Abstract
Ghrelin is a hormone that influences many physiological processes and behaviors, such as food intake, insulin and growth hormone release, and a coordinated response to chronic stress. However, little is known about the molecular pathways governing ghrelin release and ghrelin cell function. To better study ghrelin cell physiology, we have generated several transgenic mouse lines expressing humanized Renilla reniformis green fluorescent protein (hrGFP) under the control of the mouse ghrelin promoter. hrGFP expression was especially abundant in the gastric oxyntic mucosa, in a pattern mirroring that of ghrelin immunoreactivity and ghrelin mRNA. hrGFP expression also was observed in the duodenum, but not in the brain, pancreatic islet, or testis. In addition, we used fluorescent activated cell sorting (FACS) to collect and partially characterize highly enriched populations of gastric ghrelin cells. We suggest that these novel ghrelin-hrGFP transgenic mice will serve as useful tools to better understand ghrelin cell physiology.
Keywords: ghrelin, green fluorescence protein, transgenic, FACS
Introduction
The peptide hormone ghrelin was first described in 1999 following its isolation from stomach extracts [1]. Mature ghrelin is unique in that it is the only known naturally-occurring peptide to be post-translationally modified by O-acylation with octanoate, a reaction catalyzed by the recently identified ghrelin O-acyl transferase [1-3]. Ghrelin was originally identified as a potent stimulator of growth hormone secretion, although its roles as a mediator of body weight and blood glucose homeostasis have gained more attention [1, 4, 5]. Anti-depressant-like and anxiolytic-like actions, in addition to effects on gastric acid secretion, gastrointestinal motility, and memory make ghrelin a truly pleiotropic hormone, as well as an interesting, yet complicated drug target [6-10].
Several studies have confirmed that the source of most circulating ghrelin is a distinct group of endocrine cells located within the gastric oxyntic mucosa [11]. These gastric ghrelin cells are characterized by round, compact, electron-dense secretory granules [P/D(1)-type in humans, A-like-type in rodents, and X-type in dogs] that distinguish them electron-microscopically from other previously-characterized gastric endocrine cell types [11-14]. Within the stomach, ghrelin cells are of the round, closed-type variety that do not have direct contact with gastric luminal contents, and tend to cluster towards the base of the gastric mucosal glands [14, 15]. There is also good histochemical and RT-PCR evidence for ghrelin cells in much fewer numbers throughout the remainder of the gastrointestinal tract, where they exist not only as closed-type cells, but also as the more elongated, opened-type cells which do have direct contact with the intestinal lumen and thus may be regulated differently than their gastric counterparts [14, 15]. Ghrelin production also has been localized to the pancreatic islets of Langerhans of several species (where it seems particularly evident in the developing fetus), although different groups have localized ghrelin either to a distinct endocrine cell type (epsilon cell) that is devoid of the classical islet hormones or to a subset of glucagon-producing alpha cells or insulin-producing beta cells [16-20]. Ghrelin expression within the brain also has been described by several groups using RT-PCR [21], although inconsistencies exist regarding the exact localization of these brain ghrelin cells. For instance, ghrelin-immunoreactivity has been differentially localized to either the arcuate nucleus [1, 22, 23] or to a group of cells that line the 3rd ventricle and comprise portions of the capsules of the ventromedial hypothalamic nucleus and paraventricular hypothalamic nucleus [24]. Yet another study, found persistence of ghrelin-immunoreactivity in the hypothalami of ghrelin-deficient mice, and found no galactosidase-positive hypothalamic cells within ghrelin-deficient mice, as one would expect in that model in which the ghrelin coding region was replaced by the lacZ reporter gene [25]. Ghrelin expression also has been found in the testis using immunohistochemistry, and a wide variety of other tissues by RT-PCR [26, 27].
Despite an accumulating literature characterizing ghrelin action as well as a growing literature on the distribution of ghrelin cells, relatively little is known about the exact molecular pathways responsible for the biosynthesis and release of ghrelin. For instance, although we know that ghrelin levels rise before set meals and in response to acute or chronic stress, the identities of direct ghrelin secretagogues have remained elusive [6, 28-30]. Perhaps, the most informative secretagogue study to date includes a microdialysis study in which candidate compounds were infused via microdialysis probes implanted into the gastric submucosa and assessed for their ability to stimulate total ghrelin mobilization [31]. Using this method, epinephrine, norepinephrine, endothelin and secretin were found to stimulate ghrelin release while numerous other neurotransmitters, hormones and neuropeptides were shown to be ineffective. However, this microdialysis technique and others, such as an ex vivo organ culture system [32, 33], do not discriminate between direct or indirect effects of putative ghrelin secretagogues.
In order to better study the physiologic characteristics of ghrelin cells, we have generated and validated new transgenic reporter mouse lines in which humanized Renilla reniformis green fluorescent protein marks the location of ghrelin-producing cells. Furthermore, we have taken advantage of this hrGFP reporter in fluorescence activated cell sorting experiments to isolate and further characterize enriched pools of gastric ghrelin cells.
Materials and Methods
Generation of ghrelin-hrGFP BAC transgenic mice
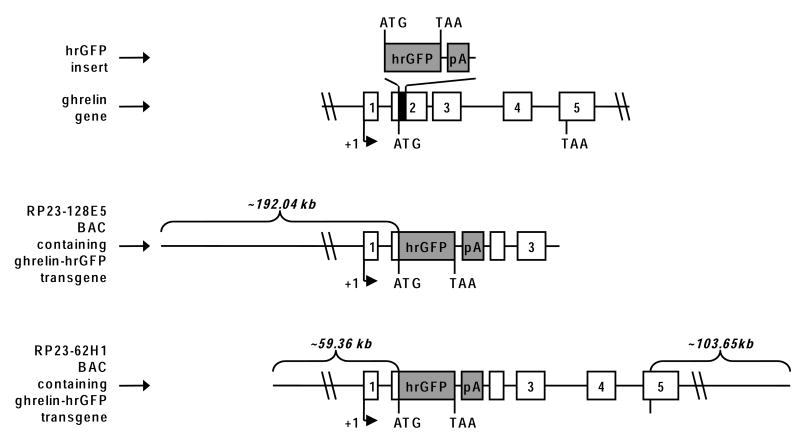
We generated several lines of transgenic mice that express humanized Renilla reniformis GFP (hrGFP) eutopically within ghrelin cells. These animals were made through the use of various ET-cloning “recombineering” technologies [34, 35]. We constructed two different ghrelin-hrGFP transgene-containing bacterial artificial chromosomes (BACs). The original ghrelin BACs were purchased from BACPAC Resources Center at Children's Hospital Oakland Research Institute and were chosen based on sequence data obtained from the Celera database. The first BAC (RP23-128E5) contained nearly 192.04 kb sequence upstream of the ghr start codon and nearly the entire coding sequence of ghrelin, ending ∼1.17 kb upstream of the ghr stop codon. The second BAC (RP23-62H1) spanned the entire coding region of ghr and contained nearly 59.36 kb sequence upstream of the start codon and ∼103.65 kb downstream of the stop codon. These BACs then were transformed into EL250 cells by electroporation. EL250 cells were provided by N. Copeland; they contain heat-inducible recE and recT recombinases, for homologous recombination, and arabinose-inducible Flp-recombinase, for site-specific recombination at frt sites [35]. Next, a fragment containing the coding sequence of hrGFP followed by an SV40 polyadenylation signal (derived from the phrGFP-1 vector, Stratagene, La Jolla) and a kanamycin resistance gene flanked by frt sites was inserted into the ghrelin BACs, at the translational start site of ghrelin, by ET-cloning. This insertion resulted in the removal of the first 29 bp of the ghrelin coding sequence. Finally, the kanamycin resistance gene was removed by arabinose induction of Flp-recombinase, and the hrGFP was sequenced to ensure that no mutations had been introduced. The hrGFP-modified ghrelin BACs were submitted to the Beth Israel Deaconess Medical Center Transgenic Facility (Boston) and the UTSW Medical Center Transgenic Core Facility for microinjection into pronuclei of fertilized one-cell stage embryos of FVB or C57Bl6/J mice. We were successful in generating 18 different potential ghrelin-hrGFP founder mice, of which seven resulted in lines with the expected hrGFP expression within the gastric mucosa. These included two lines using the RP23-128E5 BAC and five lines using the RP23-62H1 BAC. Each of the seven lines had different amounts of gastric hrGFP expression that was apparently independent of the BAC used, and none of these seven lines contained hrGFP expression within the brain. We chose to perform more detailed analyses on one line derived from each of the two BACs, using abundance of expression within the gastric mucosa as the main criterion for our choice. The line denoted as “R4” was derived from RP23-128E5 and the line denoted as “hrGFP10” was derived from RP23-62H1. The R4 mice used in this study were on a pure FVB genetic background, whereas the hrGFP10 mice used were on a pure C57Bl6/J genetic background.
