Abstract
Pairing of SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins on vesicles (v-SNAREs) and SNARE proteins on target membranes (t-SNAREs) mediates intracellular membrane fusion. VAMP3/cellubrevin is a v-SNARE that resides in recycling endosomes and endosome-derived transport vesicles. VAMP3 has been implicated in recycling of transferrin receptors, secretion of α-granules in platelets, and membrane trafficking during cell migration. Using a cell fusion assay, we examined membrane fusion capacity of the ternary complexes formed by VAMP3 and plasma membrane t-SNAREs syntaxin1, syntaxin4, SNAP-23 and SNAP-25. VAMP3 forms fusogenic pairing with t-SNARE complexes syntaxin1/SNAP-25, syntaxin1/SNAP-23 and syntaxin4/SNAP-25, but not with syntaxin4/SNAP-23. Deletion of the N-terminal domain of syntaxin4 enhanced membrane fusion more than two fold, indicating that the N-terminal domain negatively regulates membrane fusion. Differential membrane fusion capacities of the ternary v-/t-SNARE complexes suggest that transport vesicles containing VAMP3 have distinct membrane fusion kinetics with domains of the plasma membrane that present different t-SNARE proteins.
Keywords: SNARE, VAMP3, membrane fusion, vesicular trafficking
Introduction
Compartmentalization into membrane-bound organelles is a fundamental principle of eucaryotic cells. Transport vesicles deliver membrane-impermeable molecules including membrane proteins and secretory factors from one organelle to another and in and out of cells. The vesicular delivery process involves the successive steps of transport on cytoskeleton, tethering, docking, and finally membrane fusion. SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins form the core machinery for intracellular membrane fusion [1, 2].
When transport vesicles traffic to the vicinity of the target organelles, SNARE proteins on vesicles (v-SNAREs) and SNARE proteins on target membranes (t-SNAREs) form trans-SNARE complexes to draw the two membranes toward each other and drive membrane fusion. SNAREs are cytoplasmic oriented type I membrane proteins. They share one homologous sequence, the ‘SNARE motif” which contains 60–70 amino acids that include eight heptad repeats ready for coiled-coil formation. The SNARE proteins that mediate synaptic exocytosis and the fusion of synaptic vesicles with the presynaptic plasma membrane are well studied [3]. In the nerve terminal, v-SNARE vesicle-associated membrane protein 2 (VAMP2) resides in the membrane of synaptic vesicles, whereas t-SNAREs syntaxin1 and synaptosomal-associated protein of 25 kD (SNAP-25) are located in the plasma membrane. Syntaxin1 and SNAP-25 constitute an acceptor complex for VAMP2 [4]. The cytoplasmic domains of VAMP2, syntaxin1 and SNAP25 form an extremely stable complex that is resistant to sodium dodecyl sulfate (SDS) [5] and heat stable up to ~90°C [6], indicating that SNARE complex formation is thermodynamically favorable. One α-helix from VAMP2, one α-helix from syntaxin1 and two α-helices from SNAP-25 intertwine to form a four-helix bundle [7]. The parallel arrangement of the helices with all amino termini at one end of the helix bundle should bring the membrane anchors of the SNARE proteins and thus the vesicular and target membranes into close proximity. Energy made available from the assembly of the trans-SNARE complexes is used to drive the fusion of lipid bilayers [8–11]. After membrane fusion, the adapter protein α-SNAP (soluble NSF attachment protein) and the ATPase NSF (N-ethylmaleimide-sensitive factor) dissociate the cis-SNARE complexes at the expense of ATP [12, 13] to free the SNAREs for the next round of membrane fusion.
There are 36 SNAREs in humans [2, 14]. Individuals of the SNARE family localize to distinct subcellular organelles in the secretory pathway, suggesting that they have selective roles in specific intracellular trafficking steps [15]. Using yeast SNARE proteins as a model, a series of studies demonstrate that to a remarkable degree the specificity of intracellular membrane fusion can be predicted from the pattern of liposome fusion mediated by isolated v- and t-SNARE proteins [16, 17]. The cognate pairings between v- and t-SNARE proteins that mediate most intracellular trafficking events in mammalian cells remain to be functionally identified.
VAMP3/cellubrevin is a homologue of VAMP2 that has a broad tissue distribution [18]. Like VAMP2, it can be cleaved by tetanus toxin, a metallo-endoprotease which blocks synaptic exocytosis [18, 19]. VAMP3 is present in recycling endosomes and endosome-derived vesicles [18]. It colocalizes with endocytosed transferrin receptors [18] and the glucose transporter GLUT4 in adipocytes [20], and is also present in α-granules in platelets [21]. VAMP3 has been implicated in recycling of transferrin receptors to the plasma membrane [19], secretion of α-granules in platelets [21, 22], recycling of T-cell receptors to the immunological synapses [23], and membrane trafficking during cell migration [24, 25]. Surprisingly, VAMP3 knockout mice have no obvious phenotype [26, 27], which likely results from compensation by VAMP2 [28] and/or other v-SNARE proteins.
Biochemical studies show that VAMP3 binds to plasma membrane t-SNAREs syntaxin1, syntaxin4, SNAP-23 and SNAP-25 [29–31]. Syntaxin4 is a homologue of syntaxin1, whereas SNAP-23 is a homologue of SNAP-25 [32, 33]. It has also been reported that VAMP3 forms ternary complexes with syntaxin4/SNAP-25 [22] and syntaxin4/SNAP-23 [34]. To fully understand VAMP3-mediated vesicle fusion, functional studies are needed to examine the membrane fusion capacity of the ternary complexes formed by VAMP3 and the plasma membrane t-SNAREs.
Using a cell fusion assay [10], we report here that VAMP3 forms fusogenic pairing with t-SNARE complexes syntaxin1/SNAP-25, syntaxin1/SNAP-23 and syntaxin4/SNAP-25, but not with syntaxin4/SNAP-23.
Materials and methods
Construction of plasmids
To generate flipped VAMP3, the flipped syntaxin1 plasmid (pCH9) [10] was digested with XbaI and ApaI to excise the coding region of syntaxin1. The vector fragment was purified from an agarose gel. The coding sequence of mouse VAMP3 was amplified by PCR from a mouse skeletal muscle Marathon-Ready cDNA library (Clontech) with primer mVAMP3F (ATGTCTACAGGTGTGCCTTCGG) and primer mVAMP3R (TTAAGAGACACACCACACGATGATG). Using the PCR product as template, the coding sequence of VAMP3 (a.a. 2–102) was amplified by PCR with primer CHU30F (TCTGTCTAGATCTACAGGTGTGCCTTCGGGGTC) and primer CHU30R (AAACGGGCCCTTAAGAGACACACCACACGATGATG). The new PCR product was digested with XbaI and ApaI and cloned into the XbaI and ApaI sites of pCH9, resulting in plasmid flipped VAMP3 (pCHU30).
To generate flipped syntaxin4, the coding sequence of mouse syntaxin4 (a.a. 2–298) was amplified by PCR from a mouse skeletal muscle Marathon-Ready cDNA library (Clontech) with primer XbaSyn4 (CTGTCTAGACGCGACAGGACCCACGAGTTGAGG) and primer Syn4Apa (AAACGGGCCCTTATCCAACGGTTATGGTGATGCC). The PCR product was digested with XbaI and ApaI and cloned into the XbaI and ApaI sites of pCH9, resulting in plasmid flipped syntaxin4 (pCHU1). To generate flipped syntaxin4(Δ111), the coding region of syntaxin4 (a.a. 112–298) was amplified by PCR using primer CHU22F (CTGTCTAGAGATGAGAATTACAACTCAGTCAACACAAG) and primer Syn4Apa with plasmid pCHU1 as template. The PCR product was digested with XbaI and ApaI and cloned into the XbaI and ApaI sites of pCH9, resulting in plasmid flipped syntaxin4(Δ111) (pCHU22). To generate flipped syntaxin4 H3, the coding region of syntaxin4 (a.a. 193–298) was amplified by PCR using primer CHU23F (CTGTCTAGAACGCAGGTGACTCGGCAG) and primer Syn4Apa with plasmid pCHU1 as template. The PCR product was digested with XbaI and ApaI and cloned into the XbaI and ApaI sites of pCH9, resulting in plasmid flipped syntaxin4 H3 (pCHU23).
