Abstract
Most tumors are epithelial-derived, and although disruption of polarity and aberrant cellular junction formation is a poor prognosticator in human cancer, the role of polarity determinants in oncogenesis is poorly understood. Using in vivo selection, we identified a mammalian orthologue of the Drosophila polarity regulator crumbs as a gene whose loss of expression promotes tumor progression. Immortal baby mouse kidney epithelial (iBMK) cells selected in vivo to acquire tumorigenicity displayed dramatic repression of crumbs3 (crb3) expression associated with disruption of tight junction formation, apicobasal polarity, and contact-inhibited growth. Restoration of crb3 expression restored junctions, polarity and contact inhibition, while suppressing migration and metastasis. These findings suggest a role for mammalian polarity determinants in suppressing tumorigenesis that may be analogous to the well-studied polarity tumor suppressor mechanisms in Drosophila.
Keywords: Crumbs, crb3, tight junctions, polarity, metastasis, cancer
INTRODUCTION
The vast majority of human cancers comprise epithelial-derived adult solid tumors that respond poorly to the currently available treatments. Epithelial cells have distinct properties necessitating that mechanisms of oncogenesis be addressed in this physiologically relevant cell type. We sought to develop models for epithelial tumor progression utilizing primary epithelial cells, with the ability to apply genetics in a compound fashion, to identify genes that regulate tumor progression. Primary mouse kidney, prostate, mammary and ovarian surface epithelial cells (C57B/6) immortalized by cooperating oncogenes become apparent as colonies and can be cloned and expanded (1–3). Specific inactivation or circumvention of the RB pathway (by expression of adenovirus E1A, or c-myc), and inactivation of the p53 pathway, are both necessary and sufficient for generation of immortal rat and mouse epithelial cells from multiple tissue types (1). For example, immortal baby mouse kidney and mammary epithelial cells (iBMK and iMMEC, respectively) are non-tumorigenic and retain their normal epithelial character including the ability to form tight junctions and establish polarity that is often lost in advanced cancers (1). Therefore, RB and p53 pathway inactivation is sufficient for immortalization in vitro while permitting retention of epithelial characteristics, but is insufficient for tumorigenesis in vivo.
In addition to RB and p53 inactivation, the capacity for tumorigenesis additionally requires inactivation of p53-independent apoptosis, either through the loss of proapoptotic bax and bak or bim, or through the gain of anti-apoptotic bcl-2, events that can be selected for during tumorigenesis in vivo (2, 4, 5). Although inactivation of p53-independent apoptosis by modulation of Bcl-2 family members enables tumor growth, these cells retain epithelial characteristics. Activation of the Ras, MAP kinase or PI-3 kinase pathways in iBMK cells also promotes tumorigenesis, but in contrast to defects in apoptosis, this is associated with the loss of epithelial characteristics through induction of an epithelial to mesenchymal transition (EMT) (1, 5). Another mechanism to enable tumorigenesis is acquired through in vivo selection of non-tumorigenic iBMK cells, which results in highly tumorigenic iBMK cells that have enabled tumor growth by an unknown mechanism (1). This selection for tumorigenesis in vivo also selects for loss of epithelial polarity and EMT, but the cells otherwise lack the phenotypically resemblance to cells with Ras, MAP kinase or PI-3 kinase pathway activation. If or how polarity disruption and EMT contributes to tumorigenesis in this case is not yet understood. Thus there appears to be at least four independent functions that promote tumorigenesis of immortal epithelial cells: apoptosis defect, MAP kinase pathway activation, PI-3 kinase pathway activation, and in vivo selection that may involve an unidentified activity associated with disruption of polarity.
EMT, the process through which cells transition from a polarized, epithelial phenotype to a highly motile mesenchymal phenotype has emerged in recent years as an important factor in tumor progression (6). EMT causes pronounced morphological and functional changes in cells such as dissolution of epithelial tight junctions, reorganization of the actin cytoskeleton, loss of apicobasal polarity and migration through basement membranes and tissues. The molecular mechanisms governing EMT involve several signaling pathways including TGF-β, STAT, Ras, Wnt and Notch pathways (7, 8). EMTs during embryonic development are governed in part by STAT signaling pathway through the Snail family of transcription factors, first described in Drosophila (6). In some cases EMT is partly due to direct repression of E-cadherin transcription, and has been associated with Snail-induced repression of tight junction proteins such as Claudins-3, -4 and -7 and Occludin (9), in both early development and in cancer progression (10). E-cadherin repressor ZEB1 has been shown to repress cell polarity genes Crumbs3, HUGHL2 and Plas1-associated tight junction protein in colon and invasive breast cancer, promoting EMT associated with tumor progression (11). Loss of epithelial polarity is a hallmark of EMT in normal development and cancer but the role of polarity determinants in this process in cancer in mammals is poorly understood. The proteins involved in polarity complexes are highly conserved between Drosophila and mammalian cells (12, 13). In Drosophila, disruption of polarity determinants causes overproliferation and disruption of tissue architecture, hence their designation as tumor suppressors. Although the genetic interactions between these Drosophila polarity determinants have been studied extensively, no links are established to analogous roles of their mammalian homologues in human cancer.
Here we identify crb3, the mammalian orthologue of Drosophila and polarity determinant crumbs as a gene whose expression is lost in iBMK cells rendered tumorigenic through in vivo selection. We further demonstrate that Crb3 functions to maintain epithelial junction formation and apicobasal polarity required for contact-inhibited growth, and to suppress invasion and metastasis. These findings suggest the existence of a mammalian pathway for tumor suppression analogous to that long known to control tumor growth in Drosophila.
MATERIALS AND METHODS
Cell culture and transfection
Derivation of iBMK cell lines was previously described (4). Stable iBMK cells expressing human Bcl-2, or vector-only controls were derived by electroporation with pCDNA3.1-hBcl-2, or selection vector only (InVitrogen, Carlsbad, CA) respectively, followed by selection (W2, gentamicin; D3, zeocin) and ring cloning. Multiple clones from each condition were expanded and characterized. For RFP expression, cells were cotransfected with the pDsRed2-C1 (Clontech, Mountain View, CA) and pCDNA3.1zeo (W2.3.1–5, InVitrogen) or pPUR (D3.zeo-2, BD Biosciences, Rockville, MD) vectors, followed by selection with zeocin or puromycin, respectively, and ring cloning. For each cell line, three individual clones were characterized in animals for fluorescence and tumor growth, and one individual clone was selected for further analysis. All cell lines were maintained in DMEM (Gibco/InVitrogen) containing 10% fetal bovine serum (FBS). For MTT assays cells were plated in 96 well plates at a density of 8000 cells/well and treated after 24 hours with 3μM etoposide for 48 hours. MTT was added to the wells and incubated at 37°C for 3 hours followed by the aspiration of the media and addition of DMSO. The plates were read using Spectra Max 250 (Molecular Devices, Sunnyvale, CA).
Microarray analysis
Gene Expression analysis was carried out using the Affymetrix Mouse 430 A 2.0 chips (accession number GPLxxxx). Initial scaling was done using the Affymetrix Microarray Suite Expression Software version 5.0 and subsequent analysis was done using GeneSpring Software version 6.1. TDCLs were compared to duplicate samples of the parental cell line from which they were derived. Three different filtering conditions were applied: Expression Percentage Restriction, retaining only those genes that had a raw expression value of 80.0 or more in at least 1 out of the 5 samples followed by a filtering selecting for genes with a Flag value of Present (P) in all the samples being compared, and a pair-wise fold-change analysis comparing each tumor derived cell line to the parental cell-line. Genes that underwent at least 3-fold or more up-regulation or down-regulation in the tumor derived cell lines, compared to the parental cell line were retained.
