Abstract
We provide evidence that matrix metalloproteinase-7 (MMP-7) interacts with anionic, cationic and neutral lipid membranes, although it interacts strongest with anionic membranes. While the catalytic activity of the enzyme remains unaffected upon binding to neutral and negatively charged membranes, it is drastically impaired upon binding to the positively charged membranes. The structural data reveal that the origin of these features lies in the “biolar” distribution of the electrostatic surface potentials on the crystallographic structure of MMP-7.
Keywords: MMP-7, Electrostatic surface potentials, Liposomes, Lipid Membranes, Inhibition, Binding, Fluorescence spectroscopy
1. Introduction
Due to their involvement in a variety of human diseases, matrix metalloproteinases (MMPs) have emerged as highly desirable targets for drug design [1,2]. In physiological milieu, the degradation of extracellular matrix (ECM) as well as other selective proteins is accomplished by a coordinated interplay of twenty-four different MMPs [3,4] of which MMP-7 (Matrilysin-1) is structurally the simplest and among the smallest enzyme [5]. MMP-7 is predominantly expressed in epithelial cells, cleaving a variety of cell-associated proteins, such as E-cadherin, TNF-α, Fas ligand, insulin-like growth factors, among others [6,7]. Since some of these non-ECM substrates are confined to the cell membrane, it would make physiological sense if MMP-7 is recruited to the cell surface to hydrolyze its cognate protein substrates. In fact, preliminary data from other laboratories led to the suggestion that MMP-7 interacted with negatively charged lipid membranes [8,9]. This appeared surprising since most MMPs (except for the membrane bound forms) are regarded as soluble proteins that find their cognate targets via randomly diffusing through the extracellular matrix.
On consideration that the catalytic and regulatory features of MMP-7 would be fine tuned if it were to be localized on the plasma membrane, we purported to investigate its potential interaction with model biological membranes, and discern the functional consequences of such interactions. As will be detailed in the following sections, MMP-7 indeed interacts with differently charged biological membranes albeit its affinity is more pronounced when the membrane is constituted of negatively charged lipids. The most interesting feature of this investigation is that whereas the negatively charged (or neutral) membranes do not impair the catalytic activity of the enzyme, the positively charged membranes are drastically inhibitory.
2. Material and Methods
2.1. Buffers
Composition of buffers used for the different experiments is described in the Electronic Supplementary Information (ESI).
2.2. Cloning, Expression, and Purification of recombinant human MMP 7
The enzyme was cloned, expressed in E.coli BL21 (DE3) cells and purified as described by Kannan et.al. [10] with slight modifications as detailed in the ESI.
2.3. Liposome Preparation
Large Unilamellar vesicles (LUVs) containing differently charged lipids were prepared by hydration, sonication and extrusion of thin films, formed by mixing appropriate amounts of chloroformic solutions of desired lipids as previously described [11].
2.4. Determination of MMP-7- liposome interaction via fluorescence spectroscopy
The binding of MMP-7 to differently charged liposomes was determined by measuring the decrease in the protein’s intrinsic fluorescence (λex = 283 nm, λem= 343 nm) and the concurrent increase in the fluorescence of the lipid probe as a function of increasing lipid concentration. DansylPE (λem= 546 nm, Avanti® Polar Lipids, Alabaster, AL) was utilized as the liposome incorporated fluorescence reporter group [12]. All fluorescence measurements were performed using a LS-50B Perkin Elmer® spectroflourimeter. The emission spectra were corrected for the background fluorescence due to increasing dansylPE concentration by performing blank titrations of dansylPE-containing liposomes into buffer.
2.5. Effect of liposomes on the enzymatic activity of MMP-7
The effect of liposomes on MMP-7 activity was determined by measuring the initial rate of the MMP-7 catalyzed reaction using fluorogenic peptide (obtained from Calbiochem, CA) of the following composition: MCA-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2 [Where MCA and Dpa stand for (7-Methoxycoumarin-4-yl)acetyl and N-3-(2,4-dinitrophenyl)-L-2,3-diaminopropionyl, respectively] while maintaining the excitation and emission wavelengths of 335nm and 395 nm, respectively. The time course of the reaction was monitored for about 5 minutes. MMP-7 in reaction mixture was maintained at 1 μM while the liposome concentration (total lipid) varied between 0 and 250 μM.