To generate flipped SNAP-23, a FLAG tag (DYKDDDDK) was appended to the COOH-terminus of the pre-prolactin signal sequence: The signal sequence was first amplified by PCR using primer EcoRSignal (GTGGAATTCGCTTGTTCTTTTTGCAGAAGCTCAG) and primer FLAG1No2 (GTCGTCATCCTTGTAATCGGTGGAGACCACACCCTGGCAC) with plasmid pCH9 as template. The PCR product was further extended by a second round of PCR using primer EcoRSignal and primer FLAG2 (ATCTCTAGACTTATCGTCGTCATCCTTGTAATCGGTGGAG). The new PCR product was digested with EcoRI and XbaI. The coding sequence of SNAP-23 (a.a. 2–210) was amplified by PCR from a mouse skeletal muscle Marathon-Ready cDNA library (Clontech) with primer XbaSNAP (AAGTCTAGAGATAACCTGTCCCCAGAGGAAGTT) and primer SNAPApa (AACGGGCCCTTAACTATCAATGAGTTTCTTTGCTC) and digested with XbaI and ApaI. The two enzyme-digested fragments were cloned into the EcoRI and ApaI sites of pcDNA3.1(+) (Invitrogen) in a single ligation reaction, resulting in plasmid pCHU2.
To replace the five cysteines (Cys79, Cys80, Cys83, Cys85, Cys87) in flipped SNAP-23 with serines, and to mutate the three putative N-glycosylation sites (Asn3-Leu-Ser5; Asn88-Arg-Thr90; Asn154-Leu-Thr156), pCHU2 was site-directed mutagenized with primers CHU24F (CAGAACTCAACAAGAGTAGTGGCCTCTGCATCTG) and CHU24R (CAGATGCAGAGGCCACTACTCTTGTTGAGTTCTG) to replace Cys79 and Cys80 with Ser; then further mutagenized with primers CHU25F (CAAGAGTAGTGGCCTCAGCATCTGCCCTTG) and CHU25R (CAAGGGCAGATGCTGAGGCCACTACTCTTG) to replace Cys83 with Ser; then with primers CHU26F (GTGGCCTCAGCATCAGCCCTTGTAATAGGAC) and CHU26R (GTCCTATTACAAGGGCTGATGCTGAGGCCAC) to replace Cys85 with Ser; then with primers CHU27F (CAGCATCAGCCCTAGTGCTAGGACAAAGAACTTTG) and CHU27R (CAAAGTTCTTTGTCCTAGCACTAGGGCTGATGCTG) to replace Cys87 with Ser and Asn88 with Ala; then with primers CHU28F (GACGATAAGTCTAGAGATGCCCTGTCCCCAGAGG) and CHU28R (CCTCTGGGGACAGGGCATCTCTAGACTTATCGTC) to replace Asn3 with Ala; then with primers CHU29F (GAAGATGAGATGGAAGAGGCCCTGACTCAAGTGG) and CHU29R (CCACTTGAGTCAGGGCCTCTTCCATCTCATCTTC) to replace Asn154 with Ala. The final plasmid is called flipped SNAP-23 (pCHU29).
The QuikChange II site-directed mutagenesis kit (Stratagene) was used for mutagenesis. Pfu DNA polymerase (Stratagene) was used for PCR cloning. All coding sequences were confirmed by DNA sequencing.
Immunoblotting
The day before transfection, 2×105 COS-7 cells were seeded in each well of 6-well plates. For expression of the flipped v-SNARE, 1 μg of flipped VAMP3 was transfected into the COS-7 cells grown in each well. To express flipped t-SNAREs, 1 μg of flipped SNAP-23 was cotransfected with 1 μg of flipped syntaxin4, flipped syntaxin4(Δ111) or flipped syntaxin4 H3 into the COS-7 cells seeded in each well. Transfection was done with Lipofectamine according to the manufacturer’s instructions (Invitrogen). Mock controls were transfected with empty pcDNA3.1(+) vector. In some of the wells, tunicamycin (5 μg/ml) was included during the overnight incubation at 37°C. Twenty-four hours after transfection, 300 μl of SDS-PAGE sample buffer was added to each well to lyse the cells. The whole cell lysates (40 μg each lane in Fig. 2) were analyzed on 7.5% Tris-Glycine gels and immunoblotted with anti-Myc monoclonal antibody (9E10) diluted at 1:2,000 or anti-FLAG polyclonal antibody (SIGMA) diluted at 1:1,000 followed by HRP-conjugated secondary antibodies at 1:4,000 dilution. Bound antibodies were detected with ECL western blot system (GE Healthcare).
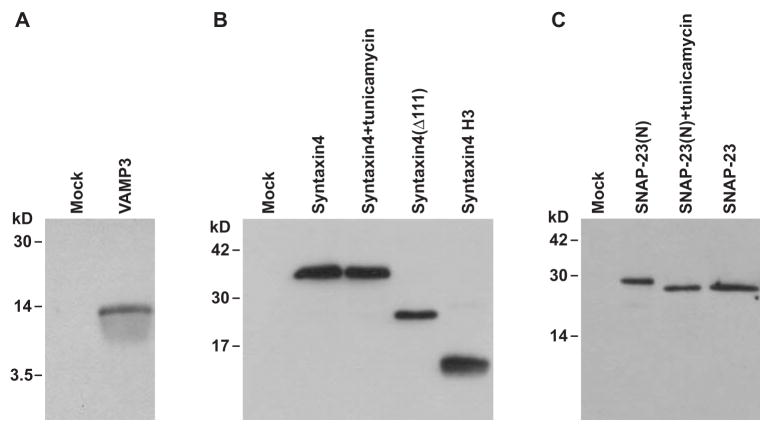
Fig. 2.
Expression of flipped SNARE proteins. (A) Twenty-four hours after transfection, whole-cell lysates of the COS-7 cells transfected with flipped VAMP3 or empty pcDNA3.1(+) vector (Mock) were immunoblotted with an antibody to Myc tag. (B) Whole cell lysates of COS-7 cells cotransfected with flipped SNAP-23 and flipped syntaxin4, flipped syntaxin4(Δ111) or flipped syntaxin4 H3 were immunoblotted with an antibody to Myc tag to detect the syntaxin4 proteins. Treatment with tunicamycin (5 μg/ml, overnight) did not change the mobility of flipped syntaxin4 proteins. (C) Whole cell lysates of COS-7 cells transfected with flipped syntaxin4 and flipped SNAP-23(N) or flipped SNAP-23 were immunoblotted with an antibody to FLAG tag to detect the SNAP-23 proteins. Flipped SNAP-23(N) is the SNAP-23 construct that contains the putative N-glycosylation sites. In flipped SNAP-23, the N-glycosylation sites have been removed by point mutations. Treatment with tunicamycin (5 μg/ml, overnight) increased the mobility of flipped SNAP-23(N) proteins to that of flipped SNAP-23 proteins.
Immunocytochemistry
The day before transfection, 3×104 COS-7 cells were seeded on sterile 12-mm glass coverslips contained in 24-well plates. For expression of flipped v-SNARE, 0.25 μg of flipped VAMP3 was transfected into the COS-7 cells grown in each well. To express flipped t-SNAREs, 0.25 μg of flipped syntaxin1 H3, flipped syntaxin4, flipped syntaxin4(Δ111) or flipped syntaxin4 H3 was cotransfected with 0.25 μg of flipped SNAP-23 or flipped SNAP-25 into the COS-7 cells grown in each well. Twenty-four hours after transfection, the COS-7 cells were fixed with 4% paraformaldehyde in PBS++ (PBS supplemented with 0.1 g/l CaCl2 and 0.1 g/l MgCl2). Primary antibodies were incubated with the cells at the following dilutions: anti-Myc monoclonal antibody, 1:300; anti-FLAG polyclonal antibody, 1:150; and anti-SNAP-25 polyclonal antibody (Synaptic Systems), 1:100. Fluorophore-conjugated secondary antibodies (Jackson Immunoresearch Laboratories) were used at a dilution of 1:500. For double staining, the cells were incubated first with a mixture of the primary antibodies, then with a mixture of the secondary antibodies. Confocal images were collected on a Zeiss 510 confocal microscope located in the Imaging Analysis Facility of West Virginia University Health Sciences Center. The images were processed with Adobe Photoshop software.
