Abstract
 Glycosylation is an essential form of post-translational modification that regulates intracellular and extracellular processes. Regrettably, conventional biochemical and genetic methods often fall short for the study of glycans, because their structures are often not precisely defined at the genetic level. To address this deficiency, chemists have developed technologies to perturb glycan biosynthesis, profile their presentation at the systems level, and perceive their spatial distribution. These tools have identified potential disease biomarkers and ways to monitor dynamic changes to the glycome in living organisms. Still, glycosylation remains the underexplored frontier of many biological systems. In this Account, we focus on research in our laboratory that seeks to transform the study of glycan function from a challenge to routine practice.
Glycosylation is an essential form of post-translational modification that regulates intracellular and extracellular processes. Regrettably, conventional biochemical and genetic methods often fall short for the study of glycans, because their structures are often not precisely defined at the genetic level. To address this deficiency, chemists have developed technologies to perturb glycan biosynthesis, profile their presentation at the systems level, and perceive their spatial distribution. These tools have identified potential disease biomarkers and ways to monitor dynamic changes to the glycome in living organisms. Still, glycosylation remains the underexplored frontier of many biological systems. In this Account, we focus on research in our laboratory that seeks to transform the study of glycan function from a challenge to routine practice.
In studies of proteins and nucleic acids, functional studies have often relied on genetic manipulations to perturb structure. Though not directly subject to mutation, we can determine glycan structure−function relationships by synthesizing defined glycoconjugates or by altering natural glycosylation pathways. Chemical syntheses of uniform glycoproteins and polymeric glycoprotein mimics have facilitated the study of individual glycoconjugates in the absence of glycan microheterogeneity. Alternatively, selective inhibition or activation of glycosyltransferases or glycosidases can define the biological roles of the corresponding glycans. Investigators have developed tools including small molecule inhibitors, decoy substrates, and engineered proteins to modify cellular glycans. Current approaches offer a precision approaching that of genetic control.
Genomic and proteomic profiling form a basis for biological discovery. Glycans also present a rich matrix of information that adapts rapidly to changing environs. Glycomic and glycoproteomic analyses via microarrays and mass spectrometry are beginning to characterize alterations in glycans that correlate with disease. These approaches have already identified several cancer biomarkers. Metabolic labeling can identify recently synthesized glycans and thus directly track glycan dynamics. This approach can highlight changes in physiology or environment and may be more informative than steady-state analyses. Together, glycomic and metabolic labeling techniques provide a comprehensive description of glycosylation as a foundation for hypothesis generation.
Direct visualization of proteins via the green fluorescent protein (GFP) and its congeners has revolutionized the field of protein dynamics. Similarly, the ability to perceive the spatial organization of glycans could transform our understanding of their role in development, infection, and disease progression. Fluorescent tagging in cultured cells and developing organisms has revealed important insights into the dynamics of these structures during growth and development. These results have highlighted the need for additional imaging probes.
Introduction
Virtually every class of biomolecule can be found in a glycosylated form. This phenomenon extends from the glycoproteins, which we now know comprise ∼50% of the total cellular proteome and >90% of the secreted proteome,1,2 to lipids, tRNA,(3) and many secondary metabolites (Figure 1). But the question, “what do the glycans do?” remains unanswered in many cases. Decades of research in the rapidly expanding field of glycobiology have provided some insights. For example, glycans have been shown to govern biological homeostasis, playing central roles in protein folding, trafficking, and stability,(4) and in organ development.(5) Inside cells, protein glycosylation is thought to play a role in signaling, perhaps in concert with phosphorylation.(6) Cell-surface glycans are poised to mediate intercellular communication,(7) including pathogen recognition,8,9 and to distinguish self from non-self immunologically.(10) In addition, the glycosylation state of both cell-surface proteins and lipids responds to external stimuli and internal cellular dysfunction. Thus, the dynamics of these molecules reflect the cell’s physiological state and can report on disease.(11)
Figure 1.
Examples of glycoconjugates. Many proteins are glycosylated at asparagine (N-linked) or serine/threonine residues (mucin-type O-linked and O-GlcNAc are shown). Lipids, secondary metabolites, and tRNA are examples of other biomolecules found in glycosylated form.
Historically, approaches to studying glycans reflected the standard tactics of biological inquiry that were developed in the context of proteins and nucleic acids: (1) alter the structure or expression level and evaluate the biological consequence (i.e., perturb); (2) define the molecular inventory as a function of physiology (i.e., profile); (3) visualize the molecule in a living system to understand its distribution and dynamics (i.e., perceive). Based primarily in genetics and biochemistry, the experimental tools used to accomplish these goals for proteins and nucleic acids did not always translate to the study of glycans. For example, perturbation of glycan structures can be achieved by genetic mutation of glycosyltransferases, but the effects of such mutations are often masked by embryonic lethality or compensatory upregulation of redundant enzymes.12,13 Lectins and antibodies with defined glycan specificities can be used to profile cell-surface glycans and to correlate global changes in their expression with developmental stages and disease.(14) Until recently, however, the available lectins and antibodies were limited in number. Finally, visualizing glycans in living systems is an unmet challenge for which no conventional experimental approach is suited. The ability to perceive these biopolymers as they undergo dynamic changes within organisms could transform our view of glycobiology.
New techniques derived from physical, analytical, and synthetic chemistry are starting to address many of the inadequacies of the conventional toolbox as applied to glycans. Several groups have contributed in important ways to the burgeoning field of chemical glycobiology. Their contributions include small molecules that interfere with glycan biosynthesis,15−17 glycopolymers that modulate carbohydrate receptor activity,(18) and synthetic methods for assembling glycoconjugates.19−22 Furthermore, analytical tools such as lectin microarrays and mass spectrometry are providing, for the first time, detailed pictures of the “glycome”.23,24 Due to restrictions of space, this Account will focus primarily on our own efforts to develop small molecules to perturb, profile, and perceive glycans.
