Abstract
V(D)J recombination is thought to be regulated by changes in the accessibility of target sites, such as modulation of methylation. To test whether demethylation of the kappa locus can activate recombination, we generated two recombinationally active B cell lines in which the gene for maintenance of genomic DNA methylation, Dnmt1, was flanked with loxP sites. Transduction with a retrovirus expressing both the cre recombinase and green fluorescent protein allowed us to purify recombinationally active cells devoid of methylation. Loss of methylation of the kappa locus was not sufficient to activate recombination, although transcription was activated in one line. It appears that demethylation of the kappa locus is not the rate-limiting step for altering accessibility and thus regulated demethylation does not generate specificity of recombination.
The V(D)J recombination machinery assembles antigen receptor genes during lymphoid development from gene segments flanked by recombination signal sequences (1). This nonhomologous site-specific recombination process is precisely controlled by a number of different regulatory mechanisms. For one thing, the coexpression of the lymphoid-specific Rag-1 and Rag-2 genes occurs restrictively in developing B and T cells, ensuring that only the appropriate cell types are recombinationally active (2). A second level of regulation also controls lineage specificity: complete Ig gene rearrangement occurs only in B cells, whereas T cell receptor genes rearrange exclusively in T cells (3). Moreover, within developing B and T cells there are temporal controls over specific gene segment usage (4, 5). Additionally, allelic exclusion ensures that each B or T cell generates only one functional antigen receptor even though two alleles exist at each locus (6).
This remarkable ability of the recombination machinery to target only a very small subset of substrates within a given cell together with the fact that a single recombinase is required for this process has led to the hypothesis that the accessibility of DNA targets to the recombination machinery is tightly controlled (7–9). Recombination loci are thought to be inaccessible until they are modified to allow the recombination machinery access to target sites. Changes in chromatin structure, transcriptional activity, or DNA methylation at each locus have been identified that correlate with recombinational activity. Many studies have attempted to define the cis-acting sequences and trans-acting factors required to regulate this process. Distinct enhancer sequences present within individual loci must be controlled by different trans-acting proteins to allow developmentally regulated changes in the accessibility of each locus to occur.
DNA methylation has been shown to regulate chromatin structure, leading to changes of gene expression (10). Additionally, methylation has been correlated with Ig gene regulation: both the μ heavy chain and κ light chain are hypermethylated in cells in which they are not recombined (11, 12). Concomitant with V(D)J rearrangement these loci become hypomethylated and transcriptionally active (13, 14). Regulated demethylation could control the specificity of the V(D)J recombinase. Consistent with such a concept, exogenously introduced hypermethylated recombination substrates including both episomes and transgenes are refractory to V(D)J recombination compared with their unmethylated counterparts (15–17). At a minimum, this suggests that methylation of target sites bound by ubiquitous trans-acting factors inhibits recombination so that demethylation of these loci should be required for the initiation of recombination.
We sought to directly test whether demethylation of an endogenous V(D)J target was sufficient to increase accessibility for recombination. If demethylation was the rate limiting step controlling the specificity of recombination, then forced demethylation should activate recombination of the unmethylated loci. To this end, we established a model system to examine the regulation of kappa recombination as a function of methylation status by using Abelson virus-transformed cell lines that were conditionally mutant for global DNA methylation. In these Abelson lines, the gene encoding the maintenance DNA methyltransferase, Dnmt1, was flanked by loxP sites allowing the excision and loss of function of the gene when the cre recombinase was provided. Using this inducible system, we were able to efficiently demethylate the kappa locus. However, this did not activate recombination. In contrast, the loss of methylation did activate transcription within the kappa locus of one of the Abelson lines. Our results suggest that demethylation of the kappa locus is not sufficient to activate V(D)J recombination, thus supporting a model whereby specific factors must be recruited to the locus to alter its accessibility for recombination independent of other properties such as transcription or methylation.
Materials and Methods
Cell Lines and Culture.
Bone marrow-derived Abelson lines from 1-month-old Dnmt2lox/DnmtN mice (mice will be described elsewhere) were established by conventional methods (18). The v-abl retrovirus used to infect bone marrow cultures was produced from a helper free retroviral packaging system (19) by using the construct pGD-v-abl (20). Abelson lines were maintained in RPMI (GIBCO/BRL) plus 10% defined FCS (HyClone), 50 μM β-mercaptoethanol (Sigma), and Pen/Strep (GIBCO/BRL). The 293 cells and derivatives were maintained in DMEM (GIBCO/BRL) plus 10% FCS (JRH Biosciences, Lenexa, KS) and Pen/Strep. Lipopolysaccharide (LPS) (Sigma) at 10 μg/ml was added to cells 24 h before collection. Cells were pelleted and resuspended in media at 107 cells/ml, sorted for live green fluorescent protein (Gfp)+ or Gfp− cells, by using a FACStar with Turbo cell sorter, and used immediately for analysis.
Retroviral Infections and Constructs.