Animals were housed under 12 hours of light/12 hours of dark per day in a temperature-controlled environment. They were fed standard chow diet (8664 F6 Rodent Diet; Harlan Teklad) and had free access to water. All animals and procedures used were in accordance with the guidelines and approval of the Beth Israel Deaconess Medical Center and UTSW Medical Center Institutional Animal Care and Use Committees.
Tissue preparation
For histological experiments, mice were deeply anesthetized with intraperitoneal injection of chloral hydrate (500mg/kg) and subsequently perfused transcardially with diethylpyrocarbonate (DEPC)-treated 0.9% saline followed by 10% neutral buffered formalin. Adult stomachs, duodenums, brains and several other organs were removed, stored in the same fixative for 4-6 hours at 4°C, immersed in 20% sucrose in DEPC-treated phosphate buffered saline (PBS), pH 7.0 at 4°C overnight, and embedded in Tissue-Tek OCT compound (Sakura Finetechnical Co., Ltd., Tokyo). Fetal pancreata (embryonic day 18.5; e18.5) were processed in the same manner. Cryosections of these peripheral organs were prepared at a 10 μm thickness using a cryostat and mounted onto SuperFrost slides (Fisher Scientific, Pittsburgh, PA). Brains were sectioned coronally, using a sliding microtome, into five equal series at a 25 μm thickness, and these sections were then mounted onto SuperFrost slides. All slides were air-dried and stored in desiccated boxes at -20°C until use. Fluorescence microscopy for endogenous hrGFP fluorescence was performed by mounting the sections in Fluoromount G (Electron Microscopy Sciences, Hatfield, PA) and then viewing them using microscopy (Zeiss, Axioskop 2).
Immunohistochemistry for ghrelin
Slides with tissue sections were washed in PBS three times, pretreated with 0.3% hydrogen peroxide in PBS for 30 minutes at room temperature and then were incubated in 3% normal donkey serum (Equitech-Bio, Kerrville, TX) with 0.25% Triton X-100 in PBS (PBT-azide) for 1 hour. Next, the slides were incubated overnight at room temperature in anti-ghrelin antiserum (Phoenix Pharmaceuticals, Belmont, CA; Code H-031-31, Lot #R451-2; 1:10,000 in PBT). Anti-ghrelin antiserum (Santa Cruz Biotechnology, Santa Cruz, CA; Code SC-10368; Lot C2503; 1:5,000 in PBT) was also used for certain analyses, with similar results. After washing in PBS, sections were incubated in Alexa Fluor 594 donkey anti-rabbit IgG (Invitrogen, Carlsbad, CA; 1:300) for 1 hour at room temperature, followed by more PBS washes. Finally, the sections were mounted in Fluoromount G (Electron Microscopy Sciences, Hatfield, PA) and viewed using microscopy (Axioskop 2, Carl Zeiss). Ghrelin-immunoreactivity within fetal pancreas was visualized using diaminobenzidine tetrahydrochloride (DAB; Sigma, St. Louis, MO), instead of with Alexa Fluor 594, as below. hrGFP fluorescence in the same sections was visualized and recorded either before (duodenum) or after (stomach) processing the sections for ghrelin-immunoreactivity. We also performed dual-label studies on representative stomach sections with DAB to visualize ghrelin-immunoreactivity on the same slides as the hrGFP endogenous fluorescence (data not shown). This alternate method of visualizing both hrGFP and ghrelin on the same sections gave similar results as when we used the red fluorophor to visualize ghrelin. Control experiments to test the specificity of the immunohistochemistry reactions included pre-adsorption of the anti-ghrelin antiserum with 50 μg/mL murine acylated ghrelin (catalog #C-et-004; Global Peptide, Fort Collins, CO; suspended in saline), and also separate reactions in which we omitted each primary antiserum. Staining was not observed in any of the pre-adsorption or primary antibody omission controls.
Immunohistochemistry for hrGFP
Sections were washed, pretreated, and incubated in normal donkey serum with Triton X-100 as described above. Next, the slides were incubated overnight at room temperature in anti-hrGFP primary antiserum (Stratagene, La Jolla, CA; Code 240 142-51, Lot #0830269; 1:10,000 in PBT). After washing in PBS, sections were incubated in biotinylated donkey antirabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, PA; 1:1,000) for 1 hour at room temperature, followed by incubation for 1 hour in a solution of avidin-biotin complex (Vectastain Elite ABC Kit, Vector Laboratories, Burlingame, CA; 1:500) diluted in PBS. The sections were next washed in PBS and incubated in a solution of 0.04% DAB and 0.01% hydrogen peroxide in PBS. After sections were washed and dried, they were dehydrated in graded ethanols, cleared in xylenes, and coverslipped with Permaslip (Alban Scientific, St. Louis, MO). Anti-hrGFP antibody was not observed to bind to any stomach cells within wild-type mice (data not shown).
Generation of ghrelin cRNA probe
The mouse ghrelin cRNA probe was generated by PCR amplification of mouse genomic (tail) DNA with the following oligonucleotides primer pairs: M296, 5′-ACAAGTAACCACGGACAGGCCTGA-3′ and M297, 5′-ATTTATTCTTAAAGTGGTAGGAGAGT-3′. The amplified 116-bp PCR product, which encompasses the last 5-bp of coding sequence, the stop codon and the first 108-bp of 3′-untranslated region, was gel-purified and then subcloned into PCR4-TOPO vector (Invitrogen), according to the manufacturer's protocol. The sequence and directionality of the insert was confirmed by DNA sequencing at the core DNA Sequencing Facility at Beth Israel Deaconess Medical Center. To generate antisense and control sense 35S-labeled cRNA to use as probes, the plasmid was linearized by restriction digestion and then subjected to in vitro transcription with either T3 or T7 RNA polymerases according to the manufacturer's protocol (Ambion, Austin, TX).
Dual-label in situ hybridization histochemistry and immunohistochemistry
In situ hybridization histochemistry was performed as reported previously [36]. Prior to hybridization, sections were fixed in 4% formaldehyde in DEPC-treated PBS, pH7.0, for 20 minutes at 4°C, dehydrated in increasing concentrations of ethanol, cleared in xylenes for 15 minutes, rehydrated in decreasing concentrations of ethanol, and placed in pre-warmed sodium citrate buffer (95-100°C, pH 6.0). While in the sodium citrate buffer, slides were placed in a commercial microwave oven for 10 minutes at 20-70% power. Afterwards, they were dehydrated, as before, in graded ethanols and air-dried. The 35S-labeled cRNA probe was diluted to 106 cpm/mL in a hybridization solution containing 50% formamide, 10mM Tris-HCl, pH7.5, 5.0 mg tRNA (Invitrogen), 100 mM dithiothreitol, 10% dextran sulfate, 0.6M NaCl, 0.5 mM EDTA, pH 8.0, and 1× Denhardt's solution. The slides with stomach sections had hybridization solution and coverslips applied and then were placed in a 57°C incubator for 12-16 hours. Next, coverslips were removed and the sections were washed with 2× SSC buffer and incubated in 0.002% RNase A (Roche Molecular Biochemicals, Indianapolis, IN) with 0.5 M NaCl, 40 mM Tris-HCl, pH 8.0, and 0.1 mM EDTA for 30 minutes, followed by a 30 minute incubation in the same buffer minus RNase. The sections were then submitted to stringency washes as follows: 2× SSC at 50°C for 1 hour; 0.2× SSC at 55°C for 1 hour; 0.2× SSC at 60°C for 1 hour. Afterwards, the sections were dehydrated in increasing concentrations of ethanol containing 0.3 M NH4OAc followed by a final submersion in 100% ethanol. Immediately following this procedure, the slides were processed for hrGFP-immunoreactivity using the protocol above, up until and including the DAB + hydrogen peroxide step. Afterwards, sections were washed and dried, then placed in X-ray film cassettes with BMR-2 film (Kodak, Rochester, NY) for 1 day. Slides were then dipped in NTB autoradiographic emulsion (Kodak), dried, and stored in desiccated, foil-wrapped boxes at 4°C for 1 day. Finally, slides were developed with Dektol developer (Kodak), dehydrated in graded ethanols, cleared in xylenes, and coverslipped with Permaslip (Alban Scientific). Our demonstration of similar results using the two different types of dual-label studies (hrGFP endogenous fluorescence/ghrelin-immunoreactivity and hrGFP-immunoreactivity/ghrelin ISHH) speaks to the reproducibility of our data and the reliability of our methods.