Perturbing Glycan Displays to Investigate Living Systems
Chemical Synthesis of Glycoconjugates
In principle, the synthesis of chemically defined glycoconjugates allows researchers to perturb biological systems in a tightly controlled fashion. Though syntheses of singly substituted glycopeptides are routinely achieved, the complexity of most glycoproteins and glycopeptides presents a formidable challenge. Full details of this approach have been recently reviewed.(25) A notable success comes from Danishefsky and colleagues who synthesized portions of the highly glycosylated protein erythropoietin A (EPO) by joining chemically synthesized glycopeptides via native chemical ligation.(26) These glycosylated peptides were more than 20 amino acids in length and contained elongated N- and O-linked glycans, thus providing a framework for the total synthesis of a complex glycoprotein. Semisynthesis of a full-length glycoprotein, GlyCAM-1, has been achieved via the combination of glycopeptide synthesis and expressed protein ligation technology. The protein included up to 13 sites of O-linked glycosylation (α-GalNAc-modified serines and threonines) located on two separate mucin domains of the protein.(27) Despite these remarkable achievements, the technical challenges associated with glycoprotein syntheses will likely prevent their routine application.
An alternative strategy to glycoprotein total synthesis is the development of synthetically tractable glycoconjugate analogs. Mucins are proteins containing repeating units of densely glycosylated domains whose biological functions often correspond to the degree of glycosylation. Recently we have reported the use of chemically defined mucin mimics to study glycans in synthetic lipid bilayers and cell surfaces.28,29 The condensation of aminooxy-sugars with polymethylvinylketone gave highly modified glycopolymers (Figure 2). These glycoconjugates resembled native mucins in size, macromolecular structure, and biophysical properties. Incorporated into synthetic membranes or cell surfaces, these glycopolymers interact with carbohydrate binding proteins (lectins) in a manner specific to the appended glycan’s composition, arrangement, and stereochemistry. The polymers also demonstrate surface two-dimensional mobilities similar to cell-surface lipid-bound mucins. These polymers are a promising alternative strategy for studying the role of glycans in both intra- and intercellular receptor-mediated signaling.
Figure 2.
Structures of mucin-type O-linked glycosylation and the structure and synthesis of a mucin mimetic. (A) Mucin-type O-linked glycosylation is characterized by densely functionalized arrays of glycans on a polypeptide backbone. (B) A derivatized polyvinylketone backbone can approximate endogenous mucins. (C) The synthesis of mucin mimics proceeds by condensing aminooxy-GalNAc with polyvinyl ketone. Extended sugars can also be incorporated.
Glycosyltransferase Inhibitors
Small molecule inhibitors of glycan biosynthesis can alter glycan structures.(30) Cell-surface glycans are constructed by the stochastic action of spatially constrained ER- and Golgi-associated glycosyltransferases as their target glycoconjugates traverse the secretory pathway. The roughly 250 glycosyltransferases predicted in mammalian genomes are attractive targets for small molecule inhibition and could help elucidate their roles in dynamic cellular processes that are opaque to course-grained genetic manipulations. To serve as useful tools for biological inquiry, glycosyltransferase inhibitors must be selective for their target, active in cells, and potentially active in living organisms. Several natural products are known inhibitors of glycosylation, including tunicamycin, which blocks the biosynthesis of N-linked glycan precursors, and deoxynojirimycin, which broadly inhibits certain glycan processing enzymes. These molecules and their relatives have been widely employed to study the effects of glycosylation on protein function, but off-target effects limit their utility.
Chemical biologists have sought to expand upon this limited toolkit with the design of inhibitors that target specific enzymes. Pioneering work by Imperiali and co-workers targeted the enzymes responsible for the initiation of N-linked glycosylation. The researchers synthesized a potent inhibitor of oligosaccharyl transferase (OST), the enzyme that transfers N-linked glycans to proteins in the ER.31,32 The compound was based on a constrained peptide that approximated the natural substrate’s ASX turn. Once optimized for cell-based environments, the compound promises to disrupt N-linked glycosylation in a more refined fashion than tunicamycin. Despite this and other notable successes in this area, the structural homology and functional overlap of the more than 200 glycosyltransferases makes inhibition of individual glycosyltransferases a challenge.
Primers of Glycosylation
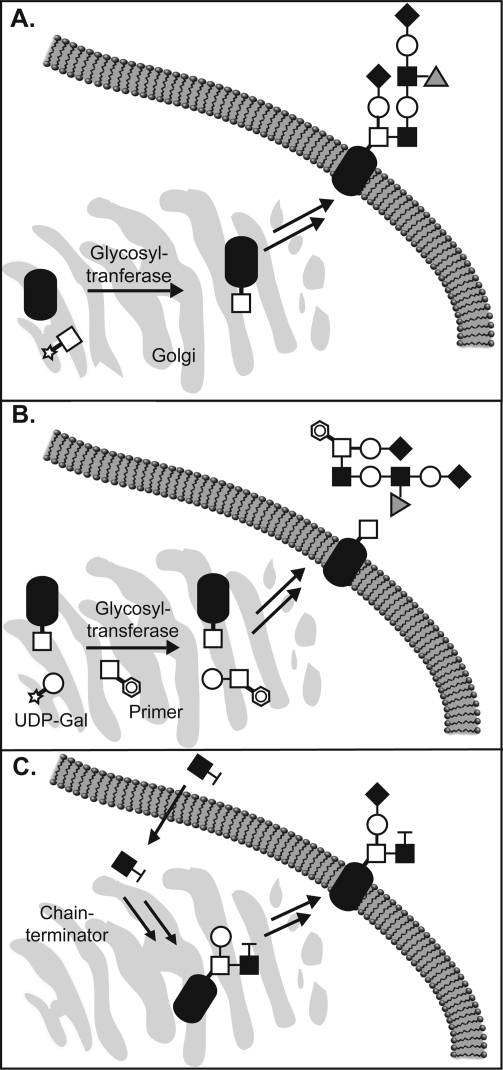
An alternative strategy for modulating cellular glycan structures involves “primers of glycosylation”. In general, these compounds are simple naturally occurring sugars adorned with hydrophobic aglycones such as benzyl, phenyl, and naphthyl groups. Including β-xylosides,(33) α-benzyl GalNAc,(34) and various substituted disaccharides, the primers act as competitive substrates for glycosyltransferases inside cells.(35) At high concentrations, the primers out compete endogenous glycans as acceptor substrates for glycosyltransferases. Thus, glycan chain elongation occurs on the soluble primers rather than on endogenous glycoproteins and glycolipids, and eventually, the modified primers are secreted from the cell (Figure 3A). In recent years, Esko and co-workers have employed a disaccharide primer, α-napthyl GlcNAcβ1−3Gal, to inhibit the elaboration of cellular N-acetyllactosamine epitopes with sialic acid (Sia) and fucose (Fuc) residues to form the sialyl Lewis X (sLeX) motif.35−38 Tumor-associated sLeX binds to P-selectin, a receptor on activated endothelial cells, and is thought to mediate the extravasion of tumor cells from the bloodstream into organs.(39) Treatment of LS180 cells with the disaccharide primer markedly decreased cell-surface expression of sLex. The cells displayed diminished capacity to colonize the lungs of nude mice, suggesting inhibition of this pathway may be a viable approach for controlling metastasis.
