Abstract
Smac mimetics (SM) have been recently reported to kill cancer cells through the extrinsic apoptosis pathway mediated by autocrine tumor necrosis factor (TNF). SM also activates nuclear factor κB (NF-κB). However, how SM induces NF-κB and the role of NF-κB in SM-induced cancer cell death has not been well elucidated. We found that effective blockage of NF-κB had no detectable effect on SM compound 3 (SMC3)-induced TNF secretion, suggesting the induction of TNF by SMC3 is independent of NF-κB. Conversely, SMC3-induced NF-κB activation was found to be mediated by autocrine TNF, because this effect of SMC3 was effectively inhibited when TNF was blocked with either a TNF neutralizing antibody or TNF siRNA. In addition, although SMC3 dramatically reduced c-IAP1 level, it had marginal effect on c-IAP2 expression, TNF-induced RIP modification, NF-κB activation, and downstream anti-apoptosis NF-κB target expression. Furthermore, blocking NF-κB by targeting IKKβ or RelA substantially potentiated SMC3-induced cytotoxicity, suggesting that the NF-κB pathway inhibits SMC3-induced apoptosis in cancer cells. Our results demonstrate that through autocrine TNF, SM induces an IKKβ-mediated NF-κB activation pathway that protects cancer cells against SM-induced apoptosis, and thus, NF-κB blockage could be an effective approach for improving the anticancer value of SM.
Keywords: NF-κB, Smac mimetic, TNF, cytotoxicity, apoptosis
Introduction
As a transcription factor that is activated by environmental stimuli and cellular stresses, NF-κB plays an important role in numerous biological and pathological processes (1). In cancer cells, NF-κB is frequently activated and contributes to proliferation, survival, invasion, and metastasis (2). NF-κB is generally considered as a cell survival factor that protect cancer cells against apoptosis because it upregulates expression of anti-apoptotic genes (3-5). However, emerging evidence from recent studies suggest that NF-κB may be pro-apoptotic because it is able to activate pro-apoptosis genes such as death receptor 5 (DR5), Bax, Fas ligand, and p73 (6-8). How the contradictory cellular outcomes by NF-κB activation are regulated has not been well understood, but they could be cell type- and stimulus-specific (9). NF-κB can be activated by many anticancer therapeutics (9-11). Because chemotherapeutics kill cancer cells through apoptosis, understanding the role of chemotherapy-induced NF-κB in apoptosis regulation would shed light on improving their anticancer efficacy.
There are two, canonical and noncanonical, pathways that lead to NF-κB activation. In most cell types the major one is the canonical pathway, which consists of IKK (IKKβ as the catalytic subunit), IκB, and NF-κB (typically a p65/p50 heterodimer) and is activated by proinflammatory cytokines such as tumor necrosis factor (TNF) and a variety of cellular stresses (3). The p65/p50 complex is retained in the cytoplasm by association with inhibitor of κB (IκB). When this pathway is turned on by binding of TNF to the TNF receptor 1(TNFR1), IKK is recruited to the TNFR1 signaling complex through TRAF2 and activated through a RIP-mediated mechanism that involves MEKK3 (12, 13). IKKβ is activated to phosphorylate IκB and trigger IκB rapid degradation. This process causes the nuclear translocation of p65/p50 to promote transcription of the NF-κB’s target genes. Notably, TNF itself is also a NF-κB target in certain cell types (14). Numerous NF-κB targets activated through this pathway, such as cellular inhibitor of apoptosis- and -2 (cIAP-1, cIAP-2), Bcl-xL, XIAP, and IEX-1L, have anti-apoptotic properties (14). Specifically, cIAP-1 and cIAP-2 function as an apoptosis brake through direct binding and suppressing the effecter caspases (15). Having an E3 ubiquitin ligase activity that modifies the key components of TNFR signaling complex, these IAP proteins participate in TNF-induced NF-κB activation (16, 17). As a critical component for TNF-induced signaling, RIP is important for transmitting signals from TNFR1 to IKK for NF-κB activation (18-20). Ubiquitination of RIP by c-IAPs was thought to be important for recruitment and activation of IKK (16, 21-25). The noncanonical pathway is initiated by NF-κB inducing kinase (NIK)-mediated activation of IKKα, which triggers processing of p100 to generate p52. Then p52 forms a functional complex with RelB and translocates to the nucleus to enhance gene expression (3). Thus, RelB and p52 represent the unique gene targets for investigation and intervention of the noncanonical pathway.
Smac (second mitochondria-derived activator of caspase, also called direct IAP binding protein with low pI, DIABLO) is a key pro-apoptotic protein released from mitochondria during the activation of the intrinsic apoptosis pathway (26-28). The pro-apoptotic activity of Smac is executed through suppression of the IAP family members c-IAP1, c-IAP2, and XIAP to release the brake for apoptosis (27, 28). Many cancer cells are insensitive to apoptosis. Thus, it is assumed that reducing the apoptosis threshold by modulating apoptosis-regulating molecules such as Smac will sensitize cells to anticancer chemotherapy (29). While other cancer cell-killing mechanisms were reported(30, 31), smac mimetics (SM)-caused apoptosis in cancer cells through induction of autocrine TNF appears to be the major SM-triggered cytotoxicity pathway (32-35). SM also activates NF-κB, which may involve both the canonical and noncanonical pathways (33, 34). Because TNF is a potential NF-κB target and NF-κB blocks TNF-induced apoptosis, there is a paradox regarding the role of NF-κB in SM-induced cytotoxicity in cancer cells.
We systematically investigated the mechanism by which SM induces NF-κB activation and the role of NF-κB in SM-induced cytotoxicity using a well characterized SM (compound 3, SMC3) (23, 32, 36). In this report, we provide evidence showing that SMC3 activates NF-κB mainly through the canonical pathway by inducing autocrine TNF and that NF-κB plays an inhibitory role in SMC3-induced cytotoxicity. The results suggest that NF-κB blockage could be an effective approach for increasing the anticancer efficacy of SM.
Materials and Methods
Reagents and antibody
SM compound 3 (SMC3) was kindly provided by Dr. Xiaodong Wang, University of Texas Southwest Medical Center. Human TNF, interleukin 1β (IL-1β) and anti-TNFR1 were purchased from R&D Systems (Minneapolis, MN). Inhibitors for and IKK (IKK inhibitor II) was from Calbiochem (San Diego, CA). Actinomycin D was from Sigma(St. Louis, MO). The following antibodies were used for Western blot: anti-cIAP2, anti-caspase-8 and -caspase-3 (Pharmingen, San Diego, CA), anti-PARP (BioSource, Camarillo, CA), anti-Bcl-XL (Cell Signaling, Beverly, MA), anti-RIP and anti-MnSOD (BD Biosciences, San Diego, CA), anti-IKKβ (Upstate, Chicago, IL), anti-ubiquitin, -IκBα, -RelA, -RelB, and anti-NF-κB p52 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-cIAP1(R&D systems), anti-β-tubulin (Sigma). Small interference RNA (siRNA) for IKKβ, RelA, RelB, and TNF, negative control siRNA were purchased from Dharmacon (Lafayette, CO). The human TNF detection ELISA kit was purchased from eBioscience (San Diego, CA).
