SUMMARY
Elongating RNA polymerases (RNAPs) can interfere with transcription from downstream promoters by inhibiting DNA binding by RNAP and activators. However, combining quantitative measurement with mathematical modelling, we show that simple RNAP elongation cannot produce the strong asymmetric interference observed between a natural face-to-face promoter pair in bacteriophage lambda. Pausing of elongating polymerases over the RNAP binding site of the downstream promoter is demonstrated in vivo, and is shown by modelling to account for the increased interference. The model successfully predicts the effects on interference of treatments increasing or reducing pausing. Gene regulation by pausing-enhanced occlusion provides a general and potentially widespread mechanism by which even weak converging or tandem transcription, either coding or non-coding, can bring about strong in cis repression.
INTRODUCTION
Widespread transcription of non-coding and/or antisense RNA has been found in the genomes of bacteria, yeast, Drosophila, Arabidopsis, mouse, and humans (Havilio et al., 2005; Johnson et al., 2005), and the thorough study of 1% of the human genome by the ENCODE pilot project has revealed that complex intercalated transcription is no rare peculiarity but the usual state of the genome (The ENCODE Project Consortium, 2007). There has also been an increasing appreciation that these transcripts possess regulatory functions (Katayama et al., 2005; Prasanth and Spector, 2007) and are often developmentally regulated (Wilhelm et al., 2008). Though a diverse range of functions and mechanisms have been uncovered, the largest class of non-coding RNAs are long transcripts of unknown function (Ponting et al., 2009), which are often antisense to coding transcripts (Katayama et al., 2005; Wilhelm et al., 2008). Transcriptional interference (TI), defined as the suppressive influence of one transcriptional process, directly and in cis, on a second transcriptional process (Shearwin et al., 2005), is a proven regulatory role for some of these transcripts, and may well be expected to explain the function of a significant fraction more (Bird et al., 2006; Callen et al., 2004; Hongay et al., 2006; Martens et al., 2004; Petruk et al., 2006).
TI can occur between convergent (face-to-face), tandem (co-directional), or overlapping arrangements of promoters, where the association and elongation of RNA Polymerases (RNAPs) from one promoter disrupts RNAPs and/or transcription factors at a second promoter. TI provides a clear evolutionary benefit by diversifying the range of functions of transcriptional regulators, permitting repressors and activators of one promoter to indirectly activate or repress a second promoter through the alleviation or enforcement of TI (Callen et al., 2004; Martens et al., 2005). Numerous studies of in cis TI in eukaryotes and prokaryotes have arrived at a model where the elongation of RNAP over a target promoter causes TI by displacing and preventing the binding of RNAP or transcription factors to the promoter (Adhya and Gottesman, 1982; Bird et al., 2006; Callen et al., 2004; Greger et al., 1998; Henderson et al., 1989; Martens et al., 2004). In our previous work we developed a mathematical model of TI by RNA polymerase traffic in E. coli, which demonstrated that ‘occlusion’, prevention of RNAP and activator binding by the passage of elongating RNAPs through a promoter, is incapable of generating a substantial quantity of TI unless either the interfering promoter initiates RNAPs at an extraordinarily rapid rate, or is at least 10× stronger than the target promoter, whose kinetic properties must be attuned to maximise vulnerability to TI (Sneppen et al., 2005). Thus, in our current mechanistic understanding of TI, substantial transcriptional interference can only arise in the action of a strong promoter upon a weak promoter.
To further study the mechanisms of action of TI, our model system is the well characterised PR and PRE promoter pair from bacteriophage λ, where a face-to-face arrangement (separated by 320bp) and preliminary measurements suggest the presence of TI (Schmeissner et al., 1980; Ward and Murray, 1979). While PR is constitutive, PRE is dependent upon the tetrameric transcriptional activator λCII. (Shih and Gussin, 1984).
In this combined in vivo and in silico study of TI between PR and PRE, experimental measurements of TI were made by β-galactosidase assays of LacZ reporter constructs, and mathematical modelling of TI was performed with stochastic simulations. We show in vivo that PR strongly interferes with PRE but not vice-versa, and that this asymmetric strong TI is inexplicable by our previous mechanistic understanding. We go on to show that this strong interference is attributable to a mechanism that can allow a weak promoter to exert TI, pausing-enhanced occlusion, where pausing of RNAP while positioned over a downstream promoter strongly inhibits transcription.
RESULTS
Induction of PRE by CII and measurement of PR and PRE transcription
Transcription from PR and PRE was assayed using a single copy, chromosomal lacZ transcriptional fusion system (Figure 1A, Supplemental Figures S1 and S2), inserting a 507 bp fragment of the λ genome in different orientations to measure either PR or PRE. This reporter system bears an RNase III cleavage site between the promoter fragment and lacZ, which has been demonstrated to ensure that lacZ message stability and translation is independent of the sequence upstream of cleavage(Linn and St Pierre, 1990). Accordingly, LacZ activity is specifically measuring interference occurring at the level of transcription.
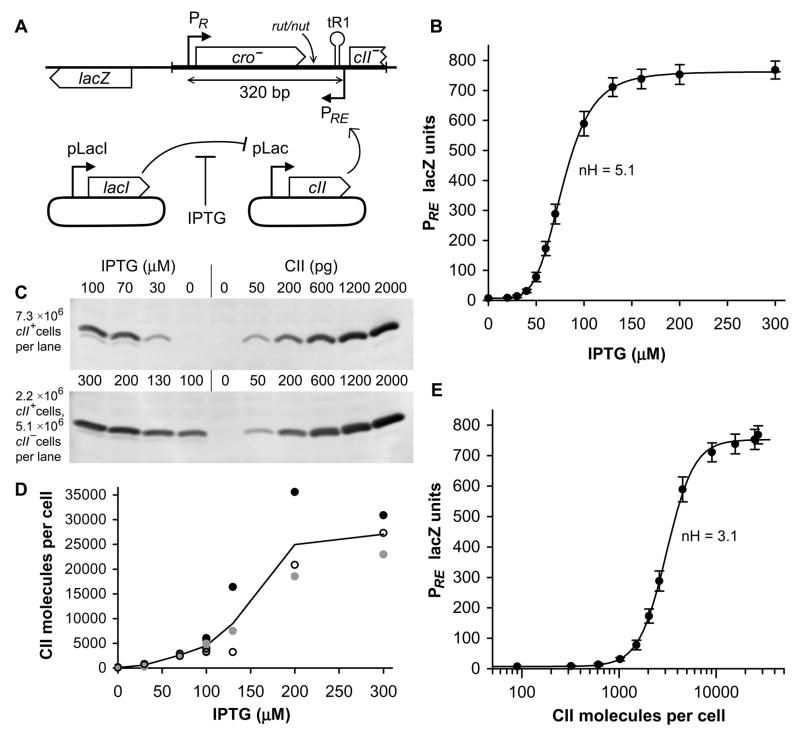
Figure 1. Activation of PRE by IPTG induction of CII.