3. Results and Discussion
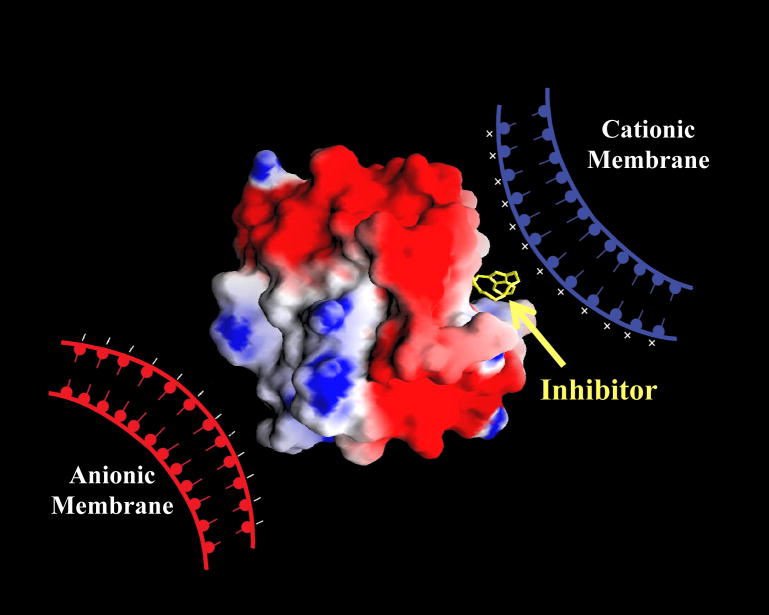
To probe whether MMP-7 exclusive interacts with the negatively charged membranes [8,9], or it also interacts with for neutral and positively charged membranes, we performed binding studies the enzyme to differently charged liposomes as the model membrane system Initially, we observed that MMP-7 strongly interacts with negatively charged liposomes containing 1-palmitoyl–2-oleoyl-sn-glycero-3-phospho-L-serine (POPS). This appeared feasible in the light of the electrostatic potentials on the surface of MMP-7 (Figure 1). Whereas the surface around the active site pocket has negative potential, the distal (opposite) end of the enzyme surface is predominantly positive. Given such a “bipolar” distribution of relative charge density on the protein surface, it appeared likely that MMP-7 would interact with both negative and positively charged lipid membranes, and the latter interaction would inhibit the enzyme activity.
Fig. 1.
Cartoon showing the electrostatic potentials on the surface MMP-7 (pdb1MMQ) containing bound hydroxamate inhibitor, and putative binding surfaces for anionic and cationic lipid membranes. The structural coordinates of MMP-7 were downloaded from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB). The red (−1.9 kcal/mol) and blue (9.8 kcal/mol) colors represent the negative and positive potentials, respectively, calculated via the aid of the GRASP software on a SGI molecular modeling workstation [14–16].
To test the above hypotheses, we determined the influence of liposomes bearing different charges on the catalytic activity of MMP-7. We prepared liposomes with zwitterionic 1-palmitoyl–2-oleoyl-sn-glycero−3-phosphocholine (POPC) as a major (75–95%) component, and other lipids (contributing different net charges) as minor components. For formulating negatively charged liposomes, we utilized anionic lipids; POPS, cholesterol 3-sulphate (CS) and 1,2-dioleoyl-sn-glycero−3-phosphoinositol–4,5-bisposphate (PIP2), which harbour −1, −1, and −3 net charges respectively, in their head groups at neutral pH. For positively charged liposomes, we utilized cationic lipid; 1-palmitoyl−2-oleoyl-sn-glycero−3-ethylphosphocholine (EPOPC), containing +1 net charge on its head group at neutral pH (for structures of these lipids, see Fig. S2 of ESI).