Cell culture
The human lung cancer cell line H23, human hepatoma cell lines HepG2 and Huh-7, and human breast cancer cell line MCF-7 were obtained from American Type Culture Collection (Manassas, VA). H23 cells were grown in RPMI 1640 with 10% fetal bovine serum, 1 mmol/L glutamate, 100 units/ml penicillin, and 100 μg/ml streptomycin. HepG2, Huh-7, and MCF-7 cells were cultured in DMEM with 4.5 g/L Glucose, 10% fetal bovine serum, 1 mmol/L glutamate, 100 units/ml penicillin, and 100 μg/ml streptomycin.
Cytotoxicity assay
Cytotoxicity was determined using a lactate dehydrogenase (LDH) release-base cytotoxicity detection kit (Promega, Madison, WI). Cells were seeded in 48-well plates at 70-80% confluence. After culture overnight, cells were treated as indicated in each figure legend. LDH release was determined and cell death was calculated as described previously (37, 38).
Measurement of autocrine TNF secretion by ELISA
Cells were plated onto 12-well plates at 70-80% confluence. After culture overnight, cells were treated as described in the figure legends. The culture media were collected and concentration of TNF was detected by ELISA analysis with the human TNF-α ELISA kit following the instruction of the manufacturer (eBioscience Inc.).
Western blot and immunoprecipitation
Cells were harvested and lysed in M2 buffer (20 mM Tris-Hcl, pH7.6, 0.5% NP-40, 250 mM Nacl, 3 mM EGTA, 3 mM EDTA, 2 mM DTT, 0.5 mM phenylmethylsulfony fluoride, 20 mM b-glycerophosphate, 1 mM sodium vanadate and 1 μg/ml leupeptin). Equal amounts of protein extracts were resolved in 12% SDS-PAGE and the proteins of interest were probed by Western blot and visualized by enhanced chemiluminescence according manufacturer’s instructions (Amersham, Piscataway, NJ) (39, 40). For immunoprecipitation, cell were cultured in 100-mm dishes, treated as indicated in figure legends, and lysed in M2 buffer. Immunoprecipitation was performed as described previously (12, 41). Briefly, 20 μl protein A agarose beads (50%) were coupled to 1 μg TNFR1 antibody in PBS for 2 h at room temperature. Then 1 mg cell lysates were added and incubated with the beads by rotating overnight at 4 °C. The beads were washed seven times with M2 buffer. The immunoprecipitants were eluted off the beads using 2× electrophoresis sample buffer. The samples were boiled for 5 minand loaded up with 12% SDS-PAGE gel. Ubiquitinated RIP was detected by Western blot with an anti-RIP antibody.
Transfection, luciferase report assay, and RNA interference
Cells grown in 24-well plates were transfected with p5×κB-Luc and pRSV-LacZ with FuGENE 6 according to manufacturer’s instruction (Roche, Indianapolis, IN). Twenty-four hours after transfection, cells were treated as indicated in each figure legend. Luciferase activity was measured using a luciferase assay kit (Promega ) and normalized to β-galactosidase activity (11, 20). siRNA was transfected with INTERFERin™ (PolyPlus-Transfection, San Marcos, CA). Forty-eight hours post-transfection, cells were treated with SMC3 as described in figure legends, then followed by Western blot or cell death assay (11).
Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Total RNA was extracted with the RNAeasy kit (Qiagen, Valencia, CA). One microgram of RNA from each sample was used as a template for cDNA synthesis with a reverse transcription kit (Promega). An equal volume of cDNA product was used in the PCR. The primers used were: MnSOD, AGTTGCTGGAAGCCATCAAACGTG and TAAGGCCTGTTGTTCCTTGCAGTG; Bcl-XL, TGGGCTCACTCTTCAGTCGGAAAT and ATGTAGTGGTTCTCCTGGTGGCAA; TNF: AATCGGCCCGACTATCTCGACTTT and TTTGAGCCAGAAGAGGTTGAGGGT; and β-actin, CCAGCCTTCCTTCCTGGGCAT and AGGAGCAATGATCTTGATCTTCATT. The reaction condition was 94°C, 45 s; 55°C, 40 s; and 72°C, 45 s. For MnSOD, Bcl-XL, TNF, and β-actin, the cycles for PCR were 25, 27, 27 and 21, respectively. PCR products were run on 2 % agarose gel with 0.5 μg/ml ethedium bromomide, visualized, and photographed.
Statistical analysis
Data are summarized with means ± standard deviations (SD). The association between design factors and cell response was examined by either one- or two-way analysis of variance. To assess the potential for interactions between factors, we included interaction terms in the models. When tests indicated an interaction, 95% confidence intervals for the differences in means were obtained to assess the magnitude of the interaction. In all analyses, P < 0.05 was considered statistically significant.
Results
Transcription-independent TNF autocrine in SMC3-induced cytotoxicity in cancer cells
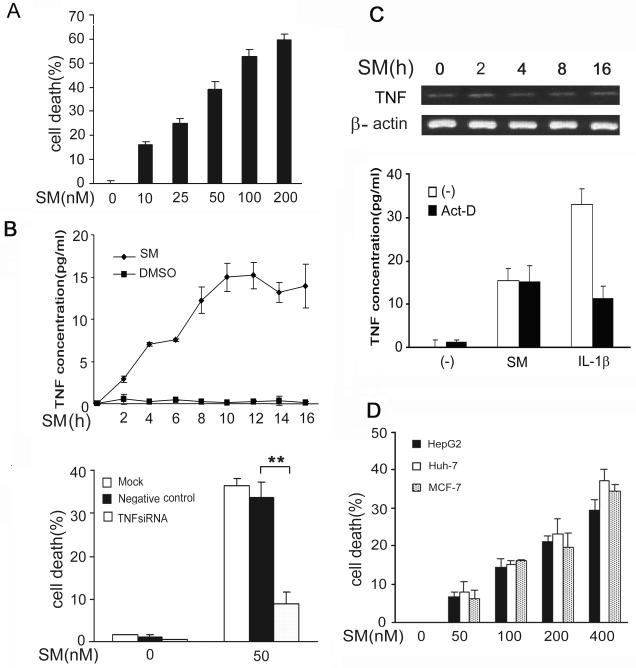
We found that the lung cancer cell line H23 is sensitive to SMC3-induced cytotoxicity. SMC3 kills this cell line in a dose-dependent manner, starting at the concentration as low as 10 nM induced cell death (Fig. 1A). The cytotoxicity induced by SMC3 was mainly apoptotic because SMC3 effectively activated caspase-8 and -3 and the cleavage of the caspase-3 substrate PARP, and the cytotoxicity was significantly suppressed by caspase inhibitors z-VAD-FMK (Fig. S1 and data not shown). The fact that the SMC3-induced apoptosis was efficiently blocked by a caspase-8 inhibitor IETD-CHO implies that the extrinsic apoptosis pathway was the main cell death pathway triggered by SMC3 (Fig. S2). We further detected SMC3-induced TNF secretion in the culture medium as early as 2 h post treatment (Fig. 1B, upper panel). The TNF siRNA that blocks TNF biosynthesis and TNF neutralizing antibody that blocks TNF binding to its receptor dramatically inhibited SMC3-induced cell death (Fig. 1B, lower panel and Fig. S3). Interestingly, SMC3 had no detectable effect on TNF message RNA expression, implying that the induction of TNF by SMC3 may not require activation of transcription on the tnf gene (Fig. 1C, upper panel). Pretreatment of the cells with the transcription inhibitor actinomycin D had no effect on SMC3-induced TNF secretion (Fig. 1C, lower panel). As a control, actinomycin D effectively blocked IL-1β-induced TNF increase in the culture medium (Fig. 1C, lower panel). These results suggest that SMC3-induced TNF autocrine is transcription-independent. The effect of TNF siRNA is likely through shutting off the constitutive TNF expression. Similar observations were made in the hepatoma cell lines HepG2 and Huh-7 and breast cancer cell line MCF-7, although the effective doses of SMC3 were much higher in these cells (Fig. 1D and data not shown). In agreement with and supplementary to previous reports (32-34), these results suggest that SMC3 induces apoptosis through TNF autocrine, which is independent of transcription, in cells derived from lung, breast and liver tumors.
