Abstract
The combined phenotypic expression of CD11clow B220+ CD122+ DX5+ has been used to define a novel cell type, termed interferon-producing killer dendritic cells (IKDC). IKDC readily produce IFN-γ and demonstrate spontaneous cytotoxic activity towards tumors, suggesting that a modulation of IKDC number may be beneficial in cancer treatment. We examined various mouse strains and found that IKDC number was highly variable between the different strains. A linkage analysis associated the distal arm of chromosome 7 with variations in IKDC number. The genetic contribution of chromosome 7 to the regulation of IKDC number was confirmed through the use of congenic mice. We further demonstrate that IKDC proportion is regulated by intrinsic hematopoietic factors. We discuss the role of various candidate genes in the regulation of this newly described cell type and its implication in therapy.
Keywords: Dendritic Cells, Natural Killer Cells, Tumor Immunity
Introduction
The idea of a dendritic cell type able to trigger cytotoxicity towards MHC-deficient cells and provide antigen presentation to T cells was first presented over ten years ago in a rat model (1). More recently, two groups clearly identified a cell type carrying phenotypic and functional properties of both NK cell and plasmacytoid dendritic cells (pDC), which was termed interferon-producing killer dendritic cells (IKDC) (2, 3). On the one hand, as for NK cells, IKDC express various NK cell surface markers such as DX5, are able to mediate cytolysis of MHC class I deficient target cells and produce vast amounts of IFN-γ (4). On the other hand, as for pDC, IKDC can be distinguished according to CD11clow B220+ expression and are able to induce T cell proliferation (2). IKDC were thus suggested to provide a direct bridge between innate and adaptive immunity through their ability to mediate both cytolysis and antigen-presentation. However, the term “IKDC” has stimulated a debate regarding the lineage relationship of this cell type to DC. Indeed, it was recently demonstrated that IKDC share more similarity with NK cells than DC (4-7). One group proposes that IKDC are merely activated NK cells (7). Regardless, recent evidence suggests that IKDC are indeed a cell type distinct from NK cells. Firstly, their bone marrow differentiation proceeds through a unique pathway (8). Secondly, their ability to respond to IL-15 through trans-presentation differs from NK cells (9). Finally, their transcriptional profile, although more similar to NK cells than DC, is clearly distinct from the former (10). Thus, although IKDC may be more closely related to NK cells than DC, they appear to define a distinct subset. For simplicity, we have opted to use “IKDC” to describe the CD11cintB220+DX5+CD122+ phenotype. Further characterization of this phenotype will be required to delineate the relationship between NK and IKDC.
Notwithstanding, the contribution of IKDC in tumor clearance has been clearly observed in various systems. Indeed, IKDC contribute to tumour immunosurveillance via secretion of high levels of IFN-γ and TRAIL-dependent lysis of tumor cells (3, 11). Therefore, modulating IKDC number may be beneficial in cancer treatment. This prompted us to examine the regulation of IKDC number. We found that each mouse strains carried a set number of IKDC. However, this number was highly variable between strains, where NOD and B10.Br mice respectively showed the lowest and highest proportion of IKDC. A linkage analysis of F2 generation mice from a B10.Br to NOD outcross associated the regulation of IKDC number to the distal arm of chromosome 7. Moreover, we demonstrate that IKDC number is regulated by intrinsic hematopoietic factors. We discuss potential genes regulating IKDC number which may contribute to the elaboration of novel cancer therapies.
Research Design and Methods
Mice
B10.Br, NOD.H2k, F1 (B10.Br x NOD.H2k) and F2 (F1 X F1) mice were maintained at Maisonneuve-Rosemont Hospital housing facility. BALB/c, C3H, C57BL/6 and NOR mice were purchased from Jackson Laboratory. NOD.Lc7 mice (formerly designated NOD.DR-4, (12)) were bred in the vivaria at the University of Virginia. 8 to 12 week-old mice were used for phenotypic analyses. F1 (B10.Br x NOD.H2k) recipients were irradiated at 11 Gy and left to reconstitute for 8 weeks with 2×106 bone marrow cells from B10.Br and NOD.H2k at 1:1 ratio. The Maisonneuve-Rosemont Hospital ethics committee overseen by the Canadian Council for Animal Protection approved all experimental procedures.