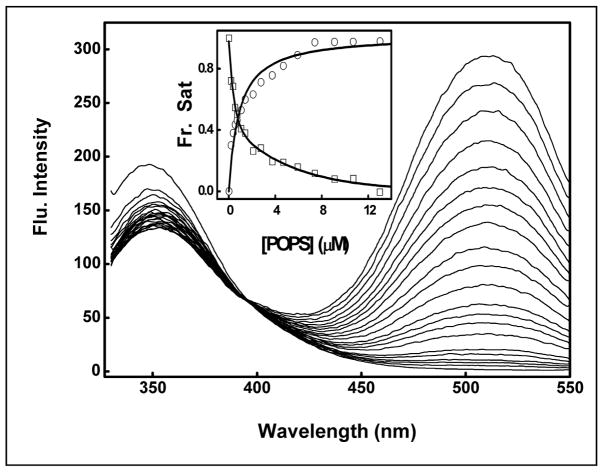
The binding of MMP-7 to liposomes was quantified by monitoring the fluorescence signals of the liposome-dansyl probe as well as that of the enzyme’s tryptophan/tyrosine residues (Fig. 2). Note the systematic decrease in the enzyme’s fluorescence emission intensity (λem = 343 nm) with concomitant increase in the intensity of the dansyl peak (at 526 nm). The overall spectral changes conform to a single isosbestic point at 397 nm, consistent with the fluorescence resonance energy transfer (FRET) from the excited state of the enzyme’s tryptophan/tyrosine residues to the liposome-resident dansyl probe, and its origin must lie in the formation of the enzyme-liposome complex [12]. In addition, the dissociation constants calculated from the liposome concentration-dependent changes in the fluorescence intensities at 343 nm (due to quenching of the enzyme’s fluorescence) and at 526 nm (due to enhancement of the dansyl fluorescence) were found to be remarkably similar, e.g., 0.73μM and 0.87μM, respectively. In case of cationic liposomes, we observed, however, that whereas the intrinsic fluorescence of MMP-7 was quenched (upon titration with the liposome), there as no significant increase in the dansyl peak at 526nm (see Fig. S1 in ESI). This was presumably due to the absence of energy transfer (due to “distance” or “orientation” factor) from the enzyme’s tryptophan/tyrosine residues to the liposome resident dansylPE probe (see Fig. 1). Hence, the Kd of the cationic liposome-MMP-7 complex was determined by measuring the intrinsic fluorescence of the enzyme at 343nm.
Fig. 2.
Fluorescence emission spectra during the course of titration of MMP-7 by anionic liposomes. MMP-7 (1 μM) was titrated with increasing concentrations of liposomes (containing POPC:POPS:dansylPE as 65:25:10 mol%), and the emission spectra (λex = 283 nm) were from 320nm to 550nm. The titration data reveals that whereas the intrinsic fluorescence of MMP-7 is quenched (λem = 343 nm), the fluorescence intensity of the liposome resident dansylPE probe (λem= 526nm; due to energy transfer from the protein) is increased. The inset shows the binding isotherms of liposome-MMP-7 complex, derived from the fluorescence intensity changes at 343nm (squares) and 526nm (circles), respectively. The solid smooth lines are the best fit of the data for the Kd values 0.73 ± 0.07 μM (343 nm peak) and 0.87 ± 0.06 μM (526 nm peak), respectively.
To probe the generality of the above findings, we determined the binding affinities of MMP-7 with liposomes containing other negatively charged lipids and with cationic and neutral liposomes. As is evident from Table 1, the negatively charged liposomes exhibit the highest binding affinity for MMP-7, and there is not much difference in the binding affinity when PIP2 or CS is substituted as the negatively charged lipid. On the other hand, the binding of MMP-7 to neutral (POPC) and positively charged (EPOPC) liposomes is relatively weak. Irrespectively, the data of Table 1 clearly shows that MMP-7 interacts with all (differently charged) liposomes. This was not surprising in light clustered positive and negative charges on the surface of MMP-7 (see Fig. 1), although the putative binding region of neutral liposomes is not clearly apparent.
Table 1.
