Abstract
Accurate DNA replication during S-phase is fundamental to maintain genome integrity. During this critical process, replication forks frequently encounter obstacles that impede their progression. While the regulatory pathways which act in response to exogenous replication stress are beginning to emerge, the mechanisms by which fork integrity is maintained at naturally occurring endogenous replication-impeding sequences remains obscure. Notably, little is known about how cells replicate through special chromosomal regions containing structured non-B DNA, e.g. G4 quartets, known to hamper fork progression or trigger chromosomal rearrangements. Here, we have investigated the role in this process of the human translesion synthesis (TLS) DNA polymerases of the Y-family (pol η, pol ι, and pol κ), specialized enzymes known to synthesize DNA through DNA damage. We show that depletion by RNA interference of expression of the genes for Pol η or Pol κ, but not Pol ι, sensitizes U2OS cells treated with the G4-tetraplex interactive compound telomestatin and triggers double-strand breaks in HeLa cells harbouring multiple copies of a G-rich sequence from the promoter region of the human c-MYC gene, chromosomally integrated as a transgene. Moreover, we found that downregulation of Pol κ only raises the level of DSB in HeLa cells containing either one of two breakage hotspot structured DNA sequences in the chromosome, the major break region (Mbr) of BCL-2 gene and the GA rich region from the far right-hand end of the genome of the Kaposi Sarcoma associated Herpesvirus. These data suggest that naturally occurring DNA structures are physiological substrates of both pol η and pol κ. We discuss these data in the light of their downregulation in human cancers.
Keywords: DNA Replication, Genetic Instability, Double Strand Breaks, non-B DNA
Introduction
Cell proliferation involves numerous processes that need to be tightly coordinated to ensure the preservation of genome integrity and to promote faithful genome propagation. Efficient and accurate DNA replication is critical for such maintenance of genome stability during the error-free duplication of chromosomes before their segregation. In eukaryotic cells, this process relies on the well-orchestrated activation of a large number of replication origins and the duplication of an enormous amount of DNA [1]. This challenging task is made more difficult by genotoxic insults and endogenous blocks, which impede fork progression and trigger chromosomal rearrangements [2]. Failure to protect stalled forks or to process the replication fork appropriately for replication restart results in the accumulation of mutations and genomic aberrations. Indeed, a variety of human genetic syndromes that lead to cancer predisposition are caused by mutations in genes that protect the genome integrity during chromosome replication [3]. While many studies using several model systems have provided new insights into how DNA replication forks are maintained and restarted when cells are transiently exposed to genotoxic drugs, much less is known about how cells tolerate fork pausing in an unchallenged S phase. Some of the endogenous sources of interference with fork progression are the naturally occurring sequences which have the potential to adopt non B-DNA conformations. These sequences are very abundant in chromosomal regions that are prone for gross rearrangement [4] and some have been associated with human diseases, such several trinucleotide repetitions [5]. Impediment of the replication fork occurs also at guanine-rich nucleic acids which have the potential to form G-quadruplex (G4) DNA, stabilized by Hoogsteen hydrogen bonding between guanine residues, which influence gene expression and genomic stability [6]. The non-B DNA structures are also associated with a variety of other types of genetic instability in vivo. Genomic regions containing fragile sites are prone to undergo DNA breaks, chromosomal translocations, deletions and amplifications [7]. Many leukemias, lymphomas, and sarcomas exhibit specific reciprocal chromosomal translocations that either result in the activation of proto-oncogenes or generate new oncogenic fusion genes. The mechanisms of instabilities occurring in these hotspot regions of the human genome are still poorly understood. Interestingly, many breakage hotspots are mapped inside or near sequences that have the potential to adopt non-B DNA structures. For example, segments of the human c-MYC gene capable of adopting non-B structures are found in the regulatory sequences of the gene [8] and overlap with the major breakage hotspots found in c-MYC-induced lymphomas or leukemias [9,10] (reviewed in [11]). An H-DNA structure also occurs at the BCL-2 gene major breakpoint region (Mbr) in follicular lymphomas [12]. Several of the fragile sites have been shown to be prone for breakage after partial inhibition of DNA synthesis, as caused by aphidicolin, an inhibitor of replicative DNA polymerases [13]. To date, the precise mechanisms maintaining genomic stability during the replication of these hotspot regions of the human genome remain poorly understood. Evidence suggests that several DNA helicases, such as the RecQ-family helicases defective in Bloom and Werner syndromes [14,15], and BRIP1 (FANCJ) [16] have roles in the resolution of aberrant DNA structures, including G quartets. Here, we investigated if the specialized enzymatic activities of the translesion (TLS) DNA polymerases could be required when the replication forks are challenged by non-B DNA to facilitate replication fork reactivation and/or to avoid fork collapses and DSB induction. TLS polymerases are specialized enzymes that present a much more open structure of their catalytic site than error-free replicative DNA polymerases (Pol delta and Pol epsilon) [17], enabling them to accommodate damaged bases in their active sites. In the translesion process, TLS polymerases take over from stalled replicative polymerases and allow synthesis through a variety of DNA lesions [17]. In human cells, many TLS DNA polymerases belong to the Y-family which includes Pol η, Pol ι and Pol κ proteins, the most well documented enzymes of this family. Pol η has the unique property of being able to synthesise past DNA containing UV-generated cyclobutane thymine dimers (CPD) with similar efficiency to that in undamaged DNA [18] and was described as the deficient protein in the variant form of Xeroderma Pigmentosum (XPV) cancer-prone syndrome [19]. Pol ι has very low processivity and is able to insert bases opposite some types of damage, but unable to extend synthesis further from the inserted base. It has a very high error rate [20] and its function remains a mystery. Pol κ is able to carry out TLS past benzo[a]pyrene-guanine and other adducts at the N2 position of guanine both in vitro and in vivo [21–23]. In addition to its role in TLS, Pol κ seems to have a role in the repair synthesis step of Nucleotide Excision Repair (NER) pathway [24].
Here, we present evidence suggesting that both Pol η and Pol κ prevent genomic instability occurring at natural DNA sequences capable of forming unusual secondary structures in human cells. We discuss the data in the scope of the frequent downregulation of these two TLS DNA polymerases in human cancers.