Flow cytometry
Spleen and lymph nodes are treated with collagenase (Collagenase of Clostridium histolyticum, Type V, Sigma-Aldrich) for 15 minutes at 37°C and passed through a cell strainer. All antibodies were purchased from Biolegend except CD11c (N418)-biotin and B220-FITC which were produced in the laboratory. Data were acquired using a FACSCalibur (BD Biosciences) and analyzed using FlowJo software (Treestar).
Linkage analysis
Genomic DNA was isolated from the tails of F2 male and female mice by using the DNeasy Blood & Tissue Kit from Qiagen and single nucleotide polymorphisms were determined using the Illumina Mouse Low Density Linkage panel serviced through The Centre for Applied Genomics at The Hospital for Sick Children, Ontario, Canada. The LOD score was obtained through a single QTL model using the library R/qtl of R software (ver 2.6.2). No impact of gender was observed (p=0.062 non-parametric, Mann-Whitney).
Results
Multiple cell surface markers have been used to define IKDC (2-4, 13). Similar to pDC, IKDC have been best defined by the expression of a low level of CD11c, along with B220 (2, 3). In addition, they express markers usually associated to NK cells, such as DX5 and CD122 (2, 5). Figure 1a shows that we can readily identify a subset of cells expressing a CD11clow B220+ DX5+ CD122+ phenotype in both spleen and lymph nodes. These plots demonstrate that not all CD11clow B220+ DX5+ cells express CD122, while all CD11clow B220+ CD122+ cells express DX5. Therefore, CD122 was routinely used in the characterization of this cell subset, as it serves as a more specific marker to define this rare cell population.
Figure 1.
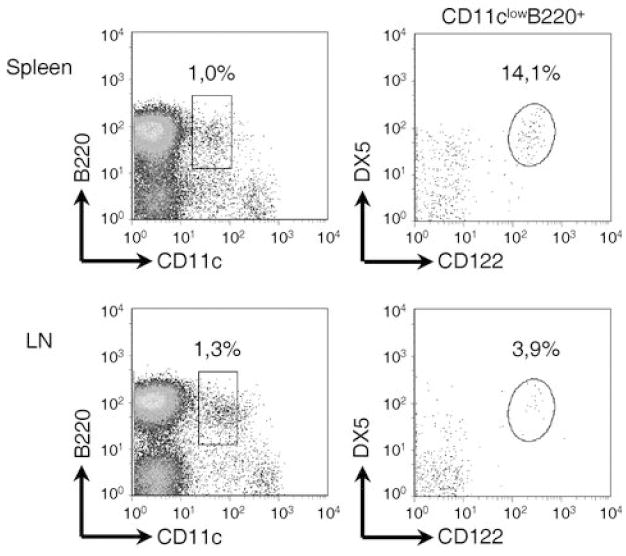
Interchangeable use of CD122 and DX5 to identify IKDC. IKDC are phenotyped according to CD11clowB220+DX5+CD122+ cells in both spleen (top) and lymph nodes (bottom).
