Abstract
Objective
To evaluate gene expression by microarray analyses of inflammatory mediators in the sinus mucosa of children with or without chronic rhinosinusitis (CRS).
Design
Prospective molecular genetics analysis of sinus mucosa from pediatric CRS and control patients.
Subjects
Eleven CRS patients who underwent endoscopic sinus surgery and ten children who underwent craniofacial resection or neurosurgical procedures.
Interventions
Gene expression levels of sinus tissue from 6 CRS and 6 control patients were analyzed on Affymetrix HGU133 plus 2.0 array chips. mRNA expression levels of upregulated inflammatory/immune response genes, as well as cytokines of interest, were further evaluated by quantitative RT-PCR.
Results
Gene expression using the Plier algorithm yielded the most consistent grouping of samples. 96 genes were significantly up-regulated more than 2 fold, and 123 genes were down-regulated by at least 50% in the CRS sinus tissues compared with controls (p<0.05). GeneSpring analysis demonstrated significant changes in several ontology categories in the CRS samples, including inflammatory/immune response genes. The chemokines CXCL13 and CXCL5, serum amyloid A, serpinB4 and defensin B1 were highly upregulated (≥ 5-fold). Increased expression of these genes was validated by quantitative RT-PCR in an independent set of tissues. Expression levels of the cytokines IL5, IL6 and IL8 were similar in both cohorts; these results were validated by RT-PCR.
Conclusions
Microarray analyses of sinus mucosa in children with CRS showed an increased expression of inflammatory genes involved in innate and adaptive immune systems. This technology can be successfully used to identify genes implicated in the pathogenesis of pediatric CRS.
INTRODUCTION
The sinus mucosa has developed both innate and adaptive immune mechanisms to protect it from pathogens and environmental irritants. Alterations in either system may make patients susceptible to chronic rhinosinusitis (CRS), which is defined as sinonasal inflammation of the paranasal sinuses manifesting with rhinorhea, nasal congestion, facial pain and headache of more than 12 weeks duration and refractory to medical management1, 2. The etiology of CRS is not well understood, however, it is generally accepted that various proinflammatory mediators and immunoreactive products play a significant role in initiating and sustaining the inflammatory response seen in these patients. Cytokines are proinflammatory mediators that function as part of the adaptive immune system via complex intercellular signals. The cytokines granulocyte macrophage-colony stimulating factor (GM-CSF) and interleukin (IL) 3 have been implicated in adults with non-allergic CRS3. These cytokines, in addition to IL4 and IL5, have been observed at high levels in adult sinus mucosa of CRS patients with allergy3, 4. The chemokine IL8, a pro-inflammatory mediator and chemoattractant produced by macrophages and by epithelial cells, is also increased in adult CRS sinus tissues and IL8 levels appear to correlate with the degree of disease severity5, 6. Interleukin 6, another proinflammatory cytokine, is also elevated in adult CRS patients6 as is RANTES, a chemotactic cytokine3, 4.
The innate immune system, another line of defense for respiratory epithelium, includes the paranasal sinus mucosa, which traps particles and pathogens and removes them from the sinus tissues through mucociliary clearance. Other mechanisms used by the innate immune system include the secretion of broad spectrum antimicrobial peptide products such as beta-defensins7, and the production of acute phase proteins like serum amyloid A (SAA), which is expressed in adult sinonasal tissues8. Properdin, complement 3, and toll like receptors have also been identified in human sinus mucosa of adult CRS and control patients8. However, little information is available on inflammatory mediators and innate immune response agents involved in the pathophysiology of pediatric CRS and these profiles may be different between adults and children.
Genome wide expression array analysis is a relatively new technology in which simultaneous analysis of mRNA expression of the > 30,000 genes in the human genome can be determined. Identification of differentially expressed genes between control and diseased tissues followed by bioinformatics analyses and integration into pathway analyses, has led to increased understanding of pathways and mechanisms wherein inflammation leads to pathology. This has been shown for several systems and diseases including muscle9 and allergy10. Furthermore, adult sinus and nasal mucosa have been evaluated by this technique11, 12. Genes associated with innate host responses, inflammation, cell activation, signal transduction and cellular proliferation were differentially expressed when the nasal polyps of 10 CRS patients (3 allergic, 5 asthmatics, and 2 aspirin sensitive patients) were compared to sphenoid sinus mucosa from 4 control patients undergoing pituitary surgery11. In another study, the inflammatory genes for IL6, IL12A, IL13, and tumor necrosis factor alpha (TNFα), were upregulated when the anterior ethmoid mucosa of 14 adult CRS patients and the nasal mucosa from 4 adult control patients were compared However, these mRNA alterations were not validated by RT-PCR in an independent set of sinus tissues12. As gene microarray analyses has the potential to direct attention to new genes of interest as well as identify new associations between established inflammatory and immune response genes that may be involved in CRS, we utilized this technique on sinus mucosa of children and adolescents with and without CRS. We evaluated the gene expression of mediators in the adaptive and innate immune system to determine whether age impacts the inflammatory and immune mediator profile in CRS.