A) Diagram of single-copy chromosomal lacZ reporter and plasmid system for IPTG regulated expression of CII. IPTG regulated expression of CII is achieved with a two plasmid system: pUHA1 contains pLacI-lacI and pZS15cII contains pLac-cII. The region of lambda used in the reporter system is shown as a thicker line. The relative locations of the Rho utilisation site (rut), N utilisation site (nut) and the tR1 terminator are shown.
(B) Activity of PRE.(PR−).lacZ (n=12) as a function of IPTG, which induces expression of CII from pLac in pZS15cII. Error bars in this and all subsequent figures are 95% confidence limits. The line connecting points is the Hill function of best fit. The average Hill coefficient is 5.1±0.4 (95% confidence limits).
(C) Western blotting of CII from E. coli pUHA1 pZS15cII grown in a range of IPTG concentrations, quantitated against a calibration curve of pure CII protein added to cII− E. coli extracts. For the IPTG concentrations 100 to 300μM (lower panel), cII+ extracts were diluted 3.3-fold into cII− extracts, to give a quantity of CII that lies within the calibration curve. The 100μM IPTG point was measured in both diluted and undiluted form, giving average measurements within 6% of one another.
(D) Quantitation of CII western blotting, showing the results of three independent sets of western blots (filled, open, and grey). Grey circles are the data of (C), and a continuous line shows the average of all data.
(E) Activity of PRE.(PR−).lacZ plotted as a function of CII molecules per cell. Using the western blotting of (C) and (D), CII molecules per cell were measured for 7 IPTG concentrations. CII molecules per cell for those IPTG concentrations not directly measured were determined by interpolation between measured data. The average Hill coefficient is 3.1±0.3 (95% confidence limits). Assuming an average cellular volume of 1.4 fL, we can determine from this data that the in vivo affinity of CII for its binding site is KD = 3.7 μM.
Between PR and PRE lies cro, a transcriptional repressor of PR. To avoid the complications of this negative feedback, the helix-turn-helix motif of cro was mutated (YQ→ER), rendering the protein unable to bind DNA (data not shown). Also located between PR and PRE is tR1, a Rho-dependent terminator of PR transcription. Rho termination factor binds to the nascent RNA at the rut sequence and leads to transcription termination at a cluster of three sites collectively termed tR1 (Banerjee et al., 2006; Lau et al., 1982). The lambda antiterminator protein, N, blocks termination at tR1 by binding to the nut site in the nascent RNA and competing with Rho binding (Vieu and Rahmouni, 2004)(Figure 1A). To investigate the termination efficiency of tR1, a shortened PR-lacZ reporter was constructed, entirely lacking cro, tR1, and PRE, such that comparison to full length reporters indicated the efficiency of termination.
The effects of TI on both PR and PRE were measured over a range of PRE activities, by the use of an IPTG inducible CII expression system. cII was encoded on a plasmid under the control of pLac, while a second plasmid expressed lacI constitutively from pLacI (Figure 1A). Measurements of non-interfered promoter activity were obtained for PRE by mutagenesis of the −10 hexamer of PR (400-fold reduction in LacZ units), and for PR by replacing CII expression vector with empty vector, leaving PRE inactive.
PRE.(PR−) demonstrates sigmoidal activation of PRE with respect IPTG (Figure 1B). Curiously, this sigmoid has a Hill coefficient of 5.1 (Figure 1B); since CII binds PRE as tetramers, this number should not exceed 4 for PRE versus CII. Western blotting of cells expressing CII under this IPTG-inducible system revealed an upward nonlinearity in CII versus IPTG (Figure 1C, D). Using this data to transform x-coordinates from IPTG to CII, revealed that the Hill coefficient with respect to CII is 3.1 (Figure 1E), within the expected range for the CII tetramer.
Transcription from PR severely interferes with PRE
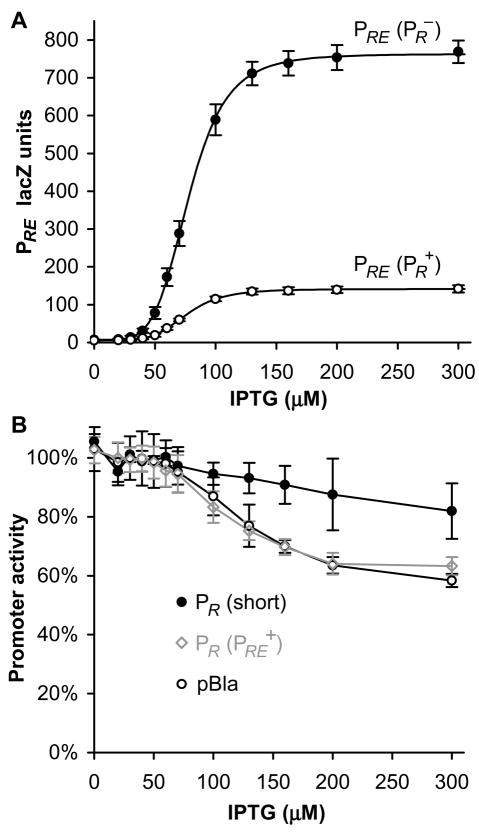
Comparison of PRE.(PR+) and PRE.(PR−) constructs demonstrated that convergent transcription from PR reduced PRE transcription 5.5-fold, while the overall shape of the sigmoidal activation curve is altered in neither Hill coefficient nor IPTG concentration required for 50% of maximum transcription (EC50) (Figure 2A).
Figure 2. Transcriptional interference between PR and PRE.
(A) Activity of PRE.(PR−).lacZ and PRE.(PR+).lacZ (n=14), demonstrating a 5.5-fold repression of PRE due to TI from PR. Lines are Hill functions of best fit; the fitted EC50s are 78μM IPTG for PRE.(PR−) and 76μM IPTG for PRE.(PR+).
(B) Activity of PR (short).lacZ (n=6), PR.(PRE+).lacZ (n=15), and pBla.lacZ (n=10), in response to IPTG induction of CII. Only PR.(PRE+).lacZ contains a CII binding site or CII-activated promoter in between the promoter and lacZ.
Transcription from PRE does not substantially interfere with PR
Measured in the absence of CII, and with lacZ positioned downstream of tR1, PR.(PRE+) produced 380 LacZ units, which by comparison to the shortened PR lacZ construct (PR (short): 1100 LacZ units), indicates 66% termination at tR1, comparable to a previous in vivo measurement of 75% (Graham, 2004).
Although transcription from PR was observed to decrease 1.5-fold with CII activation of PRE, IPTG induction of CII similarly produced a decrease in transcription from PR (short) and also a reporter of the constitutive pBla promoter (Figure 2B). As both the PR (short) and pBla constructs lack CII binding sites or a convergent promoter, it appears that CII expression indirectly influences LacZ production; this is unsurprising given the known toxic effect of CII on host DNA replication (Kedzierska et al., 2003). IPTG alone had no effect (data not shown). Whether the reduction of PR is indirect or due to PRE, the TI of PR by PRE is less than or equal to 1.5-fold, a minor effect.