Materials and Methods
Construction of non-B DNA-containing Plasmids
Natural occurring human sequences that are capable of forming non-canonical DNA structures were amplified by PCR and were cloned into plasmid pUCNIM at the same location similar as described previously [25]. pULCtrl has 600 bp control non-B DNA sequence from the GAPDH gene and is not known to form non-B DNA structures. pUMycProm has 556 bp of the human c-MYC promoter region, containing at least 3 Z-DNA forming sequences [26], one H-DNA [27], and one G-DNA forming sequence [28]. pUMBR has 520 bp from the human BCL-2 gene major break region (MBR) where several H-DNA forming sequences have been defined [12]. pUGA-rich has a GA rich region (~700 bp), a breakage hotspot region from the far right-hand end of the KSHV genome and can form an H-DNA structure.
Cell culture
U2OS cells and HeLa cells were grown in DMEM medium (Gibco) complemented with 10% of FBS (Lonza) and 80 units/mL of penicillin, and 80μg/mL of streptomycin (Gibco) for U2OS cells and 500 μg/mL neomycin only for transgenic HeLa cells containing pUCNIM derivatives that express the neomycin resistant gene.
Construction of transgenic HeLa cell lines harbouring chromosomal non-B DNA forming sequences
After the initial cloning of the naturally occurring sequences and verification by DNA sequencing, the plasmids, pULCtrl, pUMycProm, pUMBR, and pUGA-rich were digested with BsaI, and 5 μg of each linearized plasmid DNA was transfected into HeLa cells using Nucleofector kit R according to the manufacturer’s recommendations (Amaxa Biosystems, Cologne, Germany). Stably transfected cells were obtained after selecting in medium containing 500 μg/ml G418. The self-ligated episomal plasmids are not able to replicate in HeLa cells and will be loss during cell prelifereation.
The integration of plasmid fragments containing non-B DNA forming sequences into the genomic DNA were random and the copy number of the integrated vector was variable among the different cell lines. To determine the copy number of integration per genome, the genomic DNA of each transgenic cell line was isolated and subjected to quantitative real-time PCR, and the amplification of integrated fragments was compared with that from the GAPDH gene. A standard curve was derived by using serial dilutions made by mixing transgene-negative human genomic DNA with plasmid DNA at 1:1, 1:3, 1:10, 1:30 and 1:100 ratio, according to the lengths of human genome (3×109) and plasmid DNA (7×103), and was used to estimate the absolute copy number of transfected HeLa cells.
Gene silencing in U20S and HeLa cells
To stably knock down the expression of POLH, the gene encoding DNA polymerase eta, POLI, the gene encoding DNA polymerase iota and BRIP1(BACH1), the gene encoding the DNA helicase BRIP1 (BACH1/FANCJ) in U2OS cells, we used replicative short hairpin (shRNA) expressing vectors (pEBVsiRNA) which have been previously shown to impose a very strong and stable gene silencing in human cells after several months in culture [29]. siRNA design, pEBVsiRNA vector cloning and establishment of stable knockdown cells were carried out as previously described [30]. RNAi sequences for Pol η (NM 006502) extended nucleotides 5′ GTGTTGAAGTGATGGAGAT3′ in the ORF (shη 1) and 5′GCAATGAGGGCCTTGAACA3′ in the 3′UTR region (shη2). For Pol ι (NM 007195), RNAi sequences stretched nucleotides 5′CTCAGTCCTTTAGTGAAGA3′ in the ORF (shι1) and 5′GAAGTAAATTCTGGCACAA3′ in the 3′UTR region (shι2). For BACH1 (NM 032043), RNAi sequences corresponded to 5′GGAATTGCTAGATGGGAAA3′ in the ORF (shB1) and 5′TGCTTATGTTAAACTCTGT3′ in the 3′UTR region (shB2). Long term expression of shRNA in human cells was maintained by addition of 200μg/mL of Hygromycin (Invitrogen). As control, we used the pBD650 plasmid which expresses an inefficient shRNA sequence, as previously validated [29].
For transcient gene silencing in HeLa cells, two independent siRNAs were used to silence the expression of POLK, the gene encoding DNA polymerase kappa (siκ1: 5′CCAAUAGACAAGCUGUGAUdTdT3′ and siκ2: 5′CCCAGUUAAUCAACCCAAAdTdT3′) or POLH (siη1: 5′GCAATGAGGGCCTTGAACA3′ and siη2:5′GCAAACTTAGCTCTGGTTA3′). For control, siRNA against luciferase (5′CGUACGCGGAAUACUUCGAdTdT3′) [31] was used.
Western blotting
Cells were lysed in a buffer containing 50mM Tris pH 7.5, 300mM NaCl, 1% Triton, 5mM EDTA, 1mM DTT, supplemented with inhibitors. Cell lysates were cleared by centrifugation for 10 minutes at 10,000rpm. Cell extracts were boiled in loading buffer (250mM Tris pH 6.8, 5% SDS, 30% Glycerol, 20% β-Mercaptoethanol, Bromophenol blue). Proteins were separated on 7% SDS-PAGE gel during 2 hours and electrotransfered (Biorad) during 2 hours on PVDF membranes (Amersham Biosciences). Pol η was detected using DNA polymerase η polyclonal antibody ab17725 at 50μg/mL (Abcam). Pol ι antibodies were a gift from R. Woodgate (NIH). Pol κ polyclonal antibody was kindly provided by T Nohmi, Naoko Niimi, and Petr Gruz (Tokyo). BACH1/BRIP1 polyclonal antibodies were from Abcam (ab 16608). Actin and Ku70 were detected using monoclonal antibody C4 at 1:10000 (Chemicon) and monoclonal antibody N3H10 (Interchim) at 1:10000 respectively. All blots were detected by ECL Western Blotting susbtrate (PIERCE).
Colony-Forming Survival Assay after TMS treatment
500 or 1000 U2OS cells stably depleted for BRIP1 (BACH1), Pol η, or Pol ι, were seeded in triplicate 24 hours prior 5μM and 8μM treatment with telomestatin (TMS). Clones were counted after Crystal Violet coloration 8 days after treatment. Standard deviations have been calculated from three independent experiments. For Pol κ depletion experiments, 20μM of siRNA directed against Pol K were transfected 24 hours before seeding by using Lipofectamine 2000 protocol.