Materials and Methods
Tissue samples
Sinus tissues from patients who underwent craniofacial and/or neurosurgical procedures for pathologies other than sinusitis served as controls. Ten patients (4 males and 6 females), ages to 152 to 222 months were studied. Exclusion criteria for the control population included a history of sinonasal surgery, current sinonasal infection, sinonasal or allergic symptoms within the previous 3 months, and/or treatment with topical nasal steroids within one month or antihistamines within 3 months prior to surgery. A combination of computerized tomography (CT) scans and/or magnetic resonance imaging with sinonasal cuts were obtained. There was no radiographic evidence of sinusitis at the time of surgery for children in the control group. One patient had a meningioma encroaching on right frontal sinus, but it did not violate the mucosa.
Sinus tissue was obtained from CRS patients who underwent functional endoscopic sinus surgery for CRS refractory to medical management. The clinical data of eleven patients (8 males and 3 females) with CRS, ages 30 to 215 months, are shown in Table 1. Chronic rhinosinusitis was defined as persistent symptoms for more than 3 months, despite antimicrobial and topical nasal steroid therapies13. Patient criteria for entry into the study was identical to that of patients recruited for earlier studies14. Patients with sinusitis had at least two of the major signs and symptoms of chronic sinusitis: nasal congestion, rhinorhea, headache, facial pain/pressure, or change in olfaction2, 15. In addition, all these children had at least one of the following minor signs or symptoms in conjunction with a minimum of two major signs and symptoms: fever, halitosis, cough or irritability2, 15. All patients with CRS were on antibiotics and/or topical nasal steroids at the time of surgery. Patients with cystic fibrosis, ciliary dyskinesias and craniofacial abnormalities were excluded. CT scans of the sinuses were obtained on all CRS patients, evaluated and scored using the Lund-MacKay system15, 16. All CT scans received a minimum score of 8.
Table 1.
Clinical Profile of Pediatric CRS Patients
| Patient | Sex | Age (mo) | RAD* | Atopy | Analyses Performed |
|---|---|---|---|---|---|
| 1 | M | 149 | Y | N | Microarray |
| 2 | M | 94 | N | Y | Microarray |
| 3 | F | 215 | Y | Y | Microarray |
| 4 | F | 129 | Y | Y | Microarray |
| 5 | M | 89 | N | Y | Microarray |
| 6 | M | 32 | Y | N | Microarray |
| 7 | M | 76 | N | Y | Real time PCR |
| 8** | M | 30 | Y | Y | Real time PCR |
| 9 | M | 89 | N | N | Real time PCR |
| 10 | F | 213 | N | N | Real time PCR |
| 11 | M | 57 | N | N | Real time PCR |
RAD, reactive airway disease (asthma).
Patient with immunoglobin deficiency.
All patients were entered consecutively into the study after appropriate surgical and research consents (and assents when applicable) were obtained. This study was reviewed and approved by the Institutional Review Board, Children’s National Medical Center (CNMC), Washington, D.C.
Clinical Data
Patient age (months), gender, atopy, presence of reactive airway disease, immunodeficiency, and use of systemic steroids were recorded. Atopy was categorized present (yes) or absent (no) and was based on results of prick puncture or intradermal skin testing whenever possible. Immunologic status was determined from quantitative analyses of total immunoglobulins and IgG subclasses in serum, as well as antibody responses to pneumococcal vaccine challenge17. CT scans of sinuses were also reviewed.
Collection of Specimens
Using standard endoscopic instruments, mucosa from the right and left paranasal sinuses was collected separately at the time of surgery. Specimens ranged from 0.2 to 1.3 cm3. In control patients, the mucosa of the paranasal sinuses, entered as a result of the craniofacial and/or neurosurgical procedure performed, was collected in an identical manner to that of the sinusitis population. Specimens were immediately frozen in liquid nitrogen for RNA isolation.
Gene expression profiling and microarray data analysis
Sample preparation and microarray analyses were performed in the Microarray Core at CNMC, according to established protocols18 based on Affymetrix standard methodology (http://www.affymetrix.com). Total RNA from frozen mucosal samples was isolated using Trizol (Invitrogen, Carlsbad, CA) and purified with RNeasy MinElute kit (Qiagen, Valencia, CA) according to the manufacturer’s protocols. RNA was quantified on a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). Purity and integrity were assessed using the Agilent 2100 Bioanalyzer. All samples showed intact 28S and 18S ribosomal RNA bands in an approximate ratio of 2:1. Five microgram of total RNA was used to initiate the cDNA-cRNA cycle. Both cDNA synthesis and in vitro transcription were performed using GeneChip® Expression 3’Amplification One-Cycle Target Labeling and Control Reagent kit according to manufacturer’s instructions. Twenty microgram of biotin-labeled cRNA from each sample was fragmented and hybridized to Affymetrix Human Genome U133_Plus2.0 Arrays for 16 h, followed by standard washing-scanning protocols on the Affymetrix Fluidics Station 400 and incubation with phycoerythrin-streptavidin to detect bound cRNA. The signal intensity was amplified using biotin labeled anti-streptavidin antibody. Fluorescent images were captured using a Hewlett-Packard G2500A gene Array Scanner.
