Abstract
Of all the proposed causes of ageing, DNA damage remains a leading, though still debated theory. Unlike most other types of age-related cellular damage, which can hypothetically be reversed, mutations in DNA are permanent. Such errors result in the accumulation of changes to RNA and protein sequences with age, and are tightly linked to cellular senescence and overall organ dysfunction. Over the past few years, an additional, more global role has emerged for the contribution of DNA damage and genomic instability to the ageing process. We, and others have found that DNA damage and the concomitant repair process can induce genome-wide epigenetic changes, which may promote a variety of age-related transcriptional and functional changes. Here, we discuss the link between DNA damage, chromatin alterations and ageing, an interplay that explains how seemingly random DNA damage could manifest in predictable phenotypic changes that define ageing, changes that may ultimately be reversible.
Keywords: DNA damage, chromatin, sirtuin, DNA repair, ageing
Introduction
Ageing affects most eukaryotes, yet it remains unclear whether the causes of ageing are conserved. At a first glance, ageing in lower organisms seems to have little in common with ageing in mammals, yet – from an evolutionary perspective – one might not expect conservation given that ageing is not subject to natural selection. However, most life-forms share common weak spots that become increasingly susceptible to failure over time. One such “Achilles’ Heel” is the genome, a fragile and highly conserved structure that accumulates a wide range of damaging alterations with age, despite continuous surveillance and repair (Garinis et al. 2008; Lombard et al. 2005; Vijg 2004). Recent work extends the impact of genomic defects to an age-associated deregulation of the epigenome (reviewed in (Oberdoerffer and Sinclair 2007)), suggesting that the accumulation of DNA damage and genomic instability with age may be a critical contributor to the ageing process, though perhaps in a more indirect and complex way than first proposed.
The accrual of genomic defects can affect cellular function on many levels. For example, mutations in coding regions of DNA can cause abnormal protein expression or function, and chromosomal translocations and rearrangements can result in apoptosis, tumor formation or senescence (Campisi 2005). DNA damage and its repair have also been linked to wide-ranging chromatin alterations that surround the sites of damage and may affect a large number of genomic loci, including coding regions and structural components (Downs et al. 2007).
Like mutations, epigenomic changes to chromatin are a conserved hallmark of ageing (Oberdoerffer and Sinclair 2007). A major difference, though, is that epigenetic changes are theoretically reversible. This is due to the fluid nature of chromatin, a complex packaging system, in which DNA is wrapped around a protein core of four different histone dimers, forming the basic building blocks of chromatin called nucleosomes. This highly dynamic form of nuclear organization influences both DNA stability and gene-expression patterns (Cheutin et al. 2003; Grewal and Jia 2007) and its level of compaction can be modulated through a variety of reversible chemical modifications of histones or modifications of DNA itself (Kouzarides 2007). Amongst the most prominent posttranslational modifications are histone acetylation and histone or DNA methylation. The enzymes that catalyze those changes are comprehensively referred to as chromatin modifiers. Histone acetylation renders chromatin accessible for transcriptional regulators and DNA binding factors, whereas histone and DNA methylation have the opposite effect, although certain types of histone methylation are linked to active transcription (Kouzarides 2007). Highly compacted, transcriptionally silent chromatin is generally referred to as “heterochromatin”, whereas the more accessible chromatin is “euchromatin”.
The potential of DNA damage to affect cell function both through direct alterations to the DNA sequence and through indirect, epigenetic changes in chromatin structure puts it at a critical position to influence the ageing of eukaryotes. In this review we will highlight recent progress in both fields, focusing on newly discovered links between chromatin, genome instability and ageing.
Links between genomic intability and ageing
The machinery that keeps our genome stable has to counteract a wide range of potential threats, including replication errors, DNA repeat instability, telomere shortening and double-strand breaks (DSBs). DSBs are a natural aspect of lymphocyte development, but can also occur as a consequence of DNA replication and repair (Finkel et al. 2007; Lombard et al. 2005). The cell must also contend with chemical damaging agents, with reactive oxygen species (ROS) being the major source of DNA damage during physiological ageing (Bohr 2002; Bokov et al. 2004; Lu and Finkel 2008). ROS include superoxide anions, hydrogen peroxide and hydroxyl radicals, the most reactive of ROS species, and are mostly generated from within the cell as a byproduct of mitochondrial respiration, although ionizing radiation can also result in increased ROS levels (Bohr 2002).
Consistent with a role for DNA damage in ageing, a variety of specialized DNA repair pathways have been linked to the ageing process and have been reviewed extensively (see references below). A variety of distinct pathways ensure the removal of damaged bases or base adducts and the repair of the resulting single-stranded (ss) DNA lesions as well as the repair of DNA double-strand breaks (DSBs). The former is generally mediated by base excision repair (BER) or nucleotide excision repair (NER), the latter by non-homologous end joining (NHEJ) or homologous recombination (HR) (Li et al. 2008; Lombard et al. 2005; Schumacher et al. 2008a). BER and NER represent multi-step processes during which a variety of repair factors recognize, excise and “patch up” sites of damaged bases or nucleotides, respectively (Maynard et al. 2009; Schumacher et al. 2008a). NER is further divided into two subsets, global genome (GG) NER, and transcription-coupled (TC) NER. While defects in GG-NER generally result in DNA mutations, defects in TC-NER can cause cell death as a result of stalled RNA polymerase II (Garinis et al. 2009; Niedernhofer et al. 2006). Consequently, defects in GG-NER are often linked to cancer, whereas defects in TC-NER underlie a variety of progeroid syndromes, the most prominent examples include mutations in the TC-NER factors Cockayne Syndrome A and B (CSA/B) and a subset of Xeroderma pigmentosa complementation groups, such as XPB, XPD and XPF. The function of these enzymes ranges from assembly of the repair machinery upon RNA polymerase II stalling to nucleotide excision (Schumacher et al. 2008a).
With the exception of lesions in promoter regions, ssDNA lesions are generally compatible with cell division and survival even in the absence of appropriate repair (Garinis et al. 2008; Garinis et al. 2009). DSBs, on the other hand, are incompatible with DNA replication and lead to cell cycle arrest or death when unrepaired. Breaks that occur before replication (i.e. during G1 or in postmitotic cells) are generally repaired by NHEJ, during which the broken chromosome ends are directly fused. Breaks that are generated during or after DNA replication (i.e. in S phase or G2) employ the sister chromatid as a template for repair by HR (Branzei and Foiani 2008; Sung and Klein 2006). Consequently, NHEJ is error-prone and often results in small deletions or insertions, while HR is generally error-free. Recognition of the break site involves the Ku70/Ku80 heterodimer during NHEJ and the Mre11/Nbs1/Rad50 (MRN) complex during HR, although HR components can also participate in direct rejoining of broken DNA ends (Riballo et al. 2004). Break recognition triggers the activation of Ataxia Telangiectasia Mutated (ATM) and Ataxia Telangiectasia and Rad3-related (ATR) kinases during HR and DNA-PK catalytic subunit (DNA-PKcs) during NHEJ. These kinases then initiate the downstream repair process, which involves initiation of the DNA damage signaling cascade through phosphorylation of H2AX and other downstream targets including checkpoint kinases and, in the case of NHEJ, the recruitment of the Ligase IV/XRCC4 complex (Branzei and Foiani 2008). Deficiencies in any of these factors can delay or impair the DSB repair efficiency, resulting in unrepaired breaks that may invade other chromosomes and, thereby, cause chromosomal translocations and other cancer-related aberrations. Defects in DSB repair have further been linked to senescence and premature aging (see below). In this section, we will discuss the impact of DNA damage in the form of oxidative stress, DSBs and telomere instability on cellular senescence and ageing. The complex interplay between DNA lesions, DNA repair defects and their impact on cell function and physiology is summarized in Table 1.
Table I.