Retrovirus infection
The png retroviral vector and mycCRB3-png, mycCRB3ΔERLI-png and mycCRB3mutFERMΔERLI-png were generously provided by Dr. Ben Margolis (University of Michigan Medical School, Ann Arbor, MI). The production of amphotropic retroviruses and cell infection was performed as described previously (14). Selection of cells stably expressing the retroviral vector was done using selection media containing 500 μg/ml puromycin 48 hours post-infection. Cells were selected for 6 days before use, or until all non-infected cells had died.
Immunoblotting, immunofluorescence, immunohistochemistry and confocal microscopy
Immunoblotting was performed using the rabbit polyclonal antibody against Crb3 (15), a generous gift from Dr. Benjamin Margolis (University of Michigan Medical School). For immunofluorescence analysis cells were grown on glass coverslips and fixed in ice cold acetone for 10 minutes and indirect immunofluorescence was performed as described previously (16). The following antibodies were used: rabbit anti-occludin (Abcam, Cambridgeshire, UK), mouse monoclonal anti-pan-cadherin clone CH-19 (Sigma-Aldrich, St. Louis, MO), mouse monoclonal anti-β-catenin, rabbit polyclonal anti-ZO-1 (Zymed, South San Francisco, CA), mouse anti-BrdU FITC (BD Biosciences, San Jose, CA) and polyclonal rabbit anti-cleaved caspase-3 (Asp175) (Cell Signaling Technology Inc., Danvers, MA). Immunohistochemistry was performed on paraffin-embedded sections of tumors using polyclonal rabbit anti-cleaved caspase-3 (Asp175) (Cell Signaling Technology Inc., Danvers, MA). Confocal microscopy was performed using a Zeiss LSM510-META confocal microscope system at the Neuroscience Imaging Facility (W. M. Keck Center for Collaborative Neuroscience, Rutgers University).
Three-dimensional morphogenesis assay and imaging
Three-dimensional culture of TDCL 5D on a reconstituted basement membrane was performed according to the protocol previously described for the immortalized human mammary epithelial cell line MCF-10A (17). Time-lapse microscopy was performed as previously described (1). In brief, cells were cultured in the time-lapse chamber equipped with controlled environmental conditions. The time-lapse microscopy system consisted of an Olympus IX71 inverted microscope fitted with 37°C temperature and 5% CO2 controlled environmental chamber (Solent Scientific, UK) and a Coolsnap ES CCD camera. Image capturing and analysis were performed using Image-Pro Plus software (Media Cybernetics, USA). Phase contrast images (100X) at multiple fields were obtained for the indicated time period.
Tumor growth assay
Tumor formation in nude mice by subcutaneous injection using 107 cells in 0.1 mL PBS was performed essentially as described previously (4). Briefly, 107 cells were injected in each of five mice for each cell line and tumors were measured regularly. Tumor volumes were calculated using the formula length (millimeters) × width2 (millimeters)2/2. Each point represents the mean value for all five mice in each group. To visualize RFP-expressing cells in mice, animals were imaged using the Illumitool imaging and camera system (Lightools Research, Encinitas, CA).
Migration assay
The invasive capacity of TDCLs was measured utilizing an in vitro transwell assay (18). Cells, in DMEM containing 0.1% BSA, were added to the upper well of transwell chambers (Corning Inc., Corning, NY), containing an 8 μm porous membrane. Lower chambers contained 10% FBS in DMEM. Cells were cultured for 2–4 hrs before invading cells were fixed, stained with crystal violet and examined under a bright-field microscope. Cells were counted in five random fields/membrane and results presented as means ± SEM of three independent experiments.
RESULTS
Selection in vivo for acquisition of tumorigenesis
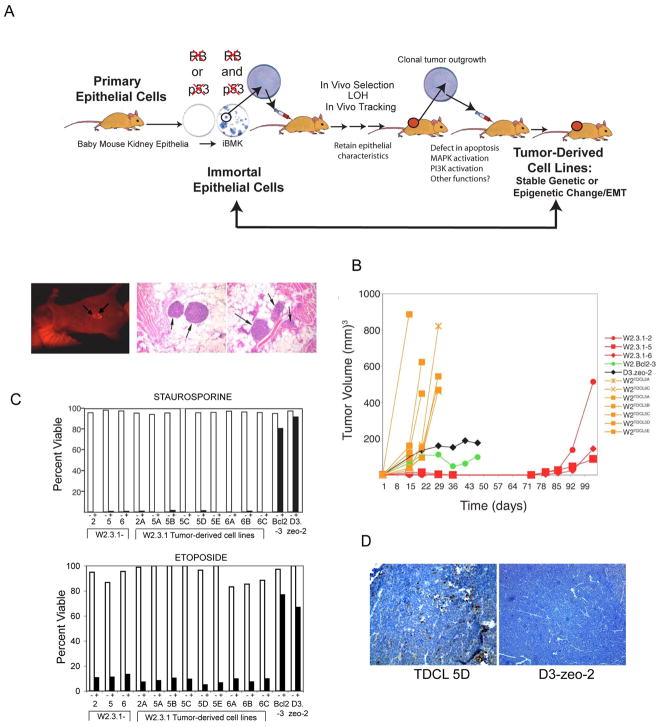
Ten million wild type non-tumorigenic W2.3.1-5 iBMK cells were introduced subcutaneously into a series of nude mice and thereby subjected to selection in vivo for tumor growth (Fig. 1A). Following two months in vivo, 1 to 10 non-tumorigenic wild type iBMK cells acquired the capacity for tumor growth, appearing initially as small clonal nodules (Fig. 1A). These clonal tumors that arise following a long latency were excised and returned to cell culture as tumor-derived cell lines (TDCLs). Seven of these TDCLs were then injected subcutaneously into nude mice along with the original parental cell line (W2.3.1) to monitor tumor growth kinetics. Despite three months of selection in vivo, TDCLs returned to cell culture readily and were profoundly tumorigenic relative to the parental, non-tumorigenic W2.3.1 cells (Fig. 1A and B). TDCLs were also more tumorigenic than iBMK cells rendered defective for apoptosis by deficiency in both bax and bak (D3) (4) or by overexpression of bcl-2 (W2 BCL2-3) (5) (Fig. 1B). As defects in apoptosis enable tumorigenesis, we determined if the TDCLs incurred mutations rendering them resistant to apoptosis by testing TDCLs for sensitivity to apoptosis-inducer staurosporine or etoposide. All TDCLs retain sensitivity to staurosporine or etoposide, and readily undergo apoptosis similarly to the parental W2.3.1-5 cell line (Fig. 1C). Moreover, the apoptotic sensitivity of TDCLs was assessed in vivo by examining the presence of cleaved, activated caspase-3 in tumor sections. Tumors from TDCL 5D displayed frequent cells positive for activated caspase-3 whereas sections from tumors derived from Bax/Bak deficient, apoptosis-defective D3 cells did not (Fig. 1D). Thus, while the TDCLs acquire a stable tumor-promoting function through in vivo selection, they did not acquire resistance to apoptosis. This suggests that TDCLs acquired stable genetic or epigenetic change(s) through selection in vivo that confers functional tumorigenic capability distinct from blockade of apoptosis.
Figure 1.