All data were analyzed using Affymetrix Microarray Analysis Software version 5.0 (MAS 5.0), dChip and Plier algorithms. Only probe sets that were statistically significant with one-way ANOVA and a p-value “cut off” less than 0.05 were used. The data set from each algorithm was loaded into a data mining program GeneSpring GX program (version 7.3.1) (Silicon Genetics) for future analysis. The results were visualized using the gene tree cluster feature of the program, which rearranges the order of the probe sets and groups them based on the similarity (Pearson’s correlation) of their expression dynamics 18. Hierarchical clustering analysis of all three algorithms data sets were performed using the Hierarchical Clustering Explorer 3.5 program19. Similarity and distance measures were assessed by Pearson Correlation Coefficient. Multivariate analyses of the expression array data set of immune and inflammatory mediator genes were performed for the control and CRS patients according to age.
Real Time RT-PCR Analysis
Sinus mucosa was homogenized in Trizol reagent (Invitrogen, Carlsbad, CA). RNA was isolated per standard techniques, followed by cleaning with RNeasy MinElute kit. Total RNA yields were measured on the NanoDrop spectrophotometer and RNA quality was visualized using agarose gel electrophoresis. Four micrograms of RNA, treated with DNAse I (Invitrogen) for 15 min at room temperature, was used to synthesize first stand cDNA with SuperScript III reverse transcription kit (Invitrogen). Real time PCR was performed with power SYBR green PCR reagent kit (Applied Biosystems, Foster City, CA) in an ABI Prism® 7900HT Sequence Detection System. mRNA for target genes and the house-keeping gene β-actin were amplified and quantified in the sample reaction plate. Primers were designed with Primer3 software to cross the intron-exon gap so as to prevent amplification of any genomic DNA contamination. Tm was between 59° – 61°C. All procedures were performed according to the manufacturer’s instructions. Standard curves were generated based on five 5-fold serial dilutions of a starting concentration of a control cDNA (150 µg/µl). PCR reactions were incubated at 50°C (2 min), 95 °C (2 min), and then 40 cycles of 95 °C (15 sec) and 60 °C (30 sec). Ct (threshold cycle) and baseline were automatically calculated using the SDS 2.2 software. A standard curve plotting CT values against the logarithm of template cDNA input was constructed for each gene. The numbers of copies in the experimental samples were calculated from the equation of the straight line for the standard curve. The final quality of each gene was normalized with housekeeping gene expression. The statistical significance of differences was determined by student’s t-test. Data were expressed as mean values ± SE. Differences were considered statistically significant at p<0.05.
RESULTS
Clinical Data
Twenty-one patients ranging in age from 30 – 220 months were studied. The median age of the pediatric patients with CRS was 94 months (range 30–215 months), compared with the median age of 204.5 months (range 152 –222 months) in the control subjects. Clinical profiles of the CRS patients are summarized in Table 1. Patient 8 had an immunoglobulin subclass deficiency. Patients with CRS had a score of 8 or more on CT scan by Lund-Mackay staging16.
Expression array analyses of sinus mucosa from control and CRS patients
To identify the differential expression of genes in the sinus mucosa of control and CRS patients, a genome-wide microarray analysis of the sinus mucosa from 12 individuals (6 CRS patients and 6 controls) was performed using Affymetrix Human Genome U133_2.0 Arrays. Data interpretation is often profoundly affected by the type of algorithm used20. Thus in order to obtain the most accurate data set of differentially expressed genes, data were analyzed separately by MAS 5.0, dChip and Plier algorithms, then loaded into GeneSpring, where the data were normalized and filtered. Hierarchical cluster analysis showed that the Plier algorithm yielded the most consistent grouping of samples in the three groups. The overall F-measure score, which can range between 0–1with the higher F-measure score reflecting better clustering results in biological samples19, showed that the Plier algorithm at F = 0.681 had the highest F-measure score. Therefore, the Plier software program was used for data presentation of each probe set in each of the twelve sinus mucosa samples for the control and CRS cohorts (Figure 1). Using a Venn Diagram, the statistically significant genes (p<0.05) from each algorithm were overlapped to yield a list of differentially expressed genes common to all algorithms (Figure 2). The data showed that 576 probe sets out of 54,000 probe sets (47,000 transcripts) were significantly differentially expressed in CRS samples (p<0.05). 96 genes were significantly up-regulated more than 2 fold and 123 genes were down-regulated by at least 50% (p<0.05), compared with control samples. We focused our initial analyses on inflammatory/immune response genes, which have been implicated in adult CRS.
Figure 1.