Cell fusion assay
The day before transfection, 1.2×106 COS-7 cells were seeded in each of 100-mm tissue culture dishes, and 5×104 COS-7 cells were seeded on sterile 12-mm glass coverslips contained in 24-well plates. For expression of flipped v-SNAREs (v-cells), 5 μg each of flipped VAMP3 and pEGFP-N3 (Clontech) were cotransfected into the cells grown in each 100-mm culture dishes. For expression of flipped t-SNAREs (t-cells), 0.25 μg each of flipped syntaxin, flipped SNAP-23 (or flipped SNAP-25) and pDsRed2-N1 (Clontech) were cotransfected into the cells seeded in each well. Twenty-four hours after transfection, the v-cells were detached from culture dishes with EDTA (Cell Dissociation Solution (SIGMA)). The detached cells were counted with a hemacytometer and resuspended in HEPES-buffered DMEM supplemented with 10% FBS and 0.5 mM DTT. v-Cells (1.2×105) were added to each coverslip already containing the t-cells. After 6 hours at 37°C in 5% CO2, the coverslips were gently washed once with PBS++, then fixed with 4% paraformaldehyde. Confocal images were collected on the Zeiss 510 confocal microscope equipped with argon (458, 488 nm), HeNe (543 nm) and HeNe (633 nm) lasers. The 488 nm argon laser line was used to excite green fluorescent protein (EGFP), which was visualized using a bandpass filter from 505 to 530 nm. The 543 nm HeNe laser line was used to excite the red fluorescent protein DsRed2 (RFP), which was visualized using a 560 nm long-pass filter. To prevent cross-contamination between EGFP and RFP, each channel was imaged sequentially using the multitrack recording module before merging the images. Before cell fusion, the cytoplasm of v-cells showed green fluorescence whereas the cytoplasm of t-cells showed red fluorescence. Fusion of v- and t-cells resulted in fused cells whose cytoplasm is yellow in the merged channel. To quantitate cell fusion, the total number of fused cells (F), the total number of unfused v-cells (V) and t-cells (T) that were in contact with each other or with fused cells in 20 to 40 random fields (images taken at ×20) were determined. The efficiency of cell fusion (%) is calculated as follows: 2F/(V+T+2F)×100.
Results
Using a cell fusion assay, previous studies showed that cells expressing v- and t-SNAREs on the cell surface fuse spontaneously, demonstrating that SNAREs are sufficient to fuse biological membranes [10]. VAMP3 binds to plasma membrane t-SNAREs syntaxin1, syntaxin4, SNAP-23 and SNAP-25 [22, 29, 30, 34]. Using the cell fusion assay in the current study, we examined the membrane fusion capacity of the ternary complexes formed by VAMP3 and the plasma membrane t-SNAREs.
Flipped SNARE constructs
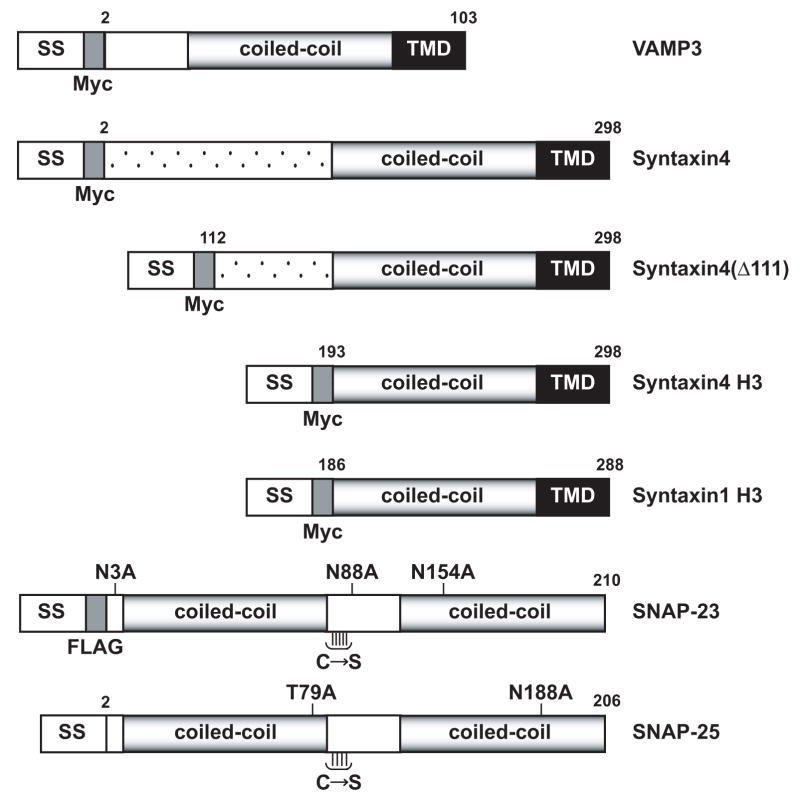
“Flipped” constructs were developed to express VAMP3, syntaxin4 and SNAP-23 on the cell surface (Fig. 1). cDNAs of VAMP3, syntaxin4 and SNAP-23 were cloned by PCR using a mouse skeletal muscle cDNA library. The signal sequence of preprolactin (SS) was fused to the N-termini of VAMP3, syntaxin4 and SNAP-23. The signal sequence directs translocation into the lumen of endoplasmic reticulum (ER) [10]. The transmembrane domains (TMD) at the C-termini of VAMP3 and syntaxin4 are expected to terminate the translocation and anchor VAMP3 proteins and syntaxin4 proteins on the cell surface after expression and intracellular transport. Like SNAP-25, SNAP-23 is anchored to the cytoplasmic side of the plasma membrane in vivo by palmitoylation of a cluster of Cys residues. Previous studies showed that replacement of the cluster of Cys residues in SNAP-25 by Ser does not interfere with its function [9, 10]. To prevent disulfide bonds that could interfere with the folding of flipped SNAP-23 proteins in the lumen of ER, we replaced all five Cys residues in SNAP-23 by Ser (Fig. 1). SNAP-23 does not contain a transmembrane domain. Flipped SNAP-23 proteins are expected to assemble with flipped syntaxin proteins in the ER and to be expressed on the cell surface after intracellular transport.
Fig. 1.
Constructs of flipped SNAREs. The pre-prolactin signal sequence (SS) was fused to the N-termini of VAMP3, syntaxin4 and SNAP-23. A Myc tag was inserted between the signal sequence and the N-termini of VAMP3 and syntaxin4. A FLAG tag was engineered between the N-terminus of SNAP-23 and the signal sequence. In flipped syntaxin4(Δ111), the first 111 amino acids of syntaxin4 were deleted. In flipped syntaxin4 H3, the first 192 amino acids of syntaxin4 were deleted. In flipped SNAP-23, the three putative N-glycosylation sites (Asn3, Asn88, and Asn154) were eliminated by point mutations to Ala. The five cysteines in SNAP-23 were replaced with serines (C→S). TMD stands for transmembrane domain. The coiled-coil segments represent the SNARE motifs. Flipped syntaxin1 H3 and flipped SNAP-25 [10] were included for comparison.
A Myc tag is inserted between the signal sequence and the N-termini of VAMP3 and syntaxin4 (Fig. 1) to detect flipped VAMP3 proteins (Fig. 2A) and flipped syntaxin4 proteins (Fig. 2B). A FLAG tag is engineered between the N-terminus of SNAP-23 and the signal sequence (Fig. 1) to detect flipped SNAP-23 proteins (Fig. 2C). Flipped syntaxin1 H3 and flipped SNAP-25 have been described in a previous publication [10]. They are included in Fig. 1 for comparison with the new constructs.
There are three putative N-glycosylation motifs (Asn-X-Ser/Thr) in mouse SNAP-23 proteins (Asn3, Asn88, Asn154). Since N-glycosylation occurs in the lumen of ER, SNAP-23 proteins are not N-glycosylated in vivo. Due to translocation into ER, flipped SNAP-23 proteins (with putative N-glycosylation sites) were N-glycosylated (flipped SNAP-23(N) in Fig. 2C). Treatment of tunicamycin, an antibiotic that inhibits N-glycosylation [35], increased the mobility of flipped SNAP-23(N) proteins (Fig. 2C). Our preliminary experiments showed that N-glycosylation disrupted the function of flipped SNAP-23(N) proteins. Therefore, in the final flipped SNAP-23 construct, we mutated the putative N-glycosylation sites by replacing the Asn residues with Ala (Fig. 1). In SDS-PAGE, flipped SNAP-23 proteins have the same mobility as the flipped SNAP-23(N) proteins after treatment with tunicamycin (Fig. 2C). Although mouse syntaxin4 proteins have one putative N-glycosylation site, treatment with tunicamycin didn’t alter the mobility of flipped syntaxin4 proteins (Fig. 2B), indicating that synaxin4 proteins were not N-glycosylated after expression.