Molecular and organismal consequences of nuclear lesions
| Nuclear lesions (associated genetic defects) | Molecular consequences | Phenotypic consequences/Diseases | Refs. |
|---|---|---|---|
| Oxidative DNA lesions(BER) | CAG repeat expansion DNA mutation Aberrant repair Gene repression |
Huntington Disease Cancer Segmental Progeria Age-associated transcriptional changes |
(Kovtun et al. 2007) (Maynard et al. 2009) (Mostoslavsky et al. 2006) (Bahar et al. 2006; Lu et al. 2004) |
| Bulky ssDNA lesions, Interstrand crosslinks(NER) | Stalled transcription (TC-NER) Mutations(GG-NER) |
Switch from growth- to somatic maintenance- associated gene expression patterns Segmental pogeria Cancer Segmental progeria |
(Garinis et al. 2009; Niedernhofer et al. 2006; Schumacher et al. 2008b) (Schumacher et al. 2008a) |
| DNA DSBs(HR, NHEJ) | Gross chromosomal rearrangements/genomic instability Redistribution of chromatin modifiers(SIRT1/HDAC1) Persistent chromatin changes at DSB sites |
Apoptosis Cancer Segmental progeria Age-associated transcriptional changes Neurodegenerative Disease Gene silencing |
(Branzei and Foiani 2008; Li et al. 2007; Li et al. 2008) (Kim et al. 2008; Oberdoerffer et al. 2008) (O’Hagan et al. 2008) |
| Telomere erosion(ATM, WRN, TERC) | Increased genomic instability Increased DDR activation |
See DSB repair defects Senescence Segmental progeria Stem cell exhaustion |
(Bohr 2008; Michishita et al. 2008; Smilenov et al. 1997) (Bell and Sharpless 2007; Campisi and d’Adda di Fagagna 2007; Choudhury et al. 2007; Rossi et al. 2007; Schaetzlein et al. 2007; Tomas-Loba et al. 2008) |
| Spindle assembly defect(BubR1, Bub1, Bub3) | Aneuploidy Activation of DDR |
Cancer Senescence/Segmental Progeria |
(Schliekelman et al. 2009) (Baker et al. 2004; Baker et al. 2006; Baker et al. 2008) |
| Disruption of nuclear lamina(Lamin A, Zmpste24) structure | Genomic instability Altered nuclear architecture/chromatin Segmental progeria structure |
See DSB repair defects Transcriptional deregulation Stem cell exhaustion Segmental Progeria | (Liu et al. 2005) (Eriksson et al. 2003; Scaffidi and Misteli 2005; Scaffidi and Misteli 2006; Scaffidi and Misteli 2008; Varela et al. 2005) |
Oxidative DNA lesions
In the “Free Radical Theory of Ageing”, ROS were proposed as a main cause of ageing (Harman 1956). This model has survived the test of time surprisingly well and remains a prominent theory, particularly with the resurging interest in mitochondria, where the majority of ROS are formed (Finley and Haigis 2009). Indeed, genetic manipulation of endogenous ROS production provides a direct link between ROS levels and tissue degeneration, underlining the importance of ROS during ageing (St-Pierre et al. 2006). ROS target a variety of macromolecules including lipids and proteins, but it is their effect on DNA that is often considered the most harmful, resulting in permanent, irreparable changes to gene sequences, the blueprint for protein and RNA-coding information (Garinis et al. 2008). One of the most common oxidative DNA lesions is 8-oxo-Guanine, which is highly mutagenic since it can mispair with adenine during DNA replication, resulting in frequent GC → TA transversions. 8-oxo-Guanine and related single-stranded DNA lesions are generally repaired by BER, while bulkier lesions are subject to NER (Maynard et al. 2009; Schumacher et al. 2008a).
Several lines of evidence link oxidative DNA lesions to ageing or age-associated degenerative diseases. Mice deficient in SIRT6, a member of the sirtuin family of NAD+-dependent protein deacetylases, exhibit a defect in the base excision repair of oxidatively damaged bases. SIRT6−/− mice die shortly after weaning and harbor a variety of symptoms consistent with a prematurely aged phenotype, including kyphosis, decreased bone density, lymphopenia and severe metabolic defects (Mostoslavsky et al. 2006).
The repair of oxidized lesions has further been directly implicated in the pathology of Huntington’s disease, an age-associated neurodegenerative disorder that involves the expansion of glutamine-encoding CAG repeats within the huntingtin (HTT) gene, resulting in extensive polyglutamine (polyQ) stretches that impair protein function. The BER enzyme OGG1 was shown to initiate a process termed “toxic oxidation”, in which the escalating excision of oxidized lesions located in CAG repeats results in age-dependent repeat expansion in a mouse model of the disease (Kovtun et al. 2007). Finally, oxidative DNA damage at promoter elements has been implicated in the repression of genes involved in synaptic plasticity, memory and inflammation in the ageing brain, as well as a stochastic deregulation of gene expression in ageing muscle tissue (Bahar et al. 2006; Lu et al. 2004). These studies suggest that oxidative stress can directly affect gene expression, and thus alter cell function (see below for a detailed discussion of age-associated gene deregulation). For an in-depth discussion of oxidative lesions, and BER in particular, in the context of ageing we refer the reader to a recent review by (Maynard et al. 2009).
DNA double strand breaks
The DNA damaging effect of oxidative stress is not limited to ssDNA lesions but can also result in DNA DSBs, either as a result of nearby ssDNA lesions or as a consequence of replication across the ssDNA lesion (Li et al. 2006). DSBs can further occur as a result of radiation, during lymphcyte development, DNA replication, or due to intrinsic genomic instability at repetitive DNA and telomeres (Lombard et al. 2005). Unrepaired DSBs are the most toxic type of DNA damage. During replication, DSBs can cause stalled replication forks, which results in cell cycle arrest, cell death or senescence. During mitosis, DSBs can cause chromosomal translocations and other gross chromosomal rearrangements (GCR) that may result in cancer and have further been linked to the ageing process (Li et al. 2008).
A causal relationship between DSBs and ageing is supported by a variety of genetic mutants. A prominent example is a defect in the human WRN protein. WRN is a member of the RecQ family of DNA helicases and is involved in several aspects of DNA metabolism, including DNA replication, maintenance of telomeres and DNA repair (Bohr 2008). WRN mutations cause a dramatic increase in DSB-related genomic instability, resulting in Werner Syndrome (WS), a progeria with onset in middle age that mimics many aspects of normal ageing including atherosclerosis, diabetes, and dramatically aged skin by age 40 (Yu et al. 1996). Interestingly, a defect in the yeast WRN ortholog SGS1 causes increased genomic instability and accelerated ageing, highlighting the evolutionarily conserved role of RecQ helicases in genome maintenance and forestalling ageing (Sinclair et al. 1997).
Recently, it has been found that age-related polyQ disorders can increase DSB formation, thereby promoting neuronal decline (Qi et al. 2007). Mutant polyQ-containing Huntingtin (HTT) or ataxin (AT1) proteins sequester and inactivate the high mobility group proteins B1 and B2 (HMGB1/2), which can otherwise recognize damaged DNA and facilitate DNA repair (Ohndorf et al. 1999; Zhang et al. 2005). Mutant HTT and AT1, as well as depletion of HMGB1, causes an increase in DSBs in primary neurons, whereas compensatory overexpression of HMGB represses genotoxic stress and ameliorates polyQ induced pathology both in vitro and in Drosophila, highlighting the potentially detrimental effect of DSBs in age-related neuropathologies (Qi et al. 2007).
Consistent with a causal role for DSBs in the ageing process, it was shown that defects in DSB repair can give rise to symptoms resembling premature ageing (Li et al. 2007; Li et al. 2008). Defects in both NHEJ and HR have been suggested to cause symptoms of premature ageing, including mutations in the HR protein Brca1 and the NHEJ factors Ku70, Ku80, Xrcc4 and DNA-PKcs (reviewed in (Li et al. 2008). While the link between HR and NHEJ repair factors and ageing is well described, recent work suggests that chromatin modifiers also contribute to DSB repair and may further impinge on the ageing process (Oberdoerffer and Sinclair 2007; van Attikum et al. 2004). Consistent with a role for chromatin modifiers in both DNA repair and degenerative processes, the class I histone deacetylase (HDAC) 1 has recently been shown to promote DSB repair in neurons in a mouse model of Alzheimer’s disease (AD). Decreased HDAC1 function, on the other hand, caused an increase in DSB formation, neuronal cell loss and accelerated neurodegeneration (Kim et al. 2008).