Figure 1. SMC3-induced transcription-independent TNF autocrine is required for SMC3-induced cytotoxicity in cancer cells.
A, H23 cells were treated with indicated concentrations of SMC3 for 36 h. Cell death was measured by LDH leakage assay. Data shown are the mean ± SD. B, H23 cells were treated with SMC3 (50 nM) or DMSO for the indicated times. The concentrations of TNF in cell culture media were measured by ELISA. C, TNF mRNA was detected by RT-PCR. β-actin was detected as an input control. D, H23 cells were pretreated with actinomycin D (10 μM) for 30 min followed by exposure to SMC3 (50 nM) or IL1β (5 ng/ml) for 8 h. TNF was detected as described in B. E, H23 cells were mock transfected or transfected with 5 nM of TNF-siRNA or negative control siRNA. Forty-eight hours after transfection, the cells were treated with SMC3 (50 nM) for 36 h or left untreated. The concentrations of TNF in cell culture media were measured As in A. **p < 0.01. F, H23 cells were pretreated with TNF neutralizing antibodies (1 μg/ml) or control antibody (1 μg/ml) for 1 h followed by SMC3 (50 nM) treatment for 36 h. Cell death was measured by LDH release assay. **p < 0.01. G, HepG2, Huh-7, and MCF-7 cells were treated with indicated concentration of SMC3 for 36 h. Cell death was measured by as described in A.
The noncanonical pathway contributes marginally to SMC3-induced NF-κB activation and is dispensable for SMC3-induced TNF secretion
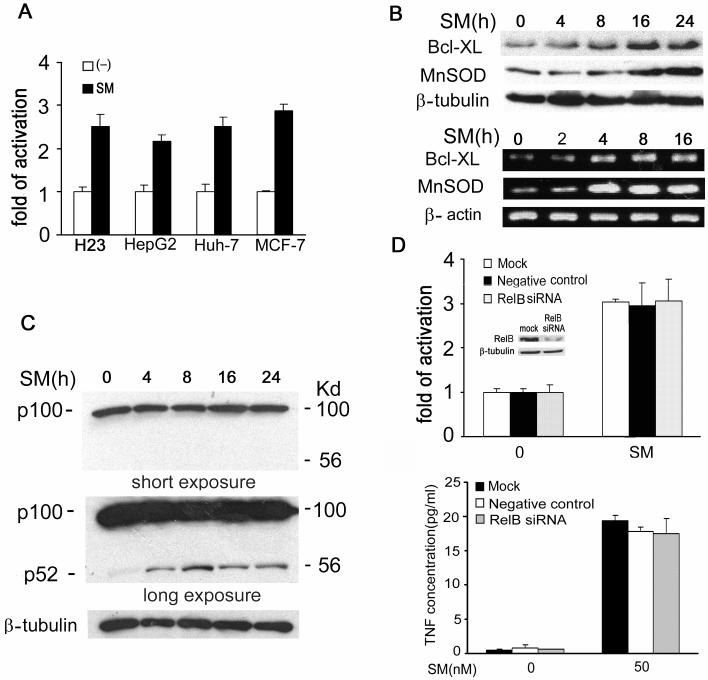
Previous reports suggested that SMC3 stimulates both the canonical and noncanonical NF-κB activation pathways (33, 34). However, the contribution of each pathway to SM-induced NF-κB activation was not determined. Thus, we sought to examine the mechanism by which SMC3 induces NF-κB activation. We confirmed that SMC3 induced NF-κB activation (Fig. 2A), and stimulated anti-apoptotic NF-κB targets’ expression at both the protein and mRNA levels in all the tested SMC3-sensitive cell lines (Fig. 2B). Consistent with previous reports, SMC3 was able to stimulate the noncanonical pathway, which was shown as generating the NF-κB p52 subunit by cleavage of the p100 precursor. The activation of the noncanonical pathway was quite moderate because no reduction of p100 was detected throughout the course of treatment and the p52 fragment could be detected only after long-time exposure (Fig. 2C, Upper and middle panels). Then we examined the contribution of the noncanonical pathway to the overall NF-κB activation by specifically blocking this pathway with siRNA, targeting the key component RelB, and with a NF-κB luciferase reporter assay that is sensitive to measure both the canonical and noncanonical pathway-mediated NF-κB activity (20, 42). The RelB siRNA efficiently blocked RelB expression (Fig. 2D upper panel, insert) but had no effect on SMC3-induced NF-κB activation (Fig. 2D). Additionally, there was no detectable effect of RelB siRNA on the SMC3-induced expression of the NF-κB target gene MnSOD (data not shown). The involvement of the noncanonical pathway in SMC3-induced TNF secretion was also tested with RelB siRNA. The results show that RelB is dispensable for SMC3-induced TNF secretion (Fig. 2D, lower panel). These results suggest that although SMC3 stimulates the processing of p100, the noncanonical pathway contributes marginally to the SMC3-induced overall NF-κB activation and TNF secretion.
Figure 2. The noncanonical pathway contributes marginally to SMC3-induced NF-κB activation and is dispensable for SMC3-induced TNF secretion.