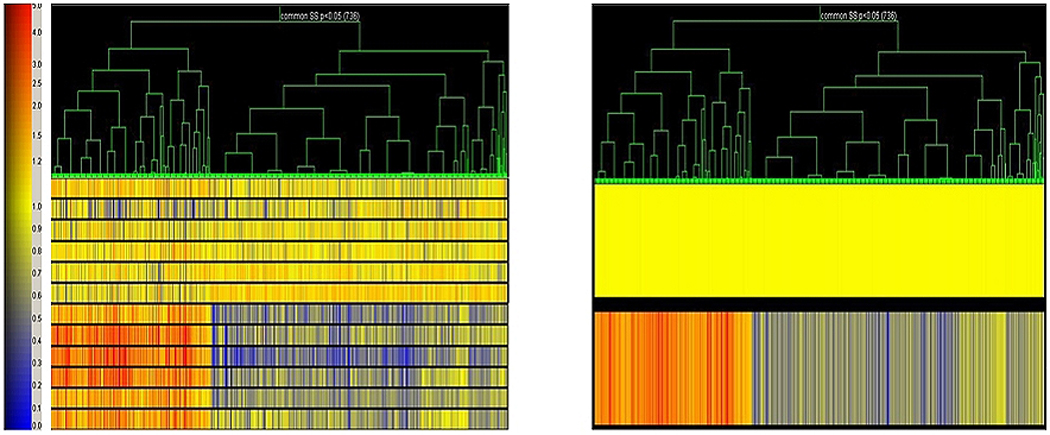
Gene tree analyses (Plier) of sinus tissues. Differential expression levels of each probe set are shown vertically for each patient sample. Red represents increased and blue decreased expression compared with control.
Figure 2.
Venn diagram showing the number of the statistically significantly altered probe sets in three algorithms: MAS5, dCHIP and Plier. The white color denotes the 576 common significantly changed probe sets in all three algorithm, p<0.05.
Microarray Analyses of Inflammatory/Immune Response Genes
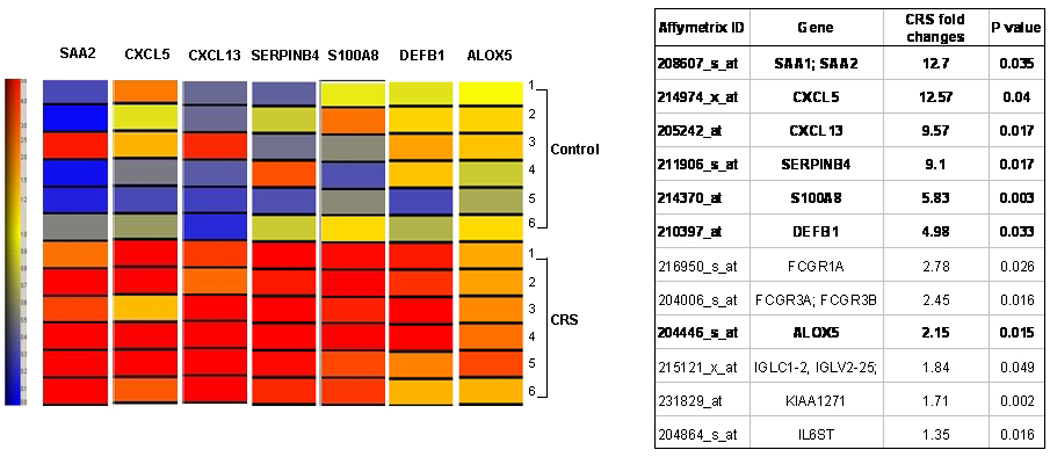
Twelve genes related to inflammatory/immune response were significantly increased (p<0.05) (Table 2). Five of these genes, including the B-cell chemoattractant chemokine (C-X-C motif) ligand 13 (CXCL13) and ligand 5 (CXCL5), serum amyloid A2 (SAA2), S100 calcium binding protein A8 (S100A8) and serpin peptidase inhibitor, member 4 (SERPINB4), were increased more than 5 fold. Beta-defensin 1 (DEFB1), arachidonate 5-lipoxygenase (ALOX5) and two IgG affinity receptors were increased more than 2 fold.
Table 2.
Up-regulated inflammatory and immune response gene list (P<0.05) identified by microarray expression profiling of sinus mucosa from 6 CRS and 6 control patients.
| Affymetrix ID | Gene | CRS fold changes |
P value | Gene Description |
|---|---|---|---|---|
| 208607_s_at | SAA1; SAA2 | 12.70 | 0.035 | Serum amyloid A1 ; Serum amyloid A2 |
| 214974_x_at | CXCL5 | 12.57 | 0.040 | Chemokine (C-X-C motif) ligand 5 |
| 205242_at | CXCL13 | 9.57 | 0.017 | Chemokine (C-X-C motif) ligand 13 (B-cell chemoattractant) |
| 211906_s_at | SERPINB4 | 9.10 | 0.017 | Serpin peptidase inhibitor, clade B (ovalbumin), member 4 |
| 214370_at | S100A8 | 5.83 | 0.003 | S100 calcium binding protein A8 (calgranulin A) |
| 210397_at | DEFB1 | 4.98 | 0.033 | Defensin, beta 1 |
| 216950_s_at | FCGR1A | 2.78 | 0.026 | Fc fragment of IgG, high affinity Ia, receptor (CD64) |
| 204006_s_at | FCGR3A; FCGR3B | 2.45 | 0.016 | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) ; Fc fragment of IgG, low affinity IIIb, receptor (CD16b) |
| 204446_s_at | ALOX5 | 2.15 | 0.015 | Arachidonate 5-lipoxygenase |
| 215121_x_at | IGLC1–2, IGLV2–25; | 1.84 | 0.049 | Immunoglobulin lambda locus ; immunoglobulin lambda constant 1 (Mcg marker) ; immunoglobulin lambda constant 2 (Kern-Oz- marker) ; immunoglobulin lambda variable 2–25 |
| 231829_at | KIAA1271 | 1.71 | 0.002 | KIAA1271 protein |
| 204864_s_at | IL6ST | 1.35 | 0.016 | Interleukin 6 signal transducer |
Note: Expression levels of genes in bolded font were also analyzed by Real time RT-PCR.