Figure 3.
Perturbing glycosylation with synthetic sugars. (A) Glycosyltransferases add activated glycan donors onto proteins in the secretory pathway to biosynthesize cell-surface glycoproteins. (B) Primers of glycosylation divert the action of glycosyltransferases away from their endogenous substrates. Instead, the primers are glycosylated at the expense of cellular glycoproteins, and elaborated primers are secreted. (C) Unnatural monosaccharides can be metabolically incorporated into glycans, wherein they prevent further elongation.
Incorporating Unnatural Monosaccharides into Glycans
Conservative modifications of monosaccharide structures are sometimes tolerated by biosynthetic enzymes, enabling the metabolic incorporation of the sugar analogs into cellular glycans (Figure 3B). The modification can disrupt recognition by downstream elaborating enzymes, or by receptors that eventually govern the glycan’s function. In a pioneering study, Reutter and co-workers demonstrated that unnatural variants of N-acetylmannosamine (ManNAc), in which the N-acyl substituent is elaborated, are recognized by the sialic acid biosynthetic machinery. Thus, treatment of cells with N-propanoylmannosamine (ManNProp) leads to replacement of a fraction of their cell surface sialic acid residues with the N-propanoyl derivative.(40) This structural perturbation interfered with sialic acid-dependent viral infection.(8)
A similar technique was employed to induce premature termination of glycan chains in neurons. Poly(sialic acid) (PSA), a homopolymer comprising 2,8-linked sialic acid residues, is a modification of the neural cell adhesion molecule (NCAM) that has been linked to neuronal plasticity.(41) Neurons grown in the presence of N-butanoyl mannosamine (ManNBut) were found to have shortened PSA chains, presumably due to inefficient extension of PSA polymers incorporating this substrate by polysialyltransferases.(42) In the future, biosynthetic modulation of PSA length using ManNBut might be applied to studies of PSA in neuronal development.
In addition to unnatural sialosides, derivatives of N-acetylglucosamine (GlcNAc) and galactose (Gal) have been incorporated into glycans using cellular machinery. Mice and human cell lines are able to incorporate 4-fluoro-GlcNAc into mucin-type O-linked glycans.43,44 Incorporation of this modification terminates further elongation of glycan chains by removal of the acceptor nucleophile, thereby diminishing the production of epitopes that bind to E-selectin, a receptor on activated endothelial cells involved in inflammation. As a consequence, T-cells do not bind to the endothelial cells and homing to the inflamed tissue is blocked.
In an application to neurobiology, chain-terminating sugars were employed to study the processes of neurite outgrowth and synapse formation. Fucα1−2Gal has long been implicated in cognitive processes.(45) Hsieh-Wilson and co-workers studied the details of this process through the use of 2-deoxy-Gal (2-d-Gal), which is incorporated into cellular glycans thereby blocking formation of the Fucα1−2Gal epitope.(46) Synapsin, a protein involved in neurite outgrowth and neurotransmitter release, was rapidly degraded by calpain-mediated proteolysis in the absence of its normal Fucα1−2Gal modification. This observation suggests a critical role for Fucα1−2Gal in neuronal plasticity.
“Switchable” Glycosyltransferases
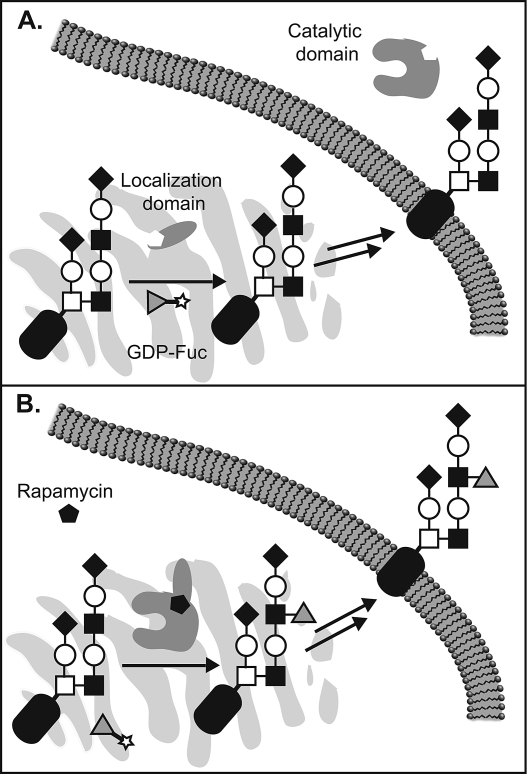
Inhibitors, primers, and unnatural substrates share the ability to downregulate glycosyation in a temporally controlled manner. However, the high degree of homology between related glycosyltransferases makes discrimination between these enzymes with specific small molecules a challenge. A complementary approach that we developed merges genetic and molecular approches to allow temporal regulation of specific glycosyltransferases using a small molecule switch. The approach exploits the conserved modular architecture of most Golgi-resident glycosyltransferases, which comprise functionally distinct catalytic (CAT) and localization (LOC) domains that must be physically associated for activity on their cellular substrates. We fused separated CAT and LOC domains to the FK506 binding protein (FKBP) and the FKB-rapamycin binding domain (FRB) of the mammalian target of rapamycin (mTOR), respectively.(47) In the absence of rapamycin, FKBP and FRB show negligible affinity for one another.(48) As a result, the LOC domain localizes normally to the Golgi membrane, while the catalytic domain is secreted from the cell, effectively limiting access to its substrates (Figure 4A). The addition of rapamycin initiates heterodimerization of FKBP and FRB, reconstituting the native localization of the CAT domain and restoring its enzymatic activity (Figure 4B). The approach was applied to glycosyltransferases47,49 and to sulfotransferases(50) that act on glycans.
Figure 4.