Expression of flipped SNARE proteins on the cell surface
Immunostaining of unpermeabilized cells showed that when COS-7 cells were transfected with flipped VAMP3, VAMP3 proteins were expressed on the cell surface (Fig. 3A). When COS-7 cells were cotransfected with flipped syntaxin4 and flipped SNAP-23, both t-SNARE proteins were expressed on the cell surface (Fig. 3B, middle row). Like flipped SNAP-25 proteins, flipped SNAP-23 proteins do not contain a transmembrane domain. They were anchored to the cell surface by forming complexes with flipped syntaxin4 proteins. When COS-7 cells were cotransfected with the same amount of DNA of flipped syntaxin1 H3 and flipped SNAP-25, lower levels of cell surface staining were detected (Fig. 3B, bottom row). In the experiments shown in Fig. 3B, both syntaxin4 and syntaxin1 H3 proteins were detected with an antibody against Myc tag. Stronger cell surface staining indicated that more syntaxin4 proteins were expressed on the cell surface than syntaxin1 H3 proteins (compare middle row and bottom row in Fig. 3B).
Fig. 3.
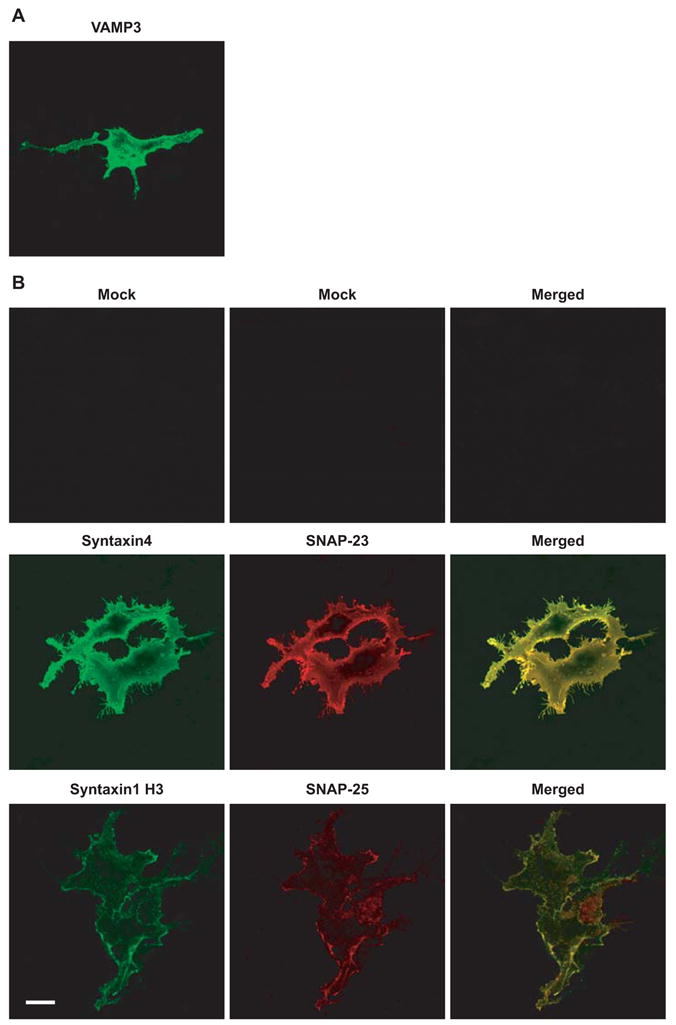
Expression of flipped SNARE proteins on the cell surface. (A) Twenty-four hours after transfection, unpermeabilized COS-7 cells transfected with flipped VAMP3 were stained with an antibody to Myc tag to detect cell surface distribution of VAMP3. (B) COS-7 cells were mock transfected (no DNA), co-transfected with flipped syntaxin4 and flipped SNAP-23, or cotransfected with flipped syntaxin1 H3 and flipped SNAP-25. Twenty-four hours after transfection, unpermeabilized COS-7 cells were dual labeled with antibodies. Syntaxin4 and syntaxin1 H3 proteins were stained with an antibody to Myc tag (green), and SNAP-23 proteins were detected with an antibody to FLAG tag (red). SNAP-25 proteins on the cell surface were detected with an antibody to SNAP-25 (red). Confocal images were collected using the same settings. Representative images of three experiments are shown. Scale bar, 20 μm.
Flipped SNAP-25 proteins do not contain a FLAG tag, which prevents the direct comparison of the cell surface expression of SNAP-25 with SNAP-23. Indirect evidence suggests that more SNAP-23 proteins were expressed on the cell surface than SNAP-25 proteins. Among the yeast SNARE proteins that mediate exocytosis, Sso1p (homologue of syntaxin1 and syntaxin4) and Sec9p (homologue of SNAP-23 and SNAP-25) form a 1:1 complex, which constitutes a three-helix bundle that serves as a binding site for Snc1/2p (homologue to VAMP2 and VAMP3) [36, 37]. It is well established that Syntaxin1 and SNAP-25 form a stoichimetric complex [4]. In addition, stoichimetric binding between syntaxin4 and SNAP-23 has been reported [38]. In the cell fusion assay, both SNAP-23 and SNAP-25 are anchored to the cell surface by forming complexes with syntaxin4 or syntaxin1 H3. Based on the biochemical evidence on the stoichimetric binding between syntaxin4 and SNAP-23 and between syntaxin1 and SNAP-25, and based on the fact that more syntaxin4 proteins were expressed on the cell surface than syntaxin1 H3 proteins, we reasoned that under the same experimental conditions, more SNAP-23 proteins were expressed on the cell surface than SNAP-25 proteins (Fig. 3B).
Cell fusion by flipped VAMP3 and flipped plasma membrane t-SNAREs
To quantitate cell fusion, we used a readout system that labels the cytoplasm of the cells that express flipped v-SNARE proteins (v-cells) differently from the cytoplasm of the cells that express flipped t-SNARE proteins (t-cells) (Fig. 4). In v-cells, flipped VAMP3 was cotransfected with a plasmid that encodes green fluorescent protein EGFP. In t-cells, the flipped t-SNARE constructs were cotransfected with a plasmid that encodes the red fluorescent protein DsRed2. Fusion between the v- and t-cells results in fused cells that contain both EGFP and DsRed2. In the merged channel, the cytoplasm of the fused cells is yellow (Fig. 4). Using this readout system, we can clearly identify those v-cells and t-cells that were in contact with each other (bound for first round of fusion), and those v-cells and t-cells that were in contact with fused cells (bound for additional rounds of fusion). Because it is difficult to distinguish whether the fused cells resulted from a single cell fusion event or from multiple cell fusion events, the extent of cell fusion (%) determined using the method described in Fig. 4 will underestimate the number of cell fusion events.
Fig. 4.