Selection scheme for TDCLs. (A) Schematic representation of mouse model used for obtaining TDCLs. Primary mouse epithelial kidney cells were immortalized by inactivation of the RB and p53 pathway (1). Wild type, non-tumorigenic W2.3.1 iBMK were injected subcutaneously in mice and tumor growth occurred with long latency (three to four months). TDCLs were returned to in vitro cell culture and reinjected into animals where tumor growth occurred within two weeks. Clonal emergence of W2.3.1-RFP expressing tumors by whole animal optical imaging after three months and hematoxylin and eosin staining of emerging clonal nodules following selection for growth for three months in vivo. (B) Tumorigenic capacity of TDCLs. Parental W2.3.1 cells (red) and TDCLs (orange) were injected subcutaneously into nude mice, and tumor volumes were measured over time. W2.Bcl2-3 (green) and D3.zeo-2 (black) cells were included as controls. (C) TDCLs retain apoptotic sensitivity. W2.3.1 and TDCLs were cultured for 24 hours with 0.4mM staurosporine or for 48 hrs with 3 μM etoposide, and viability was assessed by trypan blue exclusion for staurosporine and MTT assay for etoposide (white bars: untreated; black bars: treated). Apoptotic defective Bcl-2 expressing (W2.Bcl2-3) and Bax and Bak deficient D3.zeo-2 cells were included as controls, as these cells are known to be resistant to staurosporine-induced apoptosis. (D) TDCL 5D cells retain apoptotitc sensitivity in vivo. Paraffin-embedded sections from Bax/Bak deficient, apoptosis-defective D3 tumors and TDCL 5D tumors were subjected to immunohistochemistry for active caspase-3 showing apoptosis induction (brown staining) of TDCL 5D tumors in vivo that is absent in D3 tumors.
Acquisition of tumorigenesis results in an altered pattern of gene expression
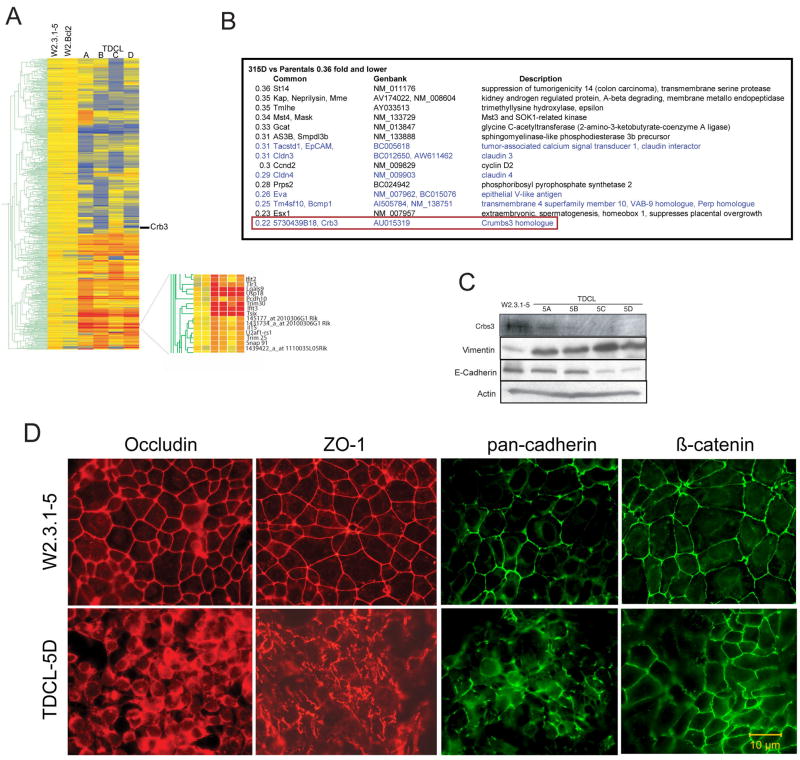
Comparison of the gene expression pattern of four independently derived TDCLs (Fig. 2A) cultured in vitro revealed a strikingly similar pattern of gene expression changes relative to the parental W2.3.1-5 cells (Fig. 2A). In contrast, TDCLs produced from W2.3.1-5 cells expressing bcl-2 did not display a significant change in the pattern of gene expression, consistent with the lack of selection pressure given that these cells readily form tumors due to defective apoptosis. Most of the upregulated genes in the TDCLs were interferon-regulated genes often referred to as the interferon cluster (Fig. 2A) (19–23) the upregulation of many was validated at the mRNA and protein levels (data not shown), and the significance of which is currently under investigation.
Figure 2.
TDCLs acquire loss of crb3 expression and defects in epithelial tight junction formation. (A) Gene tree analysis of parental W2.3.1-5 and TDCLs 5A-D. All samples were analyzed in duplicate (Affymetrix 430), accession no. GPLxxxx. (B) Crb3 expression is repressed in TDCLs. List of the 15 most repressed genes in TDCLs from the gene expression analysis. (C) Western blot showing reduced expression of Crb3 protein in TDCL A-D relative to parental W2.3.1-5 and expression of vimentin and E-cadherin in TDCL A-D relative to parental W2.3.1-5. (D) Localization of junction proteins occludin, ZO-1, cadherin, and β-catenin to intracellular junction in parental W2.3.1-5 and aberrant localization in TDCL 5D. The localization of junction proteins in TDCLs is either fragmented or localized to the cytoplasm.
Similar to the upregulated genes, there was a unifying pattern of repressed genes in all TDCLs, and these genes had the common theme of plasma membrane proteins known or predicted to play a role in epithelial adhesion and junction formation (shown in blue, Fig. 2B). Among the 15 most repressed genes in TDCL 5D (0.36 fold or lower expression relative to parental W2.3.1-5), the most repressed gene (AU015319) is crb3 (Fig. 2B), a murine homologue of the Drosophila apicobasal polarity determent Crumbs. Crb3 is the mammalian isoform of Drosophila crb that is exclusively expressed in epithelial tissues, and Western blotting for Crb3 protein expression revealed high expression in parental W2.3.1-5 cells and dramatically reduced expression in all four of the TDCLs, with undetectable expression in TDCL 5D (Fig. 2C).
In addition to crb3, another highly repressed gene in TDCLs is Tm4sf10/Bcmp1/Vab-9 (Fig. 2B), a poorly characterized adherent junction and claudin-related protein that regulates adhesion through disc-large (Dlg) in Drosophila Crumbs pathway (24). Claudins 3 and 4 were also repressed in TDCLs (Fig. 2B), and are transmembrane proteins that are the major structural elements in epithelial tight junctions (25). Another repressed gene, Tacstd1/EpCam (Fig. 2B) is a homophilic adhesion regulator and claudin interacting protein (26). It is possible that acquisition of a single defect in epithelial junction formation resulted in the coordinate down-regulation of expression of multiple cell junction components.
Acquisition of tumorigenesis is associated with EMT and deficient tight junction formation
Commonly used molecular markers for EMT include increased expression of vimentin, a mesenchymal marker, and decreased E-cadherin expression, a hallmark of metastatic carcinoma (27). In order to determine if TDCLs underwent an EMT we assayed vimentin and E-cadherin protein expression levels in comparison to the parental cell line. Western blot analysis revealed an increase in vimentin in all TDCLs compared to W2.3.1-5 and a concomitant downregulation of the E-cadherin levels in TDCL 5C and 5D (Fig. 2C). These results suggest that selection for tumorigenesis in TDCLs also selects for acquisition of properties associated with an EMT.
The repression of crb3 expression and that of other junction/adhesion regulators suggested that selection for tumorigenicity in vivo resulted in loss of epithelial junction formation. Therefore, junction formation was examined in parental W2.3.1-5 and TDCLs by immunofluorescence for the tight junction proteins occludin, ZO-1, and for the adherens junction proteins cadherins and β-catenin in culture in vitro.