Engineering glycosyltransferases for small molecule control. (A) In the absence of rapamycin, a “chemical inducer of dimerization”, catalytic and localization domains remain separate and the glycosyltransferase cannot engage its substrates. (B) In the presence of rapamycin, the glycosyltransferase domains associate and the enzyme regains activity.
Profiling Glycosylation
Decoding the information stored in the glycome is a major impetus of the field of glycobiology. Chemical approaches can vastly accelerate this process by enabling molecular-level analyses of glycans at the systems level. Furthermore, profiles of glycans associated with disease can provide new biomarkers for diagnosis and therapeutic monitoring.51,52 Three complementary approaches for profiling glycans are now assuming prominent positions: lectin/antibody arrays, mass spectrometry, and metabolic labeling.
Lectin Microarrays
Lectins are nonantibody carbohydrate binding proteins that exhibit specificity for one or more glycan-associated epitopes. Historically, lectins have been used in low-throughput agglutination assays to categorize cell surfaces.(53) Advances in microarray technology coupled with the genome-driven discovery of new lectins have enabled more rapid and detailed analyses of glycosylation. Recently, Kuno and co-workers used evanescent fluorescence to quantify the binding of diverse Cy5-labeled glycoproteins to an array of 39 different lectins.(54) Mahal and co-workers used a similar approach to characterize bacterial cell surface glycomes.(23) In an extension of this theme, Haab and co-workers used microarrayed antibodies to capture specific glycoproteins from the serum of healthy donors and cancer patients to individual array features. Through use of a type of sandwich assay, lectin binding to the captured species identified specific glycan changes unique to the cancer patients.(55) While these techniques can detect both the identity and concentration of certain glycan epitopes, not every glycan structure has a corresponding lectin and many lectins show promiscuous binding. Nonetheless, lectin arrays can provide insight into glycosylation patterns associated with disease.
Mass Spectrometry
Comprehensive structural analysis of cell-derived glycans can be achieved using mass spectrometry. For decades, mass spectrometry has been the tool of choice for sequencing the oligosaccharide components of purified glycoproteins.56,57 With the advent of ultrasensitive and ultrahigh-resolution mass spectrometry techniques, entire glycomes can now be analyzed from complex cell lysates or bodily fluids. Already, distinct glycan profiles have been identified in sera from patients with ovarian,(51) breast,(52) and prostate(58) cancers using mass spectrometry profiling techniques. These discoveries represent the first step in identifying new clinical biomarkers.
Both lectin arrays and mass spectrometric techniques provide a snapshot of glycosylation at steady state. The dynamics of glycosylation, still difficult to analyze using these techniques, may provide additional information relevant to disease monitoring or to fundamental studies of glycobiology. For instance, cancer cells are known to metabolize simple sugars more rapidly than healthy cells, a trait that has been exploited in diagnostic imaging.(59) Techniques that assess glycosylation dynamics, including monosaccharide uptake and processing, glycan biosynthesis, and membrane turnover, could provide a new dimension to glycome profiling.
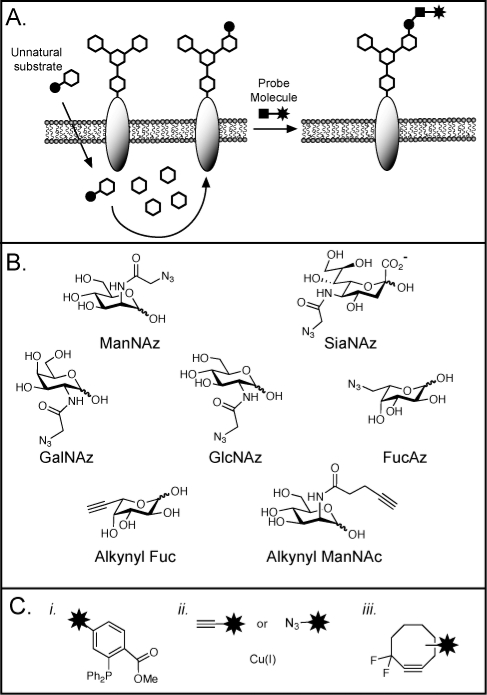
We have made use of the bioorthogonal chemical reporter strategy to specifically tag newly synthesized glycoconjugates, which can then be purified and inventoried. In this procedure, an unnatural monosaccharide containing a small, bioorthogonal functional group is incorporated into glycans using the cell’s metabolic machinery.(60) Much like a pulse−chase experiment, the incubation time and concentration of the synthetic sugar can be varied so that it labels only the most recently synthesized glycans and not the entire glycome. Subsequently, a reagent specific for the bioorthogonal functionality is used to modify the newly synthesized glycans with a probe for capture and enrichment (Figure 5A).
Figure 5.
The bioorthogonal chemical reporter strategy for profiling and visualizing glycans. (A) Metabolic labeling of cell-surface glycans with an azidosugar. The azide and alkyne functionalities can be covalently reacted with a probe that bears the functionality shown in panel C. (B) Azidosugars and alkynyl sugars that have been employed as metabolic labels. The unnatural monosaccharides are typically used in peracetylated form to facilitate cellular uptake. (C) Reagents for covalent labeling of azides (i, iii) and alkynes (ii): (i) phosphines for the Staudinger ligation; (ii) terminal alkynes or azides and copper(I) for copper-mediated azide−alkyne cylcoaddition, “click chemistry”; (iii) difluorinated cyclooctyne (DIFO) for strain-promoted cycloadditions.
The azide has proven to be the most versatile bioorthogonal functional group for metabolic labeling of glycans, because it is small, abiotic, and stable in cellular systems. Sia,(61) GalNAc,(62) GlcNAc,(63) and Fuc64,65 can all be substituted with azido analogs in cultured cells, and Sia and GalNAc can be labeled with azides in mice (Figure 5B). In contrast to chain terminators, the azide-bearing sugars are not intended to disrupt further glycan elaboration and are generally well-tolerated by cultured cells and mice.62,66,77 Once installed in a glycan, the azide can be selectively reacted with phosphines via the Staudinger ligation, with terminal alkynes via Cu-mediated “click chemistry”, or with cyclooctynes by a strain-promoted [3 + 2] cycloaddition65,67−70 (Figure 5C). These reactions allow chemical labeling of the azido glycan with any probe of choice (e.g., biotin, FLAG peptide, or fluorescent dyes). Fucose and sialic acids modified by terminal alkynes can also be incorporated onto cell surfaces and imaged via click chemistry (Figure 5B);65,71 however, the inherent toxicity of the copper reagents precludes imaging of live organisms.