Development of a mathematical model of transcriptional interference between PR and PRE
Previous work in this laboratory developed a mathematical model of TI by RNA polymerase traffic in E. coli, with analytical, stochastic, and numerical mean-field implementations (Sneppen et al., 2005). This model describes the interference of a constitutive promoter by a stronger constitutive promoter, which itself is unaffected by the interaction. In the case of PR and PRE, the similar promoter strengths and mutual interference render the analytical and mean-field models inapplicable (Sneppen et al., 2005), leaving the stochastic model, which was adapted to incorporate an activator dependent promoter (PRE) and a unidirectional terminator (tR1).
The stochastic simulation of TI is a discrete time monte carlo, with time steps dt = (RNAP elongation velocity)−1, such that elongating RNAPs advance a single base pair per time step. We model RNAP initiation with three steps: reversible binding of RNAP to DNA to form a closed complex, isomerisation of closed complexes to open complexes, and initiation of elongation by open complexes (Supplemental Figure S3). Binding of RNAP to DNA is treated as an equilibrium process acting on a faster timescale than promoter firing, and is thus governed by the equilibrium binding constant, KB, of RNAP to DNA; binding of CII to DNA is treated similarly. Isomerisation of closed to open complexes, and initiation of elongation by open complexes are governed by rate constants kco and koe, respectively. These steps received stochastic treatment; occurring with probability k. dt per time step, where k is the relevant rate constant. Parameters values are addressed in the Supplemental Data.
Before considering transcriptional interference, the model must reproduce CII activation of PRE, measured by PRE.(PR−).lacZ (Figure 1B). In vitro studies have shown CII to activate PRE by a 15-fold increase in KB and a 40-fold increase in kco (Shih and Gussin, 1984). Activation of PRE by CII was thus modelled by increasing KB and kco linearly with respect to CII occupation of PRE. Calculating CII occupation of PRE requires measurements of total CII molecules per cell as a function of IPTG (Figure 1C, D), the free energy change of CII tetramerisation, and the affinity of CII for DNA. The affinity of CII for DNA is obtained from the measurement that PRE is 50% activated at 3100 CII molecules per cell (KD = 3.7μM monomers; from Figure 1E). The free energy change of tetramerisation is the only unknown parameter, which must be fitted to produce the measured Hill coefficient: the best fit to PRE.(PR−) data is obtained at ΔG° = −22.5 kcal/mol, which compares remarkably well with an in vitro measurement of ΔG°= −23.5 kcal/mol (Ho et al., 1982).
Turning now to TI, the model contains three mechanisms (Figure 3A): occlusion, where elongating RNAPs block access to a promoter; ‘sitting duck’ interference, where initiation complexes yet to fire from a promoter are removed by elongating RNAPs; and collisions between RNAPs elongating in opposite directions. A fourth possible mechanism unique to activator dependent promoters is dislodgement of DNA-bound activator by elongating RNAP. This increases the effective dissociation rate of the activator and thereby increases the EC50 for activation, but does not alter promoter activity in the presence of saturating amounts of activator. In the event that an activator’s intrinsic dissociation rate is much faster than the rate of dislodgement by RNAP, no effect will be seen (Supplemental Figure S4). As PRE is measured to undergo no change in EC50 with TI (Figure 2A), we conclude that this mechanism is not pertinent to PRE, presumably due to rapid CII dissociation kinetics.
Figure 3. Simulations of transcriptional interference cannot explain experimental observations using existing mechanisms.
(A) Schematic of different mechanisms of transcriptional interference, in which the left (darker) promoter is interfering with the right (lighter) promoter.
(B) Alignment of experimental measurements of promoter activity with the existing mathematical model of TI. Points are experimental data and lines are the results of stochastic simulations of TI, incorporating the mechanisms of occlusion, ‘sitting duck’, and collisions.
The modelling of TI (further details in Supplemental Data) was essentially the same as in (Sneppen et al., 2005), with the exception that it was assumed here that only one RNAP, rather than both RNAPs, is lost (randomly) after a collision event. This assumption is more consistent with recent AFM imaging of collided E. coli elongation complexes suggesting that collisions induce backtracking of one RNAP (Crampton et al., 2006). The maximal activities of the PR and PRE promoters were estimated to be 0.17 and 0.12 initiations/sec, respectively, based on LacZ activities compared with a Pbla.lacZ reporter and estimates of Pbla firing rates (Liang et al., 1999). RNAP velocity was set at 60 bp/sec and termination at tR1, with probability 66%, was assumed to be instant.
A mathematical model of existing mechanisms of transcriptional interference is unable to explain the high interference of PRE by PR
Applying the above quantitative model to TI between PR and PRE, it was found that the interference of PRE by PR is inexplicably high. TI can be enhanced by the adjustment of a promoter’s kinetic parameters to maximise interference by dislodgement of open complexes; this ‘sitting duck’ interference is largest when kco and koe are similar (Sneppen et al., 2005). Upon selecting kinetic parameters for PRE that maximise TI, only 2.5 fold interference could be simulated; far short of the measured 5.5-fold interference (Figure 3B).
With the same kinetic parameters, simulated TI of PR by PRE is a reasonable match to experimental measurements. The failure of the model to explain the observed TI of PRE suggests that we are missing an asymmetric mechanism of TI, which specifically enhances the ability of PR to interfere with PRE.
RNAP from PR pause over PRE
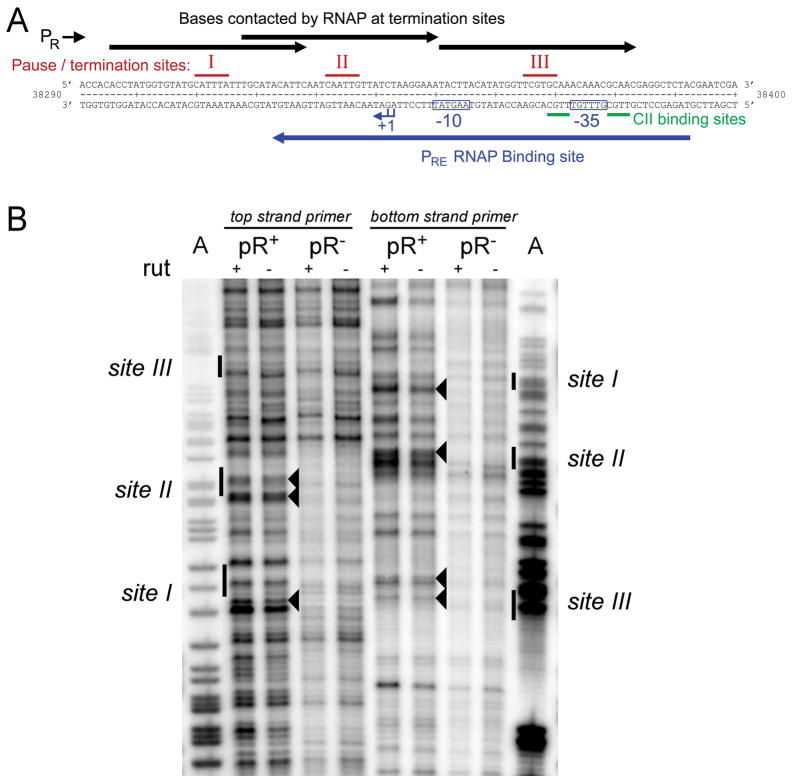
One asymmetry in the interaction of PR with PRE is the presence of the unidirectional, Rho-dependent terminator tR1. In vitro studies have found termination at tR1 to occur in three clusters located over the PRE promoter (Lau et al., 1982; Roberts et al., 1991)(Figure 4A), and also that Rho-dependent termination requires pausing at the termination site, allowing time for Rho to travel from its binding site rut along the nascent transcript to RNAP to effect termination (Lau et al., 1983; Richardson, 2002; Vieu and Rahmouni, 2004). It appeared possible then that in vivo, RNAPs from PR pause at tR1, and while so paused occlude the PRE promoter. Indeed, pausing of RNAP from PR over PRE was confirmed in vivo by potassium permanganate (KMnO4) footprinting, a technique able to detect paused elongation complexes by preferentially modifying single stranded DNA (Figure 4B). Comparison of footprints performed on PR− and PR+ templates showed a number of bands of increased intensity on the PR+ templates, at positions which correspond to the three termination sites previously identified in vitro.