Cell Preparation and Measurement of γ-H2AX by Flow Cytometry
Forty eight hours after transfection with siRNA, cells (1 × 106 cells) were harvested, fixed in ethanol 70%, washed in PBS before permeabilization for 10 min in PBS SVF 2% Triton 0.1%, centrifuged and resuspended in 100 μL of mouse monoclonal anti-phospho-histone H2AX antibody (Euromedex/Upstate Biotechnology; clone JBW301; 1:400 dilution). Samples were incubated for 1 h under constant agitation at room temperature (RT). At the end of the incubation time, samples were centrifuged, rinsed twice with FACS Buffer (PBS SVF2% Triton 0,1%) and resuspended in 100 μL of secondary antibody Alexa 488 goat antimouse IgG (H + L)F(ab′)2 fragment conjugate (Molecular Probes; 1:200 dilution) for 1h under constant agitation at RT. Samples were washed twice with FACS buffer, resuspended in PBS containing 250 μg/mL Propidium Iodide and 500 μg/mL RNAse-A before an analysis of 15,000 cells/sample with a FACScan (Becton Dickinson). Analysis of flow cytometry data were conducted using Cellquest software. Samples were gated on Propidium Iodide for DNA content and time of flight to eliminate debris and cell doublets before the analysis of γH2AX antibody staining intensity.
Processing of tissue specimen from cancer patients
Tissue specimens from 74 patients who underwent surgery for primary colorectal adenocarcinoma resection at the Toulouse University Hospital between 1999 and 2002 were selected. Exclusion criteria were patients treated with adjuvant therapy and with tumour microsatellite instability (MSI). Before RNA and DNA extraction, a frozen section was cut and stained with hematoxylin and eosin to evaluate the percentage of neoplastic cells. Except for 3 patients, malignant cells accounted for at least 50 % of the tumour involvement. The study was approved by the National Institute of Cancer (INCa) following the recommendations of the National Agency of Agreement and Evaluation for Health (ANAES). Tumour samples have been collected in agreement with the 2004 French Bioethic law.
For RNA extraction, 40 to 80 frozen sections with 5 to 10 μm thick from tumoral and adjacent normal samples were immediately homogenized in the lysis buffer of the RNeasy extraction kit and total RNA were extracted according to the manufacturer’s instruction (Qiagen). The quality of total RNA was assessed on the Agilent 2100® bioanalyser using the RNA NanoLab® chip and RNA 6000 Nano Assay® kit (Agilent Technologies). 1 to 2 μg of total RNA were reverse transcribed using the High-Capacity cDNA Archive Kit (Applied Biosystems). cDNA of Pol E and Pol H transcripts have been amplified in triplicate in tumoral and normal samples using gene expression assays (ABI cat# Hs00197814_m1 and Hs00173030_m1 respectively), TaqMan Universal PCR Master and the 7900HT fast real time PCR apparatus. Four control cDNA (18S, GAPDH, HPRT, YWHAZ) were simultaneously amplified (gene expression assays: ABI cat# Hs99999901_s1, Hs99999905_m1, Hs99999909_m1, Hs00237047_m1). For each pair of samples, the two most stable control genes were selected by qBases software (http://medgen.ugent.be/qbase/) to normalized the expression of pole and polh then ratios T (tumoral)/N(normal) between normalized values were calculated.
Results
Generation of the RNA interference-based knockdown cellular models
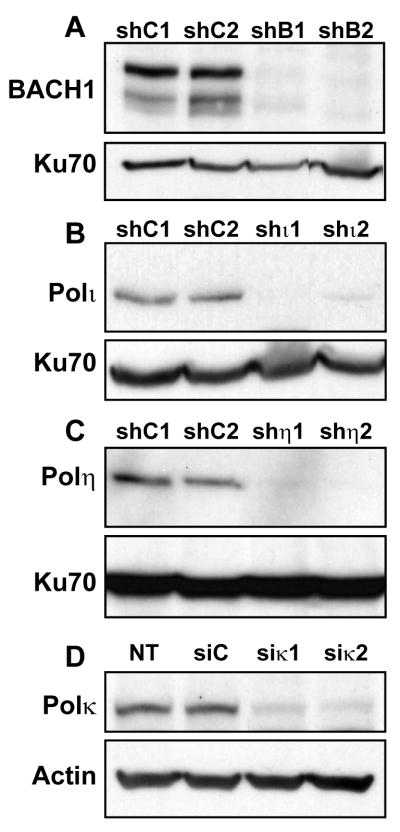
We succeeded in stably knocking down the expression of POLH, POLI, and BRIP1(BACH1) genes in U2OS cells, by using replicative short hairpin (shRNA) expressing vectors (pEBVsiRNA) which exerted a very strong and stable gene silencing after several months in culture. For POLK silencing, we failed to obtain viable long term depleted cellular clones and we decided to transiently silence the gene by using siRNA. All these siRNA/shRNA interference-based knockdown cellular clones presented a strong reduction of protein levels for Bach1, Pol η, Pol ι, and Pol κ, as evidenced by Western blotting (Fig 1). We checked by real time PCR that depletion for each TLS DNA polymerase was specific since none of replicative polymerases (pols α, δ, ε), the other Y-family polymerases, or other alternative DNA polymerases (pols β, λ, μ, and θ) were downregulated at the transcript level (data not shown).
Figure 1. Validation of the RNA interference-based knockdown cellular models (A–C).
We generated U2OS clones expressing shRNA control sequences (shC1 and shC2) and targeting BACH1 (BRIP1) (shB1 and shB2), POL I (shι1 and shι2) or POL H (shη1 and shη2). Extracts from these cells were analyzed by immunoblotting with the indicated antibodies and Ku70 as a loading control (D) U2OS cells were transfected with control siRNA (Luciferase) or two independent Pol κ siRNAs (siκ1 and siκ2). The levels of Pol κ was analyzed by Western blotting 48h after transfection and normalized against actin.