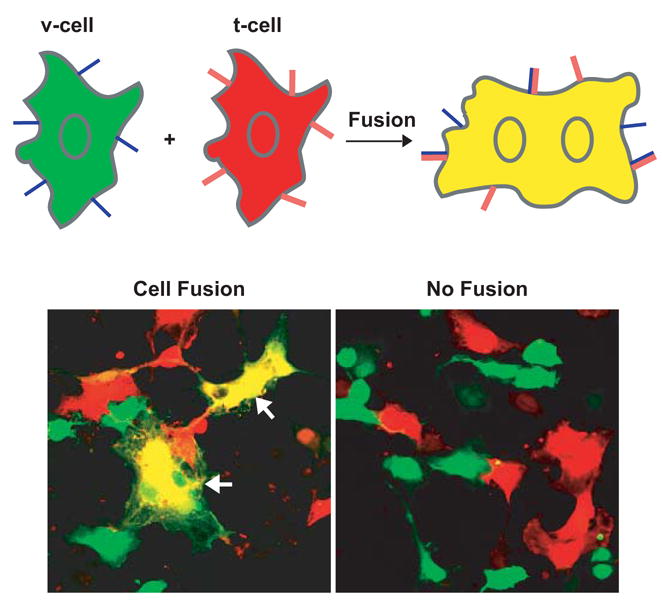
Cell fusion assay. COS-7 cells that expressed flipped v-SNAREs (v-cells) were labeled by green fluorescent protein EGFP. COS-7 cells that expressed flipped t-SNAREs (t-cells) were labeled by red fluorescent protein DsRed2 (RFP). Twenty-four hours after transfection, v-cells were detached from culture dishes, then overlaid on coverslips that already contained t-cells. Cells were fixed after 6 hours at 37°C. EGFP and RFP were imaged sequentially on a confocal microscope using the multitrack recording module before merging the images. Before cell fusion, the cytoplasm of v-cells showed green fluorescence whereas the cytoplasm of t-cells showed red fluorescence. Fusion of v- and t-cells resulted in fused cells (arrows) whose cytoplasm is yellow after the EGFP channel was merged with the RFP channel. No cell fusion was observed when either flipped v-SNAREs or flipped t-SNAREs were not expressed in COS-7 cells. To quantitate cell fusion, the total number of fused cells (F), the total number of unfused v-cells (V) and t-cells (T) that were in contact with each other or with the fused cells in 20 to 40 random fields (images taken at ×20) were determined. The extent of cell fusion (%) is calculated as 2F/(V+T+2F)×100.
Using the cell fusion assay, we determined the membrane fusion capacity of all the ternary complexes form by VAMP3 and plasma membrane t-SNAREs syntaxin1, syntaxin4, SNAP-23 and SNAP-25, i.e. VAMP3/syntaxin1/SNAP-25, VAMP3/syntaxin1/SNAP-23, VAMP3/syntaxin4/SNAP-25 and VAMP3/syntaxin4/SNAP-23. Efficient cell fusion was observed when VAMP3 partnered with syntaxin1 H3 and SNAP-25: 26% of the v- and t-cells that were in contact fused within 6 hours (Fig. 5A and B). To a lesser degree, v-cells expressing VAMP3 fused with t-cells expressing syntaxin1 H3 and SNAP-23 (5%) or t-cells expressing syntaxin4 and SNAP-25 (8 %) (Fig. 5A and B). Representative random images taken 6 hours after mixing of the v- and t-cells are shown in Fig. 5A. As expected, no cell fusion was observed between v-cells expressing VAMP3 and t-cells expressing only syntaxin1 H3 or t-cells expressing only syntaxin4 (Fig. 5B).
Fig. 5.
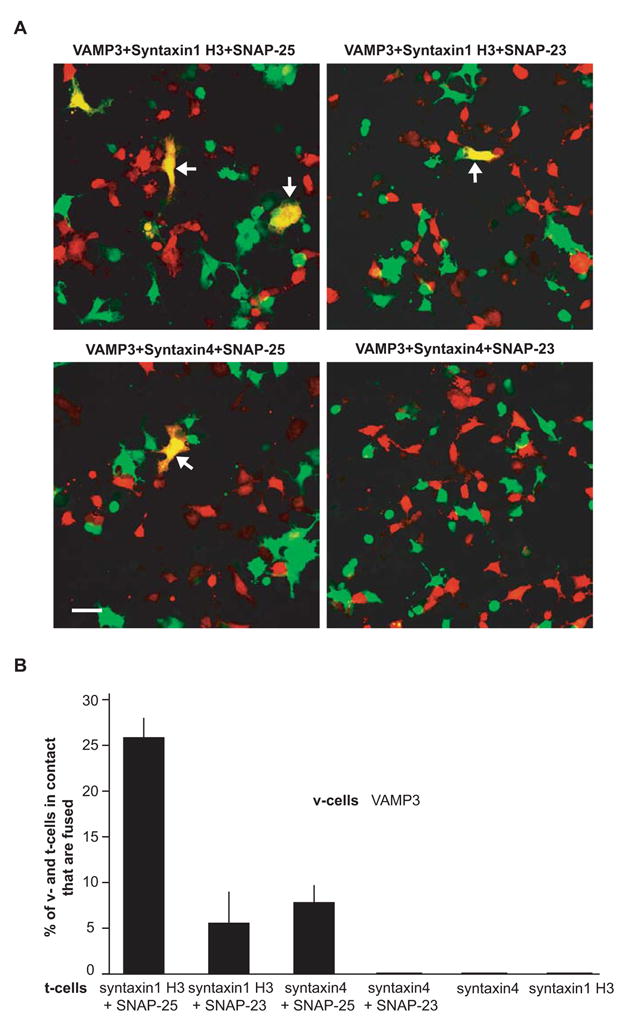
Cell fusion by flipped VAMP3 and flipped plasma membrane t-SNAREs. Twenty-four hours after transfection, v-cells (green) expressing VAMP3 were mixed with t-cells (red) expressing syntaxin1 H3 and SNAP-25, syntaxin1 H3 and SNAP-23, syntaxin4 and SNAP-25, syntaxin4 and SNAP-23, syntaxin4 alone, or syntaxin1 H3 alone for 6 hours at 37°C. (A) Representative random images were taken 6 hours after co-incubation of v-cells expressing VAMP3 and t-cells expressing the different combinations of t-SNAREs. Arrows indicate fused cells with a yellow cytoplasm. Scale bar, 50 μm. (B) Quantitation of cell fusion between v-cells expressing VAMP3 and t-cells expressing different t-SNAREs. Extent of cell fusion was determined using the method described in Fig. 4. Error bars represent SD of four independent experiments.
Surprisingly, in multiple experiments, no cell fusion was observed when v-cells expressing VAMP3 were mixed with t-cells expressing syntaxin4 and SNAP-23 (Fig. 5A and B). No cell fusion was observed even when the v- and t-cells were incubated together for 24 hours (data not shown). The lack of fusion was not due to a lower expression of syntaxin4 and SNAP-23 on the cell surface. As discussed above, immunostaining experiments (Fig. 3B) indicated that more syntaxin4 and SNAP-23 proteins were expressed on the cell surface than syntaxin1 H3 and SNAP-25 proteins. v-Cells expressing VAMP3 fused efficiently with the t-cells expressing syntaxin1 H3 and SNAP-25 (26%, with lower cell surface t-SNARE expression), but not with the t-cells expressing syntaxin4 and SNAP-23 (0%, with higher cell surface t-SNARE expression), suggesting that VAMP3 does not form fusogenic pairing with syntaxin4/SNAP-23. In summary, our cell fusion studies show that v-SNARE VAMP3 forms fusogenic pairing with plasma membrane t-SNARE complexes syntaxin1/SNAP-25, syntaxin1/SNAP-23 and syntaxin4/SNAP-25, but not with syntaxin4/SNAP-23.