Figure 4. RNAP from pR pauses at tR1.
(A) Sequence of tR1 and PRE. Above the sequence the three in vitro pause/termination sites at tR1 (Lau et al., 1982) are marked, together with the expected protected region for RNAP paused at each of these pause sites. Below the sequence is marked the binding region for RNAP at PRE, demonstrating that RNAP paused at tR1 should sterically hinder the association of RNAP to PRE.
(B) In vivo potassium permanganate footprinting was performed to identify RNAP pause sites at tR1, for rutA+ and rutA− templates. To ensure pauses were specific for transcription originating from pR, pR+ and pR− templates were compared. Footprints were obtained for both top and bottom strands, and run alongside lanes containing dideoxy sequencing reactions which had been generated using the same primers (only the A lane is shown). The bands which are the most distinctly pR specific are indicated by arrowheads. The indicated bands were observed in several (n=5) independent footprint reactions and are consistent with the known three tR1 termination sites observed in vitro (indicated at the side of figure).
A mutation which enhances pausing of RNAP over PRE increases transcriptional interference
To further investigate if pausing over PRE is a source of TI, PRE was analysed in the presence of a mutation designed to enhance pausing over PRE. Mutations of the Rho-binding site rut of the PR transcript are known to reduce termination at tR1 (Graham, 2004). Weakening the association of Rho and reducing termination at tR1 should result in prolonged occupation of pause sites by RNAP from PR, since decreased termination implies that: (1) a larger fraction of RNAP will reach the second and third pause sites, and (2) RNAP must be removed from pauses (terminated) less frequently by Rho-factor. We introduced a 3bp mutation to Rho utilisation site A (rutA−), which has been demonstrated to reduce termination through modulation of Rho-RNA interactions (Graham, 2004). By LacZ assay of PR, we confirmed that rutA− reduced termination efficiency, from 66% to 25%.
In vivo pause durations were measured by performing KMnO4 footprinting (Figure 5A, B), following the addition of rifampicin, which prevents reinitiation of RNA synthesis (Hatoum and Roberts, 2008). Pause signals decay as RNAPs leave the pause site but are not replaced, and the rate of loss of the signal gives a measure of pause duration. Scans of pixel intensity down the gel (Figure 5A) clearly show that pausing is only occurring downstream of the rutA sequence. Analysis of the most distinctly pR specific pause signals (Figures 4B, 5A) on the wild type template gave pause durations of 33, 33 and 24 seconds at sites I, II and III, which were extended approximately twofold by rutA− (Figure 5B). Analysis of a number of other bands within tR1 also gave increased pause times on the rutA− template. A second, independent time course experiment (not shown) gave similar pause durations on the wildtype template which were extended ~ 1.3 fold by rutA−. These results confirm that mutation of the rutA site, and the consequent reduction of termination efficiency, prolongs occupation of the pause sites located over PRE. The RNAP pause times observed are not unusual: a series of E. coli promoters have shown pauses of similar magnitude (Hatoum and Roberts, 2008). Comparison of PRE.(rutA− PR−) and PRE.(rutA− PR+) by LacZ assay revealed that PR now caused 21-fold TI of PRE (Figure 6A), a major increase in TI relative to PRE.(PR+).
Figure 5. A rutA mutation extends the lifetime of the pauses.
(A) Measurement of pause durations at the three tR1 sites by in vivo permanganate footprinting following addition of rifampicin. Footprints were obtained for the bottom strand on both rutA+ and rutA− templates and run alongside lanes containing dideoxy sequencing reactions which had been generated using the same primer (only the A and C lanes are shown). The appearance of a strong pR band indicates accumulation of open complexes at pR, showing that rifampicin is blocking further rounds of RNA synthesis. In contrast, the tR1 pause signals decay with time, as the paused RNAP either terminates or resumes elongation. Plots of pixel intensity down the rutA+ lanes of the gel are overlaid for the 15 (blue), 30 (green), 60 (red) and 300 (black) second time points. Black dots indicate the pR-specific bands (Figure 4B) which were used for the estimation of pause durations at sites I, II and III shown in (B).
(B) The average RNAP pause durations for rutA+ and rutA− templates at each of the three tR1 pause sites were estimated by plotting the rate of loss of signal with time following addition of rifampicin. The average pause durations, calculated as 1/slope of these plots, are indicated within each graph. The intensity of the 15 second time point was used as the initial value, in order to allow time for pR derived polymerases, which were elongating at the time of rifampicin addition, to reach tR1. Average pause durations were consistently increased on the rutA− templates.
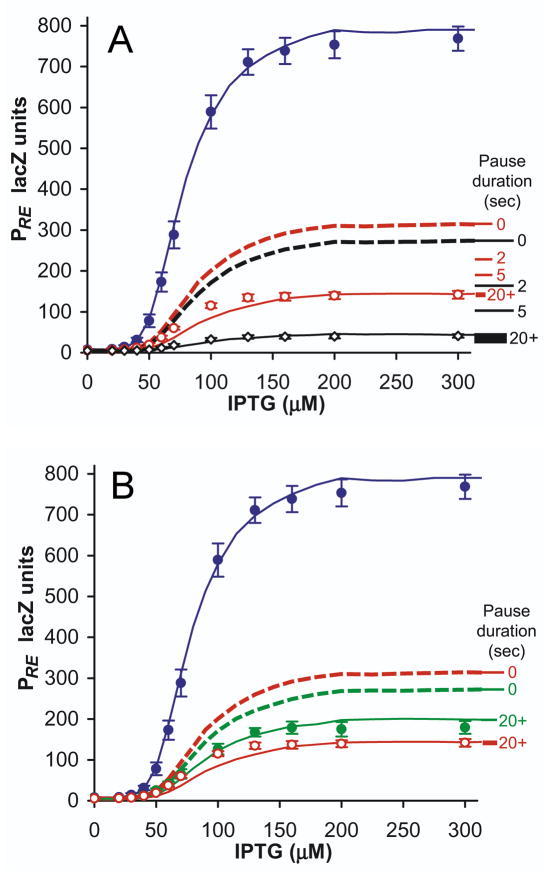
Figure 6. Occlusion by paused RNAP can explain the strong transcriptional interference of PRE.