MCiGfp was generated by inserting a Cre-SV40 nuclear localization signal fusion protein (21) into the multiple cloning site of MiGfp (22). The nonreplication competent helper plasmid pCL-eco was a gift from Inder Verma, The Salk Institute, San Diego. DNA was introduced into 293 cells by coprecipitation with calcium phosphate as described (19). The viral supernatants were used for spin infections (22) after which the viral supernatant was removed and replaced with media (day 0 postinfection).
Southern Analysis.
Cell pellets were washed once in PBS and then lysed in DNA lysis buffer plus proteinase K at 55°C overnight as described. The DNA then was phenol/chloroform-extracted, precipitated, and resuspended in TE-1 (10 mM Tris, pH 8.0/1 mM EDTA). Five micrograms of DNA was digested with the appropriate restriction enzymes and loaded on an agarose gel. The DNA was transferred to Nylon N (Hybond) and probed with random primed (Stratagene) fragments: intracisternal A-type particle (IAP) [nucleotides 1527–1690, 1712–1802, 1946–2160 from the mouse ren-2 IAP genome (23) assembled as a contig; gift from D. Humphreys, Whitehead Institute, Cambridge, MA] and CJk (24).
Recombination Assays.
One hundred nanograms of genomic DNA generated as above was used as a template for PCR. VJk PCR fragments were amplified by using the primers Vk and Jk2 for 26 cycles consisting of 1 min 94°C, 1 min at 60°C, and 1.75 min at 72°C. The final polymerization step was extended an additional 10 min, and the PCR products were detected by Southern analysis using the oligonucleotide Jk1 (25).
Reverse Transcription–PCR (RT-PCR) Analysis.
Total cellular RNA was purified from cultured cells with Trireagent (MRC, Cincinnati), DNase (Stratagene)-treated, phenol/chloroform-extracted, and precipitated. A total of 1 μg RNA was subjected to RT using Moloney murine leukemia virus reverse transcriptase (Boehringer Mannheim) and random primers. The reaction was subjected to PCR analysis as described by using the primers ko and Ck (26). The amplified products were subjected to electrophoresis on an agarose gel, transferred to a nylon membrane, and probed with Jk2 (27).
Results
Assay System.
We generated Abelson virus transformed pre-B cell lines to study the role of methylation in the regulation of V(D)J recombination (18). Abelson lines were used because they have been extensively characterized and used as a model system for the regulation of early B cell development. Abelson lines are recombinationally active and recombine their Ig loci in the same developmentally ordered program of gene segment usage as normal B cells. Abelson lines are typically D to JH rearranged on both alleles and are actively rearranging their V regions to the DJH segments. The level of recombination at the kappa locus is very low. However, recombination can be induced by treatments that alter the accessibility of the locus, allowing the retargeting of the recombinase from the heavy chain to the kappa locus. For example, treatment of Abelson lines with bacterial LPS activates both transcription and recombination of kappa. Therefore, we reasoned that Abelson lines would be a good model system to test the role of methylation in V(D)J recombination of the kappa locus.
Mice deficient for the gene that encodes the maintenance DNA methyltransferase, Dnmt1, which remethylates the hemimethylated cytidines within CpG dinucleotides generated as a product of DNA replication, die early in embryonic life. This precludes analysis of how alterations in the methylation status of recombination loci affect lymphoid development. Therefore, we generated cell lines from mice that were conditionally mutant for the Dnmt1 locus; one allele was null for the Dnmt1 gene (N allele, ref. 28) whereas the other allele contained lox P sites flanking exons required for the function of the gene (2lox, mice will be described elsewhere). Abelson lines derived from these animals are conditionally mutant for the Dnmt1 locus; upon introduction of the cre recombinase, the allele containing the lox P sites is excised, generating cells in which both alleles of the Dnmt1 gene are mutant. We generated two independent Abelson lines of the genotype Dnmt2lox/N (Dnmtcond1, Dnmtcond2) and assayed their rearrangement status. Southern blot analysis of these lines demonstrated that the heavy chain had DJH rearrangements, whereas the kappa locus was in a germ-line configuration (data not shown).
