Abstract
Numerous reports have indicated the important role of human normal flora in the prevention of microbial pathogenesis and disease. Evidence suggests that infections at mucosal surfaces result from the outgrowth of subpopulations or clusters within a microbial community and are not linked to one pathogenic organism alone. To preserve the protective normal flora while treating the majority of infective bacteria in the community, a tuneable therapeutic is necessary that can discriminate between benign bystanders and multiple pathogenic organisms. Here we describe the proof-of-principle for such a multitargeted antimicrobial: a multiple-headed specifically-targeted antimicrobial peptide (MH-STAMP). The completed MH-STAMP, M8(KH)-20, displays specific activity against targeted organisms in vitro (Pseudomonas aeruginosa and Streptococcus mutans) and can remove both species from a mixed planktonic culture with little impact against untargeted bacteria. These results demonstrate that a functional, dual-targeted molecule can be constructed from a wide-spectrum antimicrobial peptide precursor.
Keywords: Antimicrobial peptide, Targeted therapeutic, Streptococcus mutans, Pseudomonas aeruginosa, Peptide synthesis, Novel antibiotic, STAMP, Specifically-targeted antimicrobial peptide, MH-STAMP
1. Introduction
For nearly 30 years antimicrobial peptides (AMPs) have been rigorously investigated as alternatives to small molecule antibiotics and as potential solutions to the growing crisis of antibiotic-resistant bacterial infections [1,2]. Numerous reports have characterised potential AMPs from natural sources and a great body of work has been carried out designing ‘tailor-made’ AMPs owing to the approachable nature of solid-phase peptide synthesis (SPPS) [3,4]. Several examples of the latter have shown remarkable activities in vitro against fungi, Gram-positive and Gram-negative bacteria as well as some enveloped viruses [5].
Unlike small molecule antibiotics that may lose activity when their basic structures are modified even incrementally, peptides are a convenient canvas for molecular alteration. AMPs can be optimised through the incorporation of more or less hydrophobic or charged amino acids, which has been shown to affect selectivity for Gram-positive, Gram-negative or fungal membranes [6,7]. Additionally, lysine residues can be utilised to improve AMP activity per μM. In this approach, multiple AMP chains can be attached to a single peptide scaffold through branching from lysine ε-amines [8,9].
AMP activity can be specifically tuned through the attachment of a targeting peptide region, as described for a novel class of molecules, the specifically-targeted antimicrobial peptides, or STAMPs [10,11]. These chimeric molecules consist of functionally independent targeting and killing moieties within a linear peptide sequence. A pathogenic bacterium recognised (i.e. bound) by the targeting peptide can be eliminated from a multispecies community with little impact to bystander normal flora. As an extension of this concept, we hypothesised that a STAMP could be constructed with multiple targeting peptide ‘heads’ attached to a single AMP by utilising a central lysine residue branch point. Potentially, targeting ‘heads’ could be specific for the same pathogen or have different binding profiles. Utilising the former approach, microbial resistance evolution linked to a targeting peptide could be inhibited or reduced, as no single microbial population would have the genetic diversity necessary to mutate multiple discrete targeting peptide receptors in one cell [12].
Multiple-headed STAMP (MH-STAMP) molecules with differing bacterial targets may have appeal in treating polymicrobial infections or where it may be advantageous to remove a cluster of biofilm constituents without utilising several distinct molecules, for example in the simultaneous treatment of dental caries and periodontitis or in the eradication of Propionibacterium spp. and Staphylococcus spp. involved in acne and skin infections, respectively.
In this report, we present the proof-of-principle design, synthesis and in vitro activity of such a MH-STAMP, M8(KH)-20. Previously, we identified two functional STAMP targeting domains, one with specific recognition of the cariogenic pathogen Streptococcus mutans [10] and the other with Pseudomonas spp.-level selectivity [13]. Conjoined to a normally wide-spectrum linear AMP, we observed antimicrobial effects directed specifically to Pseudomonas aeruginosa and S. mutans in vitro. Additionally, treatment of mixed bacterial communities with the MH-STAMP resulted in specific eradication of the target organisms with little impact on bystander population levels.