We next undertook the challenge of defining the genetic factors regulating IKDC number. We examined the proportion and absolute number of IKDC in various mouse strains (Figure 2). Each strain showed a relatively fixed proportion and number of IKDC. However, a high variation of IKDC proportion was noted between mouse strains. For instance, B10.Br and C57BL/6 carried a high number of IKDC, while NOD mice showed a very low IKDC number (IKDC number, B10.Br vs NOD p value < 1×10-4). This variation in IKDC number between strains did not correlate with NK cell number (supplemental figure 1) and suggests that genetic factors determine IKDC number. Alternatively, the reduced IKDC number in NOD mice was possibly due to inflammation associated with autoimmune diabetes predisposition. To verify this hypothesis, we assessed the number of IKDC in NOD mice congenic for the MHC H2k locus (NOD.H2k). The MHC locus is the major susceptibility factor in NOD mice and thus NOD.H2k mice do not develop diabetes (14). Yet, NOD and NOD.H2k mice had similar numbers of IKDC (Figure 2). These results suggest that the decrease in IKDC in NOD mice is not a consequence of autoimmune diabetes development. Regardless, NOD.H2k mice still carry all non-MHC genes associated with autoimmune susceptibility, and as for NOD mice, they remain susceptible to other autoimmune diseases (15), which may provide an inflammatory state and influence IKDC number. However, the BALB/c strain is resistant to autoimmune diseases, does not show inflammation and also exhibits a relatively low number of IKDC (Figure 2). Interestingly, these results also demonstrate that IKDC proportion is not regulated by the MHC locus, as NOD and NOD.H2k mice show a comparable number of IKDC. This observation is further supported by the fact that C57BL/6 (H2b) and B10.Br (H2k), which are essentially genetically related bar the MHC, also show a comparable number of IKDC (Figure 2). Further evidence that IKDC number is regulated by genetic factors, came from the evaluation of IKDC proportion in NOR mice. NOR is a NOD-related strain with prominent non-NOD intervals on chromosomes 1, 2, 4, 5, 7, 11, 12, and 18 (16). As the proportion of IKDC is significantly different between NOD and NOR strains (p value < 1×10-3), these results suggests that at least one of the genetic intervals differing between the two strains is responsible for defining IKDC number.
Figure 2.
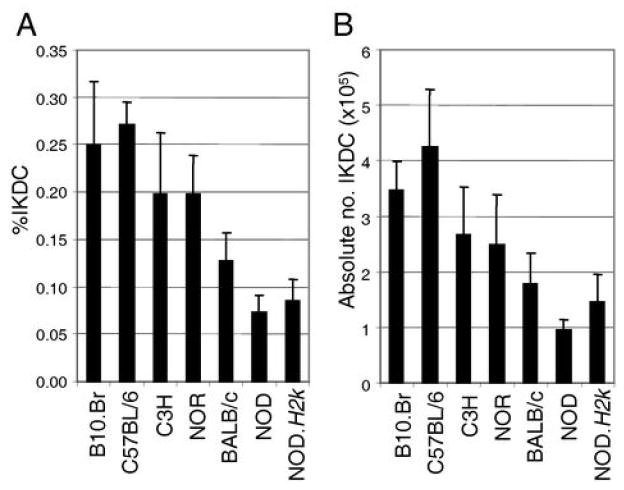
IKDC proportion and number in various mouse strains. A) The proportion and B) the absolute number of IKDC (CD11clowB220+CD122+ cells) in the spleen are shown for the indicated strains of mice. n<3.
We have determined that IKDC proportion is highest in C57BL/6 and B10.Br strains and lowest in NOD and NOD.H2k mice. For linkage analysis, we opted to perform the F2 outcross using B10.Br and NOD.H2k mice. These strains were selected for three reasons. First, they showed the highest differential in the proportion of IKDC, allowing for a better segregation of the F2 phenotypes. Second, the results would not be confounded by possible autoimmune diabetes development as NOD.H2k mice do not develop diabetes. Finally, both B10.Br and NOD.H2k strains carry the same MHC locus and our data indicate that this parameter does not regulate IKDC number.
A linkage analysis was conducted on 106 F2 mice from the B10.Br to NOD.H2k outcross. The F2 mice showed a broad distribution of IKDC frequency, suggesting that the trait is multigenic (Figure 3a). Linkage analysis using the Illumina low density platform demonstrated a suggestive linkage on chromosome 7 and a weak association with chromosome 19 (Figure 3b). To confirm the impact of chromosome 7 on the proportion of IKDC, we took advantage of congenic NOD.Lc7 mice which carry C57L alleles on the corresponding chromosome 7 interval (Figure 3c). NOD.Lc7 mice showed a high proportion of IKDC, confirming that this genetic locus regulates the proportion of these cells (Figure 3d).