A, H23, HepG2, Huh-7, and MCF-7 cells were cotransfected with p5×κB-Luc and pRSV-LacZ. Twenty-four hours after transfection the cells were treated with SMC3 (50 nM for H23 and 100 nM for the rest cells) for 24 h or left untreated. Luciferase activity was detected and normalized to β-galactosidase activity. Data shown are the mean ± SD. B, Upper panel, H23 cells were treated with SMC3 (50 nM) for the indicated times. Bcl-XL and MnSOD proteins were detected by Western blot. β-Tubulin was detected as an input control. Lower panel, Bcl-XL and MnSOD RNAs were detected by RT-PCR. β-actin was detected as an input control. C, Upper panel, H23 cells were treated with SMC3 (50nM) for indicated times. NF-κB p100 and p52 were detected by Western blot. For detecting the weak p52 signal, a long-time exposure was used (middle panel). β-Tubulin was detected as an input control. D, H23 cells were mock transfected or transfected with 5 nM of RelB-siRNA or negative control siRNA. Forty-eight h after transfection the cells were cotransfected with p5×κB-Luc and pRSV-LacZ. Twenty-four hours post-transfection the cells were treated with SMC3(50 nM) for 24 h or left untreated. Luciferase activity was detected and normalized to β-galactosidase activity. Insert, knockdown of RelB in H23 cells detected by Western blot. E, H23 cells were mock transfected or transfected with 5 nM of RelB- or negative control siRNA. Forty-eight h after transfection the cell were treated with SMC3 for 24 hours or left untreated; the TNF concentrations in the cell culture media were measured.
The canonical pathway mediates SMC3-induced NF-κB activation but is not required for SMC3-induced TNF secretion
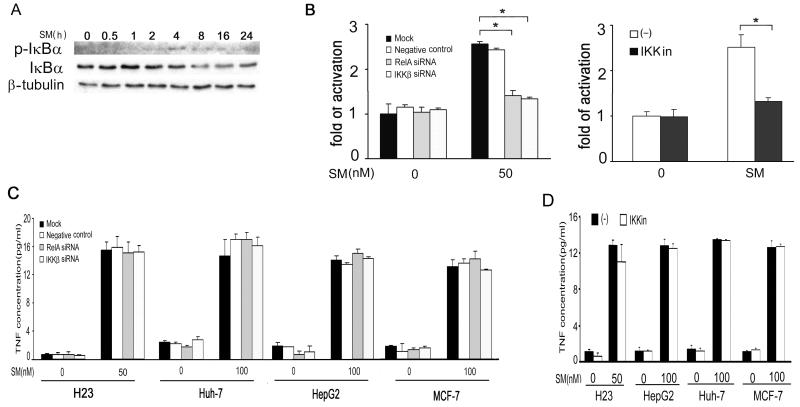
The canonical pathway was then examined by detection of the hallmark of this pathway, IκBα phosphorylation and degradation. The results showed that SMC3 triggered phosphorylation of IκBα, which was detected as early as 2 h and peaked at 4 -8 h post treatment (Fig. 3A). IκBα degradation was also seen following IκBα phosphorylation (Fig. 3A). siRNA targeting IKKβ or RelA, two key components of the canonical pathway, almost completely blocked SMC3-induced NF-κB activation (Fig. 3B). The efficiency of knockdown of IKKβ or RelA was confirmed by Western blot (Fig. S4). Together with the finding that blocking the noncanonical pathway had no effect on SMC3-induced NF-κB activity (Fig. 2D), these results suggest the canonical, but not the noncanonical, pathway is the main pathway for SMC3-induced NF-κB activation. The chemical IKK inhibitor, which blocks both the canonical and noncanonical pathways through suppressing IKKβ and IKKα, inhibited the SMC3-induced NF-κB activation in a similar efficiency as that of IKKβ or RelA siRNA (Fig. 3C), further supporting the conclusion that the canonical pathway is the main pathway for SMC3-induced overall NF-κB activation. To examined if NF-κB plays a role in SMC3-induced autocrine TNF, the H23, HepG2, Huh-7, and MCF-7 cells were transfected with siRNA targeting IKKβ or RelA to block SMC3-induced NF-κB activation and TNF in the culture media was detected by ELISA. IKKβ or RelA siRNA had no effect on SMC3-induced TNF secretion in all cell lines (Fig. 3C). The knockdown of IKKβ or RelA was confirmed by Western blot (Fig. S4) and blocking of SMC3-induced NF-κB activation was confirmed by luciferase reporter assay (Fig. 3B and data not shown). Consistent with the results with siRNA, the IKK inhibitor that blocks both the canonical and noncanonical NF-κB activation pathways did not affect autocrine TNF triggered by SMC3 (Fig. 3D). Taken together with the results that SMC3-induced TNF autocrine was independent of transcription, these experiments establish that NF-κB is dispensable for SMC3-induced autocrine TNF in cancer cells.
Figure 3. The canonical pathway mediates SMC3-induced NF-κB activation but is not required for SMC3-induced TNF secretion.
A, H23 cells were treated with SMC3 (50 nM) for various times as indicated. Phosphorylated- and total IκBα were detected by Western blot. β-Tubulin was detected as an input control. B, H23 cells were mock transfected or transfected with 5 nM of RelA-, IKKβ-, or negative control siRNA. The cells were treated with SMC3 (50 nM) for 24 h and NF-κB activity was analyzed. *p<0.05. C, H23 cells were cotransfected with p5×κB-Luc and pRSV-LacZ. Twenty-four hours post-transfection half of the cells were pretreated with IKK inhibitor II (10 μM) for 1 h followed by SMC3(50 nM) treatment for 24 h or left untreated, Luciferase activity was detected and normalized to β-galactosidase activity. *p<0.05. D, H23, HepG2, Huh-7 and MCF-7 cells were mock transfected or transfected with 5 nM of RelA-, IKKβ-, or negative control siRNA. Forty-eight hours after transfection the cells were cotransfected with p5×κB-Luc and pRSV-LacZ. Twenty-four hours post-transfection the cells were treated with indicated concentration of SMC3 for 24 h or left untreated. The concentrations of TNF in conditioned cell culture media were measured by ELISA. E, The efficiency of knockdown of RelA- and IKKβ-siRNA in H23, HepG2, Huh-7, and MCF-7 cells was detected by Western blot. β-Tubulin was detected as an input control. F, H23, HepG2, Huh-7, and MCF-7 cells were pretreated with IKK inhibitor II (10 nM) for 1 h followed by SMC3 (50 or 100 nM as indicated) treatment for 24 h or left untreated. The concentrations of TNF in conditioned cell culture media were measured by ELISA.