Pol η and Pol κ depletion sensitizes cells to the G-quadruplex ligand TMS
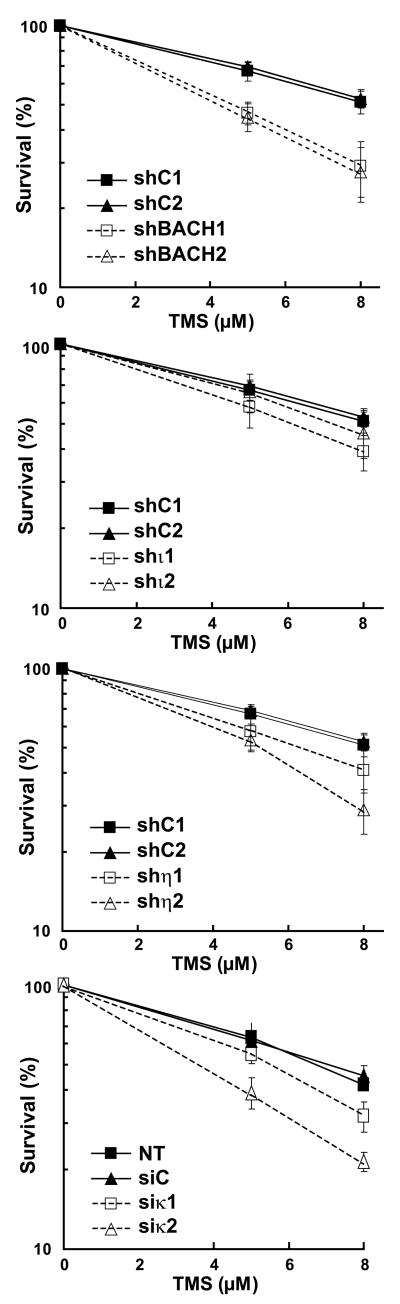
G-quadruplex structures are stabilized in vitro by small molecules called G-quadruplex ligands (G4 ligands). Although firstly described on G-quadruplex structures formed by telomeric sequence, G4 ligands are able to stabilize all G-quadruplex independently on primary DNA sequence capable of forming these secondary structures. Several classes of small molecules that bind to G-quadruplex DNA have been described. Among them, the natural compound Telomestatin (TMS) is a G4 ligand with exceptional selectivity [32]. To investigate a potential role of TLS polymerases in cellular G4 DNA metabolism, we asked if TLS pol-depleted cells were sensitive to TMS by performing clonogenic colony formation assays. As an additional reference, we used a set of isogenically cell lines deleted for the helicase BRIP1(BACH1), which has been shown to play a role in the resolution of alternate G4 DNA structures that impede DNA replication [33]. All the cell lines were exposed to 5μM and 8 μM of TMS and we determined the colony formation capability for each cell line (Fig 2). As expected, depletion of BRIP1(BACH1) with two different shRNAs sensitized cells to TMS, with only 26% of survival after 8 μM TMS treatment (as compared to 52% in control cells). We did not observe a significant impact of the Pol ι depletion on TMS sensitivity compared to control cells, supporting the notion that Pol ι is not essential in G4 DNA metabolism. In contrast, we found that TMS inhibited colony formation to a significantly greater extent among Pol η-depleted and Pol κ-depleted cells than among control cells, suggesting that both TLS DNA polymerases could have an important role in processing G4 DNA structures.
Figure 2. Pol η and Pol κ depletion sensitizes cells to TMS.
Cell survival after one week treatment with a range of TMS concentrations for U2OS cell lines depleted for BACH1, Pol η, Pol ι, or Pol κ. All data were performed in triplicate and were taken from the mean survival values obtained from three independent experiments (error bars = standard deviation).
The depletion of Pol η or Pol κ elevates DNA damage associated with non-BDNA formed at the human c-MYC promotor
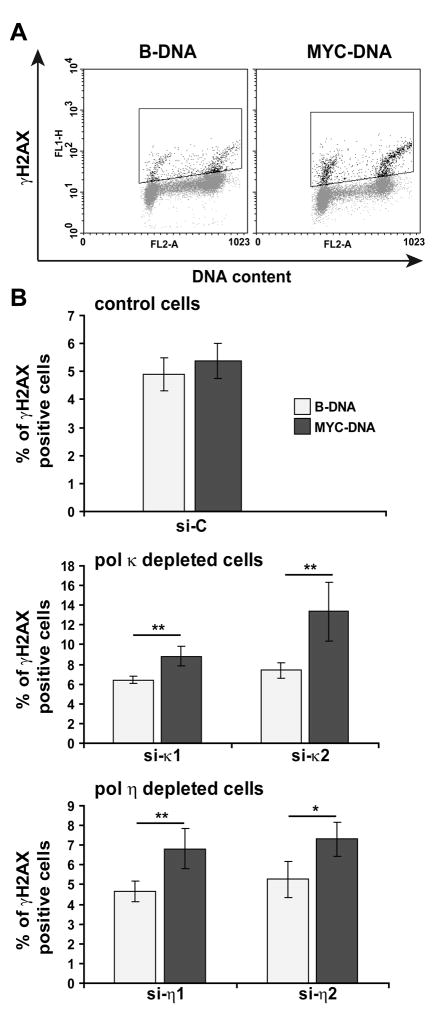
We next asked if Pol η and Pol κ depletion disrupted the cellular ability to cope with replication stress associated with the presence of non-B DNA since prolonged stalling of the replication forks at these alternate structured DNA sequences can lead to fork collapse and double-strand breaks. To assess the importance of both TLS polymerases in preventing or reducing the number of double-strand breaks that arise from the presence of non-B DNA, we established transgenic HeLa cells with approximately 8 to 12 copies (determined by quantitative real-time PCR, data not shown) of chromosomally integrated plasmid containing G-rich sequences from the human c-MYC promoter region (pUMycProm), sequences from the human BCL-2 major break region (pUMBR), GA-rich sequences from the breakage hotspot region of the KSHV genome (pUGA-rich), or a similar length (600 bp) of control B-DNA sequence from the human GAPDH gene, respectively (see Supplementary Fig 1). By using a siRNA technique, we successfully depleted Pol η or Pol κ individually in HeLa cells containing pUMycProm (MYC-DNA) or control plasmid pULCtrl (B-DNA) in the genome, as evidenced by immunoblotting of the tranfected HeLa cell lines shown in Fig 3. We then measured by Flow Cytometry (see Materials and Methods and Fig 4A) the proportion of cells positive for γ-H2AX, a well-known marker for double-strand breaks, in MYC-DNA cells and we compared to the control B-DNA cells. For the B-DNA cells, depletion of Pol η had no significant effect on γ-H2AX positive cell population, while downregulation of Pol κ seemed to slightly increase spontaneous DSB (Fig. 4B). Compared to B-DNA cells, we found that DSBs were significantly more elevated in MYC-DNA cells knockeddown for Pol η or Pol κ, while control siRNA transfection in these cells had no effect. The increased number of γ-H2AX positive cells in those populations silenced for Pol η or Pol κ compared to that in undepleted cells strongly suggests that the presence of each polymerase prevents the replicative stress associated with the non-B DNA structure (e.g. G4 DNA, H-DNA or Z-DNA), thereby decreasing the extent of DSBs. These data further support the conclusion that both polymerases contribute to the metabolism of non-B DNA forming sequences in human cells and show that these enzymes prevent genomic instability occurring at these naturally occurring sequences.