Figure 3.
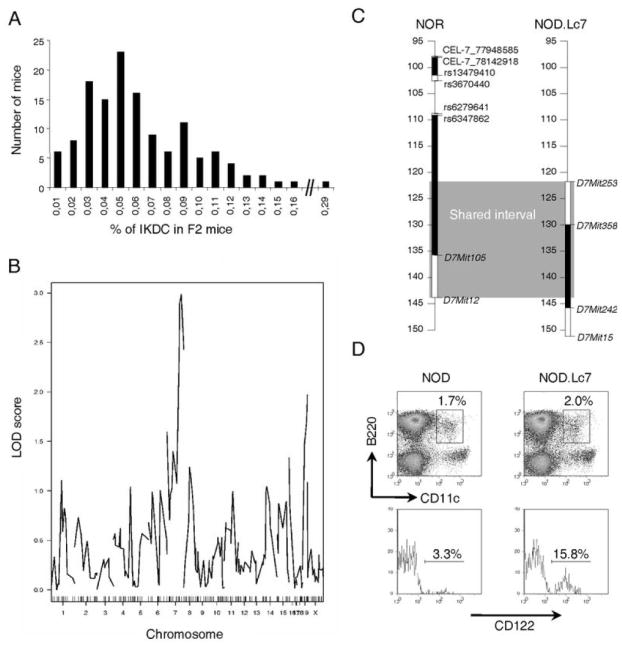
Regulation of IKDC proportion by distal region of chromosome 7. A) The proportion of IKDC was evaluated in 106 F2 mice. Shown is the distribution of mice for the %IKDC. B) Logarithm of odds plot for the %IKDC in F2 cohorts is shown. Chr 7, p < 0.01 and Chr 19, p < 0.05. C) Schematic representation of the shared diabetes-resistant intervals between NOR and NOD.Lc7 mice. The genetic region is delimited by D7Mit253 and D7Mit12. Marker placement is determined according to the NCBI m37 build. D) The proportion of IKDC is restored in NOD.Lc7 congenic mice. Data are representative of three experiments.
The interval on chromosome 7 is rather large and carries many potential candidate genes. To determine whether IKDC proportion was regulated by cell-intrinsic hematopoietic factors, which would restrict the candidate gene search, we performed bone marrow chimera experiments. B10.Br and NOD.H2k bone marrow were mixed in a 1:1 ratio in a lethally irradiated F1 (B10.Br x NOD.H2k) recipient and left to reconstitute for 8 weeks prior to analysis. Taking advantage of congenic markers for each strain, we found that the B10.Br and NOD.H2k bone marrow reconstituted the recipient in relatively equal proportions, although NOD.H2k consistently showed a slight advantage (Figure 4, left panel). However, the proportion of IKDC originating from NOD.H2k was always reduced in comparison to B10.Br (Figure 4, right panel). These results demonstrate that the proportion of IKDC is regulated by cell-intrinsic hematopoietic factors, thus greatly restricting the list of potential candidate genes.
Figure 4.