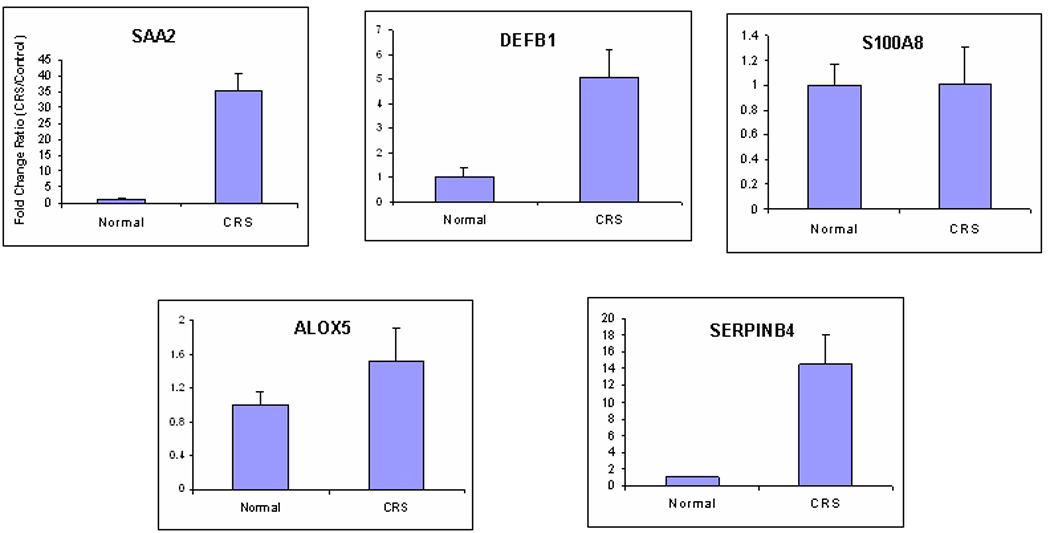
Validation of Inflammatory/Immune Response Gene mRNA Expression Levels by Real-Time PCR
The expression levels of seven inflammatory/immune response genes – CXCL5, CXCL13, SAA2, SERPINB4, S100A8, DEFB1 and ALOX5 – (bolded in Table 2) were evaluated by real-time PCR in an independent set of sinus tissues from 5 CRS patients (Table 1) and 4 control patients. Data showed that mRNA levels for CXCL13, CXCL5, SAA2, SERPINB4 and DEFB1 were increased in the sinus mucosa of CRS patients compared to sinus mucosa of control patients (Figure 3). These results are consistent with the microarray analyses (Table 2), although the rank order and magnitude of gene expression levels differed between the two methods. mRNA levels of S100A8 and ALOX5, which were significantly increased in CRS tissues by microarray analyses, were, however, not significantly increased when analyzed by real time RT-PCR (Figure 3).
Figure 3.
Quantitative RT-PCR analyses of seven up-regulated genes from microarray study. Data presented shows fold-change in transcript levels of control (n=4) and CRS (n=5) tissues +/− SE. Asterisk (*) denotes statistically significant differences (p<0.05) between the two groups.
Analyses of cytokines in the sinus mucosa of CRS and control samples
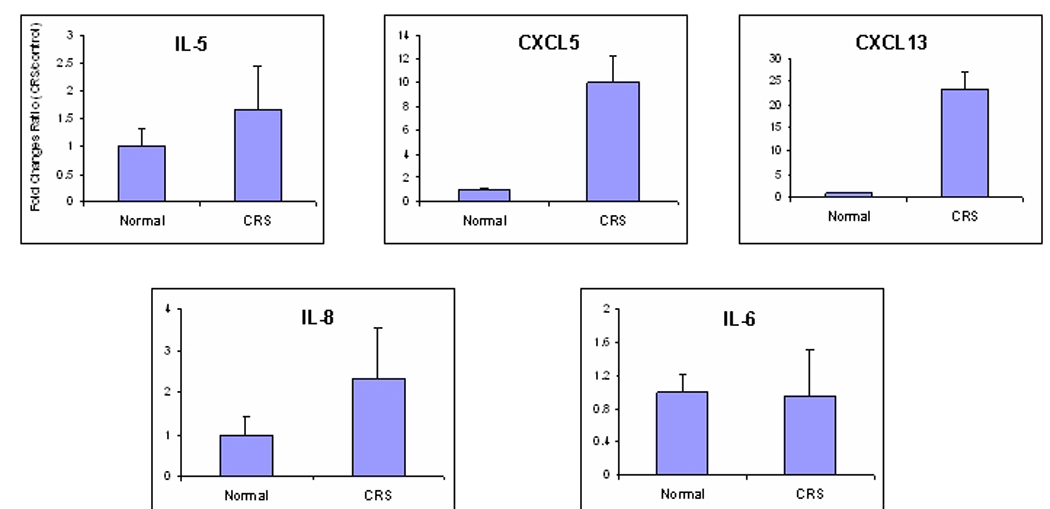
A comparison of the expression levels of most of the cytokines of interest in the CRS literature is shown in Table 3. Expression levels between the control and CRS cohorts were either similar or not statistically different for IL4, IL5, IL13, GM-CSF and various members of the TNF super family (SF), e.g. TNFSF11, TNFSF4, and TNFSF8. IL3 expression was increased in CRS, but the increase was not statistically significant. However, expression levels of IL6, IL8, IL12A, TNFSF7, TNFSF9, and TNFSF12 were significantly decreased in CRS sinus mucosa. Real-time PCR analyses of IL5, IL6 and IL8 were performed with the independent set of sinus mucosa used for validation in Figure 3. Data showed that the expression levels of these cytokines were variable, but were not significantly altered in CRS tissues (Figure 4).