Staining for the tight junction markers occludin and ZO-1 in W2.3.1-5 cells (Fig. 2D) revealed a smooth, continuous and well-defined staining at the membranes between cells that was initiated by cell-cell contact as cells began to form a monolayer. In contrast, TDCL 5D (Fig. 2D) and the other TDCLs (data not shown) displayed a diffuse staining pattern for occludin and a faint, discontinuous, saw-tooth-like staining pattern of ZO-1, even subsequent to monolayer formation, suggesting impaired tight junction formation. Staining for β-catenin and pan-cadherin revealed smooth, continuous staining at cell-cell contacts in parental W2.3.1-5 that was reduced in TDCL 5D (Fig. 2D) and the other TDCLs (data not shown). These results suggest that tight junction formation was severely impaired in TDCLs and adherens junctions are affected to a lesser degree in TDCLs compared to the parental cells. Thus essential characteristics of epithelial cell junction formation were lost in TDCLs coincident with acquisition of tumorigenicity.
Restoration of Crb3 expression in TDCL 5D corrects defective epithelial junction formation
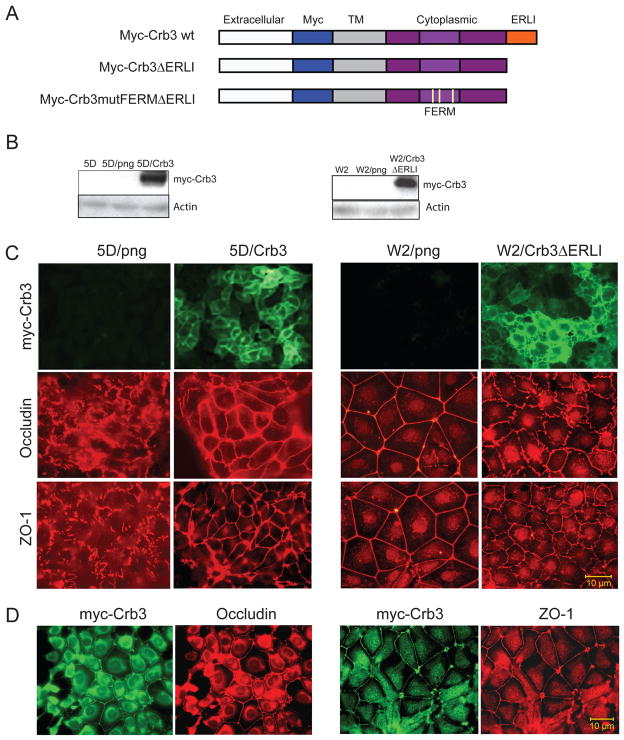
To test the hypothesis that repressed crb3 expression was responsible for defective tight junction formation in TDCLs, crb3 expression was restored in TDCL 5D where crb3 endogenous levels were undetectable. TDCL 5D was infected with either a retroviral vector alone (png) or a vector driving expression of myc-tagged Crb3 (15) and cells stably expressing Crb3 were generated (Fig. 3A and B). Importantly, Crb3 localized to the plasma membrane and substantially rescued tight junction formation in TDCL 5D as indicated by increased occludin and ZO-1 at the junctions between cells compared to the png vector control (Fig. 3C). Thus, the loss of crb3 expression in TDCL 5D is in part responsible for impaired junction formation.
Figure 3.
Role of Crb3 in controlling tight junction formation. (A) Graphic representation of wild type Crumbs 3 protein domain structure and schematic illustrating the Myc-tagged Crb3 and Crb3 mutants22. The Crb3 mutants contain a deletion of the C-terminal domain (ΔERLI) and three point mutations in the FERM domain (Crb3mutFERM). (B) Western blots illustrating the stable expression of myc-Crb3 in TDCL 5D and the mutant myc-Crb3ΔERLI in W2.3.1-5 cells. (C) Crb3 expression restores tight junction formation in TDCL 5D. TDCL 5D png vector infected or wild type Crb3 expressing vector infected cells and stained with for tight junction markers occludin, and ZO-1. Immunofluorescence showin restoration of tight junctions indicated by alterations in ZO-1 and occludin. Expressed Crb3 localized to the plasma membrane and rescued tight junction formation in TDCL 5D as indicated by increased occludin and ZO-1 at cell junctions compared to png vector control. Mutant crb3 expression impairs tight junction formation in parental W2.3.1-5 cells. W2.3.1-5 parental cells png vector infected or Crb3ΔERLI expressing vector infected were immunostained for tight junction markers occludin, and ZO-1. Immunofluorescence showing disruption of tight junctions indicated by alterations in ZO-1 and occludin. (D) Immunofluorescence showing co-localization of Crb3 with occludin and ZO-1 in TDCL 5DCrb3 cells.
Inhibition of Crb3 function prevents junction formation in W2.3.1-5 cells
Since interfering with Crb3 function disrupts junction formation (28), in a complimentary approach, we tested if expression of dominant-negative mutant of crb3 disrupted polarity in parental, non-tumorigenic W2.3.1-5 cells. The ERLI motif on Crb3 binds to the PDZ domain on PALS1 and PALS1 binds PATJ to form the Crb3/PALS1/PATJ functional protein complex. Deletion of the ERLI motif in Crb3 disrupts the Crb3/PALS1/PATJ complex, and thereby polarity and junction formation in mammalian cells (15, 28) and produces multilayer cell growth in Drosophila (29). Mutation of the FERM binding motif in Crb3 has similar effects but the interacting protein is not known (15).
Infection with retroviral vectors driving expression of dominant-negative, myc-tagged mutant forms of crb3, Crb3ΔERLI and Crb3ΔERLImutFERM was used to assess the impact on junction formation in W2.3.1-5 cells (Fig. 3A and B). Expression of either Crb3ΔERLI (Fig. 3C) or Crb3ΔERLImutFERM (data not shown) efficiently disrupted tight junction formation in W2.3.1-5 cells (Fig. 3C). Thus, as in other epithelial cell types (30, 31), Crb3 is an essential regulator of tight junction formation in iBMK cells.
