately four-times slower analgesic onset (n = 9, mean = 36.2 min; 95% CI: 13.3–59.2) compared with major homozygous (n = 61, mean = 9.4 min; 95% CI: 7.8–11.0) and heterozygotes (n = 34, mean = 9.0 min; 95% CI: 6.9–11.1) with analysis-of-variance, p = 1.8 × 10−10, df = 103, F = 28.3 (FIGURE 1C). Strong effects on analgesic onset (p = 3.5 × 10−8 to p = 2.7 × 10−7) in three different novel regions across the human genome were also detected, though Bonferroni multiple test corrections denied the significant associations for these borderline SNPs: rs7295290 (p = 3.5 × 10−8 at FDR = 4.51 × 10−3, rs10014562 (p = 2.6 × 10−7 at FDR = 2.93 × 10−2) and rs17011183 (p = 2.7 × 10−7 at FDR = 2.48 × 10−2) (FIGURE 1A).
Figure 1. Multilayer genetic association study.
(A) Plot of statistical significance (−log10P) values of genotyped SNPs across the whole genome for analgesic onset time after ketorolac administration. Reference line indicates threshold for genome wide significance (p = 4.2 × 10−7). Horizontal axis represent reference SNPs for the chromosomal location. (B) A single haploblock from HapMap containing significant associated SNPs and ZNF429. (C) Associations between four ZNF429 SNPs and analgesic onset (p = 1.8 × 10−10, degrees of freedom = 103, F = 28.3, mean ± standard error).
Table 1.
SNPs genotyped from ZNF429.
| SNP | SNP ID | Location on NCBI assembly | Nucleotide variation | Rarer allele frequency |
|---|---|---|---|---|
| 1 | rs1984771 | 21,483,637 | A/G | 0.17 |
| 2 | rs2650825 | 21,491,065 | A/G | 0.25 |
| 3 | rs1520071 | 21,493,880 | G/T | 0.17 |
| 4 | rs2650757 | 21,495,385 | A/G | 0.17 |
| 5 | rs2681365 | 21,495,804 | A/G | 0.17 |
| 6 | rs1879234 | 21,496,319 | G/T | 0.25 |
| 7 | rs11085455 | 21,496,885 | C/T | 0.17 |
| 8 | rs2173724 | 21,500,997 | A/G | 0.17 |
| 9 | rs2133816 | 21,501,221 | C/T | 0.17 |
| 10 | rs2562408 | 21,501,721 | C/G | 0.25 |
| 11 | rs2562466 | 21,506,044 | C/T | 0.25 |
NCBI: National Center for Biotechnology Information.
Table 2.
Linkage disequilibrium of SNPs genotyped from ZNF429.
| L1 | L2 | D′ | LOD | r2 | Cllow | Clhi | Dist | T-int |
|---|---|---|---|---|---|---|---|---|
| rs1984771 | rs2650825 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 7428 | 115.55 |
| rs1520071 | 1.0 | 35.53 | 1.0 | 0.94 | 1.0 | 10243 | – | |
| rs2650757 | 1.0 | 35.53 | 1.0 | 0.94 | 1.0 | 11748 | – | |
| rs2681365 | 1.0 | 35.53 | 1.0 | 0.94 | 1.0 | 12167 | – | |
| rs1879234 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 12682 | – | |
| rs11085455 | 1.0 | 30.89 | 0.921 | 0.91 | 1.0 | 13248 | – | |
| rs2173724 | 1.0 | 35.53 | 1.0 | 0.94 | 1.0 | 17360 | – | |
| rs2133816 | 1.0 | 35.53 | 1.0 | 0.94 | 1.0 | 17584 | – | |
| rs2562408 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 18084 | – | |
| rs2562466 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 22407 | – | |
| rs1520071 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 2815 | 205.86 | |
| rs2650825 | rs2650757 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 4320 | – |
| rs2681365 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 4739 | – | |
| rs1879234 | 1.0 | 46.31 | 1.0 | 0.95 | 1.0 | 5254 | – | |
| rs11085455 | 1.0 | 4.15 | 0.065 | 0.64 | 1.0 | 5820 | – | |