N-terminal domain of syntaxin4 negatively regulates membrane fusion
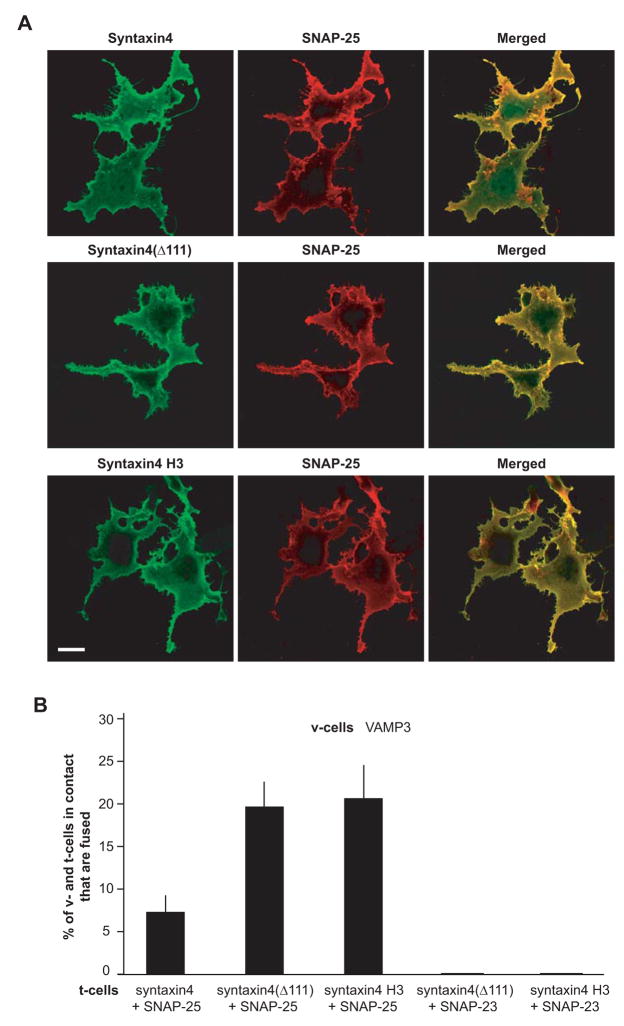
The N-terminal domain of syntaxin1 was shown to exhibit an inhibitory role in membrane fusion [39]. When co-expressed with SNAP-25, syntaxin4 was less fusogenic than syntaxin1 H3 that does not contain its N-terminal domain (compare first column and third column in Fig. 5B). To determine whether the N-terminal domain of syntaxin4 negatively regulates membrane fusion, we developed two truncation mutations of flipped syntaxin4 (Fig. 1). In syntaxin4(Δ111), the first 111 amino acids and therefore half of the N-terminal domain were deleted. In syntaxin4 H3, the entire N-terminal domain was deleted, leaving only the SNARE motif (H3 domain) and the C-terminal transmembrane domain. As expected, the truncation mutants had higher mobility than full-length syntaxin4 in SDS-PAGE (Fig 2B). When cotransfected with SNAP-25, similar amounts of syntaxin4(Δ111) proteins and syntaxin4 H3 proteins were expressed on the cell surface as compared with full-length syntaxin4 proteins (Fig. 6A). Interestingly, when co-expressed with SNAP-25, syntaxin4 (Δ111) and syntaxin4 H3 showed a more than 2-fold higher cell fusion capacity than full-length syntaxin4 (Fig. 6B), suggesting that the N-terminal domain of syntaxin4 indeed negatively regulates membrane fusion. When t-cells expressing syntaxin4 H3 and SNAP-25 were mixed with v-cells expressing VAMP3, 21% of the v- and t-cells that were in contact fused within 6 hours.
Fig. 6.
N-terminal domain of syntaxin4 negatively regulates membrane fusion. (A) Expression of syntaxin4 and truncation mutants of syntaxin4 on the cell surface. Twenty-four hours after transfection, unpermeabilized COS-7 cells cotransfected with flipped SNAP-25 and flipped syntaxin4, flipped syntaxin4(Δ111) or flipped syntaxin4 H3 were dual labeled with an antibody to Myc tag (syntaxin4 proteins, green) and an antibody to SNAP-25 (red). Confocal images were collected using the same settings. Representative images of three experiments are shown. Scale bar, 20 μm. (B) Extent of cell fusion between v-cells expressing VAMP3 and t-cells that co-expressed SNAP-25 (or SNAP-23) with syntaxin4, syntaxin4(Δ111) or syntaxin4 H3. v-Cells were incubated with t-cells for 6 hours. Extent of cell fusion was determined using the method described in Fig. 4. Error bars represent SD of three independent experiments.
Although the syntaxin4 truncation mutants showed higher membrane fusion capacity, they are fusogenic only when co-expressed with SNAP-25. When either syntaxin4 (Δ111) or syntaxin4 H3 was co-expressed with SNAP-23, no cell fusion was observed between the t-cells and the v-cells that expressed VAMP3 (Fig. 6B), further suggesting that VAMP3, syntaxin4 and SNAP-23 do not form a fusogenic pairing.
Discussion
VAMP3 is a v-SNARE protein that resides in recycling endosomes and endosome-derived transport vesicles [18]. It has been implicated in recycling of transferrin receptors to the plasma membrane [19], secretion of α-granules in platelets [21, 22], recycling of T-cell receptors to the immunological synapses [23], and membrane trafficking during cell migration [24, 25]. Using a cell fusion assay, we functionally examined the membrane fusion capacity of the ternary complexes formed by VAMP3 and plasma membrane t-SNAREs, syntaxin1, syntaxin4, SNAP-23 and SNAP-25. We showed that VAMP3 forms fusogenic pairings with syntaxin1/SNAP-25, syntaxin1/SNAP-23 and syntaxin4/SNAP-25, but not with syntaxin4/SNAP-23. The N-terminal domain of syntaxin4 was shown to negatively regulate membrane fusion. Differential membrane fusion capacities by the ternary v-/t-SNARE complexes suggest that transport vesicles containing VAMP3 have distinct membrane fusion kinetics with domains of the plasma membrane that present different t-SNARE proteins.
Both SNAP-23 and SNAP-25 bind to syntaxin1 and syntaxin4 [33]. VAMP3 forms complexes with syntaxin1, syntaxin4, SNAP-23 and SNAP-25 [29–31]. It has been reported that VAMP3 forms ternary complexes with syntaxin4/SNAP-25 [22] and syntaxin4/SNAP-23 [34]. Therefore, biochemically VAMP3 can form ternary complexes with t-SNARE complexes syntaxin1/SNAP-25, syntaxin1/SNAP-23, syntaxin4/SNAP-25 and syntaxin4/SNAP-23. In the current study, we demonstrated that the ternary combinations VAMP3/syntaxin1/SNAP-25, VAMP3/syntaxin1/SNAP-23 and VAMP3/syntaxin4/SNAP-25 are fusogenic. This result indicates that VAMP3 can interact with multiple t-SNAREs to drive the fusion of transport vesicles with the plasma membrane. Although VAMP3, syntaxin4 and SNAP-23 form a heterotrimeric complex [34], VAMP3/syntaxin4/SNAP-23 did not drive membrane fusion. Lack of fusion of the ternary combination was not due to lower expression of syntaxin4 proteins or SNAP-23 proteins on the cell surface. The lack of fusion was not due to malfunction of the flipped SNARE proteins either. Flipped VAMP3, flipped syntaxin4 or flipped SNAP-23 drive membrane fusion in other ternary combinations. Previous reports showed that syntaxin4 and SNAP-23 are phosphorylated by protein kinase C (PKC) [40] [41]. To determine if phosphorylation is required for the activation of syntaxin4/SNAP-23, we treated t-cells expressing syntaxin4 and SNAP-23 with purified PKC before mixing the t-cells with v-cells expressing VAMP3. Treatment of PKC did not trigger fusion by VAMP3/syntaxin4/SNAP-23 (data not shown), suggesting that lack of fusion was not due to the phosphorylation status of syntaxin4 and SNAP-23. Together our data strongly suggest that the VAMP3/syntaxin4/SNAP-23 ternary complex is not fusogenic. It is still possible that in vivo VAMP3/syntaxin4/SNAP-23 forms a non-efficient complex whose membrane fusion activity is below the detection limit of the cell fusion assay. It would be interesting to determine if biochemically and biophysically the VAMP3/syntaxin4/SNAP-23 complex is as stable as the other three ternary SNARE complexes that are fusogenic.
The findings of the present study may have implications for the mechanism of α-granule secretion in activated platelets. Fusion of α-granules with the plasma membrane results in the secretion of proteins that are implicated in thrombosis and wound healing, such as P-selectin, coagulation factor V and platelet-derived growth factor. Whereas syntaxin1 is not detected in platelets, VAMP3, syntaxin4 and SNAP-25 have been detected [34, 42, 43]. VAMP3 is present in α-granules [21]. Recombinant cytoplasmic domain of VAMP3 and antibodies against syntaxin4 inhibit α-granule secretion from permeabilized platelets [22, 34]. The current study showed that VAMP3 forms a fusogenic pairing with syntaxin4/SNAP-25, suggesting that VAMP3/syntaxin4/SNAP-25 can serve as a ternary SNARE complex that mediates the secretion of α-granules in platelets. Biochemical data support this hypothesis because VAMP3, syntaxin4 and SNAP-25 form SDS-resistant ternary complexes [22] and in platelets VAMP3 and SNAP-25 form complexes with syntaxin4 [43].
SNAP-23 is also expressed in platelets [34] and antibodies against SNAP-23 inhibit α-granule secretion from permeabilized platelets [44]. In platelets, VAMP3 forms ternary complexes with syntaxin4 and SNAP-23 [34]. However, in our cell fusion experiments, VAMP3 did not form a fusogenic pairing with syntaxin4/SNAP-23. The role of VAMP3/syntaxin4/SNAP-23 in mediating the secretion of α-granules warrants further examination.