Determination of plasma ghrelin levels
Fresh blood from mice with ad lib access to regular chow and from mice that had been fasted for 24 hours was collected from tails into tubes containing EDTA, between 10:00-11:00, on ice. The protease inhibitor p-hydroxymercuribenzoic acid (#12425, Sigma-Aldrich) was added to each sample to achieve a final concentration of 1mM. The samples were centrifuged and the resulting plasma was immediately treated with one-tenth volume of 1 N HCl, so as to inhibit the degradation of acylated ghrelin, and then stored at -80°C until use. Acylated ghrelin levels were determined using ghrelin (rat acylated) EIA kits according to the manufacturer's instructions (#10006307, Cayman Chemical Company, Ann Arbor, MI).
Isolation of stomach mucosal cells and fluorescence activated cell sorting (FACS)
Isolated stomach mucosal cells were prepared by the enzymatic dispersion method as previously reported [37]. Briefly, mice were sacrificed under anesthesia, and the stomachs were quickly removed and turned inside out. The stomachs were inflated and incubated in 2.4 U /mL Dispase II solution (Roche Diagnostics) for 2 hours. Next, a polyethylene transfer pipette (Fisher Scientific) filled with some of the Dispase II solution was used to aid in the mechanical release of the stomach mucosal cells from the remainder of the stomach. These released cells were then passed through a 100 μm filter and collected by centrifugation at 1500 rpm for 5 minutes. The cellular pellet was further digested in 0.25% trypsin-EDTA (Invitrogen) at 37°C for 5 minutes. After washing in PBS, cells were resuspended in FACS buffer (3% fetal bovine serum, 0.5 mM EDTA, 0.1% BSA, 10 U/mL DNaseI, 20 mg/mL glucose in Hank's Buffered Salt Solution). The cells were then analyzed and sorted with a cell sorter (MoFlo, Dakocytomation, USA) at the UTSW Medical Center Flow Cytometry Multi-user Core Facility. Cells were sorted based on size, complexity and intensity of GFP fluorescence (at 530 nm) and fluorescence at 585 nm. Sorted hrGFP-positive pools of cells were viewed using microscopy (IX51, OLYMPUS, Tokyo). Four independent FACS preparations (4-6 mice were used for each independent FACS preparation) were included in the subsequent analysis. For each preparation, those cells with the most intense fluorescence were collected as the hrGFP-enriched pool, whereas those with the least intense fluorescence were collected as the hrGFP-negative pool. Depending on the preparation, these hrGFP-enriched and hrGFP-negative pools corresponded to slightly different percentages of sorted, living cells, as follows: the hrGFP-enriched pool corresponded to 0.29%, 0.53%, 0.92%, and 0.44% of sorted, living cells in preparations 1, 2, 3, and 4, respectively; the hrGFP-negative pool corresponded to 84.1%, 92.91%, 91.48% and 93.25% of sorted, living cells in preparations 1,2,3 and 4, respectively.
RNA extraction and Quantitative RT-PCR
After FACS sorting, we adjusted the hrGFP-enriched pools and the hrGFP-negative pools to contain a matched number of cells (for instance, in the pools used for Table 2, we used 12,000 cells, 6,000 cells, 12,000 cells and 10,000 cells for preparations 1, 2, 3, and 4, respectively). The cells in each pool were collected by centrifugation at 4°C at 3000 rpm, for 10 minutes. Total RNA was extracted from these cellular pellets using the PicoPure RNA Isolation Kit (Arcturus Bioscience. Mountain View, CA), according to the manufacturer's protocol, and was stored at -80°C until use. Complementary DNA was synthesized from 25 ng total RNA using Superscript III reverse transcriptase (Invitrogen). The following primer pairs were designed to amplify ghrelin: 5′-GTCCTCACCACCAAGACCAT-3′; 5′-TGGCTTCTTGGATTCCTTTC-3′, chromogranin A: 5′-GCAGGCTACAAAGCGATCCA-3′; 5′-CTCTGTCTTTCCATCTCCATCCA-3′, prohormone convertase1/3: 5′-GGCACCTGGACATTGAAAATTAC-3′; 5′-TTCATGTGCTCTGGTTGAGAAGA-3′, prohormone convertase 2: 5′-CAAGCGGAACCAGCTTCA-3′; 5′-ATTCCAGGCCAACCCCA-3′, amylase 2: 5′-AATGGTTCTCCCAAGGAAGC-3′; 5′- ACACGGCCATTTCCAAAGTA-3′, H+/K+ ATPase β-subunit: 5′-CCCAGCTTCGGCTTCGA-3′; 5′-TGGAGACTGAAGGTGCCATTG-3′, gastric intrinsic factor: 5′-GAAAAGTGGATCTGTGCTACTTGCT-3′; 5′-AGACAATAAGGCCCCAGGATG-3′, pepsinogen F: 5′-GAAGTGGCTCTGGGTCCTT-3′; 5′-GGCTTTCCCGCAGGTTTT-3′, calpain 8: 5′-TTCTGCCTGAGGGTGTTCTC-3′; 5′- TCTTCTCCATCCATGTCACG-3′, histidine decarboxylase: 5′-GCGACCCTTCCTTCGAAATT-3′; 5′-CCTTTAACACACTTTCTGTGAGACAAT-3′, somatostatin: 5′-CCCAGACTCCGTCAGTTTCT-3′; 5′-GGGCATCATTCTCTGTCTGG-3′, tryptophan hydroxylase 1: 5′-CCTTGGAGCTTCAGAGGAGA-3′; 5′- CAGCTGTCCATCTTGTTTGC-3′, gastrin: 5′-TCCAGGGTCCTCAACACTTC-3′; 5′-CCAAAGTCCATCCATCCGTA-3′ and cyclophilin: 5′-TGGAGAGCACCAAGACAGACA -3′; 5′-TGCCGGAGTCGACAATGAT-3′. mRNA levels were measured with an ABI 7300 Real-Time PCR System (Applied Biosystems, Foster, City, CA). PCR was performed using the iTaq SYBR Supermix (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer's instructions. In brief, initial template denaturation was performed for 3 minutes at 95°C. The cycle profiles were programmed as follows: 15 seconds at 95°C (denaturation) and 45 seconds at 60°C (annealing and extension). Forty cycles of the profile were run and cyclophilin RNA was used for normalization.
Table 2.