2. Materials and methods
2.1. Bacterial strains and growth conditions
Pseudomonas aeruginosa ATCC 15692, Klebsiella pneumoniae KAY 2026 [14], Escherichia coli DH5α (pFW5, spectinomycin resistance) [15], Staphylococcus aureus Newmann [16] and Staphylococcus epidermidis ATCC 35984 were cultivated under aerobic conditions at 37°C with vigorous shaking. Aerobic Gram-negative organisms were grown in Luria–Bertani (LB) broth and Gram-positive bacteria in brain–heart infusion (BHI) broth. Streptococcus mutans JM11 (spectinomycin-resistant, UA140 background) was grown in Todd–Hewitt (TH) broth under anaerobic conditions (80% N2, 15% CO2, 5% H2) at 37°C [17]. All bacteria were grown overnight to an optical density at 600 nm (OD600) of 0.8–1.0 prior to appropriate dilution and antimicrobial testing.
2.2. Synthesis of MH-STAMPs
Conventional SPPS methodologies were utilised for the construction of all peptides shown in Fig. 1 (Symphony® synthesiser; Protein Technologies Inc., Tucson, AZ). Chemicals, amino acids and synthesis resins were purchased from AnaSpec (San José, CA). BD2.20 [FIRKFLKKWLL, amidated c-terminus, molecular weight (MW) 1491.92], an AMP developed in our laboratory with robust antimicrobial activity against a number of bacterial species (Table 1), served as the root sequence to which differing targeting peptides were attached. First, BD2.20 was synthesised by SPPS (Rink-Amide-MBHA resin, 0.015 mmol), followed by the stepwise coupling of a functionalised alkane (NH2(CH2)7COOH) and an Fmoc-protected Lys [side-chain protected with 4-methyltrityl (Mtt)] to the N-terminus. Standard SPPS methods were then employed for the step-wise addition of the S. mutans targeting peptide M8 plus a tri-Gly linker region (TFFRFLNR-GGG) to the N-terminal of the central Lys. After assembly of Fmoc-M8-GGG-K(Mtt)-(CH2)7CO-BD2.20, the Fmoc group was removed with 25% piperidine in dimethylformamide (DMF) and the N-terminal was re-protected with an acetyl group with Ac2O/DIEA (1:1, 20 molar excess) for 2 h. The Mtt-protected amino group of the central Lys was then selectively exposed with 2% trifluoroacetic acid (TFA) in dichloromethane (DCM) (1.5 mL) for 15 min (three cycles of 5 min). The resulting product was reloaded into the synthesiser and the peptide sequence built from the Lys side chain was completed with standard Fmoc SPPS methods. As shown in Fig. 1, the completed MH-STAMP M8(KH)-20 contained the side-chain peptide KH (Pseudomonas spp.-targeting, KKHRKHRKHRKH-GGG), whilst in MH-STAMP M8(BL)-20, a peptide with no bacterial binding (data not shown) BL-1 (DAANEA-GGG), was utilised. BL(KH)-20 was constructed identically to M8(KH)-20, utilising BL-1 in place of M8 (Fig. 1).
Fig. 1.
Multiple-headed specifically-targeted antimicrobial peptides (MH-STAMPs) used in this study: design, sequence and observed mass (m/z) for M8(KH)-20, BL(KH)-20 and M8(BL)-20.
Table 1.
Minimum inhibitory concentrations (MICs) of multiple-headed specifically-targeted antimicrobial peptides (MH-STAMPs) and component peptides
| MH-STAMP | MIC(μM)a |
|||||
|---|---|---|---|---|---|---|
| Pseudomonas aeruginosa | Escherichia coli | Klebsiella pneumoniae | Streptococcus mutans | Staphylococcus epidermidis | Staphylococcus aureus | |
| BD2.20 | 14.4 ± 4.40 | 5.47 ± 1.41 | 2.98 ± 0.47 | 2.86 ± 0.60 | 5.11 ± 1.58 | 5.625 ± 1.29 |
| M8(KH)-20 | 11.95 ± 3.32 | 2.72 ± 0.59 | 3.13 | 6.25 | 3.13 | 5.64 ± 1.07 |
| M8(BL)-20 | 50 | 5.97 ± 0.94 | 6.88 ± 1.98 | 6.25 | 6.25 | 18.05 ± 6.58 |
| BL(KH)-20 | 27.5 ± 7.90 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 |
Average MIC with standard deviation (n = 10 assays).