Requirement for Crb3 for establishing apicobasal polarity
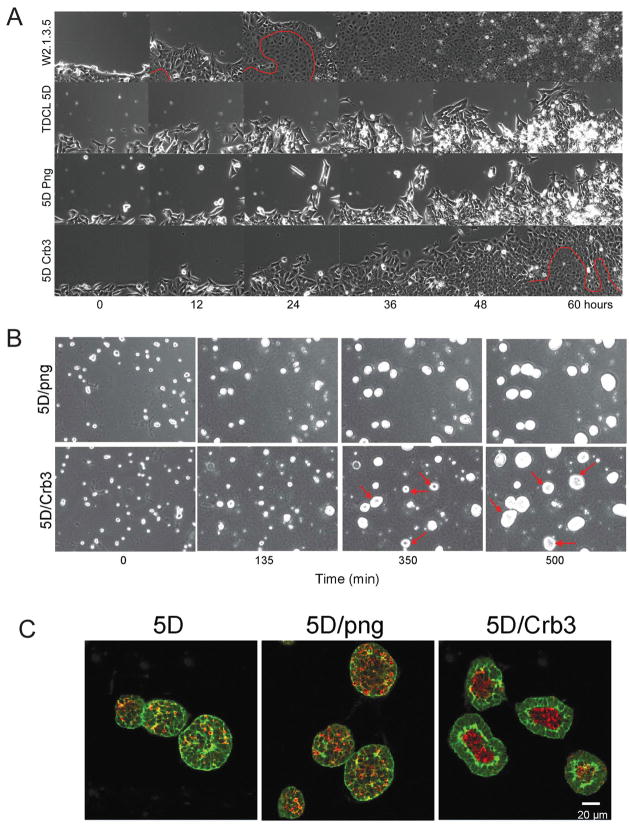
To test if loss of crb3 and junction formation caused defects polarity and possibly altered migration in TDCLs, we employed multifield time-lapse microscopy to examine a wound-healing response as a means to study directional cell migration in vitro. Parental W2.3.1-5 cells responded to the wound in vitro with a high mitotic rate and a wave of coordinated and directional migration into the wound area followed by tight junction and fusion sheet formation as indicated by the disappearance of the distinction between individual cells (Fig. 4A, below the red line, and supplemental video 1). In contrast, wounded TDCL 5D cell monolayers displayed random, uncoordinated cell movements and the cell population failed to efficiently migrate to fill the void (Fig. 4A, and supplemental video 2). There was also failure of tight junction formation (no fusion sheet), and cells piled on top of one another (multilayer growth) and succumbed to apoptosis indicated by the accumulation of highly refractile apoptotic corpses (Fig. 4A, and supplemental video 2). TDCL 5D infected with the png vector behaved similarly (Fig. 4A, and supplemental video 3). TDCL 5D expressing crb3, however, displayed a marked improvement in coordinated directional migration, junction formation (fusion sheet formation, below the red line), contact inhibited growth, and suppression of apoptosis (Fig. 4A, and supplemental video 4). These findings support a substantial role for deficiency in crb3 expression in the loss of epithelial characteristics acquired during selection for the capacity for tumor growth in vivo.
Figure 4.
Crumbs3 rescues migration, polarity and tight junction formation. (A) iBMK parental, TDCLs or derivatives infected with the retroviral vector (5D/png) or the Crb3-expressing retroviral vector (5D/Crb3) were cultured to a confluent monolayer. Scratching a line through the monolayer with a pipette tip then disrupted a small area. The open gap was then monitored as the cells migrated in and filled the void using computerized video multifield time-lapse microscopy for three days. Epithelial fusion sheet resulting from tight junction formation is indicated below the red line. (B) Restoration of Crb3 expression establishes single layer cell growth, apicobasal polarity and lumen formation in TDCL 5D spheroids. TDCL 5D uninfected or infected with the empty retroviral vector png, or the wild type Crbs3 expressing vector were cultured in Matrigel and monitored by time-lapse microscopy. (C) Spheroids were co-stained with for β-catenin (green) and active caspase-3 (red) and imaged by confocal microscopy at 14 days. Crb3 expression confers single layer cell growth and apoptotic lumen formation in three-dimensional culture. 20–30 spheroids/microscopic field were counted in five different fields for each cell line and the experiment was repeated three times (p<0.05). Greater than 90% of TDCL 5D cells expressing Crbs3 formed spheroids with hollow lumens compared to less than 10% of TDCL 5D infected with the png vector.
Crb3 is required for epithelial apicobasal polarity in three-dimensional morphogenesis
Another means to examine proper junction formation and polarity characteristics of epithelial cells is through three-dimensional growth in extracellular matrix (Matrigel) where cells form spheroids that organize into an epithelial layer with apicobasal polarity forming a lumen through apoptosis (3, 17). To test the role of crb3 in epithelial organization in three-dimensional cultures, TDCL 5D, TDL 5D png, and TDCL 5D Crb3 were grown in Matrigel and evaluated morphologically by time-lapse microscopy, and for junctions, organization and spatial localization of apoptosis by β-catenin and active caspase-3 staining by confocal microscopy.
TDCL 5D and vector png transduced derivatives form solid spheroids in Matrigel with high efficiency typified by random cellular organization and spatial localization of apoptosis, and multilayer cell growth (Fig. 4B and supplemental video 5). In contrast, TDCL 5D expressing crb3 formed spheroids composed of a single epithelial cell layer with hollow lumens (Fig. 4B, and supplemental video 6). This suggested that lack of Crb3 and tight junction formation disrupted the three-dimensional epithelial organization by preventing tight junction formation. The spheroids were stained for β-catenin (green) and active caspase-3 (red) and confocal microscopy showed that 90% of TDCL-5D cells overexpressing crb3 formed spheroids with hollow lumens compared to 10% TDCL-5D transduced with the png vector and these lumens were coincident with detection of active caspase-3 (Fig. 4C). In contrast, TDCL 5D and 5D png generated solid spheroids with random cellular organization and caspase-3 activation (Fig. 4C). Thus restoring crb3 expression in TDCL 5D cells rescued loss of specialized cell–cell contacts, polarized morphology and proper epithelial organization and function while suppressing multilayer cell growth that may impact tumorigenesis.
Role of Crb3 in regulating growth arrest through contact inhibition
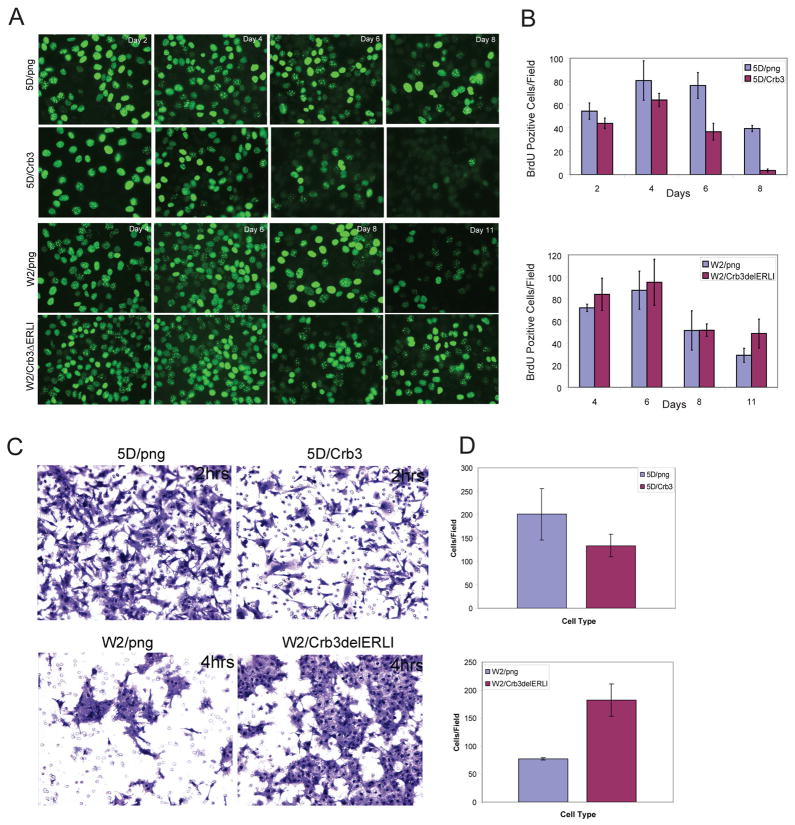
Striking evidence from the time-lapse microscopy in particular, also supported by the findings from three-dimensional culture, suggested that subconfluent crb3-expressing epithelial cells (either W2.3.1-5 or TDCL 5D Crb3) divide rapidly, but once confluence was reached and tight junctions form, mitosis was suppressed due to contact inhibition. This tight junction formation-associated suppression of cell division was absent in Crb3-deficient cells, which may contribute to multilayer cell growth. To test the role of Crb3 in mediation of contact-inhibited growth, TDCL 5D png and TDCL 5D Crb3 were followed over time and during increasing cell density for occurrence of DNA synthesis by BrdU labeling (Fig. 5B). The results indicated that TDCL 5D png continued to incorporate BrdU, even when cells were at high density and reached confluency, at which point TDCL 5D Crb3 failed to incorporate BrdU. Conversely, we tested if disruption of Crb3 function in W2.3.1-5 cells relieved contact inhibition. W2.3.1-5 cells displayed decreased BrdU incorporation and with increasing cell density (Fig. 5), as expected. Furthermore, coincident with tight junction formation, W2.3.1-5 cells expressing the dominant negative Crb3ΔERLI continued DNA synthesis even when at high cell density (Fig. 5B). These results indicate that impairing tight junction formation in iBMK cells leads to failure of contact inhibition leading to unrestrained proliferation. This is remarkably similar to the cellular overgrowth phenotype of Drosophila imaginal disc tissue with defects in junction formation (13).