Relative Expression of Various mRNAs within FACS-Separated hrGFP-Positive versus hrGFP-Negative Gastric Mucosal Cells of the hrGFP10 Line#
| hrGFP-enriched pools | hrGFP-negative pools | P value | |
|---|---|---|---|
| ghrelin | 5420.67±1097.30 | 1.36±0.24 | 0.0159* |
| chromogranin A | 76.49±16.54 | 1.86±0.73 | 0.0190* |
| prohormone convertase 1/3 | 3.84±0.99 | 0.07±0.04 | 0.0319* |
| prohormone convertase 2 | 4.48±1.21 | 0.10±0.04 | 0.0338* |
| amylase 2 | 1.64±1.26 | 0.13±0.06 | 0.2977 |
| H+/K+ ATPase β-subunit | 2.06±0.78 | 0.88±0.12 | 0.1948 |
| gastric intrinsic factor | 1.04±0.24 | 1.62±0.22 | 0.1965 |
| pepsinogen F | 0.39±0.12 | 0.52±0.21 | 0.6626 |
| calpain 8 | 0.10±0.02 | 0.07±0.02 | 0.2872 |
| histidine decarboxylase | 4.62±2.89 | 0.22±0.14 | 0.2085 |
| somatostatin | 25.70±20.07 | 5.26±2.83 | 0.3216 |
| tryptophan hydroxylase 1 | 0.12±0.05 | 0.16±0.10 | 0.7053 |
| gastrin | 14.15±3.71 | 26.97±8.34 | 0.2208 |
all values are normalized to the levels of the housekeeping gene cyclophilin
P<0.05 was considered statistically significant
Analysis of data
Tissue sections were viewed with a Zeiss Axioskop 2 microscope and photomicrographs were produced with a Zeiss digital camera attached to the microscope. To determine the degree of overlap between cells with hrGFP fluorescence and ghrelin-immunoreactivity, three different sections of gastric fundus and three different sections of duodenum, each separated by at least 100 μm along the length of each organ, were examined in each of three separate animals. Images were taken using both the green filter and the red filter at each level, and were placed side-by-side. All the cells at each level with either hrGFP fluorescence alone, ghrelin-immunoreactivity alone, or both were counted. Criteria used to determine whether an hrGFP-immunoreactive cell co-expressed ghrelin mRNA included both 1) brightfield visualization of silver granules overlying the DAB-stained cell at 5× the background density of silver granule deposition and 2) conformation of the overlying silver granules to the shape of the DAB-stained cell. An imaging editing software program, Adobe PhotoShop 7.0 (San Jose, CA) was used to adjust contrast, brightness, and color of the photomicropraphs. For analysis of the quantitative PCR data, the paired t-test was used to compare hrGFP-positive vs. hrGFP-negative values. For comparison of plasma acylated ghrelin levels in hrGFP mice vs. wild-type littermates in both the fed and fasted conditions, we used a two-way ANOVA, and there was no significant interaction between hrGFP mice and wild-type littermates. The data was then analyzed using a Student's t-test to assess the effect of food availability. P<0.05 was considered statistically significant. All of the data are expressed as mean ± SEM.
Results
Expression of hrGFP in stomach and other tissues
Transgenic mice expressing hrGFP in ghrelin cells were generated by engineering two different ghrelin bacterial artificial chromosomes (BACs) such that hrGFP is driven by ghrelin regulatory elements (Fig. 1). These ghrelin BACs each contained different amounts of genomic sequence flanking the start and stop codons of ghrelin. We examined hrGFP expression in several of the lines harboring the transgenes by both fluorescence microscopy and by use of antibodies directed against hrGFP, and chose to continue working with one line derived from each of the two BACs: R4 (derived from the RP23-128E5 ghrelin BAC) and hrGFP10 (derived from the RP23-62H1 ghrelin BAC). Fluorescence microscopy confirmed an abundant presence of hrGFP-expressing cells within the gastric oxyntic mucosa in both transgenic lines (Fig. 2A, B). Most of the hrGFP-expressing cells were found within the mucosal glandular base while far fewer hrGFP-expressing cells were observed within the glandular neck, as was expected from previously published descriptions of ghrelin distribution within the stomach [1, 15]. The total number of hrGFP-expressing cells in the stomach was larger in the hrGFP10 line than in the R4 transgenic line.
Figure 1.
Schematic diagram of the derivation of ghrelin-hrGFP mice. In order to generate the transgenes, the first 29-bp of ghrelin's coding sequence (denoted by a black rectangle) was replaced by the coding sequence of hrGFP followed by an SV40 polyadenylation signal (pA). The R4 line was derived from the RP23-128E5 ghrelin BAC, while the hrGFP10 line was derived from the RP23-62H1 ghrelin BAC. Ghrelin's and the transgenes' transcriptional start sites (+1), ghrelin's exons (denoted by numbered rectangles), and the start codons (ATG) and stop codons (TAA) for ghrelin and hrGFP are all indicated.
Figure 2.
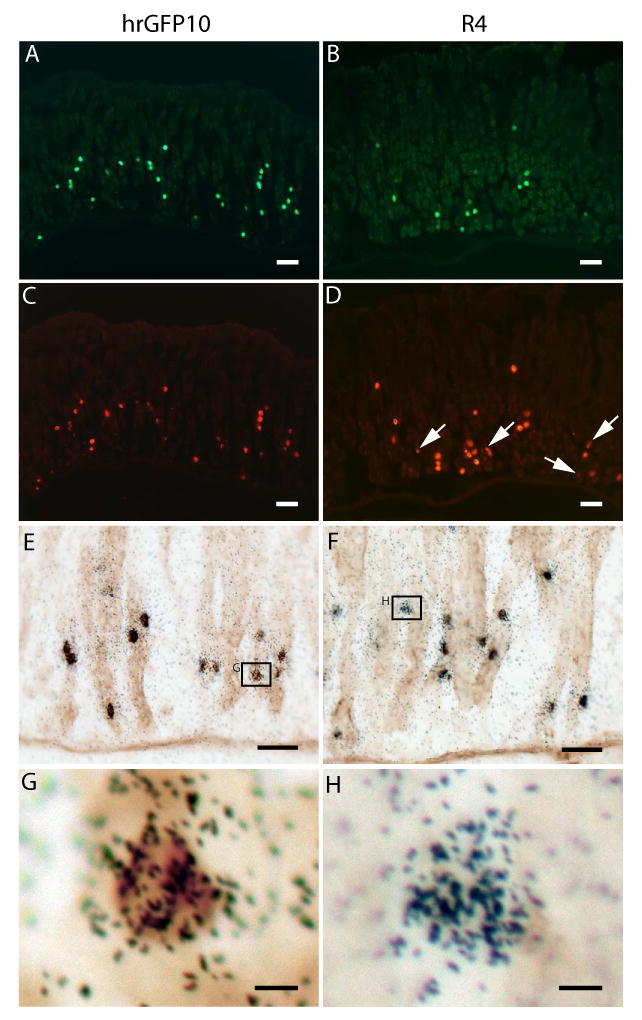
Expression of hrGFP within the gastric oxyntic mucosa of the two novel ghrelin-hrGFP transgenic mouse lines. Histochemical analyses of the hrGFP10 line appear on the left side and those of the R4 line appear on the right side. A-D) Expression of hrGFP fluorescence (A and B) within ghrelin-immunoreactive cells lining the stomach (C and D). White arrows indicate the subset of R4 ghrelin-immunoreactive cells that do not co-express hrGFP fluorescence. E-F) Expression of hrGFP (as indicated in brown) within ghrelin mRNA-containing cells (as indicated by overlying black silver granules) lining the stomach. G) High power photomicrograph of a dual-labeled cell in the hrGFP10 line. H) High power photomicrograph of one of the few ghrelin mRNA-containing gastric cells of the R4 line that does not co-express hrGFP. Scale bars = 50 μm in A-D; 20 μm in E-F; 5 μm in G-H.
To determine whether hrGFP expression was restricted to ghrelin cells, we performed several different dual-label histochemical analyses on single tissue sections. We visualized hrGFP fluorescence or hrGFP-immunoreactivity together with ghrelin-immunoreactivity or binding of ghrelin mRNA-specific riboprobe. hrGFP was visualized only within ghrelin cells in both ghrelin-hrGFP transgenic lines (Fig. 2 and Table 1). This was evidenced by both co-localization of hrGFP fluorescence with ghrelin-immunoreactivity (Fig. 2A-D) and co-localization of hrGFP-immunoreactivity with ghrelin mRNA (Fig. 2E-G). Nearly 100% of ghrelin-immunoreactive and ghrelin mRNA-containing gastric cells of the hrGFP10 line also expressed hrGFP (Fig. 2A, C, E, G and Table 1). However, only ∼66% of the gastric ghrelin cells of the R4 line co-expressed hrGFP (Fig. 2B, D, F, H and Table 1).
Table 1.
Co-expression of hrGFP and ghrelin within ghrelin-hrGFP transgenic lines
| Transgenic line | % of cells with hrGFP fluorescence co-expressing ghrelin-immunoreactivity | % of ghrelin immunoreactive cells co-expressing hrGFP fluorescence |
|---|---|---|
| hrGFP10 | 100.0 ± 0.0 | 98.6 ± 1.7 |
| R4 | 100.0 ± 0.0 | 66.1 ± 2.8 |
The data are reported as the mean percentage ± SEM for 3 animals at 3 different levels along the length of the gastric fundus for each animal, each separated by at least 100 μm.