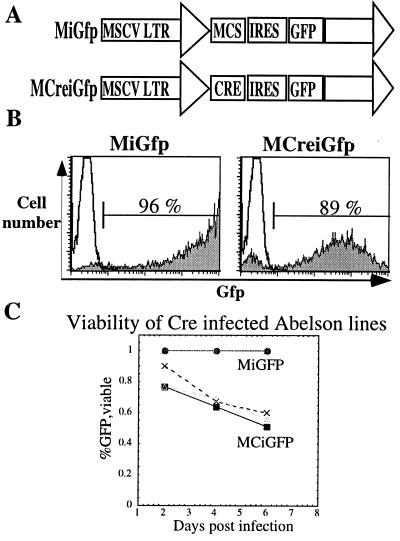
To introduce the cre recombinase efficiently into the Dnmtcond lines we used a replication incompetent retroviral construct containing the mouse stem cell virus long terminal repeat (MSCV LTR) driving the expression of Gfp as a reporter for infection (Fig. 1A; ref. 22). This retroviral vector contained a multiple cloning site (MCS) upstream of an internal ribosomal entry site (IRES) that allowed for enhanced Gfp expression (MiGfp). The experimental retrovirus had the cre recombinase gene cloned into the MCS generating a bicistronic vector (MCiGfp). This allowed us to monitor, and purify, by fluorescence-activated cell sorting the infected (Gfp+) and uninfected (Gfp−) populations of cells. In addition, given the bicistronic nature of the vector, we could be confident that the Gfp+ cells that were infected by the MCiGfp retrovirus also expressed the cre recombinase. Using a 1-h spin infection of retroviral supernatant generated by cotransfecting 293 cells with either the MiGfp or the MCiGfp vector and a replication incompetent helper plasmid, pCL-eco (29), we reproducibly infected over 70% of the cells as measured by flow cytometry (Fig. 1B).
Figure 1.
Abelson lines are efficiently infected by retroviruses. (A) Schematic diagram of retroviral vectors used in this study. The MSCV LTR drives the expression of the enhanced Gfp downstream of an IRES in the control vector, MiGfp. The cre recombinase is cloned upstream of the IRES GFP cassette and also is driven by the MSCV LTR, MCiGfp. MCS, multiple cloning site. (B) Fluorescence-activated cell sorting analysis measuring Gfp expression of the Abelson line Dnmtcond1 infected with either MiGfp or MCiGfp at 2 days postinfection gated on live cells by scatter properties. (C) Measurement of the percentage of live Gfp+ cells as a function of time postinfection either infected with MiGfp or MCiGfp as labeled. Each line represents an independent experiment.
We then assayed the viability of the cells after infection. At 2 days postinfection, we observed a similar percentage of Gfp+ viable cells that had been infected with either MiGfp or MCiGfp (Fig. 1 B and C). In contrast, by 4 days postinfection, when the cells had undergone two rounds of cell division, the number of Gfp+ cells that were infected with MCiGfp had declined whereas the number of Gfp+ cells infected with MiGfp remained stable (Fig. 1C). This finding suggests that the loss in viability of the cells might be caused by reduced genomic DNA methylation as a result of deletion of the Dnmt1 locus. This possibility is consistent with previous observations that mice deficient for Dnmt1 die early in embryogenesis (28) and mouse embryonic fibroblasts in which Dnmt1 is deleted are not viable (C.B., unpublished observation).
Cre-Mediated Loss of DNMT1.
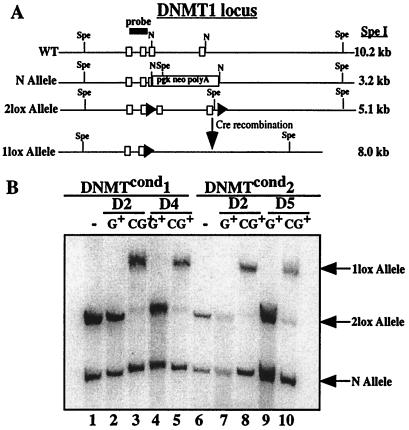
We verified that cells infected by MCiGfp had indeed deleted their Dnmt1 locus via cre-mediated recombination. Taking advantage of the incomplete infection of each cell line, we analyzed infected (Gfp+) cells separately from uninfected (Gfp−) cells that were either infected with MiGfp or MCiGfp (Fig. 1B, see gates). We purified these cells by fluorescence-activated cell sorting and performed Southern blot analysis on the genomic DNA from these purified populations. A schematic diagram of the Dnmt1 locus before and after cre-mediated recombination is shown in Fig. 2A. Southern blot analysis demonstrated that only the cells that were infected by MCiGfp and were Gfp+ had deleted their Dnmt1 locus by day 2 postinfection (Fig. 2B, lanes 3 and 8). Additionally, there was no recombination either in cells that were not infected by a cre-containing retrovirus (MiGfp) or had not been infected (Gfp−) (Fig. 2B). Over time there was an accumulation of cells that were Gfp+ but had not excised their Dnmt1 locus as seen by the presence of the 2lox band in lane 10. However, this represents less than 10% of the total cells in this fraction.
Figure 2.
Abelson lines infected with MCiGfp efficiently excise the Dnmt1 locus. (A) Schematic diagram of the genomic Dnmt1 locus. The genotype of the Abelson lines are 2lox/N before cre-mediated excision. WT, wild type. (B) Southern blot analysis of the Dnmt1 locus from purified populations of infected Dnmtcond Abelson lines infected with either MiGfp or MCiGfp and sorted for Gfp+ viable cells (G+ or CiG+, respectively). The DNA was digested with SpeI and probed with a fragment from the Dnmt1 locus. The bands are labeled on the right.
Methylation Status of the Kappa Locus.