The bioorthogonal chemical reporter strategy has been artfully employed in the proteomic analysis of protein O-GlcNAcylation, a form of glycosylation that is unique to cytosolic and nuclear proteins and thought to be a key modulator of protein function.(6) Despite its potential importance, the complete repertoire of O-GlcNAcylated proteins is not known. We demonstrated that GlcNAcylated proteins can be metabolically labeled with the azido analog N-azidoacetylglucosamine (GlcNAz), and then biotinylated by Staudinger ligation with a phosphine probe.(63) Zhao and co-workers exploited this discovery to capture O-GlcNAcylated proteins for a comprehensive proteomic analysis of cell lysates.(72)
In another application, the bioorthogonal chemical reporter method was applied to analyze mucin-type O-linked glycoproteins from cell lysates(62) and murine tissues.(73)N-Azidoacetylgalactosamine (GalNAz) was found to label the O-linked glycans in both contexts, substituting for the conserved glycan core residue GalNAc. Interestingly, the GalNAz labeling efficiency of B cells from treated mice was considerably higher than that of T cells, a result that was not predicted based on their steady-state glycomes. This observation underscores the differences in information provided by metabolic labeling versus lectin or mass spectrometry methods of glycan profiling.
Perceiving Glycans
To understand how changes in glycosylation relate to physiological changes requires a means to visualize these biopolymers in a physiologically relevant context. Tagging proteins with fluorescent-protein fusions has proven to be an invaluable technique for tracking their localization and function in living cells. Analogous tools should be sought for probing the abundance, distribution, and dynamics of glycans in vivo. The bioorthogonal chemical reporter strategy holds much promise in this regard. Glycans can be labeled with SiaNAz and GalNAz in living animals.66,73 In addition, phosphine probes injected into these animals undergo the Staudinger ligation and accumulate on cell surfaces in an azide-dependent manner. The stage has thus been set for the delivery of imaging probes that can report on the presence of specific glycan subtypes.
The development of imaging reagents that target azides is an important next step. Two recent examples include the development of phosphine- or alkyne-functionalized dyes that fluoresce only upon reaction with azides and thereby label cell surface glycans with low background staining.65,71,74 The application of these reagents in living animals, however, may be limited. The Staudinger ligation is plagued by slow kinetics and the Cu-mediated “click chemistry” requires a toxic heavy metal catalyst. To overcome these problems, we recently developed a difluorinated cyclooctyne reagent, termed DIFO (Figure 5C(iii)) that rapidly reacts with azides and is nontoxic in mice.75,76 The rapid kinetics of DIFO−azide reactions were exploited to monitor the dynamics of cellular glycosylation during zebrafish embryo development. Zebrafish embryos labeled with GalNAz were tagged with fluorescently labeled DIFO reagents to image the fish’s total O-linked glycosylation.(77) We extended this technique to monitor dynamic glycosylation by quenching unreacted azide groups on labeled cells with TCEP, pulsing the embryos with additional GalNAz, and probing the embryos with a blue-shifted DIFO conjugate (Figure 6). Areas of rapid O-linked glycan biosynthesis including the fins, jaw, and olfactory organs showed increased labeling with the blue-shifted conjugate. This quench, label, tag sequence exploits the high reactivity of DIFO to visualize glycan concentrations below the detection limit for comparable phosphine probes. In addition, these small molecule-based reagents are more accessible to the tissues than lectin or antibody-based imaging agents. Post-labeling, the zebrafish appeared to continue normal development, confirming the low toxicity of these reagents. The extension of this work to mammalian systems should enable imaging of tumors and sites of microbial infection.
Figure 6.
The quench−label−tag sequence for visualizing dynamic glycosylation. Zebrafish embryos labeled with GalNAz can be tagged with fluorescently labeled DIFO reagents to visualize total O-linked glycosylation. A dynamic picture of glycosylation can be obtained by reducing residual azides with TCEP, incubating briefly with GalNAz, and labeling the newly synthesized glycans with a blue-shifted DIFO conjugate.
Future Outlook
Chemical tools for perturbing, profiling, and perceiving glycans should reduce the barriers that have previously impeded progress in this field. Their implementation has already produced a body of knowledge that was formerly limited to the domain of proteins and nucleic acids. For example, a developing glycomic database has begun to facilitate bioinformatics studies on glycans.(78) As mentioned above, profiles of glycans from tissues and bodily fluids have produced new biomarker candidates. Many regents for perturbing glycans, including inhibitors and primers, are now commercially available. Similarly, azidosugars and both phosphine- and alkyne-based labeling reagents can now be obtained from commercial suppliers. The future is bright for use of these tools in analyzing the dynamic glycomes of normal and diseased tissues.
Acknowledgments
The authors’ work that was mentioned in this review was primarily supported by grants to C.R.B. from the National Institutes of Health (GM66047, GM58867, and GM59907).
Biographies
Nicholas J. Agard is a NIH postdoctoral fellow at the University of California, San Francisco. He completed his undergraduate degree in Chemistry from Brown University in 2002 and his Ph. D. in chemical biology from the University of California, Berkeley, in 2007 with a thesis focused on glycan perturbation and detection. Currently he is studying proteolysis in innate immunity.
Carolyn Bertozzi is a Howard Hughes Medical Investigator, Director of the Molecular Foundry, and the T. Z. and Ingrid Chu Distinguished Professor of Chemistry and Molecular and Cellular Biology at the University of California, Berkeley. She received her undergraduate degree from Harvard University in 1988 and her Ph. D. from the Univerity of California, Berkeley, in 1993. She has published more than 150 scientific articles and received numerous honors including memberships in the National Academy of Science and the American Academy of Arts and Sciences. Professor Bertozzi’s current research investigates a number of fields including glycobiology, nanomaterials, and molecular imaging.