Synthesis progression was monitored by the ninhydrin test, and completed peptides were cleaved from the resin with 95% TFA utilising appropriate scavengers and precipitated in methyl tert-butyl ether. Purification and MH-STAMP quality was confirmed by high-performance liquid chromatography (HPLC) (Waters, Milford, MA) using a linear gradient of increasing mobile phase (acetonitrile 10% to 90% in water with 0.1% TFA) and a Waters XBridge™ BEH 130 C18 column (4.6×100 mm, particle size 5 μm). Absorbance at 215 nm was utilised as the monitoring wavelength, although 260 nm and 280 nm were also collected. Liquid chromatography spectra were analysed with MassLynx Software v. 4.1 (Waters). Matrix-assisted laser desorption/ionisation mass spectroscopy (MALDI-MS) was utilised to confirm correct peptide mass (Voyager System 4291; Applied Biosystems, Foster City, CA) [18].
2.3. Minimum inhibitory concentration (MIC) assay
Peptides were evaluated for basic antimicrobial activity by broth microdilution, as described previously [10,11]. Briefly, ca. 1×105 colony-forming units (CFU)/mL bacteria were diluted in TH (S. mutans) or Mueller–Hinton (MH) broth (all other organisms) and distributed to 96-well plates. Serially-diluted (two-fold) peptides were then added and the plates were incubated at 37 °C for 18–24 h. Peptide MICs were determined as the concentration of peptide that completely inhibited organism growth when examined by eye (clear well). All experiments were conducted 10 times.
2.4. Post-antibiotic effect assay
The activity and selectivity of MH-STAMPs after 10 min incubation was determined by growth retardation experiments against targeted and untargeted bacteria in monocultures, as described previously [10,11]. Cells from overnight cultures were diluted to ca. 5×106 CFU/mL in MH broth (or TH broth with 1% sucrose for S. mutans), normalised by OD600 = 0.05–0.1, and seeded to 96-well plates. Cultures were then grown under appropriate conditions for 2 h (3 h for S. mutans) prior to the addition of peptides for 10 min. Plates were then centrifuged at 3000 ×g for 5 min, the supernatants discarded, fresh medium returned (MH or TH without sucrose for S. mutans) and incubation resumed. Bacterial growth after treatment was then monitored over time by OD600. Data were analysed for significance by an unpaired Student’s t-test.
2.5. Microbial population shift assay
Mixed planktonic populations of P. aeruginosa, E. coli, S. epidermidis and S. mutans were utilised to examine the potential of MH-STAMPs to direct species composition within a culture after treatment. Samples were prepared containing ca. 6×104 CFU/mL S. mutans, ca. 2×104 CFU/mL E. coli, ca. 2×104 CFU/mL S. epidermidis and ca. 0.5×104 CFU/mL P. aeruginosa in BHI broth (mixed immediately before peptide addition). Peptide (10 μM) or mock-treatment [1×phosphate-buffered saline (PBS)] was then added and samples were incubated at 37°C for 24 h under anaerobic conditions (80% N2, 15% CO2, 5% H2) to maintain similar growth rates between the bacterial species utilised. After incubation, samples were serially diluted (1:10) in 1×PBS and aliquots from each dilution were then spotted to agar plates selective for each species in the mixture [TH plus 800 μg/mL spectinomycin (S. mutans), LB plus 25 μg/mL ampicillin (P. aeruginosa), LB plus 200 μg/mL spectinomycin (E. coli) and mannitol salt agar (S. epidermidis)] in order to quantitate survivors from each species. Plates were then incubated 37 °C under aerobic conditions (TH plates were incubated anaerobically) and colonies were counted after 24 h to determine survivors. Expected colony morphologies were observed for each species when plated on selective media. Gram stains and direct microscopic observation (from select isolated colonies) were undertaken to confirm species identity (data not shown). The detection limit of the assay was 200 CFU/mL.