We also detected hrGFP expression within a subset of duodenal ghrelin cells of the hrGFP10 line (Fig. 3A-B) but not of the R4 line. We were unable to detect hrGFP fluorescence or hrGFP-immunoreactivity within the brain of either the hrGFP10 line (Fig. 3C-D) or the R4 line, although we also were unable to detect ghrelin-immunoreactivity or ghrelin mRNA by in situ hybridization histochemistry within the brains of these transgenic mice or wild-type mice (data not shown). Furthermore, although we were able to detect ghrelin-immunoreactivity within hrGFP10 fetal islets of Langerhans (embryonic day 18.5; Fig. 3F), no hrGFP-immunoreactivity (Fig. 3E) or fluorescence (data not shown) was observed. We failed to observe specific ghrelin-immunoreactivity or hrGFP-immunoreactivity within the testis of either transgenic line, and detected no hrGFP signal within the kidney, pituitary, ovary, neonate pancreas, adult pancreas, esophagus, jejunum, ileum, cecum, colon or rectum of the R4 line. In order to confirm that the presence of the transgene does not interfere ghrlein cell physiology, we compared plasma acylated ghrelin levels within ghrelin-hrGFP mice to those within wild-type littermates both in the fed and fasted conditions. We found no statistically significant differences in either fed or fasted levels between the two groups of mice (hrGFP mice fed: 105.1±35.2 pg/mL, fasted: 358.1±59.7 pg/mL; wild-type littermates fed: 85.1±16.2 pg/mL, fasted: 292.9±35.4 pg/mL). Furthermore, in both groups, fasting resulted in statistically significant elevations of ghrelin levels.
Figure 3.
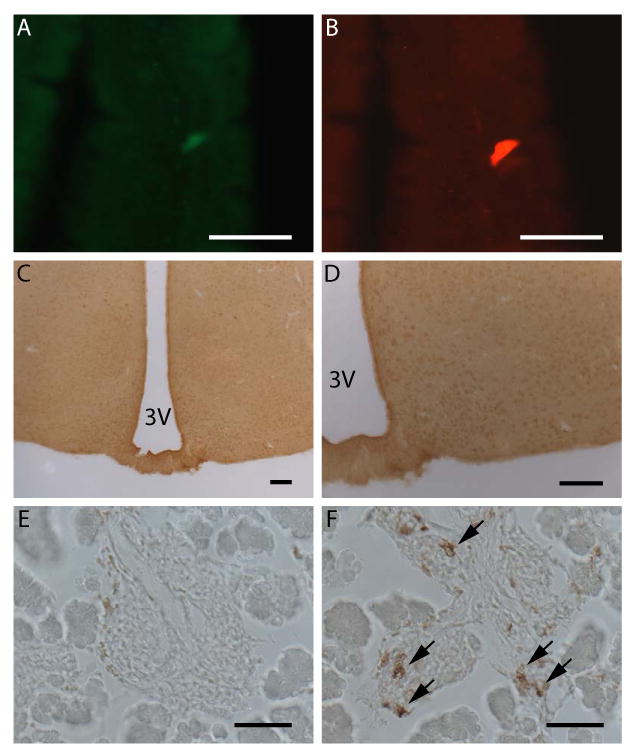
Expression of hrGFP within extra-gastric tissues. A-B) Expression of hrGFP fluorescence (A) within a ghrelin-immunoreactive cell (B) within the duodenal mucosa of the hrGFP10 line. C-D) Lack of hrGFP-immunoreactivity within the mediobasal hypothalamus of the hrGFP10 line. E-F) Lack of hrGFP-immunoreactivity within the islets of Langerhans of an e18.5 hrGFP10 mouse (E), which do contain ghrelin-immunoreactivity, as indicated by black arrows (F). Scale bars = 50 μm in A-B, E-F; 100 μm in C-D.
Fluorescence Activated Cell Sorting and Quantitative PCR
After confirming the expression of hrGFP only within ghrelin cells, we decided to take advantage of the fluorescence signal to isolate and study an enriched population of gastric ghrelin cells. Cells comprising the gastric mucosa of the hrGFP10 line were first enzymatically and mechanically dispersed and subsequently were submitted for FACS analysis to generate an enriched sample of hrGFP-positive cells (Fig. 4). This was repeated for a total of four separate preparations of gastric mucosal cells. Those cells with the most intense fluorescence were collected as the hrGFP-enriched pools, whereas those with the least intense fluorescence were collected as the hrGFP-negative pools (see Materials and Methods for details). Fluorescence microscopy of primary cultures of the isolated gastric mucosal cells prior to and following FACS analysis demonstrated this enrichment (Fig. 4B-4E).
Figure 4.
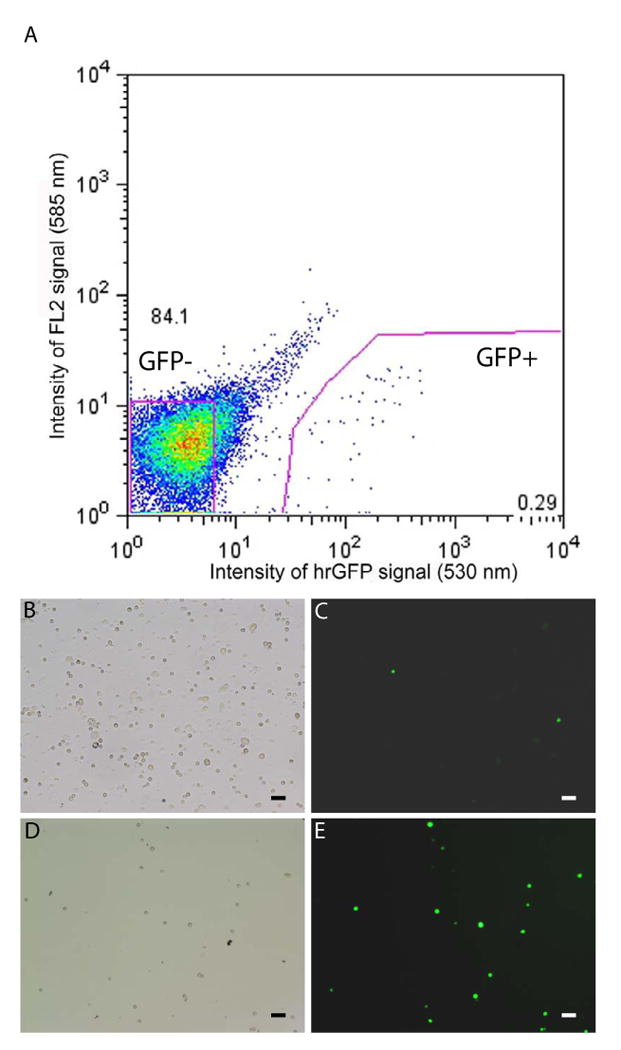
Fluorescence activated cell sorting of ghrelin-hrGFP cells. A) Graphical representation of FACS of gastric mucosal cells (preparation 1), indicating those cells collected as part of the hrGFP-enriched pool (0.29% of the total number of sorted, living cells) and those collected as part of the hrGFP-negative pool (84.1% of the total number of sorted, living cells). A-D) Photomicrographs of dispersed gastric mucosal cells prior to FACS (B-C) and after FACS (D-E), demonstrating an enrichment in the numbers of total cells (B, D) that contain hrGFP (C, E). Scale bars = 50 μm.
Characterization of Gastric Ghrelin Cells
After demonstrating our ability to isolate enriched populations of hrGFP-positive cells, we isolated mRNAs from both hrGFP-enriched pools and subsets of the remaining separated cells (the hrGFP-negative pools). These mRNAs were next submitted to quantitative RT-PCR analyses, and the mean results for the four separate sets of hrGFP-enriched vs. hrGFP-negative pools are presented in Table 2. As expected, the hrGFP transcript was only detected by RT-PCR in the hrGFP-enriched pool, and was not detected within the hrGFP-negative pool (data not shown). Ghrelin mRNA expression was several thousand times higher in the hrGFP-enriched pool than in the hrGFP-negative pool. The mRNAs for other markers of endocrine lineage – chromogranin A and prohormone convertases 1/3 and 2 – were also much higher within the hrGFP-enriched pool than within the hrGFP-negative pool. We also assayed for markers of common non-endocrine gastric mucosal cells, including amylase 2 (a marker of zymogenic/peptic/chief cells), H+/K+ ATPase β-subunit (a marker of acid-producing parietal cells), gastric intrinsic factor and pepsinogen F (markers of both zymogenic and parietal cells) and calpain-8 (a marker of mucus-secreting pit cells) [38-40]. As expected, the levels of mRNAs encoding these non-endocrine gastric mucosal cell markers were not elevated in the hrGFP-enriched pools. In addition, we assayed for markers of the other common endocrine cell types within the gastric mucosa, including histamine-containing enterochromaffin-like cells, somatostatin-containing D cells, serotonin-containing enterochromaffin cells, and gastrin-containting G cells. On average, we found no evidence of enrichment for histidine decarboxylase type (which catalyzes the rate-limiting step in the histamine biosynthetic pathway), somatostatin, tryptophan hydroxylase 1 (which catalyzes the rate-limiting step in serotonin biosynthesis) or gastrin within the hrGFP-positive pools. Similar abbreviated analyses were performed on other preparations of R4 cells, with similar results.