Figure 3. Hela cells transfected with B-DNA and non-B DNA used in this study.
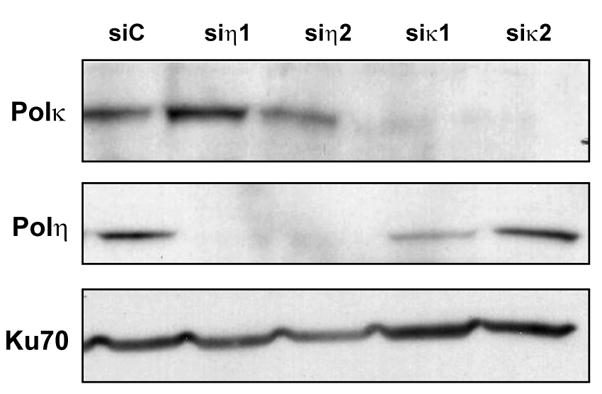
Whole cell extracts from Hela cells containing c-MYC-DNA transiently transfected with siRNA against Luciferase (siC), Pol η (siη1 and siη2) or Pol κ (siκ1 and siκ2) were probed by Western blotting with antibodies against Pol η, Pol κ, or Ku70 (as a loading control).
Figure 4. Pol H and Pol K gene silencing triggers H2AX phosphorylation in HeLa-MYC cells.
HeLa cells containing B-DNA or MYC-DNA were transfected with control siRNA (Luciferase), two independent Pol κ siRNAs (siκ1 and siκ2) or two independent Pol η siRNAs (siη1 and siη2). Quantification of γ-H2AX-positive cells in the population of each cell line was performed by Flow Cytometry Analysis as described in Materials and Methods. An example is given in (A) where a scatter plot is presented with γ-H2AX intensity on the y axis and propidium iodide intensity on the x axis for the control B-DNA cells and the MYC-DNA cells depleted for Pol κ. The cell populations used for the measurement of the γ-H2AX-positive cells were enclosed by rectangles. (B) Quantification of the FACS analysis were performed as shown in (A) with the cell lines transfected with the indicated siRNAs. The error bars indicate standard deviations of three independent experiments
Analysis of DSBs in HeLa cells transfected with Mbr-DNA and GA-DNA after depletion for Pol η or Pol κ
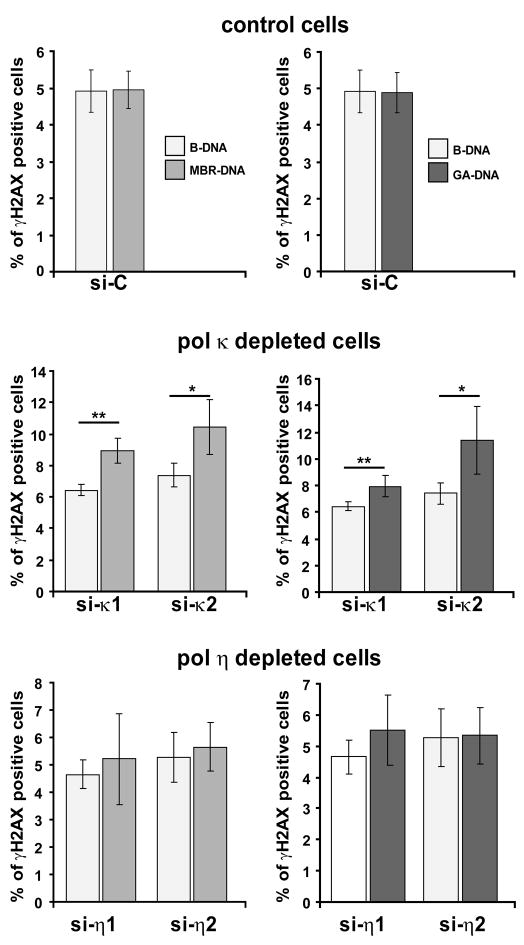
We next investigated whether the loss of Pol η or Pol κ contributes to genomic instability associated with two other naturally occurring DNA sequences capable of forming unusual secondary structures, and which have been shown to be breakage hotspots. The first one is a 520 bp sequence of the BCL-2 major break region (Mbr) within which several H-DNA forming sequences have been defined (Supplementary Fig 1). The second one is a 700 bp GA rich region from the far right-hand end of the KSHV genome (Supplementary Fig 1). HeLa cells were transfected with plasmids containing these structured regions (see Materials and Methods) and ultimately depleted for Pol η or Pol κ. The proportion of γ-H2AX positive cells was measured by Flow Cytometry and compared to that observed with control Hela cells transfected with B-DNA. We found that downregulation of Pol κ significantly enhanced the proportion of cells positive for γ-H2AX in Mbr and GA cells (Fig 5), similary to the results obtained from cells containing the c-MYC sequence. However, in contrast to what we observed with MYC-DNA, depletion of Pol η had no significant effect on γ-H2AX positive cell population for the cells carrying Mbr or GA structured sequences (Fig. 5). These data suggest that Pol κ and Pol η may play different roles in processing different types of non-B DNA structures in vivo.
Figure 5. Depletion of Pol κ, but not that of Pol η, increases H2AX phosphorylation in HeLa-Mbr and HeLa-GA cells.