3. Results
3.1. Design and synthesis of MH-STAMPs
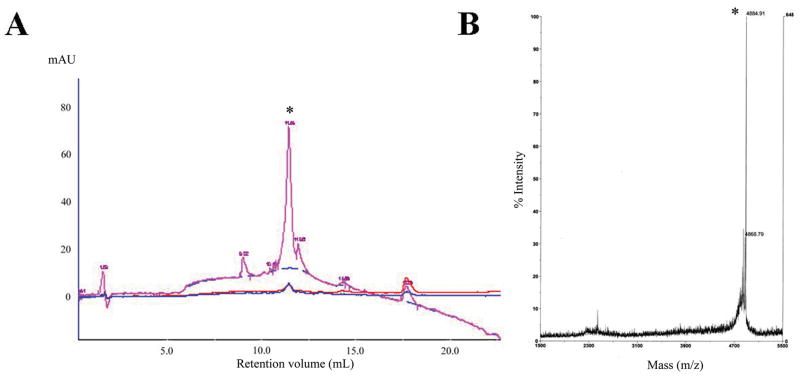
A prototype MH-STAMP was constructed from the well established targeting peptides KH (specific to Pseudomonas spp.) and M8 (specific for S. mutans). The wide-spectrum antimicrobial peptide BD2.20 was utilised as the base AMP for all MH-STAMP construction. BD2.20 is a novel synthetic AMP with a cationic and amphipathic residue arrangement, which has robust MICs against a variety of Gram-negative and Gram-positive organisms (Table 1). For the synthesis of MH-STMAP M8(KH)-20 (construct presented in Fig. 1), BD2.20 and a Lys (Mtt-protected side chain) residue were joined via an activated alkane spacer, followed by addition of the M8 targeting peptide to the N-terminus of the product. Selective deprotection of the central Lys(Mtt) side chain was then undertaken and the KH targeting peptide was attached. The correct molecular mass (4884.91) and ca. 90% purity was confirmed by HPLC and MALDI-MS (Fig. 2).
Fig. 2.
High-performance liquid chromatography (HPLC) and mass spectrometry (MS) spectra of M8(KH)-20. The quality of the completed multiple-headed specifically-targeted antimicrobial peptide (MH-STAMP) was analysed by (A) HPLC and (B) matrix-assisted laser desorption/ionisation (MALDI)-MS. At ultraviolet absorbance 215 nm (260 nm and 280 nm are also plotted), a single major product was detected by HPLC (* retention volume 11.04 mL). After fraction collection, the correct mass (m/z) for single-charged M8(KH)-20 [4884.91 (marked by *)] was observed for this peak. mAU milliabsorbance units.
The non-binding ‘blank’ targeting peptide BL-1 was incorporated into the synthesis scheme in place of KH or M8 to construct variant MH-STAMPs possessing a single functional targeting head: M8(BL)-20 and BL(KH)-20 (Fig. 1). The correct MW and acceptable purity were observed for these MH-STAMPs (data not shown).
3.2. General antimicrobial activity of multihead constructs
After synthesis, the completed MH-STAMPs were evaluated for general antimicrobial activity by MIC against a panel of bacteria. As shown in Table 1, the MH-STAMP constructs M8(KH)-20, BL(KH)-20 and M8(BL)-20 were found to have similar activity profiles to that of BD2.20 for the organisms examined (less than two titration steps difference). Additionally, a difference in general susceptibility was observed between P. aeruginosa and the other organisms tested, suggesting that this bacterium is more resistant to BD2.20. Overall, these data indicate that the addition of the targeting domains to the base sequence was tolerated and did not completely inhibit the activity of the AMP.
Peptide selectivity could not be determined utilising these methods, as STAMPs and their parent AMP molecules often display similar MICs but have radically different antimicrobial kinetics and selectivity due to increased specific killing mediated by the targeting regions [10,11]. Therefore, different experiments were performed to test for antimicrobial selectivity and functional MH-STAMP construction.