Kd values of the Liposome-MMP-7 complexes
| Type of Liposomes | Kd (μM) |
|---|---|
| POPC | 13.0 ± 1.4 |
| POPS | 0.73 ± 0.06 |
| PIP2 | 0.05 ± 0.01 |
| CS | 0.03 ± 0.01 |
| EPOPC | 5.0 ± 0.9 |
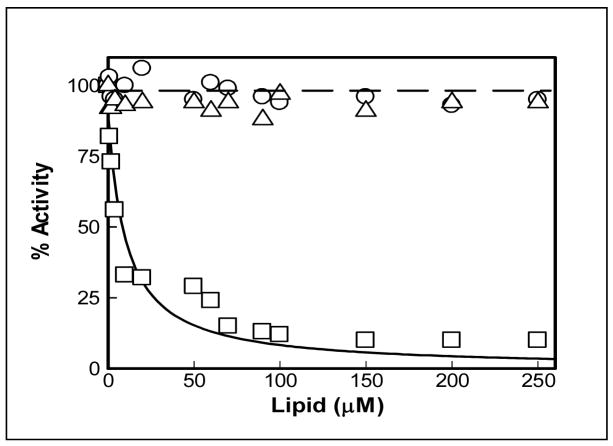
Since the negative charges appeared to be clustered in the vicinity of the active site pocket of MMP-7 (see Fig. 1), it was logical to expect that the enzyme activity would be impaired upon binding of MMP-7 to the positively (but not negatively) charged liposomes. To ascertain this, we determined the activity of MMP-7 in the presence of increasing concentrations of differently charged liposomes (Fig. 3). As expected, the data (Fig. 3) clearly shows that whereas neutral and negatively charged liposomes do not inhibit the MMP-7 catalyzed reaction, the positively charged liposomes exhibit a pronounced inhibitory effect. The analysis of the inhibition data yielded a Ki value of 10 ± 2 μM for the positively charged (EPOPC-containing)liposomes, which emerged out to be comparable to their direct binding affinity (Kd = 5 ± 0.9μM; Table 1). It should be pointed out that the above similarity leads to the suggestion that the inhibition of MMP-7 by cationic liposomes is due to their interaction. However, to exclude the possibility that the above inhibition is not due to binding of substrates to the cationic liposomes (such that the substrates are not available to the enzyme), we performed an analytical HPLC (using a C18 reverse phase column) experiment. In this experiment, we compared the elution profiles of the substrate (20 μM) in the absence and presence of cationic liposomes (200 μM) under the condition where about 90% MMP-7 was inhibited (see Figure 3). The experimental data revealed that the elution time of the substrate remained unchanged in the presence of cationic liposomes, suggesting that the substrate did not interact with the liposomes (see ESI Figure S3). In the light of these data, it appears plausible that the coulombic interaction between the negatively charged active site surface of MMP-7 and the cationic phospholipid head groups of the liposomes blocks the accessibility of substrate (to the active site pocket of the enzyme), causing the inhibition of the enzyme. Since the negatively charged liposomes are unlikely to block the accessibility of the enzyme’s active site pocket for its substrate (due to potential coulombic repulsion), they do not exhibit the inhibitory effect. The site specific binding of anionic versus cationic liposomes is also apparent from the difference in fluorescence emission profiles of the MMP-7-liposome complexes (compare Figure 2 and ESI-Fig. S 1).
Fig. 3.
Effect of differently charged liposomes on MMP-7 activity. The enzyme-catalyzed hydrolysis of the fluorogenic peptide (λex = 335nm, λ ex = 395 nm) was measured as a function of the liposome concentrations. The symbols ○- ○, △- △, and □- □ represent data obtained in the presence of anionic, neutral, and cationic liposomes, respectively. [MMP-7] = 1μM, [Substrate]= 10 μM. Note that unlike anionic and neutral liposomes, the activity of MMP-7 decreases as a function of increasing concentrations of cationic liposomes. The solid smooth line is the best fit of the data (as described by Banerjee.et.al [17]) for the Ki value of MMP-7-cationic liposome complex as being equal to 10 μM.
In summary, we report herein, for the first time, that MMP-7 not only interacts with anionic lipid membranes, but it also interacts with cationic and neutral lipid membranes. When bound to the negatively charged or neutral membranes, the active site pocket of the enzyme remains unobstructed and the enzyme stays fully active. In contrast, when MMP-7 binds to the positively charged membranes, it is drastically inhibited, and this is presumably due to the orientation of the enzyme’s active site pocket toward the positively charged membrane surface. We surmise that in the physiological milieu, the catalytic activity of MMP-7 could be fine-tuned by selective interaction with membranes of different charge and/or membrane-resident proteins, and the outcome of these studies will lead to the development of therapeutic/diagnostic tools for MMP-7, which is known to be over-expressed in a variety of pathological conditions, including cancers [13].
Supplementary Material
Acknowledgments
This work was supported by NIH and NSF grants CA113746 and DMR-0705767, respectively, to DKS and SM.