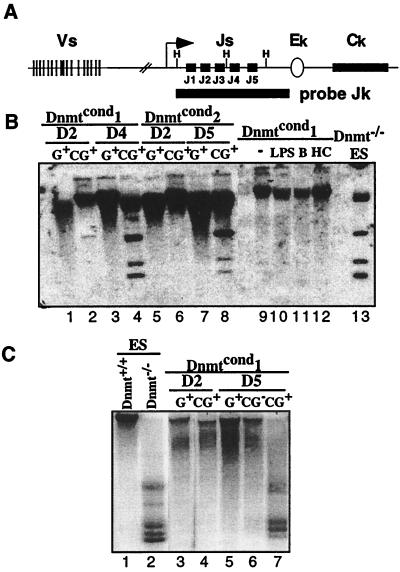
We next determined the methylation status of the Dnmtcond lines under the different conditions to access the efficiency of demethylation caused by the loss of the Dnmt1 gene. Because of the mechanism of action of the maintenance methyltransferase Dnmt1, two rounds of replication are required to generate an unmethylated genome; after one cell division both daughter cells are hemimethylated, and after the second round, two daughters are unmethylated and two daughters are hemimethylated. The original methylated parental strands of DNA are not lost, but simply diluted in the population as the cells divide. To assess whether this was apparent in the excised Dnmtcond lines we treated genomic DNA with the methylation-sensitive restriction enzyme HhaI to assay the methylation status of the Jk region of the kappa locus. We assayed this region of the kappa locus as it has been shown to be demethylated in a developmentally regulated manner that tightly correlates with the recombination potential of the region (30). A schematic diagram of the locus is shown along with the restriction sites (H) and the probe used for analysis (Fig. 3A). Dnmt1−/− embryonic stem (ES) cell DNA was used as a control for unmethylated genomic DNA (Fig. 3B, lane 13).
Figure 3.
Loss of Dnmt1 results in the loss of genomic methylation by day 4 postinfection. (A) Schematic diagram of the murine kappa locus. The map shows the V and J gene segments, constant region (Ck) and the intronic enhancer (Ek). The methylation-sensitive restrictions sites, HhaI (H), are shown as well as the probe used for analysis (Jk). The diagram is not to scale. (B) Southern blot analysis of the methylation status of the kappa locus in Abelson lines under different conditions. The DNA was digested with HhaI and probed with a Jk probe that spans the J region of the kappa locus. Lanes 1–8 are derived from Gfp+ sorted Abelson lines infected with the MiGfp (G) or MCiGfp (CG) at the time points indicated postinfection. Lanes 9–12 are generated from unsorted bulk populations of cells. Lane 9, untreated; lane 10, treated with LPS; lane 11, treated with butyrate; lane 12 treated with HC toxin. Lane 13 is derived from untreated Dnmt−/− ES cells. (C) Southern blot analysis of the methylation status of IAPs in Abelson lines. The DNA was digested with HpaII and probed with a fragment of a known, endogenous IAP. Lanes 1 and 2 are controls, derived from ES cells that are wild type or mutant for the Dnmt1 locus, respectively. Lanes 3–7 are derived from Gfp+ or Gfp− sorted Dnmtcond1 Abelson line infected with either MiGfp (G) or MCiGfp (CG) at the time points indicated. The unmethylated fragments are shown on the right.
The kappa locus of the untreated Abelson lines is fully methylated as seen by the lack of digestion of the genomic DNA by HhaI (Fig. 3B, lane 9). This was expected because the efficiency of recombination at the kappa locus is low. In contrast, cells that had been infected by MCiGfp 4 days earlier and were Gfp+ had a substantial level of unmethylated DNA (Fig. 3B, lanes 4 and 8). Genomic DNA from cells that had only been infected for 2 days or that were infected by MiGfp remained methylated (Fig. 3B, lanes 1 and 5). This is consistent with excision of the Dnmt1 gene by day 2 postinfection followed by two rounds of cell division causing the loss of DNA methylation by day 4 postinfection. The absolute level of hypomethylation is less than in cells null for the Dnmt1 locus but significant; more than 30% of the alleles are unmethylated.
We also evaluated the methylation status of IAPs because they are normally highly methylated retrotransposons present in hundreds of copies in the genome (31) and thus a sensitive readout for alterations of genomewide DNA methylation state. We digested genomic DNA with HpaII, another methylation-sensitive restriction enzyme that digests the IAP sequences a number of times, generating a diagnostic ladder of bands. Again, we used Dnmt1 null ES cell DNA as a control for unmethylated DNA (Fig. 3C, lane 2). Southern blot analysis demonstrated that the IAP sequences were highly methylated in Abelson lines (Fig. 3C, lanes 3–6). As expected, these sequences were hypomethylated in cells that were Gfp+ and were infected by MCiGfp at least 4 days earlier (Fig. 3C, lane 7). This correlates with the loss of methylation observed at the kappa locus. Once again, there is no effect on the methylation status of the IAP sequences if cells were infected by MiGfp, were Gfp−, or were only 2 days postinfection (Fig. 3C, data not shown).