Table 3.
Comparison of mRNA expression levels of differentially expressed cytokines identified by microarray expression profiling in the sinus mucosa of CRS and control patients.
| Affymetrix ID | Gene | CRS fold changes | P value |
|---|---|---|---|
| 207906_at | IL3 | 1.743 | 0.068 |
| 207538_at | IL4 | 1.044 | 0.749 |
| 207952_at | IL5 | 0.865 | 0.311 |
| 205207_at | IL6 | 0.359 | 0.001 |
| 211506_s _at | IL8 | 0.461 | 0.006 |
| 207160_at | IL12A | 0.548 | 0.001 |
| 207844_at | IL13 | 1.028 | 0.632 |
| 210643_at | TNFSF11 | 1.774 | 0.409 |
| 206508_at | TNFSF7 | 1.347 | 0.016 |
| 207426_s_at | TNFSF4 | 1.260 | 0.566 |
| 206907_at | TNFSF9 | 0.879 | 0.036 |
| 235735_at | TNFSF8 | 0.871 | 0.256 |
| 205611_at | TNFSF12 | 0.666 | 0.007 |
| 210228_at | GM-CSF | 0.999 | 0.986 |
Note: Expression levels of genes in bolded font were also analyzed by Real time RT-PCR.
Figure 4.
Quantitative RT-PCR validation of IL5, IL6, and IL8 in pediatric sinus mucosa. Data was performed and presented as in Figure 2. No statistically significant differences in expression between the control and CRS groups were observed.
Analysis of age and inflammatory gene expression
Multivariate analyses of the expression array data sets of the immune and inflammatory mediator genes shown in Table 3 were performed for the control and CRS patients according to age. The age of the control patients was significantly higher in both the microarray group (p=0.0064) and real time RT-PCR patients (p=0.032) compared with the diseased cohort. Age was not a significant factor when adjusted for disease status. Statistical analyses adjusted for age were inconclusive (data not shown).
DISCUSSION
Gene microarrays are now a standard experimental platform for investigating gene expression in different biological tissues and to identify differential gene expression in control and diseased tissues, including adult CRS tissues11, 12. This is the first study to use gene microarray technology to begin to analyze the inflammatory/immune response gene products that may be involved in the pathophysiology of pediatric CRS. However, due to the variations in clinical profiles of the disease cohort including the differences in age, medical management, length of medical management, duration of the disease, as well as presence of allergy and reactive airway disease, the results have to be interpreted somewhat cautiously because of the inability to eliminate confounding factors in the patient population. Nevertheless, the stratification of the CRS and non-CRS cohorts shown in Fig. 1 is encouraging.
In this preliminary analysis, 5 inflammatory/immune response gene products were shown by Affymetrix microarray technology and real-time RT-PCR to be highly and significantly upregulated in pediatric CRS sinus mucosa. Two of these gene products were the cytokines CXCL5 and CXCL13, both potent chemoattractants of B lymphocytes, which are cells not routinely associated with pediatric CRS 21.
Cytokines are typically small proteins that are part of the adaptive immune system and thought to play an integral role in the pathophysiology of CRS. They include interleukins, chemokines, interferons, colony stimulating factors, and tumor necrosis factors. The deregulation of cytokines is believed in large part, to be responsible for the excessive inflammation seen in the sinus mucosa of CRS patients. In the sinus mucosa of adults, elevated levels of the cytokines GM-CSF, IL3, IL4, IL5, IL6, RANTES, and IL8 have been reported3–6. IL6, IL12A, IL13, and TNF-α ave been shown by microarray analyses to be increased in the sinus mucosa of patients with non-allergic CRS without nasal polyps12. Interestingly, none of these cytokines exhibited increased expression in our microarray analyses of sinus mucosa from pediatric CRS and control patients. In fact, levels of IL6, IL8 and IL12A and several TNF factors were significantly decreased in the microarray data sets (Table 3). Additionally, IL5, IL6, and IL8 levels were not significantly altered when evaluated by real-time RT-PCR in an independent set of sinus mucosa tissues (Figure 4).