Discussion
In this paper, we describe the generation of new transgenic mouse lines expressing hrGFP under the regulation of elements from the mouse ghrelin promoter. Expression of the hrGFP reporter within these new ghrelin-hrGFP mouse lines was shown to occur only within cells co-expressing ghrelin, and was not observed in any non-ghrelin cells. The distribution of hrGFP-expressing ghrelin cells within the stomach and duodenum was consistent with the expression patterns previously described in the literature [1, 14, 15]. Yet, despite the fact that two different ghrelin BACs were used as the sources of the ghrelin “promoter” elements – each with amounts of chromosomal DNA ranging between 59.36 to 192.04 kb upstream of the ghrelin start codon and 0 to 103.65 kb downstream of the ghrelin stop codon -- we nonetheless achieved only minimal expression in extra-gastric gastrointestinal sites and did not yield any lines with pancreatic islet or testis hrGFP expression, both of which are fairly well-characterized sites of ghrelin expression [16-20, 26, 27]. We also failed to observe hrGFP, ghrelin-immunoreactivity or ghrelin mRNA expression within the brains of any of our ghrelin-hrGFP mice, although this was not totally unexpected given discrepancies among previous studies examining ghrelin expression in the brain [1, 21, 22, 24, 25]. The possible reasons for this limited hrGFP expression might include chromosomal insertion site-related effects, failure of insertion of the entire BAC, unintended effects of removing the first 29 bp of ghrelin coding sequence from the transgene, and/or lack of actual or strong ghrelin expression in one or more of the sites in question. Regarding the latter possibility, we did not observe ghrelin expression in the brain by in situ hybridization histochemistry or by histochemistry using reagents that otherwise worked remarkably well in stomach. Further studies using different BACs and/or different reporter genes may help to clarify the distribution of ghrelin cells outside of the stomach.
It is important to note that earlier this year, another group published the characterization of a separate ghrelin-GFP reporter mouse line [41]. This separate line differs from ours in that it uses enhanced GFP (eGFP) as the reporter in place of hrGFP. Also, their transgene contained an 8654-bp fragment of the ghrelin gene whereas our lines contained several thousands of basepairs of chromosomal DNA both upstream and downstream of the ghrelin start codon. This previous study examined GFP expression only within the stomach and brain, but did not discuss expression in other tissue sites. Unlike our study, in which gastric expression of GFP was observed only in ghrelin cells, the previous model does describe some eGFP-immunoreactivity within non-ghrelin-immunoreactive gastric cells, although it is suggested that these eGFP-positive cells without ghrelin-immunoreactivity might represent ghrelin cells in which all of the ghrelin had been secreted [41]. Furthermore, unlike our study in which we did not demonstrate hrGFP expression within the brain, the previous model did report eGFP expression within the arcuate nucleus. Interestingly, though, this brain expression was only detectable using wide-band spectral confocal laser microscopy to examine eGFP fluorescence; standard fluorescence microscopy and immunohistochemistry were ineffective at detecting the brain expression of eGFP [41].
Despite the lack of any substantive hrGFP expression in ghrelin cells outside of the stomach, we were able to take advantage of the highly concordant expression patterns of ghrelin and hrGFP in our transgenic mice to obtain highly enriched populations of ghrelin cells using fluorescence activated cell sorting techniques. In fact, we found that ghrelin mRNA was several thousand-fold higher in the hrGFP-enriched population of sorted cells. The endocrine nature of the hrGFP-expressing ghrelin cells was further confirmed by the finding of mRNAs encoding various markers of endocrine cells, including chromogranin A. A number of previous studies have described ghrelin and chromogranin A co-expression [11, 42]. The prohormone convertases 1/3 and 2, which are expressed in many different types of endocrine cells, also were enriched within the hrGFP pool. The enrichment for prohormone convertase 1/3 had been expected, as a previous immunohistochemical analysis had demonstrated co-localization of ghrelin with prohormone convertase1/3 in the mouse stomach [43]. Also, prohormone convertase 1/3 had been shown to cleave bacterially-produced proghrelin to its mature form, whereas deletion of the prohormone convertase1/3 gene had the effect of blocking this conversion [43, 44]. Interestingly, mature ghrelin was observed in prohormone convertase 2-deleted mice, thus corroborating in vitro data which demonstrated that prohormone convertase 2 is not involved in processing of proghrelin to ghrelin [43, 44]. Thus, the enrichment for prohormone convertase 2 that we detected within the hrGFP-positive pool was surprising and may suggest that it is involved in processing of another/other prohormone(s) synthesized in the ghrelin cell. Of mention, another study had previously searched for co-localization of prohormone convertase 2 and ghrelin, however such was observed only within the pancreas and not the stomach [45]; that study, though, failed to detect any prohormone convertase within the stomach in contradiction to other studies which had previously confirmed its gastric expression [46].
Expectedly, the hrGFP-positive pools on average were not enriched for mRNAs encoding proteins known to characterize some of the other, more abundant, non-endocrine cell types of the gastric mucosa, such as amylase 2, H+/K+ ATPase β-subunit, gastric inhrinsic factor, pepsinogen F and calpain 8. The fact that we did detect low levels of those mRNAs within the hrGFP-enriched pools likely is the result of an impure preparation of hrGFP cells. In fact, primary cultures of our FACS-separated hrGFP-enriched pool do include some non-fluorescing cells (Fig. 4D-4E). Such might be the result of insufficient dissociation of ghrelin cells from their neighbors. In addition, we also failed to observe enrichment for markers of other gastric endocrine cell types within the hrGFP-positive pools. As was observed for the markers of non-endocrine gastric mucosal cells, mRNAs encoding markers of histamine-containing enterochromaffin-like cells (histidine decarboxylase), somatostatin cells, serotonin-secreting enterochromaffin cells (tryptophan hydroxylase 1) and gastrin-secreting G cells all were found within the hrGFP-enriched pools (although, on average, their levels were not higher within the hrGFP-enriched pools as compared to the hrGFP-negative pools). Again, this most likely can be explained by impure preparations of hrGFP cells, possibly resulting from incomplete dissociation of some ghrelin cells from neighboring cells. For example, dual-label studies have shown subsets of somatostatin cells to be in close proximity to ghrelin cells [47]. Nonetheless, the possibility for co-localization of some of these other endocrine markers with ghrelin still does exist. For instance, previous immunohistochemistry studies examining co-localization of ghrelin with histamine or histidine decarboxylase within the gastric mucosa have demonstrated that the ghrelin cells are apparently distinct from histamine-producing cells [11, 48]. However, a separate study demonstrated a partial overlap between immunoreactivity for vesicular monoamine transporter 2, which is a marker of histamine-producing enterochromaffin cells, and ghrelin immunoreactivity [12, 49, 50]. Also, varying results for the co-localization of ghrelin and somatostatin have been reported in the literature [12, 42].
In summary, we have characterized newly developed transgenic mouse lines that express hrGFP within ghrelin cells located within the gastric oxyntic mucosa and duodenum. We have demonstrated the ability to generate highly enriched pools of these hrGFP-containing ghrelin cells as well as to characterize their mRNA content. Circulating levels of ghrelin and regulation of these levels with caloric restriction are unaffected by the presence of the hrGFP reporter. Thus, we believe that these new ghrelin-hrGFP transgenic mouse lines will serve as powerful tools to further investigate various aspects of ghrelin cell physiology.