References
- Apweiler R.; Hermjakob H.; Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1999, 1473, 4–8. [DOI] [PubMed] [Google Scholar]
- Jung E.; Veuthey A. L.; Gasteiger E.; Bairoch A. Annotation of glycoproteins in the SWISS-PROT database. Proteomics 2001, 1, 262–268. [DOI] [PubMed] [Google Scholar]
- Kasai H.; Nakanishi K.; Macfarlane R. D.; Torgerson D. F.; Ohashi Z.; McCloskey J. A.; Gross H. J.; Nishimura S. The structure of Q* nucleoside isolated from rabbit liver transfer ribonucleic acid. J. Am. Chem. Soc. 1976, 98, 5044–5046. [DOI] [PubMed] [Google Scholar]
- Mitra N.; Sharon N.; Surolia A. Role of N-linked glycan in the unfolding pathway of Erythrina corallodendron lectin. Biochemistry 2003, 42, 12208–12216. [DOI] [PubMed] [Google Scholar]
- Grobe K.; Ledin J.; Ringvall M.; Holmborn K.; Forsberg E.; Esko J. D.; Kjellen L. Heparan sulfate and development: Differential roles of the N-acetylglucosamine N-deacetylase N-sulfotransferase isozymes. Biochim. Biophys. Acta 2002, 1573, 209–215. [DOI] [PubMed] [Google Scholar]
- Slawson C.; Housley M. P.; Hart G. W. O-GlcNAc cycling: How a single sugar post-translational modification is changing the way we think about signaling networks. J. Cell. Biochem. 2006, 97, 71–83. [DOI] [PubMed] [Google Scholar]
- Haines N.; Irvine K. D. Glycosylation regulates Notch signalling. Nat. Rev. Mol. Cell. Biol. 2003, 4, 786–797. [DOI] [PubMed] [Google Scholar]
- Herrmann M.; von der Lieth C. W.; Stehling P.; Reutter W.; Pawlita M. Consequences of a subtle sialic acid modification on the murine polyomavirus receptor. J. Virol. 1997, 71, 5922–5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y. Sialobiology of influenza: Molecular mechanism of host range variation of influenza viruses. Biol. Pharm. Bull. 2005, 28, 399–408. [DOI] [PubMed] [Google Scholar]
- Upreti R. K.; Kumar M.; Shankar V. Bacterial glycoproteins: Functions, biosynthesis and applications. Proteomics 2003, 3, 363–379. [DOI] [PubMed] [Google Scholar]
- Dube D. H.; Bertozzi C. R. Glycans in cancer and inflammation--potential for therapeutics and diagnostics. Nat. Rev. Drug Discovery 2005, 4, 477–488. [DOI] [PubMed] [Google Scholar]
- Lowe J. B.; Marth J. D. A genetic approach to mammalian glycan function. Annu. Rev. Biochem. 2003, 72, 643–691. [DOI] [PubMed] [Google Scholar]
- Stanley P.; Ioffe E. Glycosyltransferase mutants: Key to new insights in glycobiology. FASEB J. 1995, 9, 1436–1444. [DOI] [PubMed] [Google Scholar]
- Feizi T.; Fazio F.; Chai W.; Wong C. H. Carbohydrate microarrays - a new set of technologies at the frontiers of glycomics. Curr. Opin. Struct. Biol. 2003, 13, 637–645. [DOI] [PubMed] [Google Scholar]
- Ostash B.; Walker S. Bacterial transglycosylase inhibitors. Curr. Opin. Chem. Biol. 2005, 9, 459–466. [DOI] [PubMed] [Google Scholar]
- Fuster M. M.; Esko J. D. The sweet and sour of cancer: Glycans as novel therapeutic targets. Nat. Rev. Cancer 2005, 5, 526–542. [DOI] [PubMed] [Google Scholar]
- Lairson L. L.; Withers S. G. Mechanistic analogies amongst carbohydrate modifying enzymes. Chem. Commun. 2004, 2243–2248. [DOI] [PubMed] [Google Scholar]
- Kiessling L. L.; Gestwicki J. E.; Strong L. E. Synthetic multivalent ligands in the exploration of cell-surface interactions. Curr. Opin. Chem. Biol. 2000, 4, 696–703. [DOI] [PubMed] [Google Scholar]
- Ouerfelli O.; Warren J. D.; Wilson R. M.; Danishefsky S. J. Synthetic carbohydrate-based antitumor vaccines: Challenges and opportunities. Expert Rev. Vaccines 2005, 4, 677–685. [DOI] [PubMed] [Google Scholar]
- Brik A.; Ficht S.; Wong C. H. Strategies for the preparation of homogenous glycoproteins. Curr. Opin. Chem. Biol. 2006, 10, 638–644. [DOI] [PubMed] [Google Scholar]
- Werz D. B.; Seeberger P. H. Carbohydrates as the next frontier in pharmaceutical research. Chemistry 2005, 11, 3194–3206. [DOI] [PubMed] [Google Scholar]
- Davis B. G. Synthesis of glycoproteins. Chem. Rev. 2002, 102, 579–602. [DOI] [PubMed] [Google Scholar]
- Hsu K. L.; Pilobello K. T.; Mahal L. K. Analyzing the dynamic bacterial glycome with a lectin microarray approach. Nat. Chem. Biol. 2006, 2, 153–157. [DOI] [PubMed] [Google Scholar]
- Haslam S. M.; North S. J.; Dell A. Mass spectrometric analysis of N- and O-glycosylation of tissues and cells. Curr. Opin. Struct. Biol. 2006, 16, 584–591. [DOI] [PubMed] [Google Scholar]
- Gamblin D. P.; Scanlan E. M.; Davis B. G. Glycoprotein synthesis: An update. Chem. Rev. 2009, 109, 131–163. [DOI] [PubMed] [Google Scholar]
- Wu B.; Chen J.; Warren J. D.; Chen G.; Hua Z.; Danishefsky S. J. Building complex glycopeptides: Development of a cysteine-free native chemical ligation protocol. Angew. Chem., Int. Ed. 2006, 45, 4116–4125. [DOI] [PubMed] [Google Scholar]
- Macmillan D.; Bertozzi C. R. Modular assembly of glycoproteins: Towards the synthesis of GlyCAM-1 by using expressed protein ligation. Angew. Chem., Int. Ed. 2004, 43, 1355–1359. [DOI] [PubMed] [Google Scholar]
- Rabuka D.; Forstner M. B.; Groves J. T.; Bertozzi C. R. Noncovalent cell surface engineering: Incorporation of bioactive synthetic glycopolymers into cellular membranes. J. Am. Chem. Soc. 2008, 130, 5947–5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabuka D.; Parthasarathy R.; Lee G. S.; Chen X.; Groves J. T.; Bertozzi C. R. Hierarchical assembly of model cell surfaces: Synthesis of mucin mimetic polymers and their display on supported bilayers. J. Am. Chem. Soc. 2007, 129, 5462–5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempel B. P.; Withers S. G. Covalent inhibitors of glycosidases and their applications in biochemistry and biology. Glycobiology 2008, 18, 570–586. [DOI] [PubMed] [Google Scholar]