SMC3-induced NF-κB activation in cancer cells requires TNF secretion
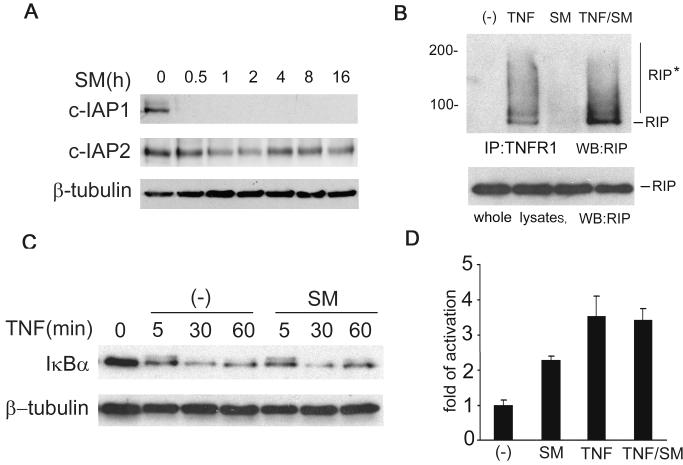
Because SMC3 induces NF-κB activation through the canonical pathway and triggers autocrine TNF, we hypothesize that SMC3 induces NF-κB activation through TNF secretion. Supporting this notion, SMC3-induced IκBα phosphorylation and degradation (4-8 h, Fig. 3A) and NF-κB targets’ expression (8-16 h, Fig. 2B) started at late time points, which was well correlated to the time course of TNF secretion (Fig. 1B). To further test this hypothesis, we first examined if the TNF-induced NF-κB activation pathway is normal in SMC3-treated cells. Consistent with the previous report (23), although SMC3 significantly suppressed the expression of c-IAP1, it only partially reduced the c-IAP2 level (Fig. 4A). Because that both c-IAP1 and c-IAP2 are involved in TNF-induced NF-κB activation through regulating RIP modification (16, 17), the retaining of c-IAP2 expression may be sufficient to allow the transmission of the NF-κB activation signal from TNFR1 to RIP. Indeed, SMC3 did not reduce recruitment of TNF-induced RIP to TNFR1 and the subsequent modification of RIP (Fig. 4B). Consistently, SMC3 had no detectable effect on TNF-induced IκBα degradation and NF-κB activation (Fig. 4C, 4D). These results suggest that the TNF-induced NF-κB activation pathway remains intact in SMC3-treated cells, allowing the secreted TNF to stimulate this pathway.
Figure 4. SMC3 does not inhibit TNF-induced NF-κB activation.
A, H23 cells were treated with SMC3 (50 nM) for the indicated times. cIAP-1 and cIAP-2 were detected by Western blot. β-Tubulin was detected as an input control. B, H23 cells were treated with SMC3 (50 nM) for 30 min followed by TNF (5 ng/ml) treatment for 10 min or left untreated. The cell lysates were immunoprecipitated with a TNFR1 antibody as described in Materials and Methods. RIP was detected by Western blot. The cell lysates (1% of the amounts used for IP) were probed for RIP as an input control. RIP*, modified RIP. C, H23 cells were treated with SMC3 (50 nM) for 30 min followed by TNF (5 ng/ml) for the indicated time points. IκBα was measured by We stern blot. β-Tubulin was detected as an input control. Note the phosphorylated IκBα shown as a weak shifted band at 5 min was comparable in samples treated with TNF alone or SMC3 plus TNF. D, H23 cells were cotransfected p5×κB-Luc and pRSV-LacZ. Twenty-four hours post-transfection the cells were treated as indicated. Luciferase activity was measured as in Fig. 2A.
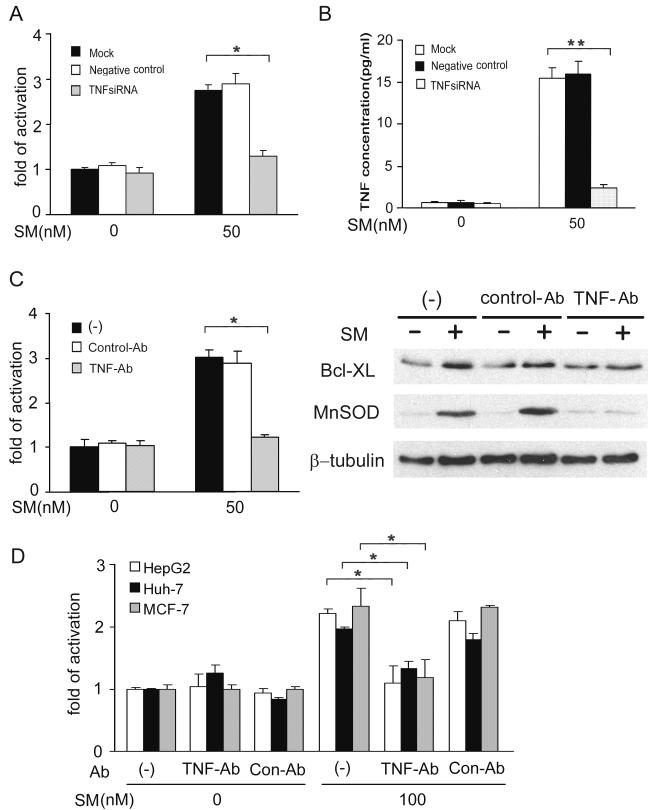
We then examined whether SMC3-induced NF-κB activation requires TNF. TNF siRNA almost completely blocked SMC3-induced NF-κB activation, which is associated with effective suppression of SMC3-triggered TNF secretion (Fig. 5A, 5B). In addition, the TNF neutralizing antibody also suppressed SMC3-induced NF-κB activation and NF-κB targets’ expression (Fig. 5C). The dependence of SMC3-induced NF-κB on TNF was also observed in HepG2, Huh-7 and MCF-7 cells (Fig. 5D). Furthermore, SMC3’s incapability of activating NF-κB was associated with its failure in TNF induction in the lung cancer cell line A549 (data not shown). Taken together, these results strongly suggest that the SMC3-induced NF-κB activation is dependent on autocrine TNF.
Fig 5. SMC3-induced NF-κB activation is TNF-dependent.
A, H23 cells were mock transfected or transfected with 5 nM of TNF-siRNA or negative control siRNA. Forty-eight h after transfection, the cells were cotransfected with p5×κB-Luc and pRSV-LacZ. After incubation for another 24 h, the cells were treated with SMC3 (50 nM) for 24 h or left untreated. Luciferase activity was detected and normalized to β-galactosidase activity. *p < 0.05. B, H23 cells were treated as in A. The concentration of TNF in culture media was determined by ELISA. **p < 0.01. C, H23 cells were cotransfected p5×κB-Luc and pRSV-LacZ. Twenty-four hours post-transfection the cells were pretreated with TNF neutralizing antibodies (1 μg/ml) or control antibody (1 μg/ml) for 1 h followed by 50 nM SMC3 treatment for 24 h. Luciferase activity was detected as in A. *p < 0.05. D, H23 cells were pretreated with TNF neutralizing antibodies (1 μg/ml) or control antibody (1 μg/ml) for 1 h followed by 50 nM SMC3 treatment for 24 h or left untreated. Bcl-XL and MnSOD were detected by Western blot. β-Tubulin was detected as an input control. E, HepG2, Huh-7, and MCF-7 cells were transfected, treated, and analyzed as described in C. *p < 0.05.