(A) Experimental and simulated TI of PRE, in rutA+ and rutA− conditions, with simulations incorporating occlusion by paused RNAP. Points are experimental data: PRE.(PR−).lacZ (blue), PRE.(PR+).lacZ (red), and PRE.(rutA− PR+).lacZ (black) (n=7). PRE.(rutA− PR+).lacZ has been scaled up to normalise PRE activity in the absence of TI, to facilitate comparison of fold-interference against rutA+ constructs; this is necessitated by rutA− causing a 27% decrease in PRE LacZ activity in the absence of TI. Lines are stochastic simulations of TI: dotted lines are simulations without RNAP pausing at tR1, and solid lines are simulations with intrinsic RNAP pause durations at tR1 of 20 seconds. Marked along the right of the graph are the predicted maximum activities of PRE.(PR−).lacZ (red) and PRE.(rutA− PR+).lacZ (black) with intrinsic pause durations of 0, 2, 5 and 20+ seconds, illustrating how pausing of RNAP progressively increases repression of PRE. The thickness of the 20+ second line spans the range of promoter activity calculated for pause durations from 20 to 1000 seconds.
(B) As for (A), showing the effects of reduced pausing due to λN in green: PRE.(PR+ N+).lacZ (n=10). Expression of λN had no influence upon PRE.(PR−).lacZ (data not shown).
‘Occlusion by pausing’ allows the mathematical model of transcriptional interference to explain the experimental data
Having experimentally confirmed that RNAPs from PR pause over PRE, and that enhanced pausing increases TI, we next examined whether occlusion by paused RNAPs can quantitatively explain the experimental data using the mathematical model. The model was modified such that RNAPs from PR pause upon arrival at each of the three tR1 pause/termination sites (Figure 4A). At each site the paused RNAPs prevent RNAP binding to PRE, and face a probability to either terminate or resume elongation at each time-step spent paused. Also included is the ability of a trailing elongating RNAP to force forward translocation of a leading paused RNAP, as has been observed in E. coli (Epshtein and Nudler, 2003). Termination probability is defined by the experimentally measured efficiency of termination for wildtype (66%) or rutA− (25%), while the probability to spontaneously resume elongation is the reciprocal of the ‘dwell time’, or the intrinsic duration of the pause: this parameter is unknown and was fitted to experimental data (below). The intrinsic duration of the pause is the average time that an RNAP would spend at a pause site in the absence of termination or trailing polymerases. In practice, RNAPs will pause for less time than this ‘intrinsic’ pause duration, as pauses will be ended prematurely by either termination or forward translocation by a trailing RNAP.
Simulated TI of PRE is seen to increase substantially with increasing intrinsic pause time, while PR.(PRE+).lacZ is a reasonable match to experiments with or without RNAP pausing (Figure 3B). The greatest increase in TI of PRE is seen in the presence of rutA−, as decreased termination efficiency means that RNAP tend to remain paused for longer. Simulated TI of PRE reaches an asymptote at large intrinsic pause duration, when the probability of spontaneous re-initiation is insignificant relative to the probability of either undergoing termination or of being ‘pushed’ by a trailing RNAP. At this asymptote, simulated TI of both PRE.(PR+).lacZ and PRE.(rutA− PR+).lacZ agree with experimental measurements (Figure 6A). That this asymptote occurs at intrinsic pause durations greater than 20 seconds is in strong agreement with experimentally measured pause durations greater than 30 seconds.
Recalling that pushing by trailing RNAPs causes pauses to be shorter than either intrinsic pause duration or pausing measured in the presence of rifampicin, simulations in the asymptotic case reveal that average RNAP pause times are only 5 seconds, distributed across all three sites. It is striking that 5 seconds of pausing per RNAP is able to quantitatively explain the 5.5-fold interference observed for PRE.(PR+).lacZ, while in the case of PRE.(rutA− PR+).lacZ, average RNAP pause times of 8 seconds can explain the measured 21-fold repression.
The introduction of occlusion by pausing enabled the model to accurately fit the TI of three data sets; PR.(PRE+), PRE.(PR+) and PRE.(rutA− PR+); by the adjustment of a single parameter: pause duration (Figure 6A). This fit is not ‘fine-tuned’, but is satisfied by any pause times greater than 20 seconds, a requirement met by experimentally measured in vivo pause durations. These conclusions are not adversely affected by a different assumption about pushing by trailing polymerases. In the absence of pushing, the model simply selects lower intrinsic pause times because the intrinsic rate of escape through the pause must increase to make up for the lack of push-through. The consequent decrease in pause half-life for a single RNAP at a specific site is compensated by the fact that when it moves on, it is replaced by the trailing RNAP.
An obvious additional experimental test of the occlusion by pausing model is provided by considering the effect of a reduction in pausing.
Reduction of pausing by λN reduces interference of PRE
The regulatory λ protein N competitively binds PR transcripts in place of Rho-factor, and renders the elongating RNAP both termination and pause resistant (Mason et al., 1992). In a model lacking occlusion by pausing, a reduction in termination by N is predicted to increase “normal” occlusion by elongating RNAP and increase TI (Figure 6B). Alternatively, if PRE is occluded by paused RNAP, then rendering RNAP from PR pause resistant should decrease the TI of PRE. To test these competing predictions, PR and PRE were measured in the presence of constitutive chromosomal expression of N. This reduced the termination of PR transcription at tR1 to 23%, from which it was inferred that approximately half of RNAP are N-modified (Supplemental Data). Measurement of PRE.(PR−) showed no influence of N upon PRE in the absence of TI, as expected. Simulations in which 50% of RNAP from PR were N-modified, and neither paused nor terminated at tR1, produced a 38% increase in PRE activity in the presence of occlusion by pausing, or a 13% decrease in PRE if occlusion by pausing was not present. Experimentally, PRE.(PR+ N+) was 27% stronger than PRE.(PR+), confirming the prediction that if paused RNAP occlude PRE, reduced pausing should reduce TI (Figure 6B).
Pausing of RNAP can generate strong occlusion of activator or RNAP binding sites
Transcription has been shown to interfere both with promoter activity and activator binding at sites distal to a promoter (Adhya and Gottesman, 1982; Bird et al., 2006; Callen et al., 2004; Greger et al., 1998; Henderson et al., 1989; Martens et al., 2004). However, strong repression by the passage of elongating RNAPs requires a very high interfering transcription rate, or RNAP flux (Callen et al., 2004; Sneppen et al., 2005). Our study of PR and PRE shows that TI can be augmented by the pausing of RNAP while passing over a promoter; a result which can in principle also apply to transcription factor binding sites.