Acknowledgments
We thank Dr. Joel Lawitts, director of the BIDMC Transgenic Facility, and Dr. Robert E. Hammer, director of the UTSW Transgenic Core Facility, for their assistance with the generation of the ghrelin-hrGFP mouse lines. We thank Syann Lee, Jen-Chieh Chuang, and Angie Bookout for many helpful discussions. This work was supported by funding from the NIH (1F32DK64564-01, K08DK068069-01A2 and R01DK71320), Takeda Pharmaceutical Company, Ltd., and a University of Texas Southwestern Medical Center Disease-Oriented Clinical Scholars Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kojima M, Hosoda H, Date Y, Nakazato M, Matsuo H, Kangawa K. Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature. 1999;402:656–60. doi: 10.1038/45230. [DOI] [PubMed] [Google Scholar]
- 2.Gutierrez JA, Solenberg PJ, Perkins DR, Willency JA, Knierman MD, Jin Z, Witcher DR, Luo S, Onyia JE, Hale JE. Ghrelin octanoylation mediated by an orphan lipid transferase. Proc Natl Acad Sci U S A. 2008;105:6320–5. doi: 10.1073/pnas.0800708105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang J, Brown MS, Liang G, Grishin NV, Goldstein JL. Identification of the acyltransferase that octanoylates ghrelin, an appetite-stimulating peptide hormone. Cell. 2008;132:387–96. doi: 10.1016/j.cell.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 4.Zigman JM, Nakano Y, Coppari R, Balthasar N, Marcus JN, Lee CE, Jones JE, Deysher AE, Waxman AR, White RD, Williams TD, Lachey JL, Seeley RJ, Lowell BB, Elmquist JK. Mice lacking ghrelin receptors resist the development of diet-induced obesity. J Clin Invest. 2005;115:3564–72. doi: 10.1172/JCI26002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun Y, Asnicar M, Saha PK, Chan L, Smith RG. Ablation of ghrelin improves the diabetic but not obese phenotype of ob/ob mice. Cell Metab. 2006;3:379–86. doi: 10.1016/j.cmet.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Lutter M, Sakata I, Osborne-Lawrence S, Rovinsky SA, Anderson JG, Jung S, Birnbaum S, Yanagisawa M, Elmquist JK, Nestler EJ, Zigman JM. The orexigenic hormone ghrelin defends against depressive symptoms of chronic stress. Nat Neurosci. 2008;11:752–3. doi: 10.1038/nn.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zigman JM, Elmquist JK. In search of an effective obesity treatment: a shot in the dark or a shot in the arm? Proc Natl Acad Sci U S A. 2006;103:12961–2. doi: 10.1073/pnas.0605959103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diano S, Farr SA, Benoit SC, McNay EC, da Silva I, Horvath B, Gaskin FS, Nonaka N, Jaeger LB, Banks WA, Morley JE, Pinto S, Sherwin RS, Xu L, Yamada KA, Sleeman MW, Tschop MH, Horvath TL. Ghrelin controls hippocampal spine synapse density and memory performance. Nat Neurosci. 2006;9:381–8. doi: 10.1038/nn1656. [DOI] [PubMed] [Google Scholar]
- 9.Asakawa A, Inui A, Kaga T, Yuzuriha H, Nagata T, Ueno N, Makino S, Fujimiya M, Niijima A, Fujino MA, Kasuga M. Ghrelin is an appetite-stimulatory signal from stomach with structural resemblance to motilin. Gastroenterology. 2001;120:337–45. doi: 10.1053/gast.2001.22158. [DOI] [PubMed] [Google Scholar]
- 10.Masuda Y, Tanaka T, Inomata N, Ohnuma N, Tanaka S, Itoh Z, Hosoda H, Kojima M, Kangawa K. Ghrelin stimulates gastric acid secretion and motility in rats. Biochem Biophys Res Commun. 2000;276:905–8. doi: 10.1006/bbrc.2000.3568. [DOI] [PubMed] [Google Scholar]
- 11.Dornonville de la Cour C, Bjorkqvist M, Sandvik AK, Bakke I, Zhao CM, Chen D, Hakanson R. A-like cells in the rat stomach contain ghrelin and do not operate under gastrin control. Regul Pept. 2001;99:141–50. doi: 10.1016/s0167-0115(01)00243-9. [DOI] [PubMed] [Google Scholar]
- 12.Rindi G, Necchi V, Savio A, Torsello A, Zoli M, Locatelli V, Raimondo F, Cocchi D, Solcia E. Characterisation of gastric ghrelin cells in man and other mammals: studies in adult and fetal tissues. Histochem Cell Biol. 2002;117:511–9. doi: 10.1007/s00418-002-0415-1. [DOI] [PubMed] [Google Scholar]
- 13.Yabuki A, Ojima T, Kojima M, Nishi Y, Mifune H, Matsumoto M, Kamimura R, Masuyama T, Suzuki S. Characterization and species differences in gastric ghrelin cells from mice, rats and hamsters. J Anat. 2004;205:239–46. doi: 10.1111/j.0021-8782.2004.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Date Y, Kojima M, Hosoda H, Sawaguchi A, Mondal MS, Suganuma T, Matsukura S, Kangawa K, Nakazato M. Ghrelin, a novel growth hormone-releasing acylated peptide, is synthesized in a distinct endocrine cell type in the gastrointestinal tracts of rats and humans. Endocrinology. 2000;141:4255–61. doi: 10.1210/endo.141.11.7757. [DOI] [PubMed] [Google Scholar]
- 15.Sakata I, Nakamura K, Yamazaki M, Matsubara M, Hayashi Y, Kangawa K, Sakai T. Ghrelin-producing cells exist as two types of cells, closed- and opened-type cells, in the rat gastrointestinal tract. Peptides. 2002;23:531–6. doi: 10.1016/s0196-9781(01)00633-7. [DOI] [PubMed] [Google Scholar]
- 16.Prado CL, Pugh-Bernard AE, Elghazi L, Sosa-Pineda B, Sussel L. Ghrelin cells replace insulin-producing beta cells in two mouse models of pancreas development. Proc Natl Acad Sci U S A. 2004;101:2924–9. doi: 10.1073/pnas.0308604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wierup N, Sundler F. Ultrastructure of islet ghrelin cells in the human fetus. Cell Tissue Res. 2005;319:423–8. doi: 10.1007/s00441-004-1044-x. [DOI] [PubMed] [Google Scholar]
- 18.Wierup N, Svensson H, Mulder H, Sundler F. The ghrelin cell: a novel developmentally regulated islet cell in the human pancreas. Regul Pept. 2002;107:63–9. doi: 10.1016/s0167-0115(02)00067-8. [DOI] [PubMed] [Google Scholar]
- 19.Date Y, Nakazato M, Hashiguchi S, Dezaki K, Mondal MS, Hosoda H, Kojima M, Kangawa K, Arima T, Matsuo H, Yada T, Matsukura S. Ghrelin is present in pancreatic alpha-cells of humans and rats and stimulates insulin secretion. Diabetes. 2002;51:124–9. doi: 10.2337/diabetes.51.1.124. [DOI] [PubMed] [Google Scholar]
- 20.Volante M, Allia E, Gugliotta P, Funaro A, Broglio F, Deghenghi R, Muccioli G, Ghigo E, Papotti M. Expression of ghrelin and of the GH secretagogue receptor by pancreatic islet cells and related endocrine tumors. J Clin Endocrinol Metab. 2002;87:1300–8. doi: 10.1210/jcem.87.3.8279. [DOI] [PubMed] [Google Scholar]
- 21.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–5. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu S, Guan JL, Wang QP, Uehara K, Yamada S, Goto N, Date Y, Nakazato M, Kojima M, Kangawa K, Shioda S. Immunocytochemical observation of ghrelin-containing neurons in the rat arcuate nucleus. Neurosci Lett. 2002;321:157–60. doi: 10.1016/s0304-3940(01)02544-7. [DOI] [PubMed] [Google Scholar]
- 23.Mondal MS, Date Y, Yamaguchi H, Toshinai K, Tsuruta T, Kangawa K, Nakazato M. Identification of ghrelin and its receptor in neurons of the rat arcuate nucleus. Regul Pept. 2005;126:55–9. doi: 10.1016/j.regpep.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 24.Cowley MA, Smith RG, Diano S, Tschop M, Pronchuk N, Grove KL, Strasburger CJ, Bidlingmaier M, Esterman M, Heiman ML, Garcia-Segura LM, Nillni EA, Mendez P, Low MJ, Sotonyi P, Friedman JM, Liu H, Pinto S, Colmers WF, Cone RD, Horvath TL. The distribution and mechanism of action of ghrelin in the CNS demonstrates a novel hypothalamic circuit regulating energy homeostasis. Neuron. 2003;37:649–61. doi: 10.1016/s0896-6273(03)00063-1. [DOI] [PubMed] [Google Scholar]