- Eason P. D.; Imperiali B. A potent oligosaccharyl transferase inhibitor that crosses the intracellular endoplasmic reticulum membrane. Biochemistry 1999, 38, 5430–5437. [DOI] [PubMed] [Google Scholar]
- Kellenberger C.; Hendrickson T. L.; Imperiali B. Structural and functional analysis of peptidyl oligosaccharyl transferase inhibitors. Biochemistry 1997, 36, 12554–12559. [DOI] [PubMed] [Google Scholar]
- Sobue M.; Habuchi H.; Ito K.; Yonekura H.; Oguri K.; Sakurai K.; Kamohara S.; Ueno Y.; Noyori R.; Suzuki S. β-d-Xylosides and their analogues as artificial initiators of glycosaminoglycan chain synthesis. Aglycone-related variation in their effectiveness in vitro and in ovo. Biochem. J. 1987, 241, 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan S. F.; Byrd J. C.; Basbaum C.; Kim Y. S. Inhibition of mucin glycosylation by aryl-N-acetyl-alpha-galactosaminides in human colon cancer cells. J. Biol. Chem. 1989, 264, 19271–19277. [PubMed] [Google Scholar]
- Sarkar A. K.; Fritz T. A.; Taylor W. H.; Esko J. D. Disaccharide uptake and priming in animal cells: inhibition of sialyl Lewis X by acetylated Galβ1→4GlcNAc β-O-naphthalenemethanol. Proc. Natl. Acad. Sci. U.S.A. 1995, 92, 3323–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. R.; Fuster M. M.; Li R.; Varki N.; Glass C. A.; Esko J. D. A disaccharide-based inhibitor of glycosylation attenuates metastatic tumor cell dissemination. Clin. Cancer Res. 2006, 12, 2894–2901. [DOI] [PubMed] [Google Scholar]
- Brown J. R.; Fuster M. M.; Whisenant T.; Esko J. D. Expression patterns of alpha 2,3-sialyltransferases and alpha 1,3-fucosyltransferases determine the mode of sialyl Lewis X inhibition by disaccharide decoys. J. Biol. Chem. 2003, 278, 23352–23359. [DOI] [PubMed] [Google Scholar]
- Fuster M. M.; Brown J. R.; Wang L.; Esko J. D. A disaccharide precursor of sialyl Lewis X inhibits metastatic potential of tumor cells. Cancer Res. 2003, 63, 2775–2781. [PubMed] [Google Scholar]
- Lowe J. B. Glycan-dependent leukocyte adhesion and recruitment in inflammation. Curr. Opin. Cell Biol. 2003, 15, 531–538. [DOI] [PubMed] [Google Scholar]
- Kayser H.; Zeitler R.; Kannicht C.; Grunow D.; Nuck R.; Reutter W. Biosynthesis of a nonphysiological sialic acid in different rat organs, using N-propanoyl-D-hexosamines as precursors. J. Biol. Chem. 1992, 267, 16934–16938. [PubMed] [Google Scholar]
- Durbec P.; Cremer H. Revisiting the function of PSA-NCAM in the nervous system. Mol. Neurobiol. 2001, 24, 53–64. [DOI] [PubMed] [Google Scholar]
- Mahal L. K.; Charter N. W.; Angata K.; Fukuda M.; Koshland D. E. Jr.; Bertozzi C. R. A small-molecule modulator of poly-alpha 2,8-sialic acid expression on cultured neurons and tumor cells. Science 2001, 294, 380–381. [DOI] [PubMed] [Google Scholar]
- Descheny L.; Gainers M. E.; Walcheck B.; Dimitroff C. J. Ameliorating skin-homing receptors on malignant T cells with a fluorosugar analog of N-acetylglucosamine: P-selectin ligand is a more sensitive target than E-selectin ligand. J. Invest. Dermatol. 2006, 126, 2065–2073. [DOI] [PubMed] [Google Scholar]
- Dimitroff C. J.; Bernacki R. J.; Sackstein R. Glycosylation-dependent inhibition of cutaneous lymphocyte-associated antigen expression: Implications in modulating lymphocyte migration to skin. Blood 2003, 101, 602–610. [DOI] [PubMed] [Google Scholar]
- Hitoshi S.; Koijima N.; Kusunoki S.; Inokuchi J.; Kanazawa I.; Tsuji S. Expression of the beta-galactoside alpha 1,2-fucosyltransferase gene suppresses axonal outgrowth of neuro2a neuroblastoma cells. J. Neurochem. 1996, 66, 1633–1640. [DOI] [PubMed] [Google Scholar]
- Murrey H. E.; Gama C. I.; Kalovidouris S. A.; Luo W. I.; Driggers E. M.; Porton B.; Hsieh-Wilson L. C. Protein fucosylation regulates synapsin Ia/Ib expression and neuronal morphology in primary hippocampal neurons. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler J. J.; Bertozzi C. R. Regulating cell surface glycosylation by small molecule control of enzyme localization. Chem. Biol. 2003, 10, 1303–1311. [DOI] [PubMed] [Google Scholar]
- Spencer D. M.; Wandless T. J.; Schreiber S. L.; Crabtree G. R. Controlling signal transduction with synthetic ligands. Science 1993, 262, 1019–1024. [DOI] [PubMed] [Google Scholar]
- Kohler J. J.; Czlapinski J. L.; Laughlin S. T.; Schelle M. W.; de Graffenried C. L.; Bertozzi C. R. Directing flux in glycan biosynthetic pathways with a small molecule switch. ChemBioChem 2004, 5, 1455–1458. [DOI] [PubMed] [Google Scholar]
- de Graffenried C. L.; Laughlin S. T.; Kohler J. J.; Bertozzi C. R. A small-molecule switch for Golgi sulfotransferases. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 16715–16720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H. J.; Miyamoto S.; Lancaster K. S.; Kirmiz C.; Li B.; Lam K. S.; Leiserowitz G. S.; Lebrilla C. B. Profiling of glycans in serum for the discovery of potential biomarkers for ovarian cancer. J. Proteome Res. 2006, 5, 1626–1635. [DOI] [PubMed] [Google Scholar]
- Kirmiz C.; Li B.; An H. J.; Clowers B. H.; Chew H. K.; Lam K. S.; Ferrige A.; Alecio R.; Borowsky A. D.; Sulaimon S.; Lebrilla C. B.; Miyamoto S. A serum glycomics approach to breast cancer biomarkers. Mol. Cell. Proteomics 2007, 6, 43–55. [DOI] [PubMed] [Google Scholar]