Using yeast SNARE proteins as a model, a series of experiments demonstrated that to a remarkable degree the specificity of intracellular membrane fusion can be predicted from the pattern of liposome fusion mediated by isolated v- and t-SNARE proteins [16, 17]. Using the cell fusion assay, it has been shown that Bet1, a non-cognate v-SNARE involved in ER-Golgi trafficking [45], does not form a fusogenic pairing with t-SNAREs syntaxin1 and SNAP-25 [10]. Our current study extends the findings that specificity is encoded in the SNARE proteins in mammalian cells. In multi-tissue organisms such as mammals, the SNARE fusion machinery has evolved to match the complex patterns of cell differentiation and cell-cell communication. In addition to the physico-chemical properties of SNARE proteins, other factors can contribute to fine tune the specificity of SNARE-mediated membrane fusion. One factor is tissue and cell-type specific expression of SNARE proteins. For example, because syntaxin1 is not expressed in platelets [42], among the three fusogenic combinations detected by the cell fusion assay, only VAMP3/syntaxin4/SNAP-25 has all the components expressed in platelets. Subcellular distribution can also limit the interaction between cognate v- and t-SNAREs. For example, in epithelial cells, syntaxin4 is restricted to the basolateral domain of plasma membrane [46], whereas syntaxin1 is delivered to both the apical and basolateral domains [47]. In epithelial cells, VAMP3 can partner with syntaxin4 to deliver cargo proteins to the basolateral side of the plasma membrane. It can partner with syntaxin1 to transport cargo proteins to both the apical and basolateral sides.
A large body of structural and biochemical data reveals that the N-terminal domains of syntaxins function as a regulatory unit [48]. The N-terminal domain of syntaxin1 folds into a three-helix bundle, containing a surface groove to which the SNARE motif of syntaxin1 may bind to form a closed conformation [49]. Upon isolation, the N-terminal domain of syntaxin1 binds to its own C-terminal SNARE motif resulting in inhibition of the interaction with VAMP2 and SNAP-25. Munc18a/nSec1, a cytosolic protein, binds to the closed form of syntaxin1 and prevents assembly of the ternary SNARE complex [50, 51]. Membrane fusion is promoted when the N-terminal domain of syntaxin1 is removed [39]. To determine the function of the N-terminal domain of syntaxin4, in the current study we developed two truncation mutants of syntaxin4 in which half or the entire N-terminal domain was removed. The membrane fusion capacities of both truncation mutants were much higher than full-length syntaxin4, indicating that the N-terminal domain of synatxin4 negatively regulates membrane fusion. Like syntaxin1, the N-terminal domain of syntaxin4 may bind to its own C-terminal SNARE motif to form a closed conformation and prevent assembly of ternary SNARE complexes.
Acknowledgments
This work was supported by a scientist development grant from the American Heart Association, a startup fund from the University of Louisville School of Medicine (to C.H.) and HL-68079 from NIH (to F.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rothman JE. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- 2.Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 3.Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. [see comments] Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 4.Fasshauer D, Margittai M. A transient N-terminal interaction of SNAP-25 and syntaxin nucleates SNARE assembly. J Biol Chem. 2004;279:7613–7621. doi: 10.1074/jbc.M312064200. [DOI] [PubMed] [Google Scholar]
- 5.Hayashi T, McMahon H, Yamasaki S, Binz T, Hata Y, Sudhof TC, Niemann H. Synaptic vesicle membrane fusion complex: action of clostridial neurotoxins on assembly. EMBO J. 1994;13:5051–5061. doi: 10.1002/j.1460-2075.1994.tb06834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang B, Gonzalez L, Jr, Prekeris R, Steegmaier M, Advani RJ, Scheller RH. SNARE interactions are not selective. Implications for membrane fusion specificity. J Biol Chem. 1999;274:5649–5653. doi: 10.1074/jbc.274.9.5649. [DOI] [PubMed] [Google Scholar]
- 7.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. [see comments] Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 8.Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 9.McNew JA, Weber T, Parlati F, Johnston RJ, Melia TJ, Sollner TH, Rothman JE. Close is not enough: SNARE-dependent membrane fusion requires an active mechanism that transduces force to membrane anchors. J Cell Biol. 2000;150:105–117. doi: 10.1083/jcb.150.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu C, Ahmed M, Melia TJ, Sollner TH, Mayer T, Rothman JE. Fusion of cells by flipped SNAREs. Science. 2003;300:1745–1749. doi: 10.1126/science.1084909. [DOI] [PubMed] [Google Scholar]
- 11.Fix M, Melia TJ, Jaiswal JK, Rappoport JZ, You D, Sollner TH, Rothman JE, Simon SM. Imaging single membrane fusion events mediated by SNARE proteins. Proc Natl Acad Sci U S A. 2004;101:7311–7316. doi: 10.1073/pnas.0401779101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sollner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- 13.Mayer A, Wickner W, Haas A. Sec18p (NSF)-driven release of Sec17p (alpha-SNAP) can precede docking and fusion of yeast vacuoles. Cell. 1996;85:83–94. doi: 10.1016/s0092-8674(00)81084-3. [DOI] [PubMed] [Google Scholar]
- 14.Bock JB, Matern HT, Peden AA, Scheller RH. A genomic perspective on membrane compartment organization. Nature. 2001;409:839–841. doi: 10.1038/35057024. [DOI] [PubMed] [Google Scholar]
- 15.Chen YA, Scheller RH. SNARE-mediated membrane fusion. Nat Rev Mol Cell Biol. 2001;2:98–106. doi: 10.1038/35052017. [DOI] [PubMed] [Google Scholar]
- 16.McNew JA, Parlati F, Fukuda R, Johnston RJ, Paz K, Paumet F, Sollner TH, Rothman JE. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. [see comments] Nature. 2000;407:153–159. doi: 10.1038/35025000. [DOI] [PubMed] [Google Scholar]
- 17.Parlati F, Varlamov O, Paz K, McNew JA, Hurtado D, Sollner TH, Rothman JE. Distinct SNARE complexes mediating membrane fusion in Golgi transport based on combinatorial specificity. Proc Natl Acad Sci U S A. 2002;99:5424–5429. doi: 10.1073/pnas.082100899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMahon HT, Ushkaryov YA, Edelmann L, Link E, Binz T, Niemann H, Jahn R, Sudhof TC. Cellubrevin is a ubiquitous tetanus-toxin substrate homologous to a putative synaptic vesicle fusion protein. Nature. 1993;364:346–349. doi: 10.1038/364346a0. [DOI] [PubMed] [Google Scholar]
- 19.Galli T, Chilcote T, Mundigl O, Binz T, Niemann H, De Camilli P. Tetanus toxin-mediated cleavage of cellubrevin impairs exocytosis of transferrin receptor-containing vesicles in CHO cells. J Cell Biol. 1994;125:1015–1024. doi: 10.1083/jcb.125.5.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Volchuk A, Sargeant R, Sumitani S, Liu Z, He L, Klip A. Cellubrevin is a resident protein of insulin-sensitive GLUT4 glucose transporter vesicles in 3T3-L1 adipocytes. J Biol Chem. 1995;270:8233–8240. doi: 10.1074/jbc.270.14.8233. [DOI] [PubMed] [Google Scholar]
- 21.Feng D, Crane K, Rozenvayn N, Dvorak AM, Flaumenhaft R. Subcellular distribution of 3 functional platelet SNARE proteins: human cellubrevin, SNAP-23, and syntaxin 2. Blood. 2002;99:4006–4014. doi: 10.1182/blood.v99.11.4006. [DOI] [PubMed] [Google Scholar]