- 25.Wortley KE, Anderson KD, Garcia K, Murray JD, Malinova L, Liu R, Moncrieffe M, Thabet K, Cox HJ, Yancopoulos GD, Wiegand SJ, Sleeman MW. Genetic deletion of ghrelin does not decrease food intake but influences metabolic fuel preference. Proc Natl Acad Sci U S A. 2004;101:8227–32. doi: 10.1073/pnas.0402763101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tena-Sempere M, Barreiro ML, Gonzalez LC, Gaytan F, Zhang FP, Caminos JE, Pinilla L, Casanueva FF, Dieguez C, Aguilar E. Novel expression and functional role of ghrelin in rat testis. Endocrinology. 2002;143:717–25. doi: 10.1210/endo.143.2.8646. [DOI] [PubMed] [Google Scholar]
- 27.Gnanapavan S, Kola B, Bustin SA, Morris DG, McGee P, Fairclough P, Bhattacharya S, Carpenter R, Grossman AB, Korbonits M. The tissue distribution of the mRNA of ghrelin and subtypes of its receptor, GHS-R, in humans. J Clin Endocrinol Metab. 2002;87:2988. doi: 10.1210/jcem.87.6.8739. [DOI] [PubMed] [Google Scholar]
- 28.Cummings DE, Purnell JQ, Frayo RS, Schmidova K, Wisse BE, Weigle DS. A preprandial rise in plasma ghrelin levels suggests a role in meal initiation in humans. Diabetes. 2001;50:1714–9. doi: 10.2337/diabetes.50.8.1714. [DOI] [PubMed] [Google Scholar]
- 29.Asakawa A, Inui A, Kaga T, Yuzuriha H, Nagata T, Fujimiya M, Katsuura G, Makino S, Fujino MA, Kasuga M. A role of ghrelin in neuroendocrine and behavioral responses to stress in mice. Neuroendocrinology. 2001;74:143–7. doi: 10.1159/000054680. [DOI] [PubMed] [Google Scholar]
- 30.Kristenssson E, Sundqvist M, Astin M, Kjerling M, Mattsson H, Dornonville de la Cour C, Hakanson R, Lindstrom E. Acute psychological stress raises plasma ghrelin in the rat. Regul Pept. 2006;134:114–7. doi: 10.1016/j.regpep.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 31.de la Cour CD, Norlen P, Hakanson R. Secretion of ghrelin from rat stomach ghrelin cells in response to local microinfusion of candidate messenger compounds: a microdialysis study. Regul Pept. 2007;143:118–26. doi: 10.1016/j.regpep.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 32.Seoane LM, Al-Massadi O, Barreiro F, Dieguez C, Casanueva FF. Growth hormone and somatostatin directly inhibit gastric ghrelin secretion. An in vitro organ culture system. J Endocrinol Invest. 2007;30:RC22–5. doi: 10.1007/BF03350806. [DOI] [PubMed] [Google Scholar]
- 33.Seoane LM, Al-Massadi O, Caminos JE, Tovar SA, Dieguez C, Casanueva FF. Sensory stimuli directly acting at the central nervous system regulate gastric ghrelin secretion. an ex vivo organ culture study. Endocrinology. 2007;148:3998–4006. doi: 10.1210/en.2007-0226. [DOI] [PubMed] [Google Scholar]
- 34.Muyrers JP, Zhang Y, Stewart AF. Techniques: Recombinogenic engineering--new options for cloning and manipulating DNA. Trends Biochem Sci. 2001;26:325–31. doi: 10.1016/s0968-0004(00)01757-6. [DOI] [PubMed] [Google Scholar]
- 35.Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 36.Zigman JM, Jones JE, Lee CE, Saper CB, Elmquist JK. Expression of ghrelin receptor mRNA in the rat and the mouse brain. J Comp Neurol. 2006;494:528–48. doi: 10.1002/cne.20823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakata I, Tanaka T, Yamazaki M, Tanizaki T, Zheng Z, Sakai T. Gastric estrogen directly induces ghrelin expression and production in the rat stomach. J Endocrinol. 2006;190:749–57. doi: 10.1677/joe.1.06808. [DOI] [PubMed] [Google Scholar]
- 38.Hata S, Koyama S, Kawahara H, Doi N, Maeda T, Toyama-Sorimachi N, Abe K, Suzuki K, Sorimachi H. Stomach-specific calpain, nCL-2, localizes in mucus cells and proteolyzes the beta-subunit of coatomer complex, beta-COP. J Biol Chem. 2006;281:11214–24. doi: 10.1074/jbc.M509244200. [DOI] [PubMed] [Google Scholar]
- 39.Mills JC, Andersson N, Stappenbeck TS, Chen CC, Gordon JI. Molecular characterization of mouse gastric zymogenic cells. J Biol Chem. 2003;278:46138–45. doi: 10.1074/jbc.M308385200. [DOI] [PubMed] [Google Scholar]
- 40.Shao JS, Schepp W, Alpers DH. Expression of intrinsic factor and pepsinogen in the rat stomach identifies a subset of parietal cells. Am J Physiol. 1998;274:G62–70. doi: 10.1152/ajpgi.1998.274.1.G62. [DOI] [PubMed] [Google Scholar]
- 41.Kageyama H, Kitamura Y, Hosono T, Kintaka Y, Seki M, Takenoya F, Hori Y, Nonaka N, Arata S, Shioda S. Visualization of ghrelin-producing neurons in the hypothalamic arcuate nucleus using ghrelin-EGFP transgenic mice. Regul Pept. 2008;145:116–21. doi: 10.1016/j.regpep.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 42.Tomasetto C, Karam SM, Ribieras S, Masson R, Lefebvre O, Staub A, Alexander G, Chenard MP, Rio MC. Identification and characterization of a novel gastric peptide hormone: the motilin-related peptide. Gastroenterology. 2000;119:395–405. doi: 10.1053/gast.2000.9371. [DOI] [PubMed] [Google Scholar]
- 43.Zhu X, Cao Y, Voogd K, Steiner DF. On the processing of proghrelin to ghrelin. J Biol Chem. 2006;281:38867–70. doi: 10.1074/jbc.M607955200. [DOI] [PubMed] [Google Scholar]
- 44.Ozawa A, Cai Y, Lindberg I. Production of bioactive peptides in an in vitro system. Anal Biochem. 2007;366:182–9. doi: 10.1016/j.ab.2007.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walia P, Asadi A, Kieffer TJ, Johnson JD, Chanoine JP. Ontogeny of Ghrelin, Obestatin, Preproghrelin, and Prohormone Convertases in Rat Pancreas and Stomach. Pediatr Res. 2008 doi: 10.1203/PDR.0b013e31818bc134. [DOI] [PubMed] [Google Scholar]
- 46.Macro JA, Dimaline R, Dockray GJ. Identification and expression of prohormone-converting enzymes in the rat stomach. Am J Physiol. 1996;270:G87–93. doi: 10.1152/ajpgi.1996.270.1.G87. [DOI] [PubMed] [Google Scholar]
- 47.Shimada M, Date Y, Mondal MS, Toshinai K, Shimbara T, Fukunaga K, Murakami N, Miyazato M, Kangawa K, Yoshimatsu H, Matsuo H, Nakazato M. Somatostatin suppresses ghrelin secretion from the rat stomach. Biochem Biophys Res Commun. 2003;302:520–5. doi: 10.1016/s0006-291x(03)00178-5. [DOI] [PubMed] [Google Scholar]
- 48.Torbergsen K, Wiksen H, Johansen K, Rahimipoor S, Falkmer UG, Zhao CM. Immunoreactivity of gastric ECL and A-like cells in fasted and fed rats and mice. Biotech Histochem. 2005;80:21–30. doi: 10.1080/10520290500051229. [DOI] [PubMed] [Google Scholar]
- 49.Erickson JD, Schafer MK, Bonner TI, Eiden LE, Weihe E. Distinct pharmacological properties and distribution in neurons and endocrine cells of two isoforms of the human vesicular monoamine transporter. Proc Natl Acad Sci U S A. 1996;93:5166–71. doi: 10.1073/pnas.93.10.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rindi G, Paolotti D, Fiocca R, Wiedenmann B, Henry JP, Solcia E. Vesicular monoamine transporter 2 as a marker of gastric enterochromaffin-like cell tumors. Virchows Arch. 2000;436:217–23. doi: 10.1007/s004280050033. [DOI] [PubMed] [Google Scholar]