In addition, the class III histone deacetlyases (sirtuins) SIRT1 and SIRT6 are also required for efficient DSB repair (McCord et al. 2009; Oberdoerffer et al. 2008; Wang et al. 2008). Lack of SIRT1 causes an increase in chromosomal aberrations in response to ROS or ionizing irradiation and mice with reduced SIRT1 levels were more tumor-prone than littermate controls, whereas increased SIRT1 levels suppress tumor formation in a mouse model of genomic instability (Oberdoerffer et al. 2008; Wang et al. 2008). These effects are at least in part due to inefficient recruitment of DSB repair factors to the DSB site, resulting in defects in HR and, to some extent in NHEJ (Oberdoerffer et al. 2008). The demonstration of a direct interaction between SIRT1 and the DSB break-associated HR repair component Nbs1 further corroborates a role for SIRT1 in DSB repair (Yuan et al. 2007). Recently, SIRT6 was shown to promote DSB repair by NHEJ, presumably through the direct modification of histones surrounding the DSB site (McCord et al. 2009). The role for sirtuins in DSB repair is particularly interesting given that mice lacking SIRT6 exhibit premature ageing phenotypes and SIRT1 has been implicated as a mediator of the health-promoting effects of caloric restriction, a dietary regimen that can extend lifespan in a variety of organisms and was shown to promote genomic stability (Chen et al. 2005; Haigis and Guarente 2006). Together, these findings suggest a causal link between the DSB repair process and ageing.
Telomeres
Although DNA damage is generally thought to occur in a random fashion, there are certain chromosomal regions that are highly susceptible to genomic instability –telomeric DNA being the most prominent example (Deng et al. 2008). Telomeres are composed of TTAGGG repeats that can expand over >10 kb at the ends of each chromosome. They are maintained by telomerase, a ribonucleoprotein complex that consists of an RNA template (TERC) and a reverse transcriptase subunit (TERT). In the absence of telomerase, chromosomes progressively shorten with each cell division, since DNA polymerase is unable to fully replicate the lagging DNA strand.
Telomere erosion is tightly linked to the accumulation of DSBs near telomeric DNA (Meier et al. 2007) and can result in detrimental chromosomal aberrations such as end-to-end fusions or fragmented chromosomes (Feldser et al. 2003). A decrease in telomere length has also been directly correlated with a loss of replicative potential in human fibroblasts (Deng et al. 2008). Given the impact of telomere maintenance on cell proliferation and genomic stability, telomeres play a central role in both tumor formation and cellular as well as organismal ageing. In the following, we will focus on recent evidence in support of the latter.
Due to their continued shortening, telomeres are particularly susceptible to DNA damage and, consequently, defects in DNA repair. It is, therefore, not surprising that individuals with defects in DNA repair and maintenance factors show pronounced defects in telomere stability as has been reported for patients with premature ageing syndromes such as WS (Bohr 2008; Multani and Chang 2007) or Ataxia-Telangiectasia (A-T). A-T patient harbor an inherited defect in the ATM protein kinase that initiates the DNA repair cascade (Smilenov et al. 1997; Verdun and Karlseder 2006). It is of note that the histone deacetylase SIRT6 has recently been shown to physically bind to human telomeres, where its histone deacetylase activity is required for the recruitment of WRN and has, thus, been proposed to help maintain telomere integrity. Cells that lack SIRT6 undergo premature cell cycle arrest and show increased end-to-end fusions (Michishita et al. 2008). Interestingly, this function of SIRT6 seems to be specific to human cells and does, therefore, not explain the premature ageing phenotype observed in mice.
One of the mechanisms by which genomic instability at telomeres is thought to promote the ageing process is by continuously activating the DNA damage response. Recent work by Rudolph and colleagues provides evidence that both telomeric DNA damage and the cellular response to telomere instability play a causal role in this process (Choudhury et al. 2007; Schaetzlein et al. 2007). Mice deficient in TERC have dysfunctional telomeres and show impaired maintenance of organs with high rates of cell turnover, resulting in symptoms of premature ageing (Blasco et al. 1997; Lee et al. 1998; Rudolph et al. 1999). Strikingly, this phenotype is reversed in telomere-deficient mice that lack the 5′-3′ exodonuclease Exo1 (Schaetzlein et al. 2007). Exo1 is a DNA repair factor that is implicated in the processing of dysfunctional telomeres and the concomitant generation of chromosomal translocations. Consistently, Exo1-deficient cells show a reduction in telomere-associated DNA damage as well as reduced ATR-mediated DNA damage signaling. Moreover, deletion of Exo1 in vivo improved organ maintenance and extended the lifespan of TERC−/− mice.
The premature ageing phenotype of TERC−/− mice could also be rescued by deleting a downstream effector of the DNA damage response, the p53 target and cell cycle inhibitor p21/CDKN1a. TERC−/−, p21−/− mice showed improved organ maintenance, in particular in the hematopoietic system and intestine, without improving telomere dysfunction (Choudhury et al. 2007). It is of note that deletion of p21 did not increase chromosomal instability and cancer formation, whereas deletion of the upstream effector p53 did (Chin et al. 1999), suggesting that p53 can activate p21-independent check-points that prevent accelerated chromosomal instability and cancer in these mice. Together, these findings illustrate that the DNA damage response can have detrimental effects on organ function under conditions of chronic telomeric DNA damage.
The dramatic functional consequence of telomere defects and concomitant genomic instability has been highlighted by an analysis of long-term reconstituting of hematopoietic stem cells (LT-HSCs) in TERC−/− mice. While TERC deficiency did not affect the frequency of LT-HSCs in the bone marrow, their functional capacity was severely affected, resulting in increased DSB formation, diminished self renewal, increased apoptosis and ultimately functional exhaustion as revealed by competitive stem cell transplantation (Rossi et al. 2007). Interestingly, mice with defects in DSB repair or nucleotide excision repair showed a similar phenotype and the frequency of DSBs increased with age in wild-type LT-HSCs. Taken together, telomere erosion and the resulting genomic instability appears to play a critical role in the regulation of mammalian lifespan.
In yeast, the situation is more complex. Yeast cells represent both germline and soma, and therefore, generally maintain telomere length with age. Similar to what has been described in mammalian cells, yeast cells with defects in telomere maintenance show progressive telomere shortening and a concomitant increase in the recruitment of DNA repair factors (Hector et al. 2007). However, there is no direct correlation between telomere shortening and lifespan reduction in budding yeast. In fact, artificially lengthened telomeres shorten lifespan, a phenomenon that has been attributed to a redistribution of the SIR silencing complex from the rDNA to telomeres (Austriaco and Guarente 1997), which increases rDNA instability and, thereby, promotes yeast ageing. The impact of the reorganization of chromatin on ageing in yeast and mammals is discussed below.
Senescence
Cellular senescence is a state of permanent cell cycle arrest that is often referred to as cellular ageing and can be induced by telomere loss, mitotic oncogenes, or exposure to genotoxic stress. The complex role of senescence during mammalian ageing has been elegantly discussed in a recent review by (Campisi and d’Adda di Fagagna 2007). Here, we will highlight recent observations that underline the impact of genomic instability on senescence and organismal lifespan.
Given the detrimental effect of telomere shortening on mammalian lifespan, it has been speculated that increasing telomerase activity in somatic tissues may be beneficial. This line of thought is complicated by the fact that continuous telomerase activity can immortalize cells and thereby predisposes them to malignant transformation in vivo (Gonzalez-Suarez et al. 2001). To bypass this limitation, Blasco and colleagues have generated TERT-overexpressing mice that harbor extra copies of three tumor suppressor genes, p53, p16 and ARF (sp53/ARF) (Tomas-Loba et al. 2008). In this tumor-resistant background, TERT overexpression was shown to significantly delay the ageing process, extending the median lifespan by 26%. While the precise mechanism for this observation is unclear, the authors observed a reduction of telomere-associated genomic instability and delayed loss of telomeres with age. Somewhat surprisingly, it was sufficient to overexpress TERT in keratinocytes to achieve this systemic anti-ageing effect, suggesting a non-cell-autonomous mechanism. In support of this hypothesis, the authors observed perturbed insulin-like growth factor 1 (IGF-1) serum levels, although it remains to be determined if and how this finding is related to the observed lifespan extension. It is of note, that, while increased telomerase activity in aged TERT transgenic sp53/ARF mice is consistent with decreased senescence due to more stable telomeres, the observed lifespan extension strictly depends on elevated levels of p53, p16 and ARF, factors that are known to promote senescence in wild-type cells (Sharpless and DePinho 2007). To understand if increased telomere stability indeed functions by delaying cellular senescence, a more detailed analysis of these animals will be critical.