3.3. Selectivity and post-antibiotic effect of MH-STAMP constructs
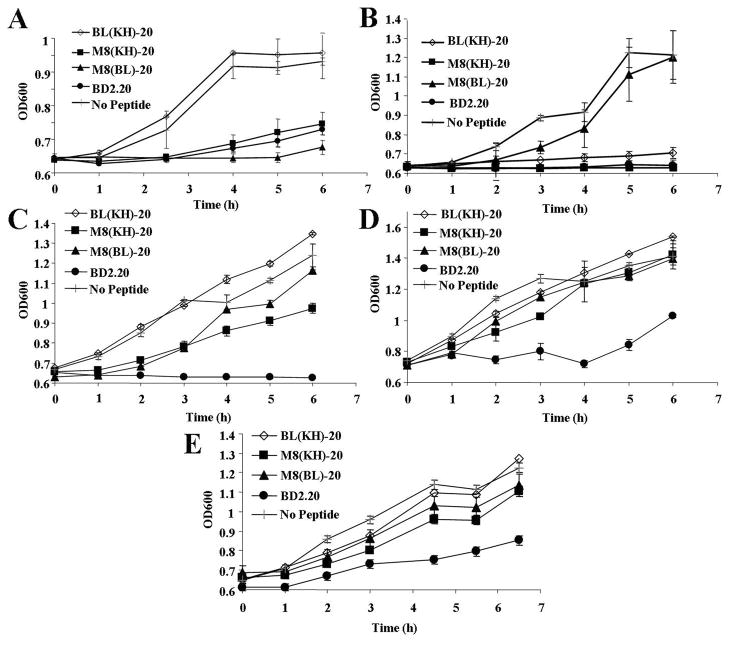
MH-STAMP antimicrobial kinetics was ascertained utilising a variation of the classical post-antibiotic effect assay, which measures the ability of an agent to affect an organism’s growth after a short exposure period. Monocultures of MH-STAMP-targeted and untargeted organisms were exposed to M8(KH)-20, M8(BL)-20, BL(KH)-20 or unmodified BD2.20 and then allowed to recover. As shown in Fig. 3A, S. mutans growth was effectively and significantly retarded as early as 2.5 h by M8-containing constructs [M8(KH)-20 and M8(BL)-20] but was not altered by a MH-STAMP construct lacking this region [BL(KH)-20] (comparison at 2.5 h, Student’s t-test, P = 0.0001). Similarly, growth of the other targeted bacterium, P. aeruginosa, was significantly inhibited in a KH-dependent manner by 4 h (Fig. 3B) [Student’s t-test, P = 0.001, comparing BL(KH)-20 or M8(KH)-20 with M8(BL)-20]. In comparison, the non-targeted bacteria E. coli, S. aureus and S. epidermidis were not inhibited by treatment with any MH-STAMP (Fig. 3C–E) and were only inhibited by the base antimicrobial peptide BD2.20, which displayed robust antimicrobial activity against all examined strains. These results indicate that MH-STAMPs containing KH or M8 targeting domains have activity against P. aeruginosa or S. mutans, respectively, and not other bacteria. Furthermore, replacement of the targeting region with a non-binding peptide abolishes specific activity.
Fig. 3.
Growth inhibitory activity of multiple-headed specifically-targeted antimicrobial peptides (MH-STAMPs). Monocultures of (A) Streptococcus mutans, (B) Pseudomonas aeruginosa, (C) Staphylococcus epidermidis, (D) Staphylococcus aureus or (E) Escherichia coli were treated with peptides for 10 min. The agent was then removed and fresh medium was returned. Culture recovery was measured over time [optical density at 600 nm (OD600)]. Plots represent the average of at least three independent experiments with standard deviations.