| rs2173724 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 9932 | – | |
| rs2133816 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 10156 | – | |
| rs2562408 | 1.0 | 46.31 | 1.0 | 0.95 | 1.0 | 10656 | – | |
| rs2562466 | 1.0 | 46.31 | 1.0 | 0.95 | 1.0 | 14979 | – | |
| rs1520071 | rs2650757 | 1.0 | 35.53 | 1.0 | 0.94 | 1.0 | 1505 | 347.82 |
| rs2681365 | 1.0 | 35.53 | 1.0 | 0.94 | 1.0 | 1924 | – | |
| rs1879234 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 2439 | – | |
| rs11085455 | 1.0 | 30.89 | 0.921 | 0.91 | 1.0 | 3005 | – | |
| rs2173724 | 1.0 | 35.53 | 1.0 | 0.94 | 1.0 | 7117 | – | |
| rs2133816 | 1.0 | 35.53 | 1.0 | 0.94 | 1.0 | 7341 | – | |
| rs2562408 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 7841 | – | |
| rs2562466 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 12164 | – | |
| rs2650757 | rs2681365 | 1.0 | 35.53 | 1.0 | 0.94 | 1.0 | 419 | 489.78 |
| rs1879234 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 934 | – | |
| rs11085455 | 1.0 | 30.89 | 0.921 | 0.91 | 1.0 | 1500 | – | |
| rs2173724 | 1.0 | 35.53 | 1.0 | 0.94 | 1.0 | 5612 | – | |
| rs2133816 | 1.0 | 35.53 | 1.0 | 0.94 | 1.0 | 5836 | – | |
| rs2562408 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 6336 | – | |
| rs2562466 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 10659 | – | |
| rs2681365 | rs1879234 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 515 | 549.37 |
| rs11085455 | 1.0 | 30.89 | 0.921 | 0.91 | 1.0 | 1081 | – | |
| rs2173724 | 1.0 | 35.53 | 1.0 | 0.94 | 1.0 | 5193 | – | |
| rs2133816 | 1.0 | 35.53 | 1.0 | 0.94 | 1.0 | 5417 | – | |
| rs2562408 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 5917 | – | |
| rs2562466 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 10240 | – | |
| rs1879234 | rs11085455 | 1.0 | 4.15 | 0.065 | 0.64 | 1.0 | 566 | 544.19 |
| rs2173724 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 4678 | – | |
| rs2133816 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 4902 | – | |
| rs2562408 | 1.0 | 46.31 | 1.0 | 0.95 | 1.0 | 5402 | – | |
| rs2562466 | 1.0 | 46.31 | 1.0 | 0.95 | 1.0 | 9725 | – | |
| rs11085455 | rs2173724 | 1.0 | 30.89 | 0.921 | 0.91 | 1.0 | 4112 | 411.72 |
| rs2133816 | 1.0 | 30.89 | 0.921 | 0.91 | 1.0 | 4336 | – | |
| rs2562408 | 1.0 | 4.15 | 0.065 | 0.64 | 1.0 | 4836 | – | |
| rs2562466 | 1.0 | 4.15 | 0.065 | 0.64 | 1.0 | 9159 | – | |
| rs2173724 | rs2133816 | 1.0 | 35.53 | 1.0 | 0.94 | 1.0 | 224 | 269.76 |
| rs2562408 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 724 | – | |
| rs2562466 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 5047 | – | |
| rs2133816 | rs2562408 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 500 | 127.8 |
| rs2562466 | 1.0 | 4.48 | 0.071 | 0.67 | 1.0 | 4823 | – | |
| rs2562408 | rs2562466 | 1.0 | 46.31 | 1.0 | 0.95 | 1.0 | 4323 | 105.73 |
L1: Loci 1; L2: Loci 2; LOD: Log of the likelihood odds ratio, a measure of confidence in the value of D′; CIlow: 95% confidence lower bound on D′; CIhi: 95% confidence upper bound on D′; Dist: Distance (in bases) between loci; T-int: Statistic used by the HapMap Project to measure the completeness of information represented by a set of markers in a region. It measures linkage disequilibrium information content in the gaps between markers.