Activation of Transcription of the Kappa Locus by Demethylation.
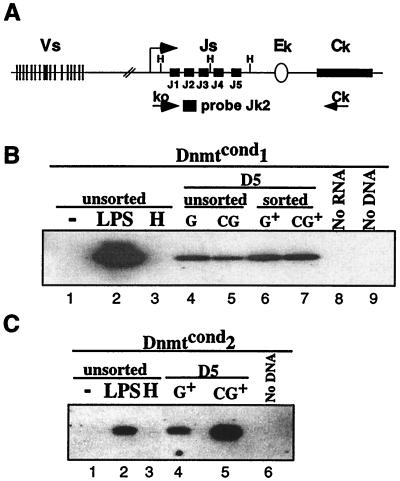
It has been demonstrated that there is a tight correlation between the transcriptional status of the antigen receptor loci and their ability to be recombined. Transcription of various Ig genes is activated concomitant with their rearrangement (7, 32, 33). Specifically, within the kappa locus the synthesis of a transcript that encompasses the Jk gene segments, k0, can be detected upon induction of kappa rearrangement (Fig. 4A; ref. 27). Additionally, hypermethylation has been associated with transcriptional silencing, and changes in methylation can alter gene expression (10). Therefore, we determined whether loss of methylation in our Abelson lines affected the expression of k0. We used a RT-PCR Southern assay on total genomic RNA randomly primed and amplified by using the primers k0 and Ck and detected by using the probe Jk2 (26) shown schematically in Fig. 4A. LPS treatment of our Abelson lines activated transcription of k0 (Fig. 4 B and C, compare lanes 1 and 2). In addition, retroviral infection increased the basal level of k0, but it remained inducible by LPS treatment (compare lanes 1 and 4 in Fig. 4 B and C).
Figure 4.
Loss of methylation at the kappa locus activates transcription in Dnmtcond2. (A) Schematic diagram of the murine kappa locus. The large arrow designates the promoter of the ko transcript. The primers ko and Ck used for PCR and the Jk2 primer used as a probe to detect the ko product are shown schematically. (B) RT-PCR analysis of the expression level of ko in the Dnmtcond1 line under different conditions. Lanes 1–3 are unsorted cells either untreated (lane 1), or treated with LPS (lane 2) or HC toxin (lane 3) for 24 h. Lanes 4–7 are derived from cells 5 days postinfection. The cells were either infected with MiGfp (G) or MCiGfp (CG). Lanes 4 and 5 are unsorted infected cells, and lanes 6 and 7 are Gfp+ sorted cells. Lane 8 is the product of an RT-PCR with no RNA template. Lane 9 is the product of a PCR assay with no DNA template. (C) RT-PCR analysis of expression of ko in the Dnmtcond2 line under different conditions. Lanes 1–3 are unsorted cells either untreated (lane 1), or treated with LPS (lane 2) or HC toxin (lane 3) for 24 h. Lanes 4 and 5 are derived from cells sorted for Gfp expression 5 days postinfection either infected with MiGfp (G) or MCiGfp (CG). Lane 6 is the product of a PCR assay with no DNA template.
In this assay the two independent Abelson lines behaved differently. Loss of methylation of Dnmtcond1 cells did not result in an increase in ko expression. Gfp+ cells infected with MCiGfp had a similar level of ko compared with cells that were infected with MiGfp at day 5 postinfection. In contrast, at day 5 postinfection Dnmtcond2 cells that were infected with MCiGfp and were Gfp+ expressed significantly more k0 as compared with Gfp+ cells infected with MiGfp. Therefore, in Dnmtcond2 cells the loss of methylation results in the activation of k0 expression, suggesting that the loss of methylation can activate transcription of the kappa locus under some conditions. In addition, hypomethylation is not a prerequisite for transcription because LPS-treated cultures that induce transcription of k0 have no detectable change in the methylation status of the kappa locus (Fig. 3B, compare lanes 9 and 10).
Recombination of the Kappa Locus.
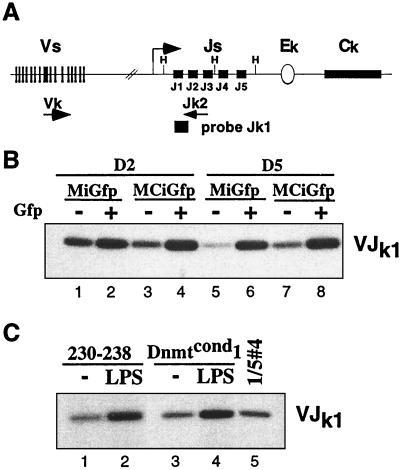
Once we generated Abelson lines that had demethylated their kappa locus, we wanted to determine the effect on the recombination efficiency of the kappa locus. We used a PCR-Southern assay that specifically amplifies the rearranged alleles by using primers Jk2 and Vk (Fig. 5A; ref. 27). Our Abelson lines have a similarly low level of recombination at the kappa locus as other previously assayed Abelson lines such as line 230–238 (Fig. 5C, compare lanes 1 and 5). The level of recombination has been estimated at 0.01%–0.001% and hypermethylation of the locus is thought to be responsible for this repression. Interestingly, we found that the Gfp+ cells infected with both the experimental and control retrovirus had an increased level of recombination as compared with their Gfp− counterparts (Fig. 5B, compare lanes 1 and 2). Nevertheless, when we compared Gfp+ cells that were either infected with MiGfp or MCiGfp, either at 2 days or 4 or 5 days postinfection, we could detect no difference in the efficiency of recombination at the kappa locus (Fig. 5B, compare lanes 2 and 4 and lanes 6 and 8). This demonstrates that demethylation of the kappa locus is not sufficient to activate recombination.