The other three genes that exhibited markedly increased upregulation in this study were SAA2, SERPINB4 and DEFB1, which genes encode for proteins that are involved in the innate immune system. SAA2 is an acute phase protein, DEFB1 is a broad spectrum antimicrobial protein and SERPINB4 neutralize serine or cysteine proteinases. Serum amyloid A has previously been shown to be expressed in sinonasal tissue of control and CRS patients22. DEFB1 has been identified in the lung7. SERPINB4 has been detected in the sinonasal squamous epithelium23. Other innate immunity mediators such as properdin, complement 3, and toll like receptors have also been shown to be expressed in the sinus mucosa of control and CRS patients8, 22 Altered expression of TLR2 was not observed in our studies, although it is reported increased in one study22 but not in another8.
The microarray gene profile generated by this study indicates that the sinus mucosa in children with CRS is immunocompetent and that both arms of the immune system are involved. Innate immunity is evolutionarily conserved and normally acts in concert with the adaptive arm of the immune system. Both adults and children are expected to utilize these functional mechanisms and upregulation of components of the innate immune system in our study is not unexpected. While there are similarities to adult patients with respect to the innate immune system especially with regard to SAA, the cytokine gene expression pattern is different, suggesting that pediatric CRS is not the same entity as adult CRS. This is not a surprising conclusion and has been supported in the literature with respect to other variables. Chan et al. have demonstrated that the sinus mucosa of children with CRS has less eosinophilic inflammation, basement membrane thickening, and mucus gland hyperplasia compared to adults with CRS 24.
Other investigators have shown that age can impact the composition and magnitude of the inflammatory mediators25. We considered whether the variation in the age of the patients studied impacted the identification of immune and inflammatory mediator genes with microarray technique Statistical analyses adjust for age were inconclusive due to the small sample size and large variations in expression levels of these gene products. Therefore, no definite conclusion can be made with respect to the impact age has on the inflammatory and immune mediators identified in this investigation.
In conclusion, this study shows that gene array profiling is a powerful technique that can be used successfully to identify the inflammatory and immune mediator profiles in children with CRS and that different mediators are seen in pediatric CRS compared to adult CRS. Validation of specific differentially expressed genes by an independent method and in an independent set of sinus mucosal tissues demonstrated that specific genes in both arms of the immune system are markedly upregulated in the sinus mucosa of CRS pediatric patients.
Acknowledgements
The authors thank Dr. Heather Gordish-Dressman for statistical analyses of inflammatory genes with regard to patient age and Drs. Diego Preciado and Dr. Hugo Escobar for their review and critique of the study.
Funding Support: This study was supported in part by NIH grants from the General Clinical Research Center (GCRC 5-MO1-RR-020359-02) and the National Center for Medical Rehabilitation Research (NCMRR 5R24HD050846-02).
Footnotes
Paper will be presented at the 2008 Annual ASPO meeting held May 2 – 4, 2008
REFERENCES
- 1.Kennedy DW. Pathogenesis of chronic rhinosinusitis. Ann Otol Rhinol Laryngol Suppl. 2004 May;193:6–9. doi: 10.1177/00034894041130s503. [DOI] [PubMed] [Google Scholar]
- 2.Lusk RP, Stankiewicz JA. Pediatric rhinosinusitis. Otolaryngol Head Neck Surg. 1997 Sep;117(3 Pt 2):S53–S57. doi: 10.1016/S0194-59989770008-1. [DOI] [PubMed] [Google Scholar]
- 3.Hamilos DL, Leung DY, Wood R, et al. Evidence for distinct cytokine expression in allergic versus nonallergic chronic sinusitis. J Allergy Clin Immunol. 1995 Oct;96(4):537–544. doi: 10.1016/s0091-6749(95)70298-9. [DOI] [PubMed] [Google Scholar]
- 4.Hamilos DL, Leung DY, Huston DP, Kamil A, Wood R, Hamid Q. GM-CSF, IL-5 and RANTES immunoreactivity and mRNA expression in chronic hyperplastic sinusitis with nasal polyposis (NP) Clin Exp Allergy. 1998 Sep;28(9):1145–1152. doi: 10.1046/j.1365-2222.1998.00380.x. [DOI] [PubMed] [Google Scholar]
- 5.Rhyoo C, Sanders SP, Leopold DA, Proud D. Sinus mucosal IL-8 gene expression in chronic rhinosinusitis. J Allergy Clin Immunol. 1999 Mar;103(3 Pt 1):395–400. doi: 10.1016/s0091-6749(99)70462-8. [DOI] [PubMed] [Google Scholar]
- 6.Lennard CM, Mann EA, Sun LL, Chang AS, Bolger WE. Interleukin-1 beta, interleukin-5, interleukin-6, interleukin-8, and tumor necrosis factor-alpha in chronic sinusitis: response to systemic corticosteroids. Am J Rhinol. 2000 Nov–Dec;14(6):367–373. doi: 10.2500/105065800779954329. [DOI] [PubMed] [Google Scholar]