- Hynes S. O.; Hirmo S.; Wadstrom T.; Moran A. P. Differentiation of Helicobacter pylori isolates based on lectin binding of cell extracts in an agglutination assay. J. Clin. Microbiol. 1999, 37, 1994–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno A.; Uchiyama N.; Koseki-Kuno S.; Ebe Y.; Takashima S.; Yamada M.; Hirabayashi J. Evanescent-field fluorescence-assisted lectin microarray: A new strategy for glycan profiling. Nat. Methods 2005, 2, 851–856. [DOI] [PubMed] [Google Scholar]
- Chen S.; Laroche T.; Hamelinck D.; Bergsma D.; Brenner D.; Simeone D.; Brand R. E.; Haab B. B. Multiplexed analysis of glycan variation on native proteins captured by antibody microarrays. Nat. Methods 2007, 4, 437–444. [DOI] [PubMed] [Google Scholar]
- Dell A.; Morris H. R. Glycoprotein structure determination by mass spectrometry. Science 2001, 291, 2351–2356. [DOI] [PubMed] [Google Scholar]
- Morelle W.; Michalski J. C. Glycomics and mass spectrometry. Curr. Pharm. Des. 2005, 11, 2615–2645. [DOI] [PubMed] [Google Scholar]
- Kyselova Z.; Mechref Y.; Al Bataineh M. M.; Dobrolecki L. E.; Hickey R. J.; Vinson J.; Sweeney C. J.; Novotny M. V. Alterations in the serum glycome due to metastatic prostate cancer. J. Proteome Res. 2007, 6, 1822–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Som P.; Atkins H. L.; Bandoypadhyay D.; Fowler J. S.; MacGregor R. R.; Matsui K.; Oster Z. H.; Sacker D. F.; Shiue C. Y.; Turner H.; Wan C. N.; Wolf A. P.; Zabinski S. V. A fluorinated glucose analog, 2-fluoro-2-deoxy-D-glucose (F-18): Nontoxic tracer for rapid tumor detection. J. Nucl. Med. 1980, 21, 670–675. [PubMed] [Google Scholar]
- Prescher J. A.; Bertozzi C. R. Chemistry in living systems. Nat. Chem. Biol. 2005, 1, 13–21. [DOI] [PubMed] [Google Scholar]
- Luchansky S. J.; Goon S.; Bertozzi C. R. Expanding the diversity of unnatural cell-surface sialic acids. ChemBioChem 2004, 5, 371–374. [DOI] [PubMed] [Google Scholar]
- Hang H. C.; Yu C.; Kato D. L.; Bertozzi C. R. A metabolic labeling approach toward proteomic analysis of mucin-type O-linked glycosylation. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 14846–14851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vocadlo D. J.; Hang H. C.; Kim E. J.; Hanover J. A.; Bertozzi C. R. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 9116–9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabuka D.; Hubbard S. C.; Laughlin S. T.; Argade S. P.; Bertozzi C. R. A chemical reporter strategy to probe glycoprotein fucosylation. J. Am. Chem. Soc. 2006, 128, 12078–12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M.; Hsu T. L.; Itoh T.; Sugiyama M.; Hanson S. R.; Vogt P. K.; Wong C. H. Glycoproteomic probes for fluorescent imaging of fucosylated glycans in vivo. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 12371–12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescher J. A.; Dube D. H.; Bertozzi C. R. Chemical remodelling of cell surfaces in living animals. Nature 2004, 430, 873–877. [DOI] [PubMed] [Google Scholar]
- Saxon E.; Bertozzi C. R. Cell surface engineering by a modified Staudinger reaction. Science 2000, 287, 2007–2010. [DOI] [PubMed] [Google Scholar]
- Agard N. J.; Baskin J. M.; Prescher J. A.; Lo A.; Bertozzi C. R. A comparative study of bioorthogonal reactions with azides. ACS Chem. Biol. 2006, 1, 644–648. [DOI] [PubMed] [Google Scholar]
- Agard N. J.; Prescher J. A.; Bertozzi C. R. A strain-promoted [3 + 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 2004, 126, 15046–15047. [DOI] [PubMed] [Google Scholar]
- Ning X.; Guo J.; Wolfert M. A.; Boons G. J. Visualizing metabolically labeled glycoconjugates of living cells by copper-free and fast Huisgen cycloadditions. Angew. Chem., Int. Ed. 2008, 47, 2253–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T. L.; Hanson S. R.; Kishikawa K.; Wang S. K.; Sawa M.; Wong C. H. Alkynyl sugar analogs for the labeling and visualization of glycoconjugates in cells. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 2614–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi A.; Sprung R.; Barma D. K.; Zhao Y.; Kim S. C.; Falck J. R.; Zhao Y. Global identification of O-GlcNAc-modified proteins. Anal. Chem. 2006, 78, 452–458. [DOI] [PubMed] [Google Scholar]
- Dube D. H.; Prescher J. A.; Quang C. N.; Bertozzi C. R. Probing mucin-type O-linked glycosylation in living animals. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 4819–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux G. A.; De Graffenried C. L.; Bertozzi C. R. A fluorogenic dye activated by the staudinger ligation. J. Am. Chem. Soc. 2003, 125, 4708–4709. [DOI] [PubMed] [Google Scholar]
- Baskin J. M.; Prescher J. A.; Laughlin S. T.; Agard N. J.; Chang P. V.; Miller I. A.; Lo A.; Codelli J. A.; Bertozzi C. R. Copper-free click chemistry for dynamic in vivo imaging. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 16793–16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codelli J. A.; Baskin J. M.; Agard N. J.; Bertozzi C. R. Second-generation difluorinated cyclooctynes for copper-free click chemistry. J. Am. Chem. Soc. 2008, 130, 11486–11493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin S. T.; Baskin J. M.; Amacher S. L.; Bertozzi C. R. In vivo imaging of membrane-associated glycans in developing zebrafish. Science 2008, 320, 664–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://www.functionalglycomics.org/fg/.