Comparison of CN across the whole genome revealed that patients with slow analgesic onset (≥30 min, n = 7, CN mean = 3.78; 95% CI: 2.3–5.3) have higher CNs than those with rapid analgesic onset (≤5 min, n = 33, CN mean = 2.06; 95% CI: 1.9–2.2) at SNP rs17011183 (p = 4.5 × 10−6, df = 38, F = 41.4), at the same SNP showed strong association above, but this effect was not significant with multiple test correction.
We also found candidate loci for the maximum post-operative pain rating (rs17122021, p = 6.9 × 10−7) and post-operative pain onset time (rs6693882, p = 2.1 × 10−6), however, correcting for multiple comparisons did not sustain these genetic associations.
Discussion
Even after the multiple test corrections, the SNP (rs2562456) located in chromosome 19 showed a significant association with the onset time of analgesia after ketorolac administration. The annotation of the genomic region around this SNP is not available yet, though it is included in the unknown gene LOC400680; the potential function of this hypothetical gene is not known at present. However, the additionally genotyped 11 SNPs neighboring around this region showed high LD values across approximately 19 kbp (data not shown). According to HapMap, this high LD extends up to 88 kbp size of a single haploblock (from rs8107940 to rs2681384, based on 95% CI) including a gene relevant to DNA binding protein: ZNF429. This gene spans approximately 32.3 kbp in the human genome with five exons and the size of mRNA is approximately 1.9 kbp. More than 20 SNPs whose heterozygosity is greater than 0.2, have been reported across the entire gene. Zinc finger proteins control the processes that modulate the frequency, rate or extent of DNA dependent transcription by binding to DNA. Except for this general function as a zinc finger protein, specific biological function of ZNF429 in analgesic drug metabolism and pain pathways has not been reported yet.
Further investigation of ZNF429 including the role of frequent SNPs in this gene revealed that genetic polymorphisms in this region are strongly associated with NSAID analgesia. We could screen SNPs from introns only because few numbers of SNPs with low minor allele frequency have been reported so far in ZNF429. It is not surprising to confirm that ZNF429 is in a strong LD as HapMap data already suggest in Caucasians. However, the GWAS missed the associations of ZNF429 SNPs with analgesic onset because we filtered out SNPs with less than ten subjects as the minimum number for one group. The number of minor homozygotes of ZNF429 was nine. It is clearly demonstrated that dense genotyping around the candidate region can reduce the risk of missing important associations.
We later analyzed analgesic drug efficacy and found that the slower analgesic onset homozygotes tend to report less analgesic effectiveness (p = 0.017, 77% pain reduction, 95% CI: 62–92%) compared with heterozygotes (89% pain reduction, 95% CI: 84–94%) and major homozygotes (90% pain reduction, 95% CI: 87–93%). Even though the statistical significance did not sustain with multiple test correction and virtually all subjects reported significant pain reduction (t-test, p = 1.75 × 10−12, VAS, mean = 23.7 mm; 95% CI: 20.3–27.2), it is noticeable that slower analgesic onset patients may also have less benefit from the analgesic drugs in terms of efficacy.
Considering overly conservative Bonferroni correction, a modified Bayesian formula with recent estimates of the total number of human genes as 20,000 has been suggested and results in a p-value threshold of approximately 4.2 × 10−7 [19]. This approach introduces three additional SNPs as candidate regions for association with analgesic onset. One of these is in chromosome 4 but this region is not annotated yet. The remaining two are:
rs7295290 located in a gene encoding ankyrin repeat domain 13A in chromosome 12. Ankyrin repeats mediate protein–protein interaction in very diverse families of proteins.
rs17011183 located in a gene encoding WDF family member 4 (WDFY4) in chromosome 10, whose functions is thought to be related to transporter activity. This was formerly known as chromosome 10 open reading frame 64 (C10orf64).