Similar to telomere erosion, defects in the mitotic spindle assembly checkpoint (SAC) can result in genomic instability that has been linked to premature cellular senescence. The SAC ensures faithful chromosome segregation and is triggered by defects in chromosome alignment or chromosomal DNA breaks during metaphase (Musacchio and Salmon 2007). SAC failure can result in aneuploidy, a deviation from normal chromosome numbers that is often linked to tumorigenesis. However, recent reports have shown that defects in the SAC components BubR1 and Bub3 can also result in premature cellular senescence and cause premature ageing in mice (Baker et al. 2004; Baker et al. 2006). Mice with a severe reduction in BubR1 (BubR1 hypomorphic mice) show progressive aneuploidy and several symptoms of premature ageing along with a significantly reduced lifespan. Consistently, aged wild-type mice have reduced BubR1 expression in multiple tissues (Baker et al. 2004). A reduction in BubR1 levels in mouse embryonic fibroblasts caused increased aneuploidy and senescence. In agreement with a role for BubR1 in senescence, BubR1 defects were recently shown to cause increased activation of the CDKN2a tumor suppressor locus, encoding the DNA damage response genes p16 and p19(ARF). However, the impact of these genes on the premature ageing phenotype is complex: p16 seems to function as an effector of premature ageing, whereas p19 attenuated premature ageing in BubR1 hypomorphic mice (Baker et al. 2008). Mice with a defect in the related SAC component Bub1 also show increased aneuploidy and premature senescence, yet these animals die perinatally (Schliekelman et al. 2009). Together, these findings suggest a causal but complex link between genomic instability, senescence and ageing.
Links between chromatin and ageing
Much like DNA itself, the eukaryotic system of DNA packaging is not immune to the ravages of time. All eukaryotes, including humans, experience changes in chromatin organization and gene-expression patterns as they age. Over a decade ago, it was proposed that changes in chromatin organization not only underlie age-related changes in gene expression but the ageing process itself (Imai and Kitano 1998; Villeponteau 1997). This hypothesis was primarily based on observations in senescent cells, which showed significant transcriptional changes (Goldstein 1990). This phenomenon has also been put forward to explain age-related decline in organ and tissue function in complex organisms. Indeed, ageing is associated with major alterations in gene expression patterns, which have been observed in a variety of tissues as well as across species (Lee et al. 1999a; Lu et al. 2004; Oberdoerffer and Sinclair 2007).
While a causal link between gene expression changes and mammalian ageing has not been established, it is clear that the long-term maintenance of a high level of chromatin organization, which is generally referred to as “nuclear architecture”, is vital for the normal functioning of cells and tissues over a lifetime. The dramatic effect of a perturbed nuclear architecture is exemplified by the Hutchinson-Gilford Progeria Syndrome (HGPS), in which a mutation that disrupts a critical component of the nuclear lamina, lamin A, leads to symptoms that resemble aspects of normal human ageing such as loss of hair, restricted joint mobility, and atherosclerosis (Hennekam 2006). Changes in chromatin organization were also found in cells that senesce due to replicative ageing in vitro (Narita et al. 2006), a process that may be responsible for the secretion of tumorigenic growth factors by these cells (Campisi 2005). Interestingly, normally aged tissues exhibit similar chromatin alterations (Herbig et al. 2006) and there are early hints that alterations to the nuclear architecture can contribute to normal human ageing (Scaffidi and Misteli 2006). For a comprehensive discussion of the role of nuclear architecture in ageing, we refer the reader to (Oberdoerffer and Sinclair 2007). In the following section, we present recent evidence linking changes in the epigenome to age-associated transcriptional deregulation and concomitant cell and organ dysfunction.
Age-associated epigenomic changes
Age-related transcriptional changes have been studied extensively over the past years. However, it is still unclear if they are a cause or merely a consequence of ageing. New clues have come from a recent gene expression profiling study in cells from patients with HGPS. HGPS cells show extensive nuclear defects including abnormal chromatin structure, which are caused by a mutant splice variant of the lamin A gene. The resulting gene product, progerin, can evoke HGPS-like changes in the nuclear architecture of human mesenchymal stem cells. These changes coincide with the transcriptional activation of major downstream effectors of the Notch signaling cascade (Scaffidi and Misteli 2008). Since upstream effectors are unaltered, the authors suggest that transcriptional regulators of downstream effectors are sequestered by the nuclear lamina, a process that may be disrupted in the presence of progerin. While the precise mechanism is unclear, deregulation of Notch targets was shown to have functional consequences, as it alters the differentiation potential of mesenchymal stem cells. Progerin can also be found at low levels during normal ageing, thus raising the possibility that the epigenetic alteration of Notch signaling may contribute to normal stem cell dysfunction with age (Scaffidi and Misteli 2006).
Changes in gene expression were also detected in ageing hematopoietic stem cells (LT-HSCs). Expression profiling revealed a systemic downregulation of genes mediating lymphoid specification in LT-HSCs from old animals, and these changes correlated with a diminished capacity of aged LT-HSCs to generate mature lymphocytes (Rossi et al. 2005). It is of note that the changes in gene expression observed in LT-HSCs are distinct from those previously reported in HGPS cells or the ageing brain (Lu et al. 2004; Scaffidi and Misteli 2008), suggesting that a variety of functionally distinct gene subsets are subject to deregulation with age, possibly in a cell type dependent manner.
Further evidence for the idea that age-associated deregulation of specific gene subsets may be a causal factor in ageing comes from a comprehensive analysis of promoter elements that become deregulated with age. This analysis revealed several cis-regulatory motifs that were overrepresented in a variety of different, aged tissues, one of the most prominent examples being NFκB (Adler et al. 2007). Indeed, overexpression of a dominant-negative NFκB subunit in mouse skin tissue was able to reverse a large fraction of NFκB-related age-associated gene expression changes. Interestingly, NFκB target genes related to inflammation and immunity were not significantly affected, suggesting a more general role for NFκB in age-related gene expression (Adler et al. 2007). It is of note that NFκB target genes are also significantly deregulated in prematurely ageing SIRT6-deficient mice and haplo-insufficiency of the NFκB subunit RelA was able to rescue early lethality in ~50% of SIRT6-deficient animals (Kawahara et al. 2009). SIRT6 was found to attenuate NFκB signaling through direct interaction with RelA and concomitant histone H3K9 deacetylation at NFκB target promoters, thus corroborating the link between transcriptional NFκB target gene deregulation and ageing.
Support for the idea that epigenetic deregulation may not be limited to specific gene subsets but rather reflect a genome-wide phenomenon comes from recent work in the nervous system, showing that global transcriptional deregulation can modulate neuron function in a mouse model of Alzheimer’s disease (Fischer et al. 2007; Kim et al. 2008). Inhibition of class I and II histone deacetylases was able to reinstate learning behavior and long term memory access in mice with advanced neurodegeneration. This effect was attributed to increased synapse formation and dendritic sprouting, although the precise mechanism for these findings is still unclear. While HDAC inhibition can affect a variety of non-histone HDAC targets, it is of note that increased histone deacetylation was observed not only upon HDAC inhibition but also in the context of environmental enrichment, a behavioral protocol that increases synaptic activity and learning similar to HDAC inhibition (Fischer et al. 2007). It will be interesting to explore how a decrease in chromatin compaction can result in functional improvement in neurons; transcription profiling is expected to provide further mechanistic insight into this phenomenon. Interestingly, using the same transgenic mouse model, HDAC1 has been shown to repress cell cycle genes, thereby suppressing aberrant cell cycle activity in neurons, a process that is generally followed by neuronal cell death (Kim et al. 2008). Consequently, gain of HDAC1 function was able to provide protection from neurotoxicity in vitro and in vivo, highlighting the complex role of chromatin modifiers during age-related epigenomic changes.
A similar beneficial HDAC effect has been described for the class III HDAC SIRT1. A neuron-specific increase in SIRT1 expression was shown to suppress age-associated deregulation of a significant fraction of SIRT1-regulated genes in mice, suggesting that increasing SIRT1 activity may help maintain youthful gene expression patterns in vivo (Oberdoerffer et al. 2008). Interestingly, age-related gene expression changes could also be suppressed in mice treated with resveratrol or on a calorically restricted diet, two interventions that increase SIRT1 activity (Barger et al. 2008; Pearson et al. 2008).