3.4. Ability of MH-STAMPs to direct a ‘population shift’ within a mixed-species population
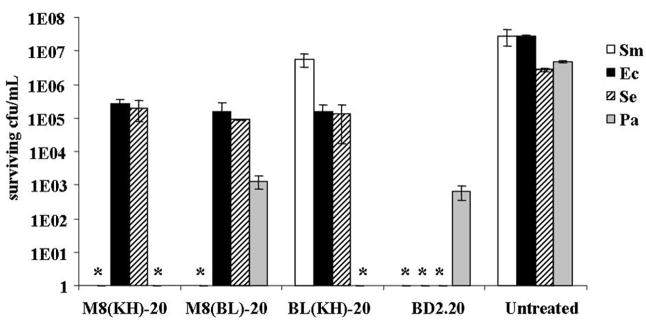
It was hypothesised that potential MH-STAMP dual functionality could affect a particular set of bacteria within a mixed population, thereby promoting the outgrowth of non-targeted organisms and ‘shifting’ the constituent make-up. To examine this possibility, defined mixed populations of planktonic cells were treated continuously and the make-up of the community was examined after 24 h. As shown in Fig. 4, treatment with the wide-spectrum antimicrobial peptide BD2.20 resulted in a significant loss of recoverable CFU/mL after 24 h from all species in the mixture. Treatment with M8(KH)-20 was found to alter this pattern; ca. 1×105 CFU/mL surviving E. coli and S. epidermidis were observed, although S. mutans and P. aeruginosa CFU/mL were not recovered. In BL(KH)-20-treated samples, P. aeruginosa CFU/mL were not observed, although higher than input CFU/mL were recovered from S. mutans and unchanged numbers of S. epidermidis and E. coli. In samples exposed to M8(BL)-20, S. mutans recoverable CFU/mL were greatly reduced compared with input CFU/mL, whilst other species were not affected or affected to a lesser extent. Interestingly, these results suggest that M8(KH)-20, M8(BL)-20 and BL(KH)-20 retain their ability to affect organisms recognised by the targeting regions present, even within a mixed population of bacteria.
Fig. 4.
Selective activity of dual-targeted and single-targeted multiple-headed specifically-targeted antimicrobial peptides (MH-STAMPs) in mixed culture. A mixture of Pseudomonas aeruginosa (Pa), Streptococcus mutans (Sm), Escherichia coli (Ec) and Staphylococcus epidermidis (Se) planktonic cells was mixed with MH-STAMPs and treated for 24 h. After incubation, colony-forming units (CFU)/mL of remaining constituent species were quantitated after plating to selective media. * Indicates <200 surviving CFU/mL recovered.
4. Discussion
Our results indicate that we have successfully constructed a STAMP with dual antimicrobial specificities controlled by the targeting peptides present in the molecule, namely KH for Pseudomonas spp. and M8 for S. mutans. In a closed multispecies system (Fig. 4), the dual specificity of M8(KH)-20 was readily discernable: the population of the culture ‘shifted’ away from targeted organisms following MH-STAMP treatment. The targeted bacteria were eliminated and the population of untargeted organisms increased, to varying degrees, above input CFU/mL. Additionally, interruption of KH or M8 in the MH-STAMP construct with the non-binding peptide BL-1 resulted in the expected elimination of only one targeted species. These results support the hypothesis that functional MH-STAMPs can be constructed from a wide-spectrum AMP base.
The emergence of metagenomics and the development of more sensitive molecular diagnostics has driven an increase in the understanding of human-associated microbial ecologies and host–microbe interactions [19–21]. At mucosal surfaces, it has become clear that human bodies harbour an abundance of residential flora that may impact innate and humoral immunity, nutrient availability, protection against pathogens and even host physiology [22–25]. Furthermore, findings have indicated that shifts in the diversity of normal flora are associated with negative clinical consequences, for example the overgrowth of S. mutans in the oral cavity during cariogenesis (linked to the uptake of sucrose) or the antibiotic-assisted colonisation of the intestine by Clostridium difficile [26,27]. Other population shifts may be linked to axilla odour (Corynebacterium spp.) [28,29] or even host obesity. Given the quantity and diversity of microbes present, pathogenesis at mucosal surfaces is not likely to be associated with the overgrowth of a single strain or species. More often, it is a population shift resulting in the predominance of two or more species, for example the persistence of Burkholderia cepacia and P. aeruginosa in cystic fibrosis airways or Treponema denticola and Porphymonas gingivalis and other ‘red cluster’ organisms in gingivitis [30,31]. In many cases (such as the latter), these species may have only distant phylogenetic relationships and display differential susceptibilities to antibiotic therapies resulting in persistent disease progression despite treatment [32,33]. Currently available treatments for infections of mucosal surfaces are largely non-specific (traditional small molecule antibiotics, mechanical removal) and thus are not effective in retaining flora or shifting the constituent balance back to a health-associated composition [34]. There is a need for a therapeutic treatment that can selectively target multiple pathogens, regardless of their phylogenetic relationship, and MH-STAMPs may help achieve this goal.