Even though we found marginal associations between these additional genetic variations and analgesic onset after ketorolac administration, it is still not clear how these genes or genetic variations influence analgesic responses. Genomic regions around those SNPs were not further investigated in this study because they did not overcome the Bonferroni multiple test corrections. However, this marginal but strong association deserves future investigation in analgesic drug responses. Overall, further investigation is needed initially to characterize the function of those genes both in animals and humans.
We did not find statistically significant association between CNVs across human genome and NSAID analgesia. SNP rs17011183 in the WDFY4 gene showed a marginal association between analgesic onset and its CNV. Considering the marginal association between analgesic onset and the genotypes of this SNP (FIGURE 1A, p = 2.7 × 10−7), genomic region of the WDFY4 gene including rs17011183 may contribute both qualitatively and quantitatively to analgesic drug responses.
Unlike NSAID analgesic onset, only weak associations were found in maximum post-operative pain rating and post-operative pain onset time but those associations were not significant with multiple test corrections. Post-operative pain is also influenced by psychological and environmental factors. It is more likely that genetic factors exert only subtle effects in more complex phenotypes. Our results imply that pharmacogenomics of analgesic responses may be a more feasible target for genetics of pain studies. Considering these weak associations for acute inflammatory pain in the well-characterized oral surgery model [20–23], finding loci responsible for multifactorial chronic pain may be very challenging. Even for a clearly heritable trait such as height, the first genetic locus reported only explains 0.3% of individual differences [24].
Whereas our results implicate a significant role of genetic factors in the responses to analgesic drugs, this study is not free from some limitation. Most of all, the sample size is relatively small for a GWAS since the sample size required is increased by the large number of hypotheses that are tested. Because we genotyped 500K SNPs from collected samples retrospectively, the sample size was not based on power analysis. We did not have required baseline data for power analysis either since GWAS related to analgesic drug efficacy has never been reported, to the best of our knowledge. Our result should be considered as preliminary because of this limitation, though our results can be used in power analysis for the future GWAS of analgesic drug responses with a larger scale of various ethnic populations. It should be also considered that current genotyping platforms represent only about two-thirds of all known common genetic variations throughout the human genome [25]. These limitations may increase the risk of false discovery. Furthermore, our results are limited to European American population only and cannot be generalized to other ethnic populations since pain responses including analgesic efficacy and genetic variations are very different among ethnic populations [26–29]. As with other genetic association studies, replications with larger sample sizes with various ethnic backgrounds by independent investigators are critical to confirm novel findings. Since the genetic association studies can only identify statistical relationships, studies are needed to characterize the underlying mechanisms. When the candidate genetic loci lack annotation, extensive additional work is needed in both animals and humans.
Even though the sample size is relatively small for a GWAS, the genome-wide significant result of this study appears robust. This study suggests that genetic variations in or near the loci encoding DNA binding proteins play a role in the individual variations in responses to analgesic drugs. These observations also suggest that inter-individual variability in analgesic drug response may be induced by variations in genes that modulate transcription in addition to genes that directly affect pharmacodynamics or pharmacokinetics.
Conclusion
A GWAS of acute clinical pain followed by additional SNP genotyping of neighboring gene suggests that genetic variations in or near the loci encoding DNA binding proteins play a role in the individual variations in responses to analgesic drugs.
Future perspective
Even with the relatively small sample size, this study provides evidence that whole-genome scale SNP genotyping is a valuable tool for hypothesis generating when searching for novel candidate genomic regions responsible for pain and analgesia in humans. It is necessary to confirm these associations with independent subjects in a bigger sample size as well as with functional genomic studies.
Executive summary.
Genome-wide association study (GWAS) of analgesic drug response
GWAS found one significant SNP associated with analgesic onset.
Three other SNPs of strong association were found but denied by Bonferroni multiple test correction.
Multilayer study
Using HapMap data, additional SNPs were genotyped in a gene with linkage disequilibrium.