SIRT1 has recently been implicated in another aspect of genome maintenance, the regulation of circadian rhythms through deacetylation of circadian promoters (Asher et al. 2008; Nakahata et al. 2008). Lack of SIRT1 was shown to result in disturbances in the circadian cycle in vitro and in a liver-specific SIRT1 knock-out mouse model (Nakahata et al. 2008). A possible explanation for this observation may be altered acetylation of histone H3 at circadian promoters as well as BMAL1, a core element of the circadian clock machinery. Direct deacetylation of a negative regulator of BMAL1, PER2, by SIRT1 has also been reported, suggesting multi-facetted regulation of circadian transcription by SIRT1 (Asher et al. 2008). Interestingly, BMAL1 deficiency causes premature ageing and age-related pathologies in mice (Kondratov et al. 2006).
Together, these results highlight the complex regulation and impact of chromatin structure on cell and organ function but also reveal the therapeutic potential that lies in manipulating the epigenome. Unlike DNA damage, changes to chromatin are highly dynamic and, at least theoretically, reversible.
A mechanistic link between genome stability, chromatin and ageing
Despite the seemingly distinct effects of ageing on genome stability and chromatin organization, it is tempting to speculate that the two are interconnected, given the intimate association between DNA and chromatin. In yeast, this connection was made more than a decade ago, and over the past few years evidence has accumulated that DNA damage can contribute to age-related epigenetic changes also in higher organisms. In the following we will summarize findings that, together, indicate that DNA-damage driven reorganization of chromatin could be an evolutionarily conserved driving force of ageing.
Lessons from yeast
The yeast “SIR” complex was first identified by virtue of its role in silencing the yeast mating type loci, HMR and HML, which are the repositories of yeast mating type information, and must be repressed if the yeast is to express only one mating type at a time and, thus, remain fertile (Klar et al. 1979; Rine and Herskowitz 1987). SIR-mediated silencing is also present at telomeres and the ribosomal DNA (rDNA). As yeast cells age, recombination between inherently unstable ribosomal DNA (rDNA) repeats results in the excision of a single circular molecule of DNA (an extra-chromosomal rDNA circle, or ERC) that is replicated during S-phase (Sinclair and Guarente 1997). The accumulation of up to 1000 ERCs eventually causes cell death, presumably by recruiting essential proteins including the SIR complex from the rest of the genome to the rDNA (Kennedy et al. 1997; Sinclair et al. 1997). The concomitant loss of SIR-mediated mating type silencing was shown to be a causal event in yeast ageing (Kaeberlein 1999). In young yeast cells, rDNA recombination is kept in check by the SIR component Sir2, which directly binds to and deacetylates the rDNA resulting in chromatin compaction and rDNA stabilization. Consistently, deleting the SIR2 gene causes a loss of rDNA silencing, elevated rDNA recombination and accelerated ageing, whereas integration of an extra copy of the SIR2 gene increases rDNA silencing, suppresses rDNA recombination, stabilizes mating type expression and extends lifespan (Kaeberlein et al. 1999).
Genomic instability at the rDNA is not the only process that causes redistribution of the Sir2 containing silencing complex. In 1999, four studies showed that a single DNA break is sufficient to illicit a DNA-damage response that releases Sir proteins from mating-type loci and telomeres and relocalizes them to the DNA break, possibly to facilitate the repair process (Lee et al. 1999b; Martin et al. 1999; McAinsh et al. 1999; Mills et al. 1999). Tyler and colleagues subsequently showed that Sir2 and other histone deacetylases are recruited to DSBs and modify chromatin that surrounds the break site in a temporally coordinated manner, a process that appears to be a prerequisite of efficient DNA repair (Tamburini and Tyler 2005). Similar to a single DSB, oxidative stress was recently shown to result in Sir2-dependent chromatin reorganization, causing a loss of mating type silencing, increased rDNA instability and a concomitant reduction in lifespan, effects that could be counteracted by increased Sir2 expression (Oberdoerffer et al. 2008).
Together, these observations demonstrate a direct link between DNA damage and global genomic changes that cause yeast ageing. We have previously termed this process the redistribution of chromatin-modifiers, or RCM response (Oberdoerffer and Sinclair 2007). We propose that RCM is a beneficial DNA damage response in young cells, and consistently, both Sir2 recruitment to DSBs and mating type derepression were shown to contribute to DSB repair (Lee et al. 1999b; Tamburini and Tyler 2005). In old cells, however, accumulating DNA damage and increased rDNA instability are chronic RCM triggers, resulting in permanent epigenomic alterations that eventually cause ageing, which in yeast is manifested as sterility.
DNA damage and the ageing epigenome
A number of recent reports indicate that a link between DNA damage can also generate chromatin alterations in higher organisms. Deficiency in the DNA repair factor ERCC1 accelerates ageing phenotypes and generates gene expression profiles reminiscent of aged animals (Niedernhofer et al. 2006), suggesting that the accumulation of DNA damage can promote global transcriptional changes. Although the mechanism behind these changes is still unclear, a subset of deregulated genes reflects a non-cell-autonomous protective response that shifts energy usage from growth to maintenance, which may reflect an attempt to provide sufficient resources for DNA repair. These changes include a dampening of insulin/IGF1 signaling, which is generally associated with longevity. This observation seems paradoxical at first, but has been suggested to reflect a “survival” response that is unable to overcome the severe defects in ERCC1-deficient mice (Garinis et al. 2008; Niedernhofer et al. 2006).
A second line of evidence for a causal link between DNA damage and chromatin reorganization was recently presented by Vijg and colleagues, who demonstrated that cells damaged by oxidative stress in vitro undergo stochastic transcriptional changes that parallel those in aged heart tissue (Bahar et al. 2006). Similarly, oxidative DNA damage at promoters was found to correlate with gene repression in the ageing human brain (Lu et al. 2004) and has been linked to both transcriptional and epigenetic changes that may contribute to Alzheimer disease (Wu et al. 2008). It is of note that systemic transcriptional changes in hematopoietic stem cells are tightly linked to functional defects in the LT-HSC pool (Rossi et al. 2005). These defects are further reminiscent of those observed in LT-HSCs from DNA repair deficient mice, although transcriptional profiling of the latter has yet to be reported (Rossi et al. 2007).
Genotoxic stress has also been implied in the deregulation of gene expression and concomitant protein secretion during senescence. Using antibody-based detection arrays, senescent cells were shown to secrete a large and functionally complex set of proteins, an effect that was due to increased protein production rather than altered secretion efficiency (Coppe et al. 2008). Remarkably, a similar secretory phenotype could be induced in normal fibroblasts when exposed to genotoxic stress, corroborating the link between genomic instability and altered gene and protein expression. It is of note that senescence-associated secreted factors were able to promote malignancy and invasiveness in premalignant tumor cell lines via a paracrine mechanism, suggesting that DNA damage-driven transcriptional changes can have non-cellautonomous effects that may promote age-related diseases such as cancer (Coppe et al. 2008).
Until recently, little was known about the mechanisms by which DNA damage might affect the expression of ostensibly undamaged genes. In yeast, this crosstalk seems to be achieved through a global reorganization of chromatin in response to rDNA instability. Given that rDNA recombination is a yeast-specific cause of ageing, it was unclear if the yeast RCM response would be relevant for mammalian ageing. However, the finding that DNA damage could cause an RCM response much like rDNA instability suggested that similar processes may be at work in higher organisms. Two recent reports provide support for this idea. In the first study, it was shown that mouse SIRT1 behaves much like its yeast ortholog Sir2 in that is recruited to DSBs in response to DNA damage (Oberdoerffer et al. 2008). SIRT1 recruitment is necessary for efficient DSB repair, a process that involves a loss of SIRT1 from other loci across the mouse genome, including a diverse set of genes and repetitive major satellite DNA. Loss of SIRT1 from these loci results in the transcriptional deregulation of repetitive DNA and a subset of genes, a phenomenon paralleled in the ageing mouse brain. Importantly, increasing SIRT1 expression in the nervous system was shown to repress these changes (Oberdoerffer et al. 2008). It is at this point unclear what triggers the redistribution of SIRT1 in mice or Sir2 in yeast, however, in both cases, recruitment to DSBs seems to depend on DNA damage signaling through ATM family kinases (Mills et al. 1999; Oberdoerffer et al. 2008). Together these findings suggest a functional link between age-dependent epigenetic changes, SIRT1 localization and DNA damage, a mechanism that appears to contribute to ageing in organisms as diverse as yeast and mammals.