Figure 5.
V(D)J recombination of the kappa locus is unaffected by loss of methylation. (A) Schematic diagram of the murine kappa locus. The degenerate primer Vk is homologous to many members of the Vk gene segment families and is used with the primer Jk2. These primers can only amplify a product once V(D)J recombination has brought the gene segments into close proximity. The product is detected by Southern blot analysis of the PCR products probed with the primer Jk1. (B) PCR-Southern analysis of genomic DNA from Dnmtcond2 infected with either MiGfp or MCiGfp and sorted at the labeled time points postinfection. The cells were sorted for both Gfp+ and Gfp− populations and assayed as described in A. (C) PCR-Southern blot of genomic DNA from unsorted Dnmtcond1 or the Abelson line 230–238 previously demonstrated to respond to LPS treatment. The cells were either untreated (lanes 1 and 3) or treated with LPS for 24 h (lanes 2 and lane 4). Lane 5 is a 5-fold dilution of the genomic DNA in lane 4 and amplified in parallel.
To verify that the V(D)J recombination of the kappa locus of the Dnmtcond Abelson lines was inducible under previously described conditions we treated them with LPS. Indeed, we found that both Dnmtcond lines had an increased level of recombination 24 h post-LPS treatment (Fig. 5C, compare lanes 3 and 4; data not shown) that was similar to the level of induction measured in the 230–238 Abelson line (Fig. 5C, lanes 1 and 2; ref. 27). We also treated the infected cultures with LPS at day 5 postinfection and found that we were able to increase the level of recombination to similar extents whether the cells were infected with MiGfp or MCiGfp (data not shown). This finding demonstrates that the demethylated cells were recombinationally active and activatable given the appropriate stimulus.
In addition, we verified that the cells expressed the V(D)J recombinase, Rag-1 and Rag-2, using the same cDNA as generated for the ko transcription assay. RT-PCR demonstrated that the expression levels of both genes were unaffected by retroviral infection with either MiGfp or MCiGfp at all time points (data not shown).
Discussion
The regulation of V(D)J recombination must be very complex to ensure the functional assembly of only one antigen receptor gene per cell and to avoid unnecessary double-stranded breaks in the process. To accomplish this, V(D)J recombination is controlled at many levels. Changes in the accessibility of target sites mediated by demethylation has been long thought to be one of the major strategies used to regulate the specificity of the recombination process. Two opposing hypotheses can account for the correlation between the activation of transcription and demethylation with recombinational activity. The first is that there is a causal relationship between recombination and transcription/hypomethylation. In this case, changes in methylation and/or transcription would directly alter the chromatin structure of target sites, thus allowing the recombinase access to previously silent loci. An alternate hypothesis is that specific changes in chromatin, mediated by trans-acting factors, allow the recombination machinery access to target sites. This change also may be permissive for transcription and/or demethylation, resulting in the simultaneous but coincidental change in these properties. Our results support the second model, which posits that specific factors are required to change the chromatin structure of target sites, allowing the recombinase access to the DNA.
Using our assay system we determined whether a hypermethylated and transcriptionally inactive kappa locus can become accessible simply by demethylating the locus. We reasoned that if DNA methylation was the rate-limiting step controlling accessibility and specificity of the recombination process, then the loss of methylation should allow the recombination machinery access to those sequences, and an increase in recombination at the kappa locus should be detectable. We found that the loss of methylation at the kappa locus did not significantly affect the efficiency of V(D)J recombination. In contrast, we found that hypomethylation activated expression of the kappa locus in one line, Dnmtcond2. This transcriptional activation was comparable to LPS-mediated activation at the kappa locus and demonstrates that hypermethylation is at least in part responsible for transcriptional repression. The finding that LPS can activate transcription without generally affecting the methylation status suggests that multiple pathways exist to activate the ko promoter. Distinct mechanistic pathways to activate the ko promoter have been demonstrated by the differential requirements for NF-κB in LPS- and interferon-mediated induction of the promoter in Abelson lines (27).