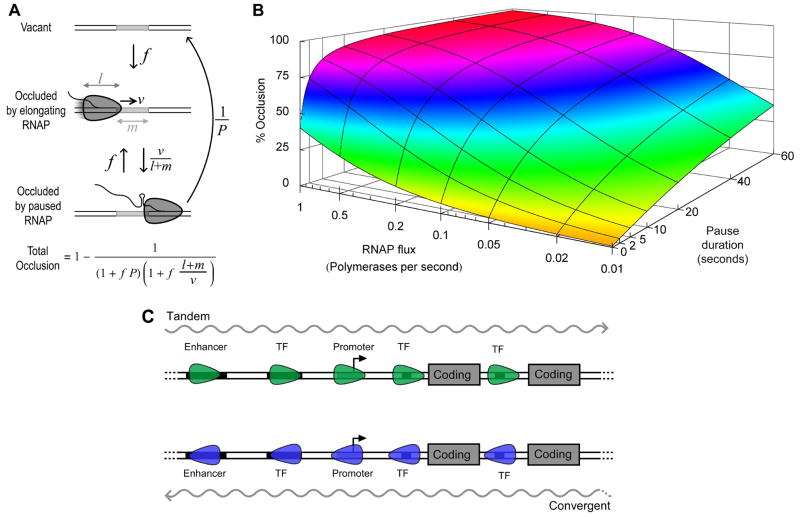
An analytical model was constructed to investigate the effect of RNAP elongation and pausing on the availability of a downstream transcription factor binding site (Figure 7A). Considering a 10 bp operator, with a 30 bp long RNAP elongating at a rate of 60 bp/sec, each RNAP occludes the operator for less than 0.7 seconds, and thus even brief pauses can substantially increase occlusion (Figure 7B). In the absence of pausing, very little occlusion (≤ 40%) can be generated even with a flux as high as 1 RNAP per second, while occlusion increases steeply with pause duration even for fluxes as low as 1 RNAP every 20 seconds. If an operator can be occluded by adjacent, non-overlapping pause sites, the fold-interference will be squared: this tactic appears to be utilised at tR1 to obtain strong repression of PRE with brief pauses.
Figure 7. Substantial occlusion can only be obtained with RNAP pausing.
(A) Analytical model of occlusion by RNAP pausing. RNAP with an occlusion length of l bp, travelling at velocity v bp/sec, arrive at an operator of length m bp, with flux f (RNAP/second). The RNAP move to the end of the operator with rate v.(l + m) −1, whereupon they reach a pause site and remain paused for an average duration P sec. Thus with rate P−1 the paused state can change to a vacant state, or with rate f another RNAP may arrive at the start of the operator. The fraction of time the operator is occluded is given by the equation shown. This result is independent of whether the pause site is positioned at the start or end of the operator.
(B) Occlusion as a function of pause duration and RNAP flux, calculated for a m = 10bp operator being occluded by l = 30bp long RNAPs which elongate at a rate of v = 60 bp/sec.
(C) Genetic arrangements where occlusion by pausing may regulate gene expression. Transcription may be tandem (upper) or convergent (lower) to the interfered promoter and may be either coding or noncoding. Pausing of RNAP over the promoter itself, or over associated elements such as transcription factor binding sites (TF) or enhancer sequences, may lead to reduction in promoter activity.
DISCUSSION
When maximally activated by CII, the λPRE promoter reaches 60% of the constitutive strength of λPR, positioned 320 bp downstream. Transcriptional interference (TI) between these convergent promoters produces no more than a 1.5-fold reduction in PR activity, but reduces transcription from PRE 5.5-fold. This strong and asymmetric interference is inconsistent with a computational model of TI which predicts moderate mutual TI by the mechanisms of (1) collisions between elongating RNAP, (2) initiation complex dislodgement and (3) promoter occlusion by elongating RNAP. Our acquisition of quantitative data and the use of modelling allowed us to identify a discrepancy between theory and experiment, which demanded a reconsideration of the mechanisms of TI. In vivo footprinting confirmed past in vitro observations: that RNAP from PR repeatedly pause over PRE in the process of Rho-dependent termination at tR1. Rho-binding site mutations were shown toenhance pausing, and dramatically increased TI of PRE to 21-fold. Expression of the λN antiterminator protein, known to reduce RNAP pausing at tR1 (Mason et al., 1992), partially alleviated TI of PRE. These findings strongly support a new mechanism of TI, occlusion by pausing, in which RNAPs pause over a promoter and sterically hinder the association of RNAP and/or transcription factors. When the experimentally measured RNAP pausing over PRE is introduced to the model, it quantitatively explains all experimental observations.
Though other biological functions for pausing have been described (Landick, 2006; Margaritis and Holstege, 2008), repression of transcription initiation is a new role. Occlusion by paused RNAP provides a mechanism by which the manner of transcription elongation, and by extension elongation regulatory factors, can directly and potently influence the expression of genes encoded on a different transcript. Theoretical analysis of other mechanisms of TI (Sneppen et al., 2005) (Figure 3A), namely collisions, occlusion by elongating RNAP, dislodgement of transcription factors and RNAP (sitting duck), indicates that: (1) adjustment of the quantity of the TI requires changes to a promoter’s activity or kinetic parameters, (2) these mechanisms have a maximal TI that depends upon the strength of the interfering promoter, and (3) extremely strong promoters are necessary to generate more than about 3-fold interference. In contrast, occlusion by pausing can potentially be tuned to any quantity of TI by adjustment of pause duration; demonstrated here by an increase from 5.5 to 21-fold repression due to a doubling of pause times. Since the quantity of repression can be widely tuned, independently of promoter activity or kinetics, occlusion by pausing is a powerful and readily evolvable mechanism.
Transcription of antisense or noncoding RNAs has been recently appreciated as a common mechanism of repression. In most eukaryotes these noncoding RNAs can repress their target transcripts with greater than 1:1 efficiency through the catalytic machinery of RNA interference (RNAi). Yet in organisms lacking RNAi such as prokaryotes and S. cerevisiae, intergenic and antisense transcription still appears to be a powerful and frequently utilised means of gene regulation, pointing to the existence of other mechanisms (Shearwin et al., 2005). Direct TI, occurring in cis by the passage of RNAP across the binding sites of activators and/or RNAP at another promoter, has been observed in bacteria, yeast, flies, HIV and mouse (Adhya and Gottesman, 1982; Bird et al., 2006; Callen et al., 2004; Greger et al., 1998; Henderson et al., 1989; Hongay et al., 2006; Martens et al., 2004; Petruk et al., 2006). We have shown here, in agreement with past work, that strong occlusion of activator and/or RNAP binding sites cannot be obtained even with a very high flux of elongating RNAP (Callen et al., 2004; Sneppen et al., 2005). However, repression of downstream promoters can be drastically improved by the pausing of RNAPs while over activator or RNAP binding sites, producing strong repression with a moderate RNAP flux.