In monoculture experiments (Fig. 3), our results suggest that inclusion of M8 or KH in the MH-STAMP drove activity towards S. mutans or P. aeruginosa, but also that the presence of a targeting domain reduced the activity of the parent AMP BD2.20 against untargeted organisms. In contrast, the results of our MIC assays (Table 1) indicate little difference in activity between BD2.20 and any MH-STAMP. Against untargeted organisms, the M8 and KH regions are likely to have a negative, but not completely inhibitory, impact on BD2.20 activity. Given the long duration of activity and the lower inoculum size in the MIC assay (compared with experiments in Fig. 3), it is likely that all BD2.20-containing peptides could reach equal levels of growth inhibition, despite large and target-specific differences in antimicrobial speed. This pattern of results was also observed when comparing the MICs of targeted and untargeted organisms utilising STAMPs against S. mutans and Pseudomonas mendocina [10,11].
Although more rigorous studies and a more medically relevant combination of pathogen targets are necessary, these findings suggest that it is possible to design an AMP-based therapeutic with multiple and defined fidelities in vitro. MH-STAMPs may help improve human health through the promotion of healthy microbial constituencies.
Acknowledgments
The authors are grateful for the technical contributions of A. Wang as well as the helpful discussions with R. Lux, C. Kaplan, L. Sim and L. Li.
Funding: National Center on Minority Health and Health Disparities [STTR Grant (US): 2R42MD001831-02].
Footnotes
Competing interests: None declared.
Ethical approval: Not required.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–20. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 2.Hancock RE, Lehrer R. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 1998;16:82–8. doi: 10.1016/s0167-7799(97)01156-6. [DOI] [PubMed] [Google Scholar]
- 3.Genco CA, Maloy WL, Kari UP, Motley M. Antimicrobial activity of magainin analogues against anaerobic oral pathogens. Int J Antimicrob Agents. 2003;21:75–8. doi: 10.1016/s0924-8579(02)00275-3. [DOI] [PubMed] [Google Scholar]
- 4.He J, Eckert R, Pharm T, Simanian MD, Hu C, Yarbrough DK, et al. Novel synthetic antimicrobial peptides against Streptococcus mutans. Antimicrob Agents Chemother. 2007;51:1351–8. doi: 10.1128/AAC.01270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–50. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 6.Muhle SA, Tam JP. Design of Gram-negative selective antimicrobial peptides. Biochemistry. 2001;40:5777–85. doi: 10.1021/bi0100384. [DOI] [PubMed] [Google Scholar]
- 7.Tossi A, Sandri L, Giangaspero A. Amphipathic, alpha-helical antimicrobial peptides. Biopolymers. 2000;55:4–30. doi: 10.1002/1097-0282(2000)55:1<4::AID-BIP30>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 8.Tam JP, Lu YA, Yang YL. Antimicrobial dendrimeric peptides. Eur J Biochem. 2002;269:923–32. doi: 10.1046/j.0014-2956.2001.02728.x. [DOI] [PubMed] [Google Scholar]
- 9.Pini A, Giuliani A, Falciani C, Runci Y, Ricci C, Lelli B, et al. Antimicrobial activity of novel dendrimeric peptides obtained by phage display selection and rational modification. Antimicrob Agents Chemother. 2005;49:2665–72. doi: 10.1128/AAC.49.7.2665-2672.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckert R, He J, Yarbrough DK, Qi F, Anderson MH, Shi W. Targeted killing of Streptococcus mutans by a pheromone-guided ‘smart’ antimicrobial peptide. Antimicrob Agents Chemother. 2006;50:3651–7. doi: 10.1128/AAC.00622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckert R, Qi F, Yarbrough DK, He J, Anderson MH, Shi W. Adding selectivity to antimicrobial peptides: rational design of a multidomain peptide against Pseudomonas spp. Antimicrob Agents Chemother. 2006;50:1480–8. doi: 10.1128/AAC.50.4.1480-1488.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–86. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckert R, Brady KM, Greenberg EP, Qi F, Yarbrough DK, He J, et al. Enhancement of antimicrobial activity against Pseudomonas aeruginosa by coadministration of G10KHc and tobramycin. Antimicrob Agents Chemother. 2006;50:3833–8. doi: 10.1128/AAC.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sprenger GA, Lengeler JW. L-Sorbose metabolism in Klebsiella pneumoniae and Sor+ derivatives of Escherichia coli K-12 and chemotaxis toward sorbose. J Bacteriol. 1984;157:39–45. doi: 10.1128/jb.157.1.39-45.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Podbielski A, Spellerberg B, Woischnik M, Pohl B, Lütticken R. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS) Gene. 1996;177:137–47. doi: 10.1016/0378-1119(96)84178-3. [DOI] [PubMed] [Google Scholar]