The second study shows a similar dual function for the Class I histone deacetylase HDAC1. Using p25/Cdk5 transgenic mice, which model aspects of Alzheimers disease, Tsai and colleagues show that p25 directly interferes with HDAC1 activity, resulting in aberrant cell cycle activity and increased DSB induction (Kim et al. 2008). HDAC1 overexpression provided protection from these events in vitro and in vivo. The observed cell cycle reentry of post-mitotic neurons was a result of aberrant expression of cell cycle regulators, including a variety of cyclins, p21 and E2F1, a phenomenon that has been previously reported to result in neuronal cell death, thereby contributing to the pathology of many neurodegenerative disorders (Busser et al. 1998). The authors confirmed previous findings that HDAC1 can directly suppress cell cycle genes, an effect that is abrogated in the presence of p25 (Kim et al. 2008). The observation that HDAC1 is also linked to DSB repair suggests a mechanism similar to the redistribution of SIRT1, by which HDAC1 could promote DSB repair in p25 transgenic mice by temporarily leaving its post at the promoters of cell cycle genes. However, it is unclear how HDAC1 protects from DNA damage and if these two processes are functionally linked or independent consequences of p25-mediated inhibition of HDAC1 function. Nevertheless, these two studies identify chromatin modifiers as a mechanistic link between DNA damage and transcriptional deregulation. Interestingly, the observations that SIRT6 can mediate DSB repair in mice but is also required to suppress NFκB target genes at the promoter level is consistent with the idea that DSB-mediated redistribution of SIRT6 may account for the observed age-related changes in the NFκB gene signature (Kawahara et al. 2009; McCord et al. 2009). Importantly, increased HDAC activity was able to revert the observed epigenetic changes in all cases, suggesting that DNA damage-mediated chromatin reorganization is a dynamic process that may be targeted for therapeutic intervention.
A unifying theory of nuclear ageing
It is tempting to speculate that RCM may, in part, account for the various age-associated transcriptional changes described above and that it may do so in a tissue or cell-type dependent manner. The subset of genes affected by the redistribution of chromatin modifiers such as SIRT1 or HDAC1 is expected to vary between tissues, depending on the silencing status of individual genes and the chromatin modifiers involved. Thus, a seemingly random distribution of DNA damage across the genome could result in what appears to be a programmed and cell-type specific deregulation of gene expression with age. RCM could also provide a mechanistic basis for the deregulation of ostensibly undamaged genes, whereas until recently, the only direct link between DNA damage and gene expression was through silencing of damaged promoter regions (Lu et al. 2004; O’Hagan et al. 2008).
While RCM may explain the epigenetic deregulation of large and functionally diverse gene sets, there is accumulating evidence for gene-specific transcriptional changes that may be part of an orchestrated DNA damage driven “survival” response, causing a shift in gene expression from growth to somatic maintenance (see above, (Garinis et al. 2008)). At this point, little is known about the mechanism behind this response and it will be interesting to investigate whether the transcriptional changes that are a part of this “survival” program reflect a programmed response that is entirely distinct from RCM. A comparison of gene expression patterns during both processes should help resolve this question. It is of note that changes in the IGF-1/insulin signaling axis appear to be limited to DNA repair defects that affect transcription-coupled nucleotide excision repair, whereas mouse mutants with DSB repair defects do not show the same transcriptional changes despite displaying symptoms of premature ageing (Garinis et al. 2008; Li et al. 2007; Niedernhofer et al. 2006).
Reconciling the theories that describe the relationship between (epi)genomic integrity and ageing will be a challenge for future research (Garinis et al. 2008; Li et al. 2008; Oberdoerffer and Sinclair 2007; Vijg and Dolle 2007). Given the pleiotropic nature of the ageing process, it is possible that different DNA repair pathways may result in distinct molecular changes that can promote ageing independently (Li et al. 2008). Nevertheless, the fundamental role of DNA damage and the maintenance of chromatin in ageing across species is becoming increasingly evident and can be held accountable for a variety of ageing phenotypes, including senescence, loss of proper cell function as well as genomic aberrations (see Figure 1). It is conceivable that these changes make a cell even more susceptible to DNA damage, thus engaging in a self-amplifying loop of cellular decline. A number of reports have demonstrated non-cellautonomous effects of these epigenetic changes, demonstrating the potential for systemic age-related decline (Coppe et al. 2008; Niedernhofer et al. 2006). However, the reversible nature of epigenetic modifications provides, at least in theory, an opportunity for therapeutic intervention, and is in a certain way reminiscent of a “reset button” that may eventually allow to restore cells and tissues to a more youthful epigenomic profile.
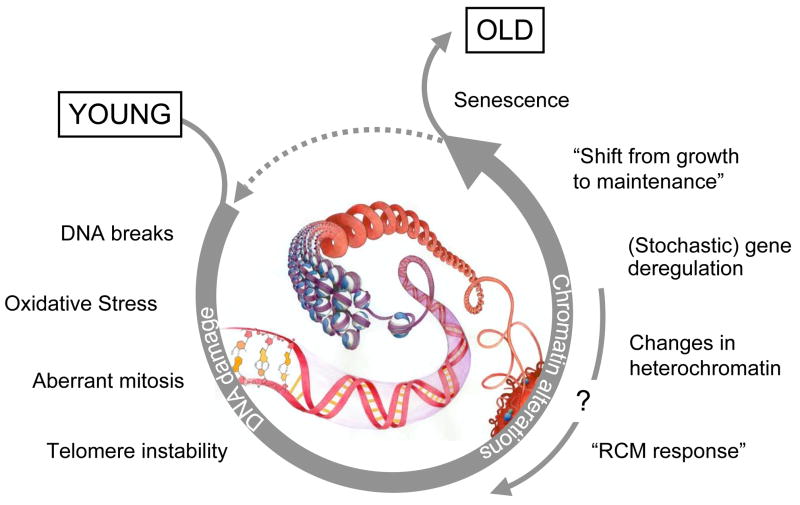
Figure 1. A “nucleocentric” view of ageing.
DNA damage drives chromatin alterations that eventually cause senescence and promote ageing. Chromatin changes may render cells more susceptible to further cycles of damage and epigenetic alterations, thereby amplifying the effect. Changes to chromatin that do not encompass genomic aberrations are theoretically reversible and may allow to “reset” the epigenome to a more youthful state.