How likely is it that occlusion by pausing, which in contrast to direct TI can produce strong repression with a moderate RNAP flux, is a widespread mechanism of gene control in higher organisms? Several lines of evidence suggest that this may the case. There are many common genetic contexts which are potentially susceptible to occlusion by pausing (Figure 7C). Both basal promoters and their associated proximal or distal transcription factor binding sites can be influenced by polymerase pausing, as can enhancer sequences, where pausing could cause disruption of complexes that activate or repress distant promoters. The transcription which ‘delivers’ the polymerase may be either convergent or tandem to the interfered promoter, and significantly, this transcription may be either coding or non-coding (Ponting et al., 2009; Shearwin et al., 2005). Secondly, several studies have shown that at least 85% of eukaryotic genomes are transcribed (Birney et al., 2007; David et al., 2006) and that there is a large degree of overlapping transcription (Katayama et al., 2005), suggesting that promoters and their associated elements are likely to be subject to considerable passing polymerase traffic. Both single molecule (Adelman et al., 2002; Neuman et al., 2003) and bulk in vitro studies suggest that there is an abundance of RNAP pause sites in transcribed DNA (Glover-Cutter et al., 2008) and a study of RNA polymerase II dynamics in live human cells in culture indicated that a significant fraction of elongating polymerases pause for cumulatively long periods (>1 minute) (Darzacq et al., 2007). Given the simple evolutionary adjustment required to appropriately locate a pause site, it seems probable that many instances of in cis transcriptional interference will feature occlusion by RNAP pausing as a primary mechanism of repression.
EXPERIMENTAL PROCEDURES
Strains and media
NK7049 (ΔlacIZYA)X74 galOP308 StrR Su− from R. Simons (Simons et al., 1987) was the host for all LacZ assays. DH5α and MC1061 were hosts for recombinant DNA work. Cells were grown at 37°C in Luria Broth with the addition of carbenicillin (100 μg/mL for pZS15) and kanamycin (50 μg/mL for pUHA1).
Constructs
Fusions of the region of λ containing PR and PRE (λ: 37954–38461)(Supplemental Figure S1) were first formed in the LacZ reporter plasmid pTL61T (Linn and St Pierre, 1990), and then transferred to the LacZ reporter phage λRS45ΔYA for insertion in single copy into the E. coli chromosome, as described in (Dodd et al., 2001) and Supplemental Figure S2. The pBla promoter was amplified from plasmid pBR322, in a product extending from −171 to +9, as per (Liang et al., 1999). This reporter system bears an RNase III cleavage site between the promoter fragment and lacZ, which reduces contextual differences in LacZ translation, and should prevent any antisense RNA interactions between PR and PRE transcripts from influencing LacZ activity. When preparing reporters of weakly terminated PR.lacZ (PR (short), rutA− and N+), shuttle strains expressing λCI protein were used to repress strong LacZ transcription. PR−, cro−, and rutA− mutations were constructed using Quikchange oligonucleotide mutagenesis (Stratagene).
N expression system
λN was expressed from single copy in the E. coli chromosome in the integrating plasmid pAH162 (Haldimann and Wanner, 2001), with the bacteriophage 186pR promoter (186: 2656–2784) transcribing N (λ: 35383–35016).
CII expression system
λCII was expressed from the wild-type pLac promoter on the ampicillin resistant plasmid pZS15, which is pZE15 (Dodd et al., 2001) with the colE1 origin replaced by the pSC101 origin (Lutz and Bujard, 1997). The wild-type λcII gene (λ: 38357–38662) was inserted into pZS15 downstream of pLac, with the intervening sequence containing lacZ up to stop codons at aa 20 and 21, followed after 15bp by the pET ribosome binding site AGGAGA to drive efficient CII translation. To control CII expression from pLac on pZS15cII, Lac repressor was supplied by pUHA1, a p15A plasmid encoding kanamycin resistance and carrying the wild-type lacI gene and promoter, obtained from H. Bujard (Heidelberg University, Germany).
LacZ assays
Kinetic LacZ assays were performed in 96-well microtitre plates by a protocol modified from (Dodd et al., 2001). Fresh colonies on selective LB plates were resuspended in LB and used to inoculate 200μL of selective LB in a 96-well microtitre plate, sealed and incubated overnight at 37°C without shaking. Overnight cultures were diluted into LB in proportion to their density, approximately 3-fold, before further diluting 2μL into 98μL of fresh selective LB plus IPTG in a microtitre plate. Cultures were incubated with rotation at 37°C until OD600 reached 0.65 – 0.75 (log phase) and were then assayed for LacZ activity (Dodd et al., 2001).
Western blotting of CII
Cultures of NK7049 pUHA1 pZS15cII were grown to log phase by the same protocol as for LacZ assays. Cultures were resuspended in 1/25th volume B-PER lysis reagent (Pierce) supplemented with 0.2 mg/mL fresh lysozyme and 2.5 units of Benzonase (Merck), incubated on ice for 1 hour, diluted 1:1 with 2× Novex Tricine SDS sample buffer (Invitrogen), and heated to 85°C for 2 minutes. Pure CII protein was supplied by Pradeep Parrack (Bose Institute, India). For consistency of background bands upon western blotting, pure CII protein was added to cII− extract prepared as per cII+ extracts, such that for both pure CII and cII+ extracts 5μL of cell extract was loaded in a net volume of 10μL for each lane during electrophoresis. For quantitation of IPTG concentrations 100–300μM, cII+ extracts were diluted 1.5μL into 3.5μL of cII− extract.
Membranes were scanned and images analysed by a Typhoon Trio and ImageQuant (Amersham). Numbers of E. coli cells per sample were determined by measurement of culture density prior to harvesting and comparison to colony forming assays.
In vivo permanganate footprinting
In vivo permanganate footprinting was performed on E. coli strain NK7049 carrying pTL61T based LacZ reporter plasmids. PRE.(PR−) and PRE.(PR+) constructs, containing either a wild type or mutated rutA site were used. There was no source of CII in these strains, in order that there was minimal transcription from PRE. To determine the location of the presumed pause site(s) of RNAP transcribing from PR, plasmid DNA prepared from KMnO4 treated cultures were subject to primer extension analysis, as detailed in Supplemental Data.
Stochastic simulations of transcriptional interference
The essential mechanics of the stochastic simulation of TI are described in Results with further details in the Supplemental Data. Simulated promoter activities in Figures 2 and 3 are the result of simulating 200 hours of transcription at each point. The program, written in FORTRAN, is available on request.