Blockage of NF-κB activation potentiates SMC3-induced cytotoxicity in cancer cells
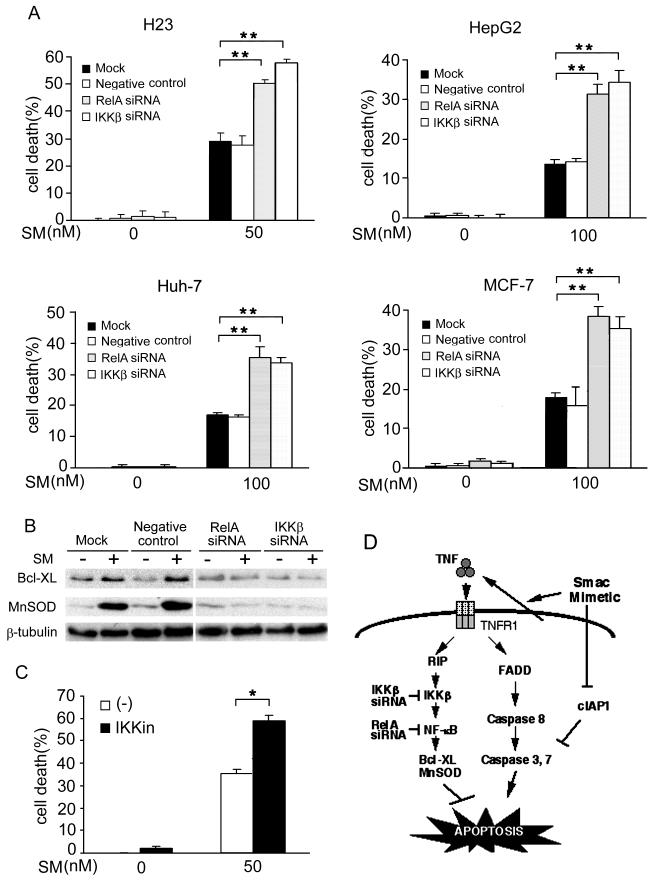
Because SMC3-induced cancer cell death occurs mainly through autocrine TNF, TNF-induced NF-κB activation remains normal in SMC3-treated cells, and NF-κB blocks TNF-induced apoptosis, we hypothesize that NF-κB plays an inhibitory role in SMC3-induced cancer cell death. To test this hypothesis IKK inhibitor and siRNA targeting IKKβ or RelA were employed to block SMC3-induced NF-κB activation and the effect of these reagents on SMC3-induced cell death was examined. Indeed, blockage of NF-κB by either siRNA or the IKK inhibitor significantly sensitized all the tested cells to SMC3-induced cytotoxicity (Fig. 6A, 6C). The substantial increase in cell death when either siRNA or the IKK inhibitor was paired with SMC3 was significantly higher than would be expected under an additive model, indicating significant synergy (p < 0.0001 for all cell lines). The sensitization of cell death was associated with suppression of SMC3-induced anti-apoptosis gene expression (Fig. 6B). These results suggest that SMC3-induced activation of NF-κB attenuates SMC3-induced cancer cell apoptosis and therefore blockage of NF-κB could be an effective approach in sensitizing SMC3’s anticancer efficacy.
Figure 6. Blocking NF-κB sensitizes SMC3-induced cytotoxicity in cancer cells.
A, H23, HepG2, Huh-7, and MCF-7 cells were mock transfected or transfected with 5 nM of RelA-, IKKβ-, or negative control siRNA. Forty-eight hours after transfection the cells were treated as indicated. Cell death was measured as described in Fig. 1A. **p < 0.01. B, H23 cells were transfected and treated as described in A. BCL-XL and MnSOD were measured by Western blot. β-Tubulin was detected as an input control. C, H23 cells were pretreated with IKK inhibitor II (10 μM) for 1 h followed by SMC3 (50 nM) treatment for 24 h or left untreated. Cell death was measured as in Fig. 1A. *p < 0.05. D, Model of SM-induced NF-κB activation and apoptosis. SM induces apoptosis through suppressing c-IAP1 expression and autocrine TNF. The SM-induced NF-κB activation is mainly through autocrine TNF, which blocks apoptosis. Blockage of the NF-κB pathway by targeting IKKβ or RelA sensitizes SM-induced apoptosis.
Discussion
This report delineates our systematic investigation into the mechanism of NF-κB activation by SM and its role in SM-induced cancer cell death. The results show that although SMC3 can stimulate a slight processing of p100, the noncanonical pathway marginally contributes to the overall NF-κB activity induced by SMC3. The NF-κB activation by SMC3 is mainly through the canonical pathway that is dependent on autocrine TNF. Although SMC3 suppresses c-IAP1 expression, it does not interfere with TNF-induced NF-κB activation or expression of anti-apoptotic NF-κB targets. SMC3-induced autocrine TNF is NF-κB independent and blockade of NF-κB efficiently sensitized SMC3-induced cytotoxicity in different cancer cell types. Thus, blockage of NF-κB activation suppresses cell survival while does not compromise the apoptosis pathway, shifting the outcome of the SM-responding cells to death (Fig. 6D).
Consistent with recent reports, SMC3 stimulated both the canonical and noncanonical NF-κB activation pathways (33, 34). However, we found that the noncanonical pathway was only moderately activated and contributed marginally to the overall NF-κB activity, which was demonstrated by specific blockage of this pathway with a RelB siRNA and an IKK inhibitor. The result is in agreement with the observations that the canonical NF-κB pathway is the main pathway in cancer cells (43, 44). Thus, the pathophysiological role of the noncanonical pathway in a cell’s response to SM remains to be determined. Although the activation of the noncanonical NF-κB pathway was determined to be through suppression of c-IAP1-mediated degradation of NIK (33), how the canonical NF-κB activation pathway is activated by SM has not been determined. Our results demonstrate that activation of the canonical NF-κB pathway by SMC3 is achieved by induction of autocrine TNF, because inhibition of TNF by either a neutralizing antibody or siRNA against TNF effectively blocked SMC3-induced NF-κB activity.