Figure 5.
Role of Crb3 in regulation of growth arrest and in vitro migration. (A) Immunofluorescence staining showing BrdU incorporation, as a measure of proliferation in TDCL 5D png and 5D Crb3 cells or W2.3.1-5 png and W2.3.1-5 Crb3ΔERLI with increasing cell density. Crb3 deficient TDCL 5D png with impaired tight junction formation continued to incorporate BrdU even after the cells reached confluency, whereas the TDCL 5D Crb3 suppressed proliferation in response to tight junction formation, contact inhibition. (B) Graphs summarizing the number of positive BrdU cells with increasing time and cell density. 50–100 cells/microscopic field were counted in four different fields for each cell line and the experiment was repeated three times. Results are presented as means ± SD of the number of cells/field, p<0.05 (p= 0.0348). (C) TDCL 5D png and 5D Crb3 and (B) W2.3.1-5 png and W2.3.1-5 Crb3ΔERLI cells were cultured to confluency in the upper well of transwell chambers. After 24 hrs the medium was changed in the upper chamber to serum depleted and cells passing through the filter were fixed, stained, and counted. Cells were cultured in triplicate wells/experiment and the experiment was replicated three times. Representative microscopic fields of crystal violet stained cells are shown. (D) Graphs summarizing the results of in vitro migration assay. Results are presented as means ± SD of the number of cells/field, evaluated in five-fields/membrane, p<0.05 (p=0.037).
Role of Crb3 in suppressing in vitro migration
As TDCL 5D displayed a profound defect in migration as a sheet in the wound-healing assay caused by defective tight junction formation, we also assessed the migratory ability of individual cells. In the process of migration as a sheet, epithelial cells must form biological barriers in which individual cells associate with each other through tight junctions and migrate as a cohesive unit. This process is distinct from migration of independent, single, unattached cells that measures the ability of individual cells to move through porous membranes by a process that may be linked to increased intravasation, invasion, and capacity for metastasis. The invasive behavior of TDCL 5D png and TDCL 5D Crb3 was assessed in parallel with W2.3.1-5 png and W2.3.1-5 Crb3ΔERLI using an in vitro migration assay. As illustrated in Fig. 5C and D, png vector transduced TDCL 5D efficiently migrated as individual cells across a porous membrane whereas restoring crb3 expression in TDCL 5D cells suppressed migration capabilities. Parental W2.3.1-5 cells displayed reduced migration likely due to proper tight junction formation often appearing to transverse the pores as continuous sheets of cells (Fig. 5C and D). Expressing mutant Crb3ΔERLI in W2.3.1-5 cells greatly increased the migratory capacity, although the cells still retained some capacity to form cell-cell contacts (Fig. 5C and D). Thus restoring Crb3 expression in TDCLs repressed migration through a porous membrane, which may have been impaired by tight junction formation, while disrupting Crb3 function in W2.3.1-5 cells repressed junction formation and stimulated migration. This suggested that tight junction formation dependent on Crb3 reduced the potential for cell migration most likely by sustaining intercellular interactions.
Crb3 expression suppresses metastatic potential
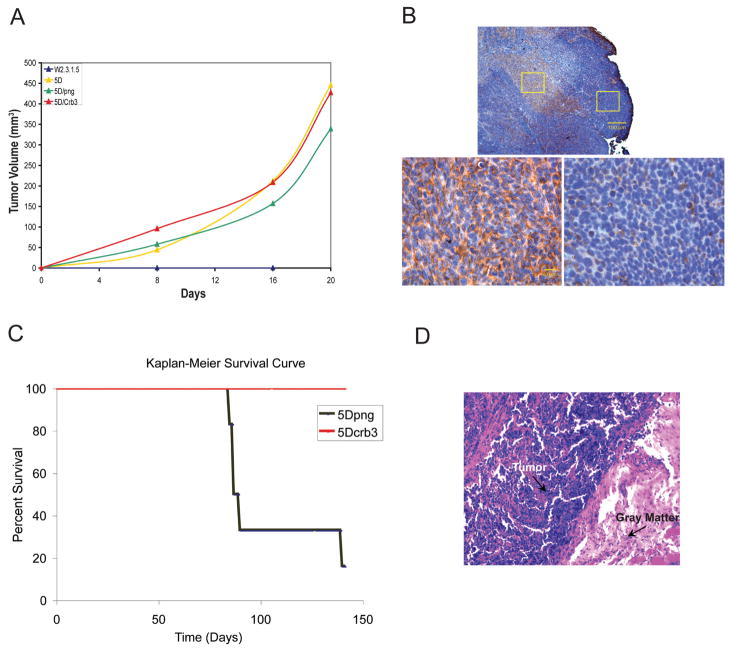
To examine the role of Crb3 in suppressing tumor growth, the tumorigenicity of TDCL 5D png and TDCL 5D Crb3 was evaluated following subcutaneous injection. Surprisingly, both vector and Crb3 transduced TDCLs formed tumors at similar rates (Fig. 6A). However, immunohistochemistry for myc-Crb3 expression, revealed a striking loss of Crb3 staining in the expanding growth regions at the perimeter of TDCL 5D Crb3 tumors (Fig. 6B). Even though all the cells were positive for Crb3 at the start of tumor formation, three weeks later most were negative for Crb3 expression in tumors in vivo. Furthermore, loss of transgene expression is not observed under conditions where expression is required for tumor growth (5) (data not shown). These findings indicate selection for loss of Crb3 expression during tumor expansion, suggesting that its expression imparts a growth disadvantage in tumors in vivo. Moreover, expression of Crb3ΔERLI in W2.3.1 cells and disruption of Crb3 function was not sufficient to confer tumorigenic capacity (data not shown) indicating that impairing tight junction formation may not be sufficient for tumorigenesis. Thus, other genetic or epigenetic changes in TDCLs are responsible for enabling tumor growth independent of the role of the loss of Crb3 in disrupting junctions, polarity and enabling proliferation at high cell density.
Figure 6.