Acknowledgments
We thank Raul Mostoslavsky, Brian North and Ralph Scully for discussion and comments. P.O. was supported by a fellowship from the National Space Biomedical Research Institute (grant PF00903). D. A. S. is supported by NIH grants RO1GM068072 and R01AG19719, The Ellison Medical Foundation and the Glenn Foundation for Medical Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adler AS, Sinha S, Kawahara TL, Zhang JY, Segal E, Chang HY. Motif module map reveals enforcement of aging by continual NF-kappaB activity. Genes Dev. 2007;21 (24):3244–3257. doi: 10.1101/gad.1588507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134 (2):317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Austriaco NR, Jr, Guarente LP. Changes of telomere length cause reciprocal changes in the lifespan of mother cells in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1997;94 (18):9768–9772. doi: 10.1073/pnas.94.18.9768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dolle ME, Calder RB, Chisholm GB, Pollock BH, Klein CA, Vijg J. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441 (7096):1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, van Deursen JM. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36 (7):744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Jeganathan KB, Malureanu L, Perez-Terzic C, Terzic A, van Deursen JM. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J Cell Biol. 2006;172 (4):529–540. doi: 10.1083/jcb.200507081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Perez-Terzic C, Jin F, Pitel K, Niederlander NJ, Jeganathan K, Yamada S, Reyes S, Rowe L, Hiddinga HJ, Eberhardt NL, Terzic A, van Deursen JM. Opposing roles for p16Ink4a and p19Arf in senescence and ageing caused by BubR1 insufficiency. Nat Cell Biol. 2008;10 (7):825–836. doi: 10.1038/ncb1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barger JL, Kayo T, Vann JM, Arias EB, Wang J, Hacker TA, Wang Y, Raederstorff D, Morrow JD, Leeuwenburgh C, Allison DB, Saupe KW, Cartee GD, Weindruch R, Prolla TA. A low dose of dietary resveratrol partially mimics caloric restriction and retards aging parameters in mice. PLoS ONE. 2008;3 (6):e2264. doi: 10.1371/journal.pone.0002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JF, Sharpless NE. Telomeres, p21 and the cancer-aging hypothesis. Nat Genet. 2007;39 (1):11–12. doi: 10.1038/ng0107-11. [DOI] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91 (1):25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Bohr VA. Repair of oxidative DNA damage in nuclear and mitochondrial DNA, and some changes with aging in mammalian cells. Free Radic Biol Med. 2002;32 (9):804–812. doi: 10.1016/s0891-5849(02)00787-6. [DOI] [PubMed] [Google Scholar]
- Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem Sci. 2008;33 (12):609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokov A, Chaudhuri A, Richardson A. The role of oxidative damage and stress in aging. Mech Ageing Dev. 2004;125 (10–11):811–826. doi: 10.1016/j.mad.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9 (4):297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- Busser J, Geldmacher DS, Herrup K. Ectopic cell cycle proteins predict the sites of neuronal cell death in Alzheimer’s disease brain. J Neurosci. 1998;18 (8):2801–2807. doi: 10.1523/JNEUROSCI.18-08-02801.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120 (4):513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8 (9):729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310 (5754):1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299 (5607):721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- Chin L, Artandi SE, Shen Q, Tam A, Lee SL, Gottlieb GJ, Greider CW, DePinho RA. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97 (4):527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- Choudhury AR, Ju Z, Djojosubroto MW, Schienke A, Lechel A, Schaetzlein S, Jiang H, Stepczynska A, Wang C, Buer J, Lee HW, von Zglinicki T, Ganser A, Schirmacher P, Nakauchi H, Rudolph KL. Cdkn1a deletion improves stem cell function and lifespan of mice with dysfunctional telomeres without accelerating cancer formation. Nat Genet. 2007;39 (1):99–105. doi: 10.1038/ng1937. [DOI] [PubMed] [Google Scholar]
- Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6 (12):2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Chan SS, Chang S. Telomere dysfunction and tumour suppression: the senescence connection. Nat Rev Cancer. 2008;8 (6):450–458. doi: 10.1038/nrc2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JA, Nussenzweig MC, Nussenzweig A. Chromatin dynamics and the preservation of genetic information. Nature. 2007;447 (7147):951–958. doi: 10.1038/nature05980. [DOI] [PubMed] [Google Scholar]
- Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, Dutra A, Pak E, Durkin S, Csoka AB, Boehnke M, Glover TW, Collins FS. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423 (6937):293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldser DM, Hackett JA, Greider CW. Telomere dysfunction and the initiation of genome instability. Nat Rev Cancer. 2003;3 (8):623–627. doi: 10.1038/nrc1142. [DOI] [PubMed] [Google Scholar]
- Finkel T, Serrano M, Blasco MA. The common biology of cancer and ageing. Nature. 2007;448 (7155):767–774. doi: 10.1038/nature05985. [DOI] [PubMed] [Google Scholar]
- Finley LWS, Haigis MC. The coordination of nuclear and mitochondrial communication during aging and CR. Ageing Res Rev this issue. 2009 doi: 10.1016/j.arr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447 (7141):178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- Garinis GA, van der Horst GT, Vijg J, Hoeijmakers JH. DNA damage and ageing: new-age ideas for an age-old problem. Nat Cell Biol. 2008;10 (11):1241–1247. doi: 10.1038/ncb1108-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garinis GA, Uittenboogaard LM, Stachelscheid H, Fousteri M, van Ijcken W, Breit TM, van Steeg H, Mullenders LH, van der Horst GT, Bruning JC, Niessen CM, Hoeijmakers JH, Schumacher B. Persistent transcription-blocking DNA lesions trigger somatic growth attenuation associated with longevity. Nat Cell Biol. 2009 doi: 10.1038/ncb1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein S. Replicative senescence: the human fibroblast comes of age. Science. 1990;249 (4973):1129–1133. doi: 10.1126/science.2204114. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Suarez E, Samper E, Ramirez A, Flores JM, Martin-Caballero J, Jorcano JL, Blasco MA. Increased epidermal tumors and increased skin wound healing in transgenic mice overexpressing the catalytic subunit of telomerase, mTERT, in basal keratinocytes. Embo J. 2001;20 (11):2619–2630. doi: 10.1093/emboj/20.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8(1):35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20 (21):2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11 (3):298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Hector RE, Shtofman RL, Ray A, Chen BR, Nyun T, Berkner KL, Runge KW. Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol Cell. 2007;27 (5):851–858. doi: 10.1016/j.molcel.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Hennekam RC. Hutchinson-Gilford progeria syndrome: review of the phenotype. Am J Med Genet A. 2006;140 (23):2603–2624. doi: 10.1002/ajmg.a.31346. [DOI] [PubMed] [Google Scholar]
- Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM. Cellular senescence in aging primates. Science. 2006;311 (5765):1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- Imai S, Kitano H. Heterochromatin islands and their dynamic reorganization: a hypothesis for three distinctive features of cellular aging. Exp Gerontol. 1998;33 (6):555–570. doi: 10.1016/s0531-5565(98)00037-0. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13 (19):2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY, Chua KF. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136 (1):62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Gotta M, Sinclair DA, Mills K, McNabb DS, Murthy M, Pak SM, Laroche T, Gasser SM, Guarente L. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell. 1997;89 (3):381–391. doi: 10.1016/s0092-8674(00)80219-6. [DOI] [PubMed] [Google Scholar]
- Kim D, Frank CL, Dobbin MM, Tsunemoto RK, Tu W, Peng PL, Guan JS, Lee BH, Moy LY, Giusti P, Broodie N, Mazitschek R, Delalle I, Haggarty SJ, Neve RL, Lu Y, Tsai LH. Deregulation of HDAC1 by p25/Cdk5 in neurotoxicity. Neuron. 2008;60 (5):803–817. doi: 10.1016/j.neuron.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar AJ, Fogel S, Macleod K. MAR1-a Regulator of the HMa and HMalpha Loci in SACCHAROMYCES CEREVISIAE. Genetics. 1979;93 (1):37–50. doi: 10.1093/genetics/93.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20 (14):1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128 (4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007 doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Klopp RG, Weindruch R, Prolla TA. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999a;285 (5432):1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- Lee HW, Blasco MA, Gottlieb GJ, Horner JW, 2nd, Greider CW, DePinho RA. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392 (6676):569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- Lee SE, Paques F, Sylvan J, Haber JE. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr Biol. 1999b;9 (14):767–770. doi: 10.1016/s0960-9822(99)80339-x. [DOI] [PubMed] [Google Scholar]