In contrast to demethylation, LPS treatment activated recombination of the kappa locus in these lines (27). These two findings at first seem contradictory: both LPS and hypomethylation can lead to transcription of the locus, yet, only LPS treatment activates recombination. These data can be explained by the requirement for specific factors to alter the accessibility of the target sites for the recombination machinery, at least one of which can be induced by LPS, resulting in these properties being regulated independently. Therefore, demethylation of the kappa locus is not the regulatory mechanism used by the cell to control the specificity of locus choice. Moreover, that LPS activates recombination in the absence of detectable demethylation suggests that demethylation may be irrelevant to the recombination process. Nevertheless, other processes must exist to control target site selection for the V(D)J recombinase.
Previous studies using exogenous substrates and genetically modified endogenous loci largely demonstrated a direct relationship between transcription, hypomethylation, and recombination, leading to the suggestion that changes in these properties were required for recombination. However, these studies were unable to directly modulate transcription or methylation but instead relied on alterations of the substrate and subsequent analysis of transcription, methylation, and recombination. Some substrates did have discordant effects on these properties; whereas some substrates were transcriptionally active but recombinationally silent (34, 35), others were transcriptionally silent but recombinationally active (36). Additionally, a genetically modified heavy chain locus was developmentally demethylated but recombinationally silent (37). Therefore, transcription and demethylation per se are not simply permissive for rearrangement, but specific enhancer sequences and cognate binding factors must be critical. This supports our findings at the kappa locus. Moreover, recent studies using an episomal recombination substrate demonstrated that the repression of recombination by methylation could be overcome, at least in part, by changes in chromatin structure, suggesting a role for specific factors in remodeling chromatin for recombination (37).
Because demethylation was not sufficient for the activation of recombination at the kappa locus, other factors, acting independently of methylation, must exist to alter the accessibility of the kappa locus for recombination. Recent data suggests that nucleosomal remodeling may be the critical change required to allow the V(D)J recombinase access to target sites (15, 38, 39). The role of demethylation may be to alter the chromatin structure of the kappa locus, providing a permanent change in the accessibility of the locus for transcription. This locus must remain transcriptionally active for the lifespan of the B cell because loss of expression in a mature lymphocyte leads to apoptosis (40). Methylation also may serve a negative role by repressing the basal level of recombination throughout the loci and cryptic genomic sites. Previous studies have demonstrated that methylated recombination substrates are less recombinationally active (15–17). Further studies are required to isolate the factors that confer specificity to the recombination process and to determine the mechanism by which they alter the accessibility of loci for recombination.
Acknowledgments
We thank members of the Jaenisch and Baltimore labs for advice and discussions. We are grateful to Dr. Luk van Parijs for advice on retroviral infections; Dr. Christopher Roman for assistance in generating Abelson lines and for discussions and comments on the manuscript; and Glen Paradis for fluorescence-activated cell sorting and technical assistance. D.B. is supported by a National Institutes of Health grant.
Abbreviations
- LPS
lipopolysaccharide
- RT-PCR
reverse transcription–PCR
- MSCV LTR
mouse stem cell virus long terminal repeat
- Gfp
green fluorescent protein
- IRES
internal ribosomal entry site
- ES
embryonic stem
- IAP
intracisternal A-type particle
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.150218497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.150218497
References
- 1.Lewis S M. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 2.Schatz D G, Oettinger M A, Schlissel M S. Annu Rev Immunol. 1992;10:359–383. doi: 10.1146/annurev.iy.10.040192.002043. [DOI] [PubMed] [Google Scholar]
- 3.Willerford D M, Swat W, Alt F W. Curr Opin Genet Dev. 1996;6:603–609. doi: 10.1016/s0959-437x(96)80090-6. [DOI] [PubMed] [Google Scholar]
- 4.Alt F, Rosenberg N, Lewis S, Thomas E, Baltimore D. Cell. 1981;27:381–390. doi: 10.1016/0092-8674(81)90421-9. [DOI] [PubMed] [Google Scholar]