Abbreviations
- MMP-7
matrix metalloproteinase-7
- POPC
1-palmitoyl–2-oleoyl-sn-glycero-3- phosphocholine
- dansyl-PE
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N (5-dimethylamino-1-naphthalenesulfonyl) lipid
- POPS
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine
- PIP2
1,2-dioleoyl-sn-glycero-3-phosphoinositol–4,5-bisposphate
- CS
and cholesterol 3-sulphate
- EPOPC
1-palmitoyl–2-oleoyl-sn-glycero-3-ethylphosphocholine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Matter H, Schudok M. Recent advances in the design of matrix metalloprotease inhibitors. Curr Opin Drug Discov Devel. 2004;7:513–535. [PubMed] [Google Scholar]
- 2.Overall CM, Kleifeld O. Tumor microenvironment - opinion: Validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat Rev Cancer. 2006;6:227–239. doi: 10.1038/nrc1821. [DOI] [PubMed] [Google Scholar]
- 3.Vartak DG, Gemeinhart RA. Matrix metalloproteases: underutilized targets for drug delivery. J Drug Target. 2007;15:1–20. doi: 10.1080/10611860600968967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lauer-Fields JL, Juska D, Fields GB. Matrix metalloproteinases and collagen catabolism. Biopolymers. 2002;66:19–32. doi: 10.1002/bip.10201. [DOI] [PubMed] [Google Scholar]
- 5.Yu WH, Woessner FJ., Jr Heparan sulphate proteoglycans as extracellular docking molecules for matrilysin (matrix metalloproteinase 7) J Biol Chem. 2000;275:4183–4191. doi: 10.1074/jbc.275.6.4183. [DOI] [PubMed] [Google Scholar]
- 6.Ii M, Yamamoto H, Adachi Y, Maruyama Y, Shinomura Y. Role of matrix metalloproteinase-7 (matrilysin) in human cancer invasion, apoptosis, growth, and angiogenesis. Exp Biol Med (Maywood) 2006;231:20–27. doi: 10.1177/153537020623100103. [DOI] [PubMed] [Google Scholar]
- 7.Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Ann Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berton A, Selvais C, Lemoine P, Henriet P, Courtoy PJ, Marbaix E, Emonard H. Binding of matrilysin-1 to human epithelial cells promotes its activity. Cell Mol Life Sci. 2007;64:610–620. doi: 10.1007/s00018-007-6415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamamoto K, Higashi S, Kioi M, Tsunezumi J, Honke K, Miyazaki K. Binding of active matrilysin to cell surface cholesterol sulfate is essential for its membrane associated proteolytic action and induction of homotypic cell adhesion. J Biol Chem. 2006;281:9170–9180. doi: 10.1074/jbc.M510377200. [DOI] [PubMed] [Google Scholar]
- 10.Kannan R, Ruff M, Kochins JG, Manly SP, Stoll I, Fahime ME, Noel A, Foidart JM, Rio MC, Dive V, Basset P. Purification of active matrix metalloproteinase catalytic domains and its use for screening of specific stromelysin-3 inhibitors. Protein Expr Purif. 1999;16:76–83. doi: 10.1006/prep.1999.1068. [DOI] [PubMed] [Google Scholar]
- 11.Elegbede AI, Haldar MK, Manokaran S, Kooren J, Roy BC, Mallik S, Srivastava DK. A Strategy for designing “multi-prong” enzyme inhibitors by incorporating selective ligands to the liposomal surface. Chem Commun. 2007;32:3377–3379. doi: 10.1039/b707141h. [DOI] [PubMed] [Google Scholar]
- 12.Vaz WL, Kaufmann K, Nicksch A. Use of energy transfer to assay the association of proteins with lipid membranes. Anal Biochem. 1977;83:385–393. doi: 10.1016/0003-2697(77)90047-1. [DOI] [PubMed] [Google Scholar]
- 13.Ii M, Yamamoto H, Adachi Y, Maruyama Y, Shinomura Y. Role of Matrix Metalloproteinase-7 (Matrilysin) in Human Cancer Invasion, Apoptosis, Growth, and Angiogenesis. Exp Biol Med (Maywood) 2006;231:20–27. doi: 10.1177/153537020623100103. [DOI] [PubMed] [Google Scholar]
- 14.Browner MF, Smith WW, Castelhano AL. Matrilysin- inhibitor complexes: common themes among metalloproteases. Biochemistry. 1995;34:6602–6610. doi: 10.1021/bi00020a004. [DOI] [PubMed] [Google Scholar]
- 15.Johnson LN, Barford D. Electrostatic effects in the control of glycogen phosphorylase by phosphorylation. Protein Sci. 1994;3:1726–1730. doi: 10.1002/pro.5560031011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 17.Banerjee AL, Swanson M, Roy BC, Jia X, Haldar MK, Mallik S, Srivastava DK. Protein surface –assisted enhacement in the binding affinity of an inhibitor for recombinant human carbonic anhydrase –II. J Am Chem Soc. 2004;126:10875–10883. doi: 10.1021/ja047557p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.