- 7.Singh PK, Jia HP, Wiles K, et al. Production of beta-defensins by human airway epithelia. Proc Natl Acad Sci U S A. 1998 Dec 8;95(25):14961–14966. doi: 10.1073/pnas.95.25.14961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lane AP, Truong-Tran QA, Myers A, Bickel C, Schleimer RP. Serum amyloid A, properdin, complement 3, and toll-like receptors are expressed locally in human sinonasal tissue. Am J Rhinol. 2006 Jan–Feb;20(1):117–123. [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao P, Seo J, Wang Z, Wang Y, Shneiderman B, Hoffman EP. In vivo filtering of in vitro expression data reveals MyoD targets. C R Biol. 2003 Oct–Nov;326(10–11):1049–1065. doi: 10.1016/j.crvi.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 10.Benson M, Jansson L, Adner M, Luts A, Uddman R, Cardell LO. Gene profiling reveals decreased expression of uteroglobin and other anti-inflammatory genes in nasal fluid cells from patients with intermittent allergic rhinitis. Clin Exp Allergy. 2005 Apr;35(4):473–478. doi: 10.1111/j.1365-2222.2005.02206.x. [DOI] [PubMed] [Google Scholar]
- 11.Liu Z, Kim J, Sypek JP, et al. Gene expression profiles in human nasal polyp tissues studied by means of DNA microarray. J Allergy Clin Immunol. 2004 Oct;114(4):783–790. doi: 10.1016/j.jaci.2004.04.052. [DOI] [PubMed] [Google Scholar]
- 12.Anand VK, Kacker A, Orjuela AF, Huang C, Manarey C, Xiang J. Inflammatory pathway gene expression in chronic rhinosinusitis. Am J Rhinol. 2006 Jul–Aug;20(4):471–476. doi: 10.2500/ajr.2006.20.2891. [DOI] [PubMed] [Google Scholar]
- 13.Gandhi A, Brodsky L, Ballow M. Benefits of antibiotic prophylaxis in children with chronic sinusitis: assessment of outcome predictors. Allergy Proc. 1993 Jan–Feb;14(1):37–43. doi: 10.2500/108854193778816833. [DOI] [PubMed] [Google Scholar]
- 14.Pena MT, Aujla PK, Patel KM, Zalzal GH, Rose MC. Immunohistochemical analyses of MUC5AC mucin expression in sinus mucosa of children with sinusitis and controls. Ann Otol Rhinol Laryngol. 2005 Dec;114(12):958–965. doi: 10.1177/000348940511401212. [DOI] [PubMed] [Google Scholar]
- 15.Lund VJ, Kennedy DW The Staging and Therapy Group. Quantification for staging sinusitis. Ann Otol Rhinol Laryngol Suppl. 1995 Oct;167:17–21. [PubMed] [Google Scholar]
- 16.Lund VJ, Mackay IS. Staging in rhinosinusitus. Rhinology. 1993 Dec;31(4):183–184. [PubMed] [Google Scholar]
- 17.Hilleman MR, Carlson AJ, Jr, McLean AA, Vella PP, Weibel RE, Woodhour AF. Streptococcus pneumoniae polysaccharide vaccine: age and dose responses, safety, persistence of antibody, revaccination, and simultaneous administration of pneumococcal and influenza vaccines. Rev Infect Dis. 1981 Mar–Apr;(3 Suppl):S31–S42. doi: 10.1093/clinids/3.supplement_1.s31. [DOI] [PubMed] [Google Scholar]
- 18.Almon RR, DuBois DC, Yao Z, Hoffman EP, Ghimbovschi S, Jusko WJ. Microarray analysis of the temporal response of skeletal muscle to methylprednisolone: comparative analysis of two dosing regimens. Physiol Genomics. 2007 Aug 20;30(3):282–299. doi: 10.1152/physiolgenomics.00242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seo J, Gordish-Dressman H, Hoffman EP. An interactive power analysis tool for microarray hypothesis testing and generation. Bioinformatics. 2006 Apr 1;22(7):808–814. doi: 10.1093/bioinformatics/btk052. [DOI] [PubMed] [Google Scholar]
- 20.Seo J, Hoffman EP. Probe set algorithms: is there a rational best bet? BMC Bioinformatics. 2006;7:395. doi: 10.1186/1471-2105-7-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peña MT, Brodsky L, Gorfien J, Noble B. Immunohistochemical Analysis of Mononuclear Inflammatory Cells in Nasal and Sinus Epithelium in Children With Sinusitis. American Journal of Rhinology. 1996;10:149–159. [Google Scholar]
- 22.Lane AP, Truong-Tran QA, Schleimer RP. Altered expression of genes associated with innate immunity and inflammation in recalcitrant rhinosinusitis with polyps. Am J Rhinol. 2006 Mar–Apr;20(2):138–144. [PMC free article] [PubMed] [Google Scholar]
- 23.Yasumatsu R, Nakashima T, Kuratomi Y, et al. Serum squamous cell carcinoma antigen is a useful biologic marker in patients with inverted papillomas of the sinonasal tract. Cancer. 2002 Jan 1;94(1):152–158. doi: 10.1002/cncr.10144. [DOI] [PubMed] [Google Scholar]
- 24.Chan KHAM, Coffinet L, Simoes EA, Cool C, Liu AH. Chronic rhinosinusitis in young children differs from adults: a histopathology study. Journal of Pediatrics. 2004;144:206–212. doi: 10.1016/j.jpeds.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 25.Unden AL, Andreasson A, Elofsson S, et al. Inflammatory cytokines, behaviour and age as determinants of self-rated health in women. Clin Sci (Lond) 2007 Jun;112(6):363–373. doi: 10.1042/CS20060128. [DOI] [PubMed] [Google Scholar]