We provide evidence generated with different approaches showing this process does not require NF-κB. First, the IKK inhibitor II, which blocks both the canonical and noncanonical pathways through inhibiting IKKα and IKKβ, had no effect on SMC3-induced TNF secretion. Second, although it effectively suppressed the SMC3-induced NF-κB activation, siRNA targeting either IKKβ or RelA did not show a detectable effect on SMC3-triggered TNF secretion. Third, the RelB siRNA, which blocks the noncanonical pathway, had no effect on autocrine TNF. Additionally, there was no detectable change of TNF mRNA following SMC3 treatment (Fig. 1C) and suppression of transcription did not inhibit SMC3-induced TNF secretion (Fig. 1D), further supporting that transcription of TNF gene is not involved in SMC3-induced TNF autocrine. This is inconsistent with a recent report showing that the TNF protein expression in rhabdomyosarcoma (Kym1) and ovarian cancer (SKOV3) cells induced by a SM was dependent on NF-κB (34). The discrepancy is likely due to different SMs used (34, 36), because similar as in other cell lines we tested, NF-κB blockage had no effect on SMC3-induced TNF autocrine while significantly enhanced SMC3-induced cytotoxicity in SKOV3 cells (Fig S5). As it is does not involve transcriptional regulation, SMC3-induced TNF autocrine is likely through stimulating TNF from the cell membrane because protein synthesis inhibitor cycloheximide had no effect on SMC3-indcued TNF secretion while TACE inhibitor, which specifically suppresses TNF release, dramatically suppressed this effect of SMC3 (Fig. S6).
We found that while SMC3 induced effective cytotoxicity, it had no inhibitory effect on TNF-induced NF-κB activation, which is consistent with a recent report using the same compound by Wang and colleagues (23). Indeed, although SMC3 caused dramatic downregulation of c-IAP1, it had minimal effect on c-IAP2 expression. The modification of RIP and activation of the NF-κB targets were retained intact in SMC3-treated cells, allowing the activation of NF-κB by TNF (Fig. 4). Additionally, modification of RIP was found in cells after long time (8h) SMC3 exposure (data not shown), presumably through autocrine TNF. This is not contradictory to the reports that high doses of SM cause extensive RIP deubiquitination and switch the TNF-induced signaling from RIP-mediated NF-κB activation to caspase-8 activation that results in apoptosis (23, 35). It is reasonable to speculate that under our conditions apoptosis caused by SMC3 occurs mainly through the c-FLIP-regulated caspase-8 activation pathway, because RIP modification was retained and NF-κB activation was unaffected (23).
We further determined the effect of NF-κB blockage on SMC3-induced cancer cell death. This is based on the fact that SMC3 kills cancer cells mainly through autocrine TNF (32-34), NF-κB is activated by SMC3 through TNF, and NF-κB blocks TNF-induced apoptosis (5, 37, 45, 46). The results demonstrated a substantial potentiation of SMC3-induced cell death by blocking the canonical NF-κB activation pathway with either chemical IKK inhibitor or siRNA targeting IKKβ or RelA. This potentiation is well correlated to the suppression of SMC3-induced expression of the anti-apoptotic NF-κB targets. Thus, combination of NF-κB-blocking means that target IKKβ or RelA and SM could be a useful strategy to improve the anticancer efficacy of SM. This approach may also reduce the adverse effects of SM by reducing the SM dosages without lowering the cancer cell-killing activity. Further in vivo studies are needed to verify the usefulness of NF-κB blockage in improving SM’s anticancer efficacy.
Supplementary Material
Acknowledgements
We thank Drs. Xiaodong Wang for providing the Smac mimetic compound 3 and Chris Stidley for help in statistical analyses, respectively, and V. Fisher for editing the manuscript. This study was partly supported by a grant from NCI/NIH (R03CA125796).
Abbreviations
- IAP
inhibitor of apoptosis protein
- NF-κB
nuclear factor κB
- NIK
NF-κB inducing kinase
- Smac
second mitochondria-derived activator of caspase
- SM
Smac mimetics
- SMC3
SM compound 3
- TNF
tumor necrosis factor
- TNFR1
TNF receptor 1
References
- 1.Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol. 2003;3:745–56. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- 2.Karin M. The IkappaB kinase - a bridge between inflammation and cancer. Cell Res. 2008;18:334–42. doi: 10.1038/cr.2008.30. [DOI] [PubMed] [Google Scholar]
- 3.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–62. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 4.Kamata H, Honda S, Maeda S, Chang L, Hirata H, Karin M. Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell. 2005;120:649–61. doi: 10.1016/j.cell.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Ju W, Renouard J, Aden J, Belinsky SA, Lin Y. 17-allylamino-17-demethoxygeldanamycin synergistically potentiates tumor necrosis factor-induced lung cancer cell death by blocking the nuclear factor-kappaB pathway. Cancer Res. 2006;66:1089–95. doi: 10.1158/0008-5472.CAN-05-2698. [DOI] [PubMed] [Google Scholar]
- 6.Singh NP, Nagarkatti M, Nagarkatti PS. Role of dioxin response element and nuclear factor-kappaB motifs in 2,3,7,8-tetrachlorodibenzo-p-dioxin-mediated regulation of Fas and Fas ligand expression. Mol Pharmacol. 2007;71:145–57. doi: 10.1124/mol.106.028365. [DOI] [PubMed] [Google Scholar]
- 7.Shou Y, Li N, Li L, Borowitz JL, Isom GE. NF-kappaB-mediated up-regulation of Bcl-X(S) and Bax contributes to cytochrome c release in cyanide-induced apoptosis. J Neurochem. 2002;81:842–52. doi: 10.1046/j.1471-4159.2002.00880.x. [DOI] [PubMed] [Google Scholar]
- 8.Kikuchi H, Ozaki T, Furuya K, et al. NF-kappaB regulates the stability and activity of p73 by inducing its proteolytic degradation through a ubiquitin-dependent proteasome pathway. Oncogene. 2006;25:7608–17. doi: 10.1038/sj.onc.1209748. [DOI] [PubMed] [Google Scholar]
- 9.Janssens S, Tschopp J. Signals from within: the DNA-damage-induced NF-kappaB response. Cell Death Differ. 2006;13:773–84. doi: 10.1038/sj.cdd.4401843. [DOI] [PubMed] [Google Scholar]
- 10.Hur GM, Lewis J, Yang Q, et al. The death domain kinase RIP has an essential role in DNA damage-induced NF-kappa B activation. Genes Dev. 2003;17:873–82. doi: 10.1101/gad.1062403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W, Wang X, Bai L, Liang X, Zhuang J, Lin Y. Blockage of NF-kappaB by IKKbeta- or RelA-siRNA rather than the NF-kappaB super-suppressor IkappaBalpha mutant potentiates adriamycin-induced cytotoxicity in lung cancer cells. J Cell Biochem. 2008;105:554–61. doi: 10.1002/jcb.21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devin A, Cook A, Lin Y, Rodriguez Y, Kelliher M, Liu Z. The distinct roles of TRAF2 and RIP in IKK activation by TNF-R1: TRAF2 recruits IKK to TNF-R1 while RIP mediates IKK activation. Immunity. 2000;12:419–29. doi: 10.1016/s1074-7613(00)80194-6. [DOI] [PubMed] [Google Scholar]