This multilayer design successfully found a candidate gene, ZNF429.
ZNF429
ZNF429 is a zinc finger protein that can influence the expression of various types of genes.
Acknowledgments
This study was supported by the Division of Intramural Research, National Institute of Nursing Research and National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD 20892, USA.
Appendix
The p-value, which is labeled ‘p =’ in a tree node display, can be calculated as follows.
Suppose there are n observations to be split into k subgroups with D unique values of the continuous or ordinal predictor (not counting a possible missing value). Let F0 be the sum of squared deviations from the mean over all responses and Fk be the sum over all segments of the squared deviations from the mean responses of each respective segment. Then F-statistic can be defined as:
The p-value can be obtained from performing an F-test with k-1 and n-k degrees of freedom. The optimal set of segments is found with the smallest p-value and such sets are constructed by recursive partitioning.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Woodcock J, Witter J, Dionne RA. Stimulating the development of mechanism-based, individualized pain therapies. Nat. Rev. Drug Discov. 2007;6(9):703–710. doi: 10.1038/nrd2335. [DOI] [PubMed] [Google Scholar]
- 2. Zubieta JK, Heitzeg MM, Smith YR, et al. COMT val158met genotype affects µ-opioid neurotransmitter responses to a pain stressor. Science. 2003;299(5610):1240–1243. doi: 10.1126/science.1078546. ▪ First report of association between COMT and acute experimental pain.
- 3.Klepstad P, Rakvag TT, Kaasa S, et al. The 118 A > G polymorphism in the human µ-opioid receptor gene may increase morphine requirements in patients with pain caused by malignant disease. Acta Anaesthesiol. Scand. 2004;48(10):1232–1239. doi: 10.1111/j.1399-6576.2004.00517.x. [DOI] [PubMed] [Google Scholar]
- 4.Janicki PK, Schuler G, Francis D, et al. A genetic association study of the functional A118G polymorphism of the human µ-opioid receptor gene in patients with acute and chronic pain. Anesth. Analg. 2006;103(4):1011–1017. doi: 10.1213/01.ane.0000231634.20341.88. [DOI] [PubMed] [Google Scholar]
- 5. Tegeder I, Costigan M, Griffn RS, et al. GTP cyclohydrolase and tetrahydrobiopterin regulate pain sensitivity and persistence. Nat. Med. 2006;12(11):1269–1277. doi: 10.1038/nm1490. ▪ Reported the role of GCH1 genetic variations in acute experimental and chronic clinical pains from multiple clinics. However, population stratification of human samples may cause false-positive findings.
- 6. Kim H, Dionne RA. Lack of influence of GTP cyclohydrolase gene (GCH1) variations on pain sensitivity in humans. Mol. Pain. 2007;3:6. doi: 10.1186/1744-8069-3-6. ▪ Reported failure to repeat the findings of Tegeder et al. [5] in larger samples.
- 7. Reyes-Gibby CC, Shete S, Rakvag T, et al. Exploring joint effects of genes and the clinical efficacy of morphine for cancer pain: OPRM1 and COMT gene. Pain. 2007;130(1–2):25–30. doi: 10.1016/j.pain.2006.10.023. ▪ First report of joining effect of multiple genetic variations. However, the result shows opposite effect of COMT compared to Zubieta et al. [2].