- 16.Duthie ES, Lorenz LL. Staphylococcal coagulase; mode of action and antigenicity. J Gen Microbiol. 1952;6:95–107. doi: 10.1099/00221287-6-1-2-95. [DOI] [PubMed] [Google Scholar]
- 17.Merritt J, Kreth J, Qi F, Sullivan R, Shi W. Non-disruptive, real-time analyses of the metabolic status and viability of Streptococcus mutans cells in response to antimicrobial treatments. J Microbiol Methods. 2005;61:161–70. doi: 10.1016/j.mimet.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 18.Anderson RC, Rehders M, Yu PL. Antimicrobial fragments of the pro-region of cathelicidins and other immune peptides. Biotechnol Lett. 2008;30:813–8. doi: 10.1007/s10529-007-9628-7. [DOI] [PubMed] [Google Scholar]
- 19.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–32. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boman HG. Innate immunity and the normal microflora. Immunol Rev. 2000;173:5–16. doi: 10.1034/j.1600-065x.2000.917301.x. [DOI] [PubMed] [Google Scholar]
- 21.Kreth J, Merritt J, Shi W, Qi F. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol. 2005;187:7193–203. doi: 10.1128/JB.187.21.7193-7203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metges CC. Contribution of microbial amino acids to amino acid homeostasis of the host. J Nutr. 2000;130:1857S–64S. doi: 10.1093/jn/130.7.1857S. [DOI] [PubMed] [Google Scholar]
- 23.Sears CL. A dynamic partnership: celebrating our gut flora. Anaerobe. 2005;11:247–51. doi: 10.1016/j.anaerobe.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Lievin-Le Moal V, Servin AL. The front line of enteric host defense against unwelcome intrusion of harmful microorganisms: mucins, antimicrobial peptides, and microbiota. Clin Microbiol Rev. 2006;19:315–37. doi: 10.1128/CMR.19.2.315-337.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DiBaise JK, Zhang H, Crowell MD, Krajmalnik-Brown R, Deckert GA, Rittmann BE. Gut microbiota and its possible relationship with obesity. Mayo Clin Proc. 2008;83:460–9. doi: 10.4065/83.4.460. [DOI] [PubMed] [Google Scholar]
- 26.Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–80. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gould CV, McDonald LC. Bench-to-bedside review: Clostridium difficile colitis. Crit Care. 2008;12:203. doi: 10.1186/cc6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leyden JJ, McGinley KJ, Holzle E, Labows JN, Kligman AM. The microbiology of the human axilla and its relationship to axillary odor. J Invest Dermatol. 1981;77:413–6. doi: 10.1111/1523-1747.ep12494624. [DOI] [PubMed] [Google Scholar]
- 29.Elsner P. Antimicrobials and the skin physiological and pathological flora. Curr Probl Dermatol. 2006;33:35–41. doi: 10.1159/000093929. [DOI] [PubMed] [Google Scholar]
- 30.Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–74. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–83. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlessinger D. Failure of aminoglycoside antibiotics to kill anaerobic, low-pH, and resistant cultures. Clin Microbiol Rev. 1988;1:54–9. doi: 10.1128/cmr.1.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tresse O, Jouenne T, Junter G. Antibacterial efficacy of tobramycin against anaerobic Escherichia coli cultures in the presence of electron acceptors. J Antimicrob Chemother. 1997;40:419–21. doi: 10.1093/jac/40.3.419. [DOI] [PubMed] [Google Scholar]
- 34.Keene HJ, Shklair IL. Relationship of Streptococcus mutans carrier status to the development of carious lesions in initially cariesfree recruits. J Dent Res. 1974;53:1295. doi: 10.1177/00220345740530053801. [DOI] [PubMed] [Google Scholar]