- 5.Hardy R R, Carmack C E, Shinton S A, Kemp J D, Hayakawa K. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajewsky K. Nature (London) 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 7.Yancopoulos G D, Alt F W. Cell. 1985;40:271–281. [PubMed] [Google Scholar]
- 8.Yancopoulos G D, Blackwell T K, Suh H, Hood L, Alt F W. Cell. 1986;44:251–259. doi: 10.1016/0092-8674(86)90759-2. [DOI] [PubMed] [Google Scholar]
- 9.Alt F W, Blackwell T K, Yancopoulos G D. Science. 1987;238:1079–1087. doi: 10.1126/science.3317825. [DOI] [PubMed] [Google Scholar]
- 10.Yeivin A, Razin A. In: DNA Methylation: Molecular Biology and Biological Significance. Jost J P, Saluz H P, editors. Basel: Birkhauser; 1993. pp. 523–568. [Google Scholar]
- 11.Mather E L, Perry R P. Proc Natl Acad Sci USA. 1983;80:4689–4693. doi: 10.1073/pnas.80.15.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Storb U, Arp B. Proc Natl Acad Sci USA. 1983;80:6642–6646. doi: 10.1073/pnas.80.21.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelley D E, Pollok B A, Atchison M L, Perry R P. Mol Cell Biol. 1988;8:930–937. doi: 10.1128/mcb.8.2.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodhardt M, Cavelier P, Doyen N, Kallenbach S, Babinet C, Rougeon F. Eur J Immunol. 1993;23:1789–1795. doi: 10.1002/eji.1830230809. [DOI] [PubMed] [Google Scholar]
- 15.Cherry S R, Baltimore D. Proc Natl Acad Sci USA. 1999;96:10788–10793. doi: 10.1073/pnas.96.19.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engler P, Haasch D, Pinkert C A, Doglio L, Glymour M, Brinster R, Storb U. Cell. 1991;65:939–947. doi: 10.1016/0092-8674(91)90546-b. [DOI] [PubMed] [Google Scholar]
- 17.Hsieh C-L, Lieber M R. EMBO J. 1992;11:315–325. doi: 10.1002/j.1460-2075.1992.tb05054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenberg N, Baltimore D, Scher C D. Proc Natl Acad Sci USA. 1975;72:1932–1936. doi: 10.1073/pnas.72.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott M L, Van Etten R A, Daley G Q, Baltimore D. Proc Natl Acad Sci USA. 1991;88:6506–6510. doi: 10.1073/pnas.88.15.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacob J, Baltimore D. Nature (London) 1999;399:593–597. doi: 10.1038/21208. [DOI] [PubMed] [Google Scholar]
- 22.Refaeli Y, Van Parijs L, London C A, Tschopp J, Abbas A K. Immunity. 1998;8:615–623. doi: 10.1016/s1074-7613(00)80566-x. [DOI] [PubMed] [Google Scholar]
- 23.Burt D W, Reith A D, Brammar W J. Nucleic Acids Res. 1984;12:8579–8593. doi: 10.1093/nar/12.22.8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin D J, van Ness B G. Mol Cell Biol. 1990;10:1950–1958. doi: 10.1128/mcb.10.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ten Boekel E, Melchers F, Rolink A. Int Immunol. 1995;7:1013–1019. doi: 10.1093/intimm/7.6.1013. [DOI] [PubMed] [Google Scholar]
- 26.Grawunder U, Rolink A, Melchers F. Int Immunol. 1995;7:1915–1925. doi: 10.1093/intimm/7.12.1915. [DOI] [PubMed] [Google Scholar]
- 27.Schlissel M S, Baltimore D. Cell. 1989;58:1001–1007. doi: 10.1016/0092-8674(89)90951-3. [DOI] [PubMed] [Google Scholar]
- 28.Li E, Bestor T H, Jaenisch R. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 29.Naviaux R K, Costanzi E, Haas M, Verma I M. J Virol. 1996;70:5701–5705. doi: 10.1128/jvi.70.8.5701-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mostoslavsky R, Singh N, Kirillov A, Pelanda R, Cedar H, Chess A, Bergman Y. Genes Dev. 1998;12:1801–1811. doi: 10.1101/gad.12.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walsh C P, Bestor T H. Genes Dev. 1999;13:26–34. doi: 10.1101/gad.13.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Ness B G, Weigert M, Coleclough C, Mather E L, Kelley D E, Perry R P. Cell. 1981;27:593–602. doi: 10.1016/0092-8674(81)90401-3. [DOI] [PubMed] [Google Scholar]
- 33.Blackwell T K, Moore M W, Yancopoulos G D, Suh H, Lutzker S, Selsing E, Alt F W. Nature (London) 1986;324:585–589. doi: 10.1038/324585a0. [DOI] [PubMed] [Google Scholar]
- 34.Capone M, Watrin F, Fernex C, Horvat B, Krippl B, Wu L, Scollay R, Ferrier P. EMBO J. 1993;12:4335–4346. doi: 10.1002/j.1460-2075.1993.tb06118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okada A, Mendelsohn M, Alt F. J Exp Med. 1994;180:261–272. doi: 10.1084/jem.180.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kallenbach S, Babinet C, Pournin S, Cavelier P, Goodhardt M, Rougeon F. Eur J Immunol. 1993;23:1917–1921. doi: 10.1002/eji.1830230828. [DOI] [PubMed] [Google Scholar]
- 37.Chen J, Young F, Bottaro A, Stewart V, Smith R K, Alt F W. EMBO J. 1993;12:4635–4645. doi: 10.1002/j.1460-2075.1993.tb06152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Golding A, Chandler S, Ballestar E, Wolffe A P, Schlissel M S. EMBO J. 1999;18:3712–3723. doi: 10.1093/emboj/18.13.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwon J, Imbalzano A N, Matthews A, Oettinger M A. Mol Cell. 1998;2:829–839. doi: 10.1016/s1097-2765(00)80297-x. [DOI] [PubMed] [Google Scholar]
- 40.Lam K P, Kuhn R, Rajewsky K. Cell. 1997;90:1073–1083. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]