- 22.Polgar J, Chung SH, Reed GL. Vesicle-associated membrane protein 3 (VAMP-3) and VAMP-8 are present in human platelets and are required for granule secretion. Blood. 2002;100:1081–1083. doi: 10.1182/blood.v100.3.1081. [DOI] [PubMed] [Google Scholar]
- 23.Das V, Nal B, Dujeancourt A, Thoulouze MI, Galli T, Roux P, Dautry-Varsat A, Alcover A. Activation-induced polarized recycling targets T cell antigen receptors to the immunological synapse; involvement of SNARE complexes. Immunity. 2004;20:577–588. doi: 10.1016/s1074-7613(04)00106-2. [DOI] [PubMed] [Google Scholar]
- 24.Tayeb MA, Skalski M, Cha MC, Kean MJ, Scaife M, Coppolino MG. Inhibition of SNARE-mediated membrane traffic impairs cell migration. Exp Cell Res. 2005;305:63–73. doi: 10.1016/j.yexcr.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 25.Proux-Gillardeaux V, Gavard J, Irinopoulou T, Mege RM, Galli T. Tetanus neurotoxin-mediated cleavage of cellubrevin impairs epithelial cell migration and integrin-dependent cell adhesion. Proc Natl Acad Sci U S A. 2005;102:6362–6367. doi: 10.1073/pnas.0409613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang C, Mora S, Ryder JW, Coker KJ, Hansen P, Allen LA, Pessin JE. VAMP3 null mice display normal constitutive, insulin- and exercise-regulated vesicle trafficking. Mol Cell Biol. 2001;21:1573–1580. doi: 10.1128/MCB.21.5.1573-1580.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schraw TD, Rutledge TW, Crawford GL, Bernstein AM, Kalen AL, Pessin JE, Whiteheart SW. Granule stores from cellubrevin/VAMP-3 null mouse platelets exhibit normal stimulus-induced release. Blood. 2003;102:1716–1722. doi: 10.1182/blood-2003-01-0331. [DOI] [PubMed] [Google Scholar]
- 28.Borisovska M, Zhao Y, Tsytsyura Y, Glyvuk N, Takamori S, Matti U, Rettig J, Sudhof T, Bruns D. v-SNAREs control exocytosis of vesicles from priming to fusion. EMBO J. 2005;24:2114–2126. doi: 10.1038/sj.emboj.7600696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chilcote TJ, Galli T, Mundigl O, Edelmann L, McPherson PS, Takei K, De Camilli P. Cellubrevin and synaptobrevins: similar subcellular localization and biochemical properties in PC12 cells. J Cell Biol. 1995;129:219–231. doi: 10.1083/jcb.129.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray RZ, Kay JG, Sangermani DG, Stow JL. A role for the phagosome in cytokine secretion. Science. 2005;310:1492–1495. doi: 10.1126/science.1120225. [DOI] [PubMed] [Google Scholar]
- 31.Schraw TD, Lemons PP, Dean WL, Whiteheart SW. A role for Sec1/Munc18 proteins in platelet exocytosis. Biochem J. 2003;374:207–217. doi: 10.1042/BJ20030610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bennett MK, Garcia-Arraras JE, Elferink LA, Peterson K, Fleming AM, Hazuka CD, Scheller RH. The syntaxin family of vesicular transport receptors. Cell. 1993;74:863–873. doi: 10.1016/0092-8674(93)90466-4. [DOI] [PubMed] [Google Scholar]
- 33.Ravichandran V, Chawla A, Roche PA. Identification of a novel syntaxin- and synaptobrevin/VAMP-binding protein, SNAP-23, expressed in non-neuronal tissues. J Biol Chem. 1996;271:13300–13303. doi: 10.1074/jbc.271.23.13300. [DOI] [PubMed] [Google Scholar]
- 34.Flaumenhaft R, Croce K, Chen E, Furie B, Furie BC. Proteins of the exocytotic core complex mediate platelet alpha-granule secretion. Roles of vesicle-associated membrane protein, SNAP-23, and syntaxin 4. J Biol Chem. 1999;274:2492–2501. doi: 10.1074/jbc.274.4.2492. [DOI] [PubMed] [Google Scholar]
- 35.Elbein AD. Inhibitors of the biosynthesis and processing of N-linked oligosaccharides. CRC Crit Rev Biochem. 1984;16:21–49. doi: 10.3109/10409238409102805. [DOI] [PubMed] [Google Scholar]
- 36.Nicholson KL, Munson M, Miller RB, Filip TJ, Fairman R, Hughson FM. Regulation of SNARE complex assembly by an N-terminal domain of the t-SNARE Sso1p. Nat Struct Biol. 1998;5:793–802. doi: 10.1038/1834. [DOI] [PubMed] [Google Scholar]
- 37.Fiebig KM, Rice LM, Pollock E, Brunger AT. Folding intermediates of SNARE complex assembly. Nat Struct Biol. 1999;6:117–123. doi: 10.1038/5803. [DOI] [PubMed] [Google Scholar]
- 38.Buxton P, Zhang XM, Walsh B, Sriratana A, Schenberg I, Manickam E, Rowe T. Identification and characterization of Snapin as a ubiquitously expressed SNARE-binding protein that interacts with SNAP23 in non-neuronal cells. Biochem J. 2003;375:433–440. doi: 10.1042/BJ20030427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parlati F, Weber T, McNew JA, Westermann B, Sollner TH, Rothman JE. Rapid and efficient fusion of phospholipid vesicles by the alpha-helical core of a SNARE complex in the absence of an N-terminal regulatory domain. [see comments] Proc Natl Acad Sci U S A. 1999;96:12565–12570. doi: 10.1073/pnas.96.22.12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung SH, Polgar J, Reed GL. Protein kinase C phosphorylation of syntaxin 4 in thrombin-activated human platelets. J Biol Chem. 2000;275:25286–25291. doi: 10.1074/jbc.M004204200. [DOI] [PubMed] [Google Scholar]
- 41.Hepp R, Puri N, Hohenstein AC, Crawford GL, Whiteheart SW, Roche PA. Phosphorylation of SNAP-23 regulates exocytosis from mast cells. J Biol Chem. 2005;280:6610–6620. doi: 10.1074/jbc.M412126200. [DOI] [PubMed] [Google Scholar]
- 42.Lemons PP, Chen D, Bernstein AM, Bennett MK, Whiteheart SW. Regulated secretion in platelets: identification of elements of the platelet exocytosis machinery. Blood. 1997;90:1490–1500. [PubMed] [Google Scholar]
- 43.Reed GL, Houng AK, Fitzgerald ML. Human platelets contain SNARE proteins and a Sec1p homologue that interacts with syntaxin 4 and is phosphorylated after thrombin activation: implications for platelet secretion. Blood. 1999;93:2617–2626. [PubMed] [Google Scholar]
- 44.Lemons PP, Chen D, Whiteheart SW. Molecular mechanisms of platelet exocytosis: requirements for alpha-granule release. Biochem Biophys Res Commun. 2000;267:875–880. doi: 10.1006/bbrc.1999.2039. [DOI] [PubMed] [Google Scholar]
- 45.Jahn R, Sudhof TC. Membrane fusion and exocytosis. Annu Rev Biochem. 1999;68:863–911. doi: 10.1146/annurev.biochem.68.1.863. [DOI] [PubMed] [Google Scholar]
- 46.Li X, Low SH, Miura M, Weimbs T. SNARE expression and localization in renal epithelial cells suggest mechanism for variability of trafficking phenotypes. Am J Physiol Renal Physiol. 2002;283:F1111–1122. doi: 10.1152/ajprenal.00185.2002. [DOI] [PubMed] [Google Scholar]
- 47.Rowe J, Calegari F, Taverna E, Longhi R, Rosa P. Syntaxin 1A is delivered to the apical and basolateral domains of epithelial cells: the role of munc-18 proteins. J Cell Sci. 2001;114:3323–3332. doi: 10.1242/jcs.114.18.3323. [DOI] [PubMed] [Google Scholar]
- 48.Dietrich LE, Boeddinghaus C, LaGrassa TJ, Ungermann C. Control of eukaryotic membrane fusion by N-terminal domains of SNARE proteins. Biochim Biophys Acta. 2003;1641:111–119. doi: 10.1016/s0167-4889(03)00094-6. [DOI] [PubMed] [Google Scholar]
- 49.Fernandez I, Ubach J, Dulubova I, Zhang X, Sudhof TC, Rizo J. Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell. 1998;94:841–849. doi: 10.1016/s0092-8674(00)81742-0. [DOI] [PubMed] [Google Scholar]
- 50.Yang B, Steegmaier M, Gonzalez LC, Jr, Scheller RH. nSec1 binds a closed conformation of syntaxin1A. J Cell Biol. 2000;148:247–252. doi: 10.1083/jcb.148.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Misura KM, Scheller RH, Weis WI. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. [see comments] Nature. 2000;404:355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]