Supplementary Material
Acknowledgments
We thank Kim Sneppen for advice and support, and Pradeep Parrack and Amos Oppenheim for their gifts of pure CII protein and anti-CII antibody. Research in the Shearwin laboratory is supported by the U.S. NIH (GM062976) and the Australian Research Council.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adelman K, La Porta A, Santangelo TJ, Lis JT, Roberts JW, Wang MD. Single molecule analysis of RNA polymerase elongation reveals uniform kinetic behavior. Proc Natl Acad Sci U S A. 2002;99:13538–13543. doi: 10.1073/pnas.212358999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhya S, Gottesman M. Promoter occlusion: transcription through a promoter may inhibit its activity. Cell. 1982;29:939–944. doi: 10.1016/0092-8674(82)90456-1. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Chalissery J, Bandey I, Sen R. Rho-dependent transcription termination: more questions than answers. J Microbiol. 2006;44:11–22. [PMC free article] [PubMed] [Google Scholar]
- Bird AJ, Gordon M, Eide DJ, Winge DR. Repression of ADH1 and ADH3 during zinc deficiency by Zap1-induced intergenic RNA transcripts. Embo J. 2006;25:5726–5734. doi: 10.1038/sj.emboj.7601453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen BP, Shearwin KE, Egan JB. Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol Cell. 2004;14:647–656. doi: 10.1016/j.molcel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Crampton N, Bonass WA, Kirkham J, Rivetti C, Thomson NH. Collision events between RNA polymerases in convergent transcription studied by atomic force microscopy. Nucleic Acids Res. 2006;34:5416–5425. doi: 10.1093/nar/gkl668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci U S A. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd IB, Perkins AJ, Tsemitsidis D, Egan JB. Octamerization of lambda CI repressor is needed for effective repression of P(RM) and efficient switching from lysogeny. Genes Dev. 2001;15:3013–3022. doi: 10.1101/gad.937301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epshtein V, Nudler E. Cooperation between RNA polymerase molecules in transcription elongation. Science. 2003;300:801–805. doi: 10.1126/science.1083219. [DOI] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JE. Sequence-specific Rho-RNA interactions in transcription termination. Nucleic Acids Res. 2004;32:3093–3100. doi: 10.1093/nar/gkh630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger IH, Demarchi F, Giacca M, Proudfoot NJ. Transcriptional interference perturbs the binding of Sp1 to the HIV-1 promoter. Nucleic Acids Res. 1998;26:1294–1301. doi: 10.1093/nar/26.5.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldimann A, Wanner BL. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol. 2001;183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum A, Roberts J. Prevalence of RNA polymerase stalling at Escherichia coli promoters after open complex formation. Mol Microbiol. 2008;68:17–28. doi: 10.1111/j.1365-2958.2008.06138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havilio M, Levanon EY, Lerman G, Kupiec M, Eisenberg E. Evidence for abundant transcription of non-coding regions in the Saccharomyces cerevisiae genome. BMC Genomics. 2005;6:93. doi: 10.1186/1471-2164-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson SL, Ryan K, Sollner-Webb B. The promoter-proximal rDNA terminator augments initiation by preventing disruption of the stable transcription complex caused by polymerase read-in. Genes Dev. 1989;3:212–223. doi: 10.1101/gad.3.2.212. [DOI] [PubMed] [Google Scholar]
- Ho Y, Lewis M, Rosenberg M. Purification and properties of a transcriptional activator. The cII protein of phage lambda. J Biol Chem. 1982;257:9128–9134. [PubMed] [Google Scholar]
- Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Edwards S, Shoemaker D, Schadt EE. Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 2005;21:93–102. doi: 10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kedzierska B, Glinkowska M, Iwanicki A, Obuchowski M, Sojka P, Thomas MS, Wegrzyn G. Toxicity of the bacteriophage lambda cII gene product to Escherichia coli arises from inhibition of host cell DNA replication. Virology. 2003;313:622–628. doi: 10.1016/s0042-6822(03)00376-3. [DOI] [PubMed] [Google Scholar]
- Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem Soc Trans. 2006;34:1062–1066. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- Lau LF, Roberts JW, Wu R. Transcription terminates at lambda tR1 in three clusters. Proc Natl Acad Sci U S A. 1982;79:6171–6175. doi: 10.1073/pnas.79.20.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau LF, Roberts JW, Wu R. RNA polymerase pausing and transcript release at the lambda tR1 terminator in vitro. J Biol Chem. 1983;258:9391–9397. [PubMed] [Google Scholar]
- Liang S, Bipatnath M, Xu Y, Chen S, Dennis P, Ehrenberg M, Bremer H. Activities of constitutive promoters in Escherichia coli. J Mol Biol. 1999;292:19–37. doi: 10.1006/jmbi.1999.3056. [DOI] [PubMed] [Google Scholar]
- Linn T, St Pierre R. Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ. J Bacteriol. 1990;172:1077–1084. doi: 10.1128/jb.172.2.1077-1084.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margaritis T, Holstege FC. Poised RNA polymerase II gives pause for thought. Cell. 2008;133:581–584. doi: 10.1016/j.cell.2008.04.027. [DOI] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- Martens JA, Wu PY, Winston F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 2005;19:2695–2704. doi: 10.1101/gad.1367605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason SW, Li J, Greenblatt J. Host factor requirements for processive antitermination of transcription and suppression of pausing by the N protein of bacteriophage lambda. J Biol Chem. 1992;267:19418–19426. [PubMed] [Google Scholar]
- Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and Functions of Long Noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- Richardson JP. Rho-dependent termination and ATPases in transcript termination. Biochim Biophys Acta. 2002;1577:251–260. doi: 10.1016/s0167-4781(02)00456-6. [DOI] [PubMed] [Google Scholar]
- Roberts EA, Eisenbraun TL, Andrews CL, Bear DG. 3′-end formation at the phage lambda tR1 rho-dependent transcription termination site. Biochemistry. 1991;30:5429–5437. doi: 10.1021/bi00236a015. [DOI] [PubMed] [Google Scholar]
- Schmeissner U, Court D, Shimatake H, Rosenberg M. Promoter for the establishment of repressor synthesis in bacteriophage lambda. Proc Natl Acad Sci U S A. 1980;77:3191–3195. doi: 10.1073/pnas.77.6.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearwin KE, Callen BP, Egan JB. Transcriptional interference--a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih MC, Gussin GN. Role of cII protein in stimulating transcription initiation at the lambda PRE promoter. Enhanced formation and stabilization of open complexes. J Mol Biol. 1984;172:489–506. doi: 10.1016/s0022-2836(84)80019-4. [DOI] [PubMed] [Google Scholar]
- Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Sneppen K, Dodd IB, Shearwin KE, Palmer AC, Schubert RA, Callen BP, Egan JB. A mathematical model for transcriptional interference by RNA polymerase traffic in Escherichia coli. J Mol Biol. 2005;346:399–409. doi: 10.1016/j.jmb.2004.11.075. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieu E, Rahmouni AR. Dual role of boxB RNA motif in the mechanisms of termination/antitermination at the lambda tR1 terminator revealed in vivo. J Mol Biol. 2004;339:1077–1087. doi: 10.1016/j.jmb.2004.04.022. [DOI] [PubMed] [Google Scholar]
- Ward DF, Murray NE. Convergent transcription in bacteriophage lambda: interference with gene expression. J Mol Biol. 1979;133:249–266. doi: 10.1016/0022-2836(79)90533-3. [DOI] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.