HeLa cells containing B-DNA, Mbr-DNA or GA-DNA were transfected with control siRNA (Luciferase), two independent Pol κ siRNAs (siκ1 and siκ2) or two independent Pol η siRNAs (siη1 and siη2). Quantification of γ-H2AX-positive cells was achieved as in Fig 4. The error bars indicate standard deviations of three independent experiments
Concomitant down-regulation of Pol η and Pol κ in colorectal cancer
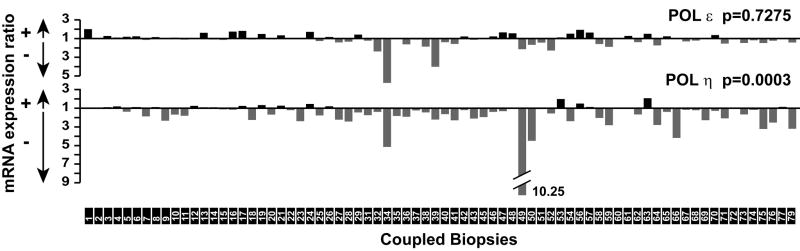
Recent studies have emphasized the existence of a relationship between replication stress and tumor progression [34,35]. Since loss of Pol η or Pol κ expression results in increased DSBs linked to natural structured DNA sequences which interfere with the progression of the replication fork during S phase, down-regulation of these TLS DNA polymerases could be an important contributor for endogenous replication stress and genetic instability, as it occurs in tumours. We previously reported that levels of expression of POL K were significantly lower in tumors than in adjacent tissues in a human colon cancer cohort including 74 coupled primary colorectal carcinomas at different stages of development [36]. Using the same cohort, we analyzed here by real-time PCR-based Micro Fluidic Cards the POL H expression profiles (Fig 6). Unlike Pol E, which encodes the replicative Pol ε and whose expression was not significantly affected, we found that, like POL K, levels of expression of POL H were strongly affected in tumors than in adjacent tissues (Fig 6). We next explored if the deregulations of both DNA polymerases gene expression were concomitant in cancer tissues by performing a non parametric Spearman’s rank-correlation test to analyze gene/gene correlations in the cohort. The closer to 1 the Spearman coefficient (rho), the more associated the expression of the two genes would be. Interestingly, we found that expression defects of genes encoding POL H and POL K were the most correlated (Table 1).
Figure 6. mRNA expression of Pol η in the 74 colorectal-adenocarcinomas.
Expressions of Pol η as well as the replicative DNA polymerase Pol ε have been measured in the matched human tumoral and normal samples. T (tumoral)/N (normal) mRNA expression ratios between normalized values were calculated. T/N values lower than 1 were transformed to the inverse value N/T. + and − indicate a higher or lower expression respectively in the tumour versus adjacent control tissue. Black boxes above the x axis point out the sample numbers. P-values from bilateral exact binomial test are given uncorrected, the significance level is evaluated using the Benjamini Krieger Yekutieli 2001 procedure for and overall FDR of 0.05. All computations were performed using Stata 9.0 SE (Tiermips).
Table 1.
Downregulation of Pol η and Pol κ are significantly correlated in a cohort of colorectal tumours
| Pol κ | Pol ε | Pol δ | |
|---|---|---|---|
| Pol η | 0.7145 | 0.6532 | 0.4610 |
Discussion
S phase is a period of great vulnerability for the genome of eukaryotic cells. Many complicated processes are undertaken during this critical phase of the cell cycle, including the unwinding together with the duplication of enormously complex DNA molecules. During this process, replication forks frequently encounter obstacles that impede their progression. Arrested forks are unstable structures that have to be stabilized and restarted in order to prevent the formation of double-strand breaks, unscheduled homologous recombination, or possibly NHEJ, and consequently genomic instability. To this aim, cells have evolved complex surveillance mechanisms sensing DNA damage and replication stress [37]. The past decade has seen a dramatic advance in our understanding of how these regulatory pathways act in response to exogenous replication stress. However, the mechanisms by which genome stability is conserved at natural replication-impeding sequences remain obscure. Alternate DNA structures capable of forming unusual secondary structures in human cells such as G4 structures, H-DNA or Z-DNA have been implicated in critical biological processes and can interfere with normal cellular DNA transactions such as the advancement of the replication fork during S phase [11]. Based on the ability of TLS DNA polymerases to facilitate the progression of the replication fork through external replicative barriers such bulky adducts [17], we have examined in this study the role of these specialized enzymes for preventing the replicational stress response associated to these alternate non-B DNA structures. The results of our investigations suggest that Pol η and Pol κ help prevent genomic instability occurring at such natural DNA sequences.
To probe the functional importance of TLS DNA polymerases in G4 metabolism in vivo, we examined the effects of TMS, a selective quadruplex binding agent, on cell survival in Pol η-, Pol κ-, and Pol ι-depleted versus control cells. Although TMS was originally identified as a potent telomerase inhibitor by virtue of its ability to stabilize telomeric quadruplex DNA and inhibit the polymerase reaction, results from recent studies suggest that TMS exert cytotoxic effects through its action on nontelomere G4 targets [38–40]. The selectivity of TMS for the G-quadruplex structure over a single-stranded or duplex DNA structure has been demonstrated [41]. This compound does not make covalent bonds with DNA and just inserts in G-quadruplex with stacking effect on the G-quartet sequence. We hypothesized that cells depleted for TLS DNA polymerases would be hypersensitive to the biological effects of TMS. Indeed, this turned out to be the case for Pol η and Pol κ, and much less for Pol ι. TMS was observed to significantly impair cell proliferation and growth in Pol η- and Pol κ-depleted cells, suggesting that both TLS DNA polymerases operate in G4 metabolism. This was further supported by showing increased DSB induction after Pol κ or Pol η depletion in HeLa cells transfected by G-rich MYC-DNA. Thus, our work provides the first description of DNA polymerases required specifically for the maintenance of guanine-rich DNA in vivo. Recent evidence showed a functional cooperation between the WRN helicase, known to unwind G-quadruplex DNA, and the Y-family TLS DNA polymerases Pols η and κ for lesion bypass [42]. It would be of interest to investigate if such interaction could facilitate the replication of tetraplex DNA structures.
Interestingly, elevated DNA breakage associated with other alternative DNA structures than G4, such Mbr-DNA or repetitive GA-DNA, was observed only after Pol κ downregulation, and not Pol η, supporting the idea that Pol κ may have a more general role for genomic maintenance during unperturbed S-phase.