- 13.Yang J, Lin Y, Guo Z, et al. The essential role of MEKK3 in TNF-induced NF-kappaB activation. Nat Immunol. 2001;2:620–4. doi: 10.1038/89769. [DOI] [PubMed] [Google Scholar]
- 14.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–59. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 15.Roy N, Deveraux QL, Takahashi R, Salvesen GS, Reed JC. The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J. 1997;16:6914–25. doi: 10.1093/emboj/16.23.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varfolomeev E, Goncharov T, Fedorova AV, et al. c-IAP1 and c-IAP2 are critical mediators of tumor necrosis factor alpha (TNFalpha)-induced NF-kappaB activation. J Biol Chem. 2008;283:24295–9. doi: 10.1074/jbc.C800128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahoney DJ, Cheung HH, Mrad RL, et al. Both cIAP1 and cIAP2 regulate TNFalpha-mediated NF-kappaB activation. Proc Natl Acad Sci U S A. 2008;105:11778–83. doi: 10.1073/pnas.0711122105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ting AT, Pimentel-Muinos FX, Seed B. RIP mediates tumor necrosis factor receptor 1 activation of NF-kappaB but not Fas/APO-1-initiated apoptosis. Embo J. 1996;15:6189–96. [PMC free article] [PubMed] [Google Scholar]
- 19.Kelliher MA, Grimm S, Ishida Y, Kuo F, Stanger BZ, Leder P. The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity. 1998;8:297–303. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- 20.Lin Y, Devin A, Rodriguez Y, Liu ZG. Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev. 1999;13:2514–26. doi: 10.1101/gad.13.19.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–57. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 22.Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol. 2005;7:758–65. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Kobayashi M, Blonska M, You Y, Lin X. Ubiquitination of RIP is required for tumor necrosis factor alpha-induced NF-kappaB activation. J Biol Chem. 2006;281:13636–43. doi: 10.1074/jbc.M600620200. [DOI] [PubMed] [Google Scholar]
- 25.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected] Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 26.Verhagen AM, Ekert PG, Pakusch M, et al. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 2000;102:43–53. doi: 10.1016/s0092-8674(00)00009-x. [DOI] [PubMed] [Google Scholar]
- 27.Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 28.Chai J, Du C, Wu JW, Kyin S, Wang X, Shi Y. Structural and biochemical basis of apoptotic activation by Smac/DIABLO. Nature. 2000;406:855–62. doi: 10.1038/35022514. [DOI] [PubMed] [Google Scholar]
- 29.Wu H, Tschopp J, Lin SC. Smac mimetics and TNFalpha: a dangerous liaison? Cell. 2007;131:655–8. doi: 10.1016/j.cell.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu J, Bai L, Sun H, et al. SM-164: a novel, bivalent Smac mimetic that induces apoptosis and tumor regression by concurrent removal of the blockade of cIAP-1/2 and XIAP. Cancer Res. 2008;68:9384–93. doi: 10.1158/0008-5472.CAN-08-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bank A, Wang P, Du C, Yu J, Zhang L. SMAC mimetics sensitize nonsteroidal anti-inflammatory drug-induced apoptosis by promoting caspase-3-mediated cytochrome c release. Cancer Res. 2008;68:276–84. doi: 10.1158/0008-5472.CAN-07-5242. [DOI] [PubMed] [Google Scholar]
- 32.Petersen SL, Wang L, Yalcin-Chin A, et al. Autocrine TNFalpha Signaling Renders Human Cancer Cells Susceptible to Smac-Mimetic-Induced Apoptosis. Cancer Cell. 2007;12:445–56. doi: 10.1016/j.ccr.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varfolomeev E, Blankenship JW, Wayson SM, et al. IAP Antagonists Induce Autoubiquitination of c-IAPs, NF-kappaB Activation, and TNFalpha-Dependent Apoptosis. Cell. 2007;131:669–81. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 34.Vince JE, Wong WW, Khan N, et al. IAP Antagonists Target cIAP1 to Induce TNFalpha-Dependent Apoptosis. Cell. 2007;131:682–93. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 35.Bertrand MJ, Milutinovic S, Dickson KM, et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 36.Li L, Thomas RM, Suzuki H, De Brabander JK, Wang X, Harran PG. A small molecule Smac mimic potentiates TRAIL- and TNFalpha-mediated cell death. Science. 2004;305:1471–4. doi: 10.1126/science.1098231. [DOI] [PubMed] [Google Scholar]
- 37.Ju W, Wang X, Shi H, Chen W, Belinsky SA, Lin Y. A critical role of luteolin-induced reactive oxygen species in blockage of tumor necrosis factor-activated nuclear factor-kappaB pathway and sensitization of apoptosis in lung cancer cells. Mol Pharmacol. 2007;71:1381–8. doi: 10.1124/mol.106.032185. [DOI] [PubMed] [Google Scholar]
- 38.Chen W, Wang X, Zhuang J, Zhang L, Lin Y. Induction of death receptor 5 and suppression of survivin contribute to sensitization of TRAIL-induced cytotoxicity by quercetin in non-small cell lung cancer cells. Carcinogenesis. 2007;28:2114–21. doi: 10.1093/carcin/bgm133. [DOI] [PubMed] [Google Scholar]
- 39.Wang X, Chen W, Lin Y. Sensitization of TNF-induced cytotoxicity in lung cancer cells by concurrent suppression of the NF-kappaB and Akt pathways. Biochem Biophys Res Commun. 2007;355:807–12. doi: 10.1016/j.bbrc.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 40.Wang X, Chen W, Zeng W, et al. Akt-mediated eminent expression of c-FLIP and Mcl-1 confers acquired resistance to TRAIL-induced cytotoxicity to lung cancer cells. Mol Cancer Ther. 2008;7:1156–63. doi: 10.1158/1535-7163.MCT-07-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin Y, Yang Q, Wang X, Liu ZG. The essential role of the death domain kinase receptor-interacting protein in insulin growth factor-I-induced c-Jun N-terminal kinase activation. J Biol Chem. 2006;281:23525–32. doi: 10.1074/jbc.M601487200. [DOI] [PubMed] [Google Scholar]
- 42.Kim YS, Nedospasov SA, Liu ZG. TRAF2 plays a key, nonredundant role in LIGHT-lymphotoxin beta receptor signaling. Mol Cell Biol. 2005;25:2130–7. doi: 10.1128/MCB.25.6.2130-2137.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121:977–90. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 44.Greten FR, Eckmann L, Greten TF, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–96. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 45.Liu ZG, Hsu H, Goeddel DV, Karin M. Dissection of TNF receptor 1 effector functions: JNK activation is not linked to apoptosis while NF-kappaB activation prevents cell death. Cell. 1996;87:565–76. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 46.Wang X, Lin Y. Tumor necrosis factor and cancer, buddies or foes? Acta Pharmacol Sin. 2008;29:1275–88. doi: 10.1111/j.1745-7254.2008.00889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.