Crb3 expression suppresses metastasis. (A) Tumorigenic capacity of TDCL 5D and png and Crb3 transfectants compared to parental W2.3.1-5 cells. Animals were injected subcutaneously with 107 cells and tumor formation was monitored over time. The parental W2.3.1-5 cells failed to form tumors by 20 days post injection while the TDCL 5D formed subcutaneous tumors as expected. Expression of Crb3 in TDCL 5D did not suppress subcutaneous tumor growth. (B) Subcutaneous tumor growth of TDCL 5D Crb3 is associated with the loss of Crb3 expression. TDCL 5D Crb3 tumors that formed 3 weeks post injection were examined by immunohistochemistry for Myc-Crb3 using the Myc tag. Crb3 positive cells with Crb3 localized to cellular junctions (left) were found in central (top) but not expanding, peripheral regions (right) of the tumor. (C) Crb3 expression suppresses metastasis. Kaplan-Meier survival curve of mice injected (tail vein) with 5×105 cells TDCL 5D png (black, n=6) and TDCL 5D Crb3 (red, n=6). Most (5/6) of the TDCL 5D png injected animals succumbed to multiple bone and kidney metastases by 10 weeks whereas all of the TDCL 5D Crb3 injected animals remained tumor free. (D) Immunohistochemistry showing metastasis to the spinal cord, illustrated by purple cells invading the gray matter (pink area) in animals injected with TDCL 5D png cells.
Since loss of Crb3 expression was associated with tumor expansion, we also evaluated the requirement for Crb3 loss in a model for metastatic growth. 5×105 TDCL 5D png and TDCL 5D Crb3 cells were injected into the tail vein of nude mice that were monitored over time for tumor growth and for intravasation and colonization of organs. Multiple tumors from TDCL 5D injected cells were found extensively colonizing the kidney and bone, especially the legs and spinal cord in five of six injected mice (Fig. 6C and D). In contrast, all of the TDCL 5D Crb3 injected animals remained tumor free (Fig. 6C). This suggested that restoring epithelial junction formation by expressing crb3 limits invasiveness and metastatic potential. Increased intercellular adhesion and attachment among tumor cells may be expected to diminish migration, intravasation and invasiveness, in addition to its role in mediating contact-inhibited growth. Thus loss of crb3 expression in tumors in vivo may be one means to overcome a barrier to tumor progression.
DISCUSSION
One of the primary diagnostic features in malignant tumors of epithelial origin is a profound disruption of cell architecture and organization. Even though the link between loss of epithelial organization and tumor progression has been previously made, an important unanswered question remains: is it correlative or does the loss of architecture contribute to progression toward malignancy in mammalian tumors? In Drosophila, however, most of the genes associated with tumor suppression function to establish and maintain epithelial cell junctions and polarity, the loss of which disrupts tissue organization and produces an overgrowth phenotype. If this process is evolutionarily conserved, there may be a role for the mammalian homologues of nTSGs in mammalian cancer that has yet to be elucidated.
The tumor suppressors identified in Drosophila are prominently comprised of genes that regulate epithelial junctions and polarity determination. In the Drosophila Crumbs pathway the polarity regulators Crumbs, Scribble (Scrib), Lethal giant larvae (Lgl), and Discs large (Dlg) function to suppress overgrowth and tumorigenesis. Perturbation in the function of Drosophila crumbs, scrib, lgl or dlg leads to disruption of polarity and to cell overgrowth in the epithelial tissue of imaginal discs(12–14, 32, 33). There are three junction complexes conserved in invertebrates and mammals that act together to establish apicobasal polarity and junction formation (12, 14): mammalian Crb3/PALS1/PATJ or Drosophila Crumbs/Stardust/dPATJ; mammalian Par-3/Par-6/aPKC or Drosophila Baz/Par-6/aPKC; mammalian hScrib/Dlg1 or Drosophila Scrib/Dlg and mammalian Scrib/Dlg/Lgl or Drosophila Scrib/Dlg/Lgl. Direct interaction between these proteins and protein complexes and their localization to the cellular membranes establishes junction formation and apicobasal polarity.
It is not surprising that a tumor suppression mechanism identified in Drosophila, such as polarity regulation, is conserved in mammals. Nonetheless establishing that this is the case is important and supported by the location of crb3 at 19p13.3, which is frequently deleted in carcinomas and where multiple tumor suppressor genes reside (34). Advanced cancers often have undergone a partial or full EMT that disrupts epithelial junction formation that may contribute to invasion and metastasis. Previous studies have shown that mutations in polarity genes in Drosophila lead to uncontrolled proliferation and abnormal tissue architecture (12). An EMT can be achieved by epithelial tumor cells reactivating one of the developmental EMT programs controlled by TGF-β, RTK/RAS, Wnt, Notch, Hedgehog, or NF-κB (8). Alternatively, in mammalian cells a partial or full EMT can be achieved by oncogenic activation of signal transduction pathways such as the MAP and PI-3 kinase pathways. Increased levels of PI-3 kinase contribute to loss of polarity and increased cell proliferation through its downstream effectors Akt and Rac1 in human mammary tumors (35). The ability of STAT pathways to disrupt polarity by direct repression of E-cadherin by Snail or Twist is also documented (8). STAT3 activation by β4 integrin in conjunction with Erb2 signaling contributes to disruption of epithelial polarity and overproliferation in aggressive breast tumors (36). Moreover, activation of Erb2 signaling disrupts apicobasal polarity by associating with PAR-6-aPKC components of the Par polarity complex (37). Our data suggests that direct perturbation of the downstream effectors of polarity, such as the polarity determinant Crb3, can also lead to phenotypic changes that facilitate tumor progression.
Comparing the global expression profiles of four independent TDCLs with the non-tumorigenic parental cell line from which they were derived, showed striking downregulation of Crb3, an important determinant of tight junction formation (15) along with coordinate downregulation of other known or putative cell junction/adhesion proteins. Previous studies have shown a correlation between tumor progression and the reduction in tight junctions suggesting their importance as key aspect that cancer cells must overcome in order to metastasize (38). We established that TDCLs have lost the capacity for contact-inhibited growth and expressing Crb3 substantially restored proper tight junction formation leading to inhibition of cell proliferation. Thus, the loss of Crb3 expression in TDCLs plays a significant role in altering epithelial three-dimensional organization by conferring multilayer cell growth associated with absent contact inhibition that may contribute to tumorigenesis.
Previous studies have shown a correlation between tumor metastasis and a reduction in tight junctions (38). Our studies showed that restoring crb3 expression in TDCL cells reduced the migratory capacity and limited the invasiveness and metastatic potential of TDCLs. The dramatic loss of Crb3 expression in tumors in vivo also suggests that it is a growth disadvantage. Taken together, these results establish that Crb3 plays an important role in maintaining the epithelial phenotype, and downregulation or loss of function of Crb3 contributes to tumorigenic progression. Our data suggest that Crb3 is required for tight junction formation, establishment of polarity and contact inhibited growth, and suppression of migration, which suppresses metastasis. Thus, in mammalian epithelial tumors, as in Drosophila, similar mechanisms may control tumor growth. It will be of great interest to examine the status of Crb3 in human tumors and the possible contribution of Crb3 deregulation to the tumor-promoting functions of developmental and oncogenic pathways that modulate polarity in human cancers.
Acknowledgments
We thank Dr. Benjamin Margolis for the generous gift of Crb3 and Crb3 mutants retroviral expression vectors, and Crb3 antibody, Tissue Analytic Services and, the Gene Expression Core Facilities at the Cancer Institute of New Jersey for tissue preparation and microarray analysis, respectively. The Howard Hughes Medical Institute and a grant from the National Institute of Health (R37CA53370) to Dr. Eileen White supported this work.