How could depletion of a TLS DNA polymerase enhance genomic instability at naturally occurring structured DNA? One possibility would be that downregulation of Pol η or Pol κ may affect normal DNA replication per se through alternate structured DNA domains. Non-B DNA structures such as G4 and H-DNA have been shown to affect the progress of some DNA polymerases [43,44] and RNA polymerases [45–47]. So it is possible that TLS pol η or pol κ is required for overcoming the structural barrier at a stalled replication fork. Knocking down the expression of these polymerases might result in a failure to rescue the stalled replication fork and thus result in DNA breakage. Alternatively, their depletion may compromise the repair processes of replication-induced DSB at these non-B DNA sites. Recent evidence has suggested that, outside its role in TLS, Pol η may be required in the Homologous Recombination Repair pathway (HRR) in chicken DT40 cells during the generation of antibody diversity of Ig genes [48] and in vitro for the extension of the invading strand in a D-loop structure [49], facilitating the RAD52-mediated second-end capture during the repair process [50]. Therefore, we cannot rule out the possible involvement of Pol η in recombinational events associated with G4-induced DSB repair. Further investigations will be required to understand by which mechanisms Pol η and Pol κ could act and eventually cooperate to prevent genomic instability at structured DNA in absence of external stress. Notably, it would be of interest to look at the posttranslational processing of the polymerases as well as PCNA in relation to their action at non-canonical DNA structure encumbrances compared to DNA lesions that interfere with DNA replication. Whether these TLS polymerases contribute to the normal genome replication is an attractive possibility that deserves further exploration.
Interestingly, we have uncovered unexpected alterations in the expression pattern of these two TLS DNA polymerases in a human colorectal cancer collection. Indeed, we have found that they are down-regulated as compared to the normal adjacent tissues while the expression of the replicative DNA polymerases δ and ε was not affected. These data may help to understand how cancer cells use this altered expression pattern of TLS DNA polymerases to promote genomic instability in absence of external stress.
Supplementary Material
DNA sequences used in this study: Control B-DNA, c-MYC-DNA, Mbr-DNA and GA-DNA.
Acknowledgments
We thank T Nohmi, Naoko Niimi, and Petr Gruz (Tokyo) for the Pol κ antibodies and R Woodgate (NIH) for the Pol ι antibodies. We thank R Guimbaud and K Gordien (Inserm/CHU Purpan, Toulouse) for collection and annotation of the tumors, PA Gourraud for statistical analysis, F Viala (IPBS Toulouse) for iconography assistance, D Gomez (IPBS Toulouse) for helpful comments, as well as JJ Maoret (IFR31 Toulouse) from the plateforme de “Genomique et Biologie Moleculaire” for his technical help on the 7900HT fast real time PCR systems.
Grant Supports: This work was supported by INCa (“Checkpol” projet libre 2007 to JSH), the Association pour la Recherche sur le Cancer (N° 4887 to JSH), the Canceropole Grand Sud Ouest grant 2004–2009 (to CC), Electricite de France (EDF) (to DB), and by NIH, NCI grant (CA093729) TO KMV.
Abbreviations
- Pol
DNA polymerase
- TLS
translesion synthesis
- DSB
double-strand breaks
- NER
nucleotide excision repair
- HRR
homologous recombination repair
- NHEJ
non homologous end joining
- Mbr
major breakpoint region
- TMS
Telomestatin
- G4 DNA
Guanine-quadruplex DNA
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- KSHV
Kaposi Sarcoma associated Herpesvirus
References
- 1.Diffley JF. Regulation of early events in chromosome replication. Curr Biol. 2004;14(18):R778–786. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 2.Kolodner RD, Putnam CD, Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297(5581):552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- 3.Burhans WC, Carr AM, Wahl GM. DNA Replication and Cancer. In: DePamphilis ML, editor. DNA Replication and Human Disease. Cold Spring Harbor Laboratory press; 2006. pp. 481–500. [Google Scholar]
- 4.Bacolla A, Wojciechowska M, Kosmider B, Larson JE, Wells RD. The involvement of non-B DNA structures in gross chromosomal rearrangements. DNA Repair (Amst) 2006;5(9–10):1161–1170. doi: 10.1016/j.dnarep.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 5.Bowater RP, Wells RD. The intrinsically unstable life of DNA triplet repeats associated with human hereditary disorders. Prog Nucleic Acid Res Mol Biol. 2001;66:159–202. doi: 10.1016/s0079-6603(00)66029-4. [DOI] [PubMed] [Google Scholar]
- 6.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat Struct Mol Biol. 2006;13(12):1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 7.Popescu NC. Genetic alterations in cancer as a result of breakage at fragile sites. Cancer Lett. 2003;192(1):1–17. doi: 10.1016/s0304-3835(02)00596-7. [DOI] [PubMed] [Google Scholar]
- 8.Michelotti GA, Michelotti EF, Pullner A, Duncan RC, Eick D, Levens D. Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol Cell Biol. 1996;16(6):2656–2669. doi: 10.1128/mcb.16.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joos S, Haluska FG, Falk MH, et al. Mapping chromosomal breakpoints of Burkitt’s t(8;14) translocations far upstream of c-myc. Cancer Res. 1992;52(23):6547–6552. [PubMed] [Google Scholar]
- 10.Care A, Cianetti L, Giampaolo A, et al. Translocation of c-myc into the immunoglobulin heavy-chain locus in human acute B-cell leukemia. A molecular analysis. Embo J. 1986;5(5):905–911. doi: 10.1002/j.1460-2075.1986.tb04302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability. Mutat Res. 2006;598(1–2):103–119. doi: 10.1016/j.mrfmmm.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 12.Raghavan SC, Chastain P, Lee JS, et al. Evidence for a triplex DNA conformation at the bcl-2 major breakpoint region of the t(14;18) translocation. J Biol Chem. 2005;280(24):22749–22760. doi: 10.1074/jbc.M502952200. [DOI] [PubMed] [Google Scholar]
- 13.Durkin SG, Glover TW. Chromosome Fragile Sites. Annu Rev Genet. 2007 doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 14.Huber MD, Duquette ML, Shiels JC, Maizels N. A conserved G4 DNA binding domain in RecQ family helicases. J Mol Biol. 2006;358(4):1071–1080. doi: 10.1016/j.jmb.2006.01.077. [DOI] [PubMed] [Google Scholar]
- 15.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306(5703):1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 16.Wu Y, Shin-ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol. 2008;28(12):4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 18.McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA. Preferential cis-syn thymine dimer bypass by DNA polymerase eta occurs with biased fidelity. Nature. 2004;428(6978):97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 19.Masutani C, Kusumoto R, Yamada A, et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399(6737):700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 20.Tissier A, McDonald JP, Frank EG, Woodgate R. poliota, a remarkably error-prone human DNA polymerase. Genes Dev. 2000;14(13):1642–1650. [PMC free article] [PubMed] [Google Scholar]