- 8.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3(4):285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 9.Mattick JS. The functional genomics of noncoding RNA. Science. 2005;309(5740):1527–1528. doi: 10.1126/science.1117806. [DOI] [PubMed] [Google Scholar]
- 10.Scuteri A, Sanna S, Chen WM, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(7):E115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salonen JT, Uimari P, Aalto JM, et al. Type 2 diabetes whole-genome association study in four populations: the DiaGen consortium. Am. J. Hum. Genet. 2007;81(2):338–345. doi: 10.1086/520599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plenge RM, Seielstad M, Padyukov L, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis – a genomewide study. N. Engl. J. Med. 2007;357(12):1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shifman S, Bhomra A, Smiley S, et al. A whole genome association study of neuroticism using DNA pooling. Mol. Psychiatry. 2007;13(3):302–321. doi: 10.1038/sj.mp.4002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444(7118):444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat. Rev. Genet. 2006;7(2):85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez E, Kulkarni H, Bolivar H, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307(5714):1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 17.Matsushima S, Ide T, Yamato M, et al. Overexpression of mitochondrial peroxiredoxin-3 prevents left ventricular remodeling and failure after myocardial infarction in mice. Circulation. 2006;113(14):1779–1786. doi: 10.1161/CIRCULATIONAHA.105.582239. [DOI] [PubMed] [Google Scholar]
- 18.Ikeuchi M, Matsusaka H, Kang D, et al. Overexpression of mitochondrial transcription factor A ameliorates mitochondrial deficiencies and cardiac failure after myocardial infarction. Circulation. 2005;112(5):683–690. doi: 10.1161/CIRCULATIONAHA.104.524835. [DOI] [PubMed] [Google Scholar]
- 19.Lencz T, Morgan TV, Athanasiou M, et al. Converging evidence for a pseudoautosomal cytokine receptor gene locus in schizophrenia. Mol. Psychiatry. 2007;12(6):572–580. doi: 10.1038/sj.mp.4001983. [DOI] [PubMed] [Google Scholar]
- 20.Hargreaves K, Dionne R. Evaluating endogenous mediators of pain and analgesia in clinical studies. In: Max M, Portenoy R, Laska E, editors. Advances in Pain Research and Therapy. Volume 18. New York, NY, USA: Raven Press; 1991. pp. 579–598. [Google Scholar]
- 21.Gordon SM, Brahim JS, Rowan J, Kent A, Dionne RA. Peripheral prostanoid levels and nonsteroidal anti-inflammatory drug analgesia: replicate clinical trials in a tissue injury model. Clin. Pharmacol. Ther. 2002;72(2):175–183. doi: 10.1067/mcp.2002.126501. [DOI] [PubMed] [Google Scholar]
- 22.Cooper SA, Beaver WT. A model to evaluate mild analgesics in oral surgery outpatients. Clin. Pharmacol. Ther. 1976;20(2):241–250. doi: 10.1002/cpt1976202241. [DOI] [PubMed] [Google Scholar]
- 23.Hargreaves KM, Schmidt EA, Mueller GP, Dionne RA. Dexamethasone alters plasma levels of β-endorphin and postoperative pain. Clin. Pharmacol. Ther. 1987;42(6):601–607. doi: 10.1038/clpt.1987.206. [DOI] [PubMed] [Google Scholar]
- 24.Weedon MN, Lettre G, Freathy RM, et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat. Genet. 2007;39(10):1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pe’er I, de Bakker PI, Maller J, et al. Evaluating and improving power in whole-genome association studies using fixed marker sets. Nat. Genet. 2006;38(6):663–667. doi: 10.1038/ng1816. ▪ Emphasizes that even in the highly heritable trait such as height, mostly significant SNP from GWAS can only explain small portion of individual differences.
- 26.Edwards C, Fillingim R, Keefe F. Race, ethnicity and pain. Pain. 2001;94(2):133–137. doi: 10.1016/S0304-3959(01)00408-0. [DOI] [PubMed] [Google Scholar]
- 27.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 28.Green CR, Ndao-Brumblay SK, Nagrant AM, Baker TA, Rothman E. Race, age, and gender influences among clusters of African American and white patients with chronic pain. J. Pain. 2004;5(3):171–182. doi: 10.1016/j.jpain.2004.02.227. [DOI] [PubMed] [Google Scholar]
- 29.Kim H, Dionne RA. Comment on Diatchenko et al. Catechol-O-methyltransferase gene polymorphisms are associated with multiple pain-evoking stimuli. Pain. 2007;129(3):365–366. doi: 10.1016/j.pain.2007.02.011. author reply 366–370. [DOI] [PubMed] [Google Scholar]