References
- 1.Degenhardt K, White E. A mouse model system to genetically dissect the molecular mechanisms regulating tumorigenesis. Clin Cancer Res. 2006;12:5298–304. doi: 10.1158/1078-0432.CCR-06-0439. [DOI] [PubMed] [Google Scholar]
- 2.Tan TT, Degenhardt K, Nelson DA, et al. Key roles of BIM-driven apoptosis in epithelial tumors and rational chemotherapy. Cancer Cell. 2005;7:227–38. doi: 10.1016/j.ccr.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Karantza-Wadsworth V, Patel S, Kravchuk O, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–35. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Degenhardt K, Chen G, Lindsten T, White E. BAX and BAK mediate p53-independent suppression of tumorigenesis. Cancer Cell. 2002;2:193–203. doi: 10.1016/s1535-6108(02)00126-5. [DOI] [PubMed] [Google Scholar]
- 5.Nelson DA, Tan TT, Rabson AB, Anderson D, Degenhardt K, White E. Hypoxia and defective apoptosis drive genomic instability and tumorigenesis. Genes Dev. 2004;18:2095–107. doi: 10.1101/gad.1204904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christiansen JJ, Rajasekaran AK. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res. 2006;66:8319–26. doi: 10.1158/0008-5472.CAN-06-0410. [DOI] [PubMed] [Google Scholar]
- 7.Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–74. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 8.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–58. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Ikenouchi J, Matsuda M, Furuse M, Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J Cell Sci. 2003;116:1959–67. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- 10.Cano A, Perez-Moreno MA, Rodrigo I, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 11.Aigner K, Dampier B, Descovich L, et al. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26:6979–88. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bilder D. Epithelial polarity and proliferation control: links from the Drosophila neoplastic tumor suppressors. Genes Dev. 2004;18:1909–25. doi: 10.1101/gad.1211604. [DOI] [PubMed] [Google Scholar]
- 13.Lu H, Bilder D. Endocytic control of epithelial polarity and proliferation in Drosophila. Nat Cell Biol. 2005;7:1232–9. doi: 10.1038/ncb1324. [DOI] [PubMed] [Google Scholar]
- 14.Margolis B, Borg JP. Apicobasal polarity complexes. J Cell Sci. 2005;118:5157–9. doi: 10.1242/jcs.02597. [DOI] [PubMed] [Google Scholar]
- 15.Fogg VC, Liu CJ, Margolis B. Multiple regions of Crumbs3 are required for tight junction formation in MCF10A cells. J Cell Sci. 2005;118:2859–69. doi: 10.1242/jcs.02412. [DOI] [PubMed] [Google Scholar]
- 16.Perez D, White E. E1B 19K inhibits Fas-mediated apoptosis through FADD-dependent sequestration of FLICE. J Cell Biol. 1998;141:1255–66. doi: 10.1083/jcb.141.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–68. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 18.Albini A, Iwamoto Y, Kleinman HK, et al. A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res. 1987;47:3239–45. [PubMed] [Google Scholar]
- 19.Chang YH, Chao Y, Hsieh SL, Lin WW. Mechanism of LIGHT/interferon-gamma-induced cell death in HT-29 cells. J Cell Biochem. 2004;93:1188–202. doi: 10.1002/jcb.20282. [DOI] [PubMed] [Google Scholar]
- 20.Klampfer L, Huang J, Swaby LA, Augenlicht L. Requirement of histone deacetylase activity for signaling by STAT1. J Biol Chem. 2004;279:30358–68. doi: 10.1074/jbc.M401359200. [DOI] [PubMed] [Google Scholar]
- 21.Kulaeva OI, Draghici S, Tang L, Kraniak JM, Land SJ, Tainsky MA. Epigenetic silencing of multiple interferon pathway genes after cellular immortalization. Oncogene. 2003;22:4118–27. doi: 10.1038/sj.onc.1206594. [DOI] [PubMed] [Google Scholar]
- 22.Nusinzon I, Horvath CM. Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc Natl Acad Sci U S A. 2003;100:14742–7. doi: 10.1073/pnas.2433987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakamoto S, Potla R, Larner AC. Histone deacetylase activity is required to recruit RNA polymerase II to the promoters of selected interferon-stimulated early response genes. J Biol Chem. 2004;279:40362–7. doi: 10.1074/jbc.M406400200. [DOI] [PubMed] [Google Scholar]
- 24.Simske JS, Koppen M, Sims P, Hodgkin J, Yonkof A, Hardin J. The cell junction protein VAB-9 regulates adhesion and epidermal morphology in C. elegans Nat Cell Biol. 2003;5:619–25. doi: 10.1038/ncb1002. [DOI] [PubMed] [Google Scholar]
- 25.Swisshelm K, Macek R, Kubbies M. Role of claudins in tumorigenesis. Adv Drug Deliv Rev. 2005;57:919–28. doi: 10.1016/j.addr.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Ladwein M, Pape UF, Schmidt DS, et al. The cell-cell adhesion molecule EpCAM interacts directly with the tight junction protein claudin-7. Exp Cell Res. 2005;309:345–57. doi: 10.1016/j.yexcr.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 27.Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat Rev Cancer. 2004;4:118–32. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- 28.Roh MH, Fan S, Liu CJ, Margolis B. The Crumbs3-Pals1 complex participates in the establishment of polarity in mammalian epithelial cells. J Cell Sci. 2003;116:2895–906. doi: 10.1242/jcs.00500. [DOI] [PubMed] [Google Scholar]
- 29.Klebes A, Knust E. A conserved motif in Crumbs is required for E-cadherin localisation and zonula adherens formation in Drosophila. Curr Biol. 2000;10:76–85. doi: 10.1016/s0960-9822(99)00277-8. [DOI] [PubMed] [Google Scholar]
- 30.Lemmers C, Michel D, Lane-Guermonprez L, et al. CRB3 binds directly to Par6 and regulates the morphogenesis of the tight junctions in mammalian epithelial cells. Mol Biol Cell. 2004;15:1324–33. doi: 10.1091/mbc.E03-04-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurd TW, Gao L, Roh MH, Macara IG, Margolis B. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat Cell Biol. 2003;5:137–42. doi: 10.1038/ncb923. [DOI] [PubMed] [Google Scholar]
- 32.Giebel B, Wodarz A. Tumor suppressors: control of signaling by endocytosis. Curr Biol. 2006;16:R91–2. doi: 10.1016/j.cub.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 33.Wodarz A, Hinz U, Engelbert M, Knust E. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell. 1995;82:67–76. doi: 10.1016/0092-8674(95)90053-5. [DOI] [PubMed] [Google Scholar]
- 34.Yang M, Nelson D, Funakoshi Y, Padgett RW. Genome-wide microarray analysis of TGFbeta signaling in the Drosophila brain. BMC Dev Biol. 2004;4:14. doi: 10.1186/1471-213X-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu H, Radisky DC, Wang F, Bissell MJ. Polarity and proliferation are controlled by distinct signaling pathways downstream of PI3-kinase in breast epithelial tumor cells. J Cell Biol. 2004;164:603–12. doi: 10.1083/jcb.200306090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo W, Pylayeva Y, Pepe A, et al. Beta 4 integrin amplifies ErbB2 signaling to promote mammary tumorigenesis. Cell. 2006;126:489–502. doi: 10.1016/j.cell.2006.05.047. [DOI] [PubMed] [Google Scholar]
- 37.Aranda V, Haire T, Nolan ME, et al. Par6-aPKC uncouples ErbB2 induced disruption of polarized epithelial organization from proliferation control. Nat Cell Biol. 2006;8:1235–45. doi: 10.1038/ncb1485. [DOI] [PubMed] [Google Scholar]
- 38.Martin TA, Jiang WG. Tight junctions and their role in cancer metastasis. Histol Histopathol. 2001;16:1183–95. doi: 10.14670/HH-16.1183. [DOI] [PubMed] [Google Scholar]