- Li H, Vogel H, Holcomb VB, Gu Y, Hasty P. Deletion of Ku70, Ku80, or both causes early aging without substantially increased cancer. Mol Cell Biol. 2007;27 (23):8205–8214. doi: 10.1128/MCB.00785-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Mitchell JR, Hasty P. DNA double-strand breaks: a potential causative factor for mammalian aging? Mech Ageing Dev. 2008;129 (7–8):416–424. doi: 10.1016/j.mad.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Yang J, Huang H. Oxidative stress induces H2AX phosphorylation in human spermatozoa. FEBS Lett. 2006;580 (26):6161–6168. doi: 10.1016/j.febslet.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Liu B, Wang J, Chan KM, Tjia WM, Deng W, Guan X, Huang JD, Li KM, Chau PY, Chen DJ, Pei D, Pendas AM, Cadinanos J, Lopez-Otin C, Tse HF, Hutchison C, Chen J, Cao Y, Cheah KS, Tryggvason K, Zhou Z. Genomic instability in laminopathy-based premature aging. Nat Med. 2005;11 (7):780–785. doi: 10.1038/nm1266. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120 (4):497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429 (6994):883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- Lu T, Finkel T. Free radicals and senescence. Exp Cell Res. 2008;314 (9):1918–1922. doi: 10.1016/j.yexcr.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97 (5):621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC, Bohr VA. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30 (1):2–10. doi: 10.1093/carcin/bgn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh AD, Scott-Drew S, Murray JA, Jackson SP. DNA damage triggers disruption of telomeric silencing and Mec1p-dependent relocation of Sir3p. Curr Biol. 1999;9 (17):963–966. doi: 10.1016/s0960-9822(99)80424-2. [DOI] [PubMed] [Google Scholar]
- McCord RA, Michishita E, Hong T, Berber E, Boxer LD, Kusumoto R, Guan S, Shi X, Gozani O, Burlingame AL, Bohr VA, Chua KF. SIRT6 stabilizes DNA-dependent Protein Kinase at chromatin for DNA double-strand break repair. Aging. 2009;1 (1):109–121. doi: 10.18632/aging.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier A, Fiegler H, Munoz P, Ellis P, Rigler D, Langford C, Blasco MA, Carter N, Jackson SP. Spreading of mammalian DNA-damage response factors studied by ChIP-chip at damaged telomeres. Embo J. 2007;26 (11):2707–2718. doi: 10.1038/sj.emboj.7601719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452 (7186):492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills KD, Sinclair DA, Guarente L. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 1999;97 (5):609–620. doi: 10.1016/s0092-8674(00)80772-2. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124 (2):315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Multani AS, Chang S. WRN at telomeres: implications for aging and cancer. J Cell Sci. 2007;120 (Pt 5):713–721. doi: 10.1242/jcs.03397. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8 (5):379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134 (2):329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Krizhanovsky V, Nunez S, Chicas A, Hearn SA, Myers MP, Lowe SW. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell. 2006;126 (3):503–514. doi: 10.1016/j.cell.2006.05.052. [DOI] [PubMed] [Google Scholar]
- Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, van Leeuwen W, Theil AF, Vermeulen W, van der Horst GT, Meinecke P, Kleijer WJ, Vijg J, Jaspers NG, Hoeijmakers JH. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444 (7122):1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- O’Hagan HM, Mohammad HP, Baylin SB. Double Strand Breaks Can Initiate Gene Silencing and SIRT1-Dependent Onset of DNA Methylation in an Exogenous Promoter CpG Island. PLoS Genet. 2008;4 (8):e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer P, Sinclair DA. The role of nuclear architecture in genomic instability and ageing. Nat Rev Mol Cell Biol. 2007;8 (9):692–702. doi: 10.1038/nrm2238. [DOI] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, Bonni A, Yankner BA, Scully R, Prolla TA, Alt FW, Sinclair DA. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135 (5):907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohndorf UM, Rould MA, He Q, Pabo CO, Lippard SJ. Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature. 1999;399 (6737):708–712. doi: 10.1038/21460. [DOI] [PubMed] [Google Scholar]
- Pearson KJ, Baur JA, Lewis KN, Peshkin L, Price NL, Labinskyy N, Swindell WR, Kamara D, Minor RK, Perez E, Jamieson HA, Zhang Y, Dunn SR, Sharma K, Pleshko N, Woollett LA, Csiszar A, Ikeno Y, Le Couteur D, Elliott PJ, Becker KG, Navas P, Ingram DK, Wolf NS, Ungvari Z, Sinclair DA, de Cabo R. Resveratrol delays age-related deterioration and mimics transcriptional aspects of dietary restriction without extending life span. Cell Metab. 2008;8 (2):157–168. doi: 10.1016/j.cmet.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi ML, Tagawa K, Enokido Y, Yoshimura N, Wada Y, Watase K, Ishiura S, Kanazawa I, Botas J, Saitoe M, Wanker EE, Okazawa H. Proteome analysis of soluble nuclear proteins reveals that HMGB1/2 suppress genotoxic stress in polyglutamine diseases. Nat Cell Biol. 2007;9 (4):402–414. doi: 10.1038/ncb1553. [DOI] [PubMed] [Google Scholar]
- Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, Parker AR, Jackson SP, Gennery A, Jeggo PA, Lobrich M. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell. 2004;16 (5):715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116 (1):9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi DJ, Bryder D, Zahn JM, Ahlenius H, Sonu R, Wagers AJ, Weissman IL. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc Natl Acad Sci U S A. 2005;102 (26):9194–9199. doi: 10.1073/pnas.0503280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447 (7145):725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- Rudolph KL, Chang S, Lee HW, Blasco M, Gottlieb GJ, Greider C, DePinho RA. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell. 1999;96 (5):701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat Med. 2005;11 (4):440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312 (5776):1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat Cell Biol. 2008;10 (4):452–459. doi: 10.1038/ncb1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaetzlein S, Kodandaramireddy NR, Ju Z, Lechel A, Stepczynska A, Lilli DR, Clark AB, Rudolph C, Kuhnel F, Wei K, Schlegelberger B, Schirmacher P, Kunkel TA, Greenberg RA, Edelmann W, Rudolph KL. Exonuclease-1 deletion impairs DNA damage signaling and prolongs lifespan of telomere-dysfunctional mice. Cell. 2007;130 (5):863–877. doi: 10.1016/j.cell.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliekelman M, Cowley DO, O’Quinn R, Oliver TG, Lu L, Salmon ED, Van Dyke T. Impaired Bub1 function in vivo compromises tension-dependent checkpoint function leading to aneuploidy and tumorigenesis. Cancer Res. 2009;69 (1):45–54. doi: 10.1158/0008-5472.CAN-07-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B, Garinis GA, Hoeijmakers JH. Age to survive: DNA damage and aging. Trends Genet. 2008a;24 (2):77–85. doi: 10.1016/j.tig.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Schumacher B, van der Pluijm I, Moorhouse MJ, Kosteas T, Robinson AR, Suh Y, Breit TM, van Steeg H, Niedernhofer LJ, van Ijcken W, Bartke A, Spindler SR, Hoeijmakers JH, van der Horst GT, Garinis GA. Delayed and accelerated aging share common longevity assurance mechanisms. PLoS Genet. 2008b;4 (8):e1000161. doi: 10.1371/journal.pgen.1000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat Rev Mol Cell Biol. 2007;8 (9):703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91 (7):1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Mills K, Guarente L. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science. 1997;277 (5330):1313–1316. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- Smilenov LB, Morgan SE, Mellado W, Sawant SG, Kastan MB, Pandita TK. Influence of ATM function on telomere metabolism. Oncogene. 1997;15 (22):2659–2665. doi: 10.1038/sj.onc.1201449. [DOI] [PubMed] [Google Scholar]
- St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, Simon DK, Bachoo R, Spiegelman BM. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127 (2):397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7 (10):739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25 (12):4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas-Loba A, Flores I, Fernandez-Marcos PJ, Cayuela ML, Maraver A, Tejera A, Borras C, Matheu A, Klatt P, Flores JM, Vina J, Serrano M, Blasco MA. Telomerase reverse transcriptase delays aging in cancer-resistant mice. Cell. 2008;135 (4):609–622. doi: 10.1016/j.cell.2008.09.034. [DOI] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119 (6):777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- Varela I, Cadinanos J, Pendas AM, Gutierrez-Fernandez A, Folgueras AR, Sanchez LM, Zhou Z, Rodriguez FJ, Stewart CL, Vega JA, Tryggvason K, Freije JM, Lopez-Otin C. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature. 2005;437 (7058):564–568. doi: 10.1038/nature04019. [DOI] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127 (4):709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- Vijg J. Impact of genome instability on transcription regulation of aging and senescence. Mech Ageing Dev. 2004;125 (10–11):747–753. doi: 10.1016/j.mad.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Vijg J, Dolle ME. Genome instability: cancer or aging? Mech Ageing Dev. 2007;128 (7–8):466–468. doi: 10.1016/j.mad.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeponteau B. The heterochromatin loss model of aging. Exp Gerontol. 1997;32 (4–5):383–394. doi: 10.1016/s0531-5565(96)00155-6. [DOI] [PubMed] [Google Scholar]
- Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, Jia R, Zheng ZM, Appella E, Wang XW, Ried T, Deng CX. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14 (4):312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Basha MR, Zawia NH. The environment, epigenetics and amyloidogenesis. J Mol Neurosci. 2008;34 (1):1–7. doi: 10.1007/s12031-007-0009-4. [DOI] [PubMed] [Google Scholar]
- Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner’s syndrome gene. Science. 1996;272 (5259):258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol Cell. 2007;27 (1):149–162. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li GM. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122 (5):693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]