- 21.Ogi T, Shinkai Y, Tanaka K, Ohmori H. Polkappa protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc Natl Acad Sci U S A. 2002;99(24):15548–15553. doi: 10.1073/pnas.222377899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, Livneh Z. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells: the role of DNA polymerase kappa. J Biol Chem. 2004;279(51):53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- 23.Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439(7073):225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- 24.Ogi T, Lehmann AR. The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat Cell Biol. 2006;8(6):640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- 25.Wang G, Christensen LA, Vasquez KM. Z-DNA-forming sequences generate large-scale deletions in mammalian cells. Proc Natl Acad Sci U S A. 2006;103(8):2677–2682. doi: 10.1073/pnas.0511084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wittig B, Wolfl S, Dorbic T, Vahrson W, Rich A. Transcription of human c-myc in permeabilized nuclei is associated with formation of Z-DNA in three discrete regions of the gene. Embo J. 1992;11(12):4653–4663. doi: 10.1002/j.1460-2075.1992.tb05567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinniburgh AJ. A cis-acting transcription element of the c-myc gene can assume an H-DNA conformation. Nucleic Acids Res. 1989;17(19):7771–7778. doi: 10.1093/nar/17.19.7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci U S A. 2002;99(18):11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biard DS. Untangling the relationships between DNA repair pathways by silencing more than 20 DNA repair genes in human stable clones. Nucleic Acids Res. 2007;35(11):3535–3550. doi: 10.1093/nar/gkm195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biard DS, Despras E, Sarasin A, Angulo JF. Development of new EBV-based vectors for stable expression of small interfering RNA to mimick human syndromes: application to NER gene silencing. Mol Cancer Res. 2005;3(9):519–529. doi: 10.1158/1541-7786.MCR-05-0044. [DOI] [PubMed] [Google Scholar]
- 31.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 32.Lemarteleur T, Gomez D, Paterski R, Mandine E, Mailliet P, Riou JF. Stabilization of the c-myc gene promoter quadruplex by specific ligands’ inhibitors of telomerase. Biochem Biophys Res Commun. 2004;323(3):802–808. doi: 10.1016/j.bbrc.2004.08.150. [DOI] [PubMed] [Google Scholar]
- 33.Maizels N. Genomic stability: FANCJ-dependent G4 DNA repair. Curr Biol. 2008;18(14):R613–614. doi: 10.1016/j.cub.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartkova J, Horejsi Z, Koed K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434(7035):864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 35.Gorgoulis VG, Vassiliou LV, Karakaidos P, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434(7035):907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 36.Lemee F, Bavoux C, Pillaire MJ, et al. Characterization of promoter regulatory elements involved in downexpression of the DNA polymerase kappa in colorectal cancer. Oncogene. 2007;26(23):3387–3394. doi: 10.1038/sj.onc.1210116. [DOI] [PubMed] [Google Scholar]
- 37.Tourriere H, Pasero P. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair (Amst) 2007 doi: 10.1016/j.dnarep.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 38.De Cian A, Cristofari G, Reichenbach P, et al. Reevaluation of telomerase inhibition by quadruplex ligands and their mechanisms of action. Proc Natl Acad Sci U S A. 2007;104(44):17347–17352. doi: 10.1073/pnas.0707365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gomez D, Wenner T, Brassart B, et al. Telomestatin-induced telomere uncapping is modulated by POT1 through G-overhang extension in HT1080 human tumor cells. J Biol Chem. 2006;281(50):38721–38729. doi: 10.1074/jbc.M605828200. [DOI] [PubMed] [Google Scholar]
- 40.Gomez D, O’Donohue MF, Wenner T, et al. The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Res. 2006;66(14):6908–6912. doi: 10.1158/0008-5472.CAN-06-1581. [DOI] [PubMed] [Google Scholar]
- 41.Kim MY, Gleason-Guzman M, Izbicka E, Nishioka D, Hurley LH. The different biological effects of telomestatin and TMPyP4 can be attributed to their selectivity for interaction with intramolecular or intermolecular G-quadruplex structures. Cancer Res. 2003;63(12):3247–3256. [PubMed] [Google Scholar]
- 42.Kamath-Loeb AS, Lan L, Nakajima S, Yasui A, Loeb LA. Werner syndrome protein interacts functionally with translesion DNA polymerases. Proc Natl Acad Sci U S A. 2007;104(25):10394–10399. doi: 10.1073/pnas.0702513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hile SE, Eckert KA. Positive correlation between DNA polymerase alpha-primase pausing and mutagenesis within polypyrimidine/polypurine microsatellite sequences. J Mol Biol. 2004;335(3):745–759. doi: 10.1016/j.jmb.2003.10.075. [DOI] [PubMed] [Google Scholar]
- 44.Kamath-Loeb AS, Loeb LA, Johansson E, Burgers PM, Fry M. Interactions between the Werner syndrome helicase and DNA polymerase delta specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J Biol Chem. 2001;276(19):16439–16446. doi: 10.1074/jbc.M100253200. [DOI] [PubMed] [Google Scholar]
- 45.Belotserkovskii BP, De Silva E, Tornaletti S, Wang G, Vasquez KM, Hanawalt PC. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J Biol Chem. 2007;282(44):32433–32441. doi: 10.1074/jbc.M704618200. [DOI] [PubMed] [Google Scholar]
- 46.Ditlevson JV, Tornaletti S, Belotserkovskii BP, et al. Inhibitory effect of a short Z-DNA forming sequence on transcription elongation by T7 RNA polymerase. Nucleic Acids Res. 2008;36(10):3163–3170. doi: 10.1093/nar/gkn136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tornaletti S, Park-Snyder S, Hanawalt PC. G4-forming sequences in the non-transcribed DNA strand pose blocks to T7 RNA polymerase and mammalian RNA polymerase II. J Biol Chem. 2008;283(19):12756–12762. doi: 10.1074/jbc.M705003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawamoto T, Araki K, Sonoda E, et al. Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Mol Cell. 2005;20(5):793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 49.McIlwraith MJ, Vaisman A, Liu Y, Fanning E, Woodgate R, West SC. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol Cell. 2005;20(5):783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 50.McIlwraith MJ, West SC. DNA repair synthesis facilitates RAD52-mediated second-end capture during DSB repair. Mol Cell. 2008;29(4):510–516. doi: 10.1016/j.molcel.2007.11.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA sequences used in this study: Control B-DNA, c-MYC-DNA, Mbr-DNA and GA-DNA.