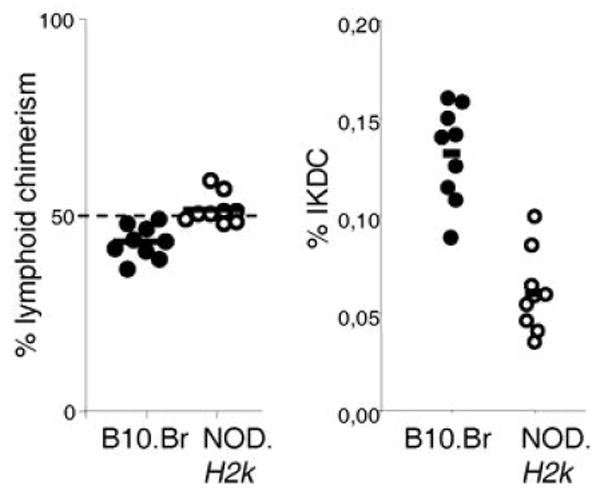
IKDC proportion is dependent on hematopoietic-intrinsic factors. CD45.1 and CD45.2 were respectively used to establish the level of bone marrow reconstitution from NOD.H2k and B10.Br mice in the spleen of an F1 recipient (left panel). The proportion of IKDC among all B10.Br or NOD.H2k cells was calculated by gating on CD45.1- or CD45.2- cells, respectively (right panel).
Discussion
IKDC have a defined tumoricidal activity (3, 11) and modulation of their numbers may yield important clinical applications. Here, we performed a linkage analysis to define the genetic parameters regulating IKDC number. We have found that the distal portion of chromosome 7 strikingly impacts on the proportion of IKDC. Indeed, both NOD.Lc7 and NOR mice, which are congenic for a C57 genetic region on chromosome 7, show a high proportion of IKDC. Moreover, the proportion of IKDC is regulated within the hematopoietic compartment, greatly restricting the potential candidate gene search. Nevertheless, IL4ra, IL21r, IL27, CD19, LAT and Mapk3 form only a short list of the potential candidate genes included within the interval defined by the loci delimiting the congenic interval regulating IKDC proportion in NOD.Lc7: D7Mit253 and D7Mit12. For example, LAT adaptative protein increases human NK cell cytotoxicity (17) and IL21 is known to promote pathogenic Th17 responses as well as to enhance both CD8 and NK cell cytotoxic activity specifically against tumor cells (18). Comparative gene profiling of IKDC from NOD and NOD.Lc7 will facilitate the identification of the genetic elements controlling this phenotype.
The NOD.Lc7 IKDC number is quite compelling and confirms the linkage results suggesting that the chromosome 7 interval provides the major genetic control of IKDC proportion. However, the broad distribution of IKDC frequencies in F2 mice illustrates that the proportion of IKDC is regulated as a multigenic trait. Moreover, it revealed a suggestive linkage on chromosome 19. Surprisingly, both the chromosome 7 and 19 intervals associate with accrued T cell IL-4 secretion (19). As IKDC rapidly produce high levels of IFN-γ upon activation, it is unlikely that they promote IL-4 production. Rather, this observation may point to the fact that the relatively large genetic regions identified may not only contribute to IKDC proportion: they may impact various other pathways including resistance to oxidative stress (20). Moreover, in NOD mice, other genetic regions of chromosome 7 have been shown to impact the inflammatory response (21). The contribution of these multiple genetic regions towards immune regulation remains to be thoroughly addressed.
In conclusion, NOD mice demonstrate a severe reduction in IKDC number, which can be restored by genetic elements contained within the distal arm of chromosome 7. Interestingly, NOD.SCID or RAG-deficient mice develop thymomas while the SCID and RAG deficiency on other genetic backgrounds does not promote thymic tumor development (22). It remains to be determined whether restoration of IKDC number in NOD.SCID mice will impact on thymomagenesis. Future work will address whether NOD mice are generally more susceptible to developing multiple types of cancers. Understanding the genetic control of IKDC proportion may help define susceptibility to cancer as well as facilitate the development of novel therapeutic targets.
Acknowledgments
The authors wish to thank Edward S. Weiss for invaluable help in the interpretations of single nucleotide polymorphism data and Nathalie Labrecque for critical review on the manuscript.
This project was supported by Diabète Québec and La Fondation de l’Hôpital Maisonneuve-Rosemont. S.L. is a recipient of a Junior 1 scholarship from Fonds de la recherché en santé du Québec. FGD is a recipient of scholarships from La Fondation du Dr. Georges Phénix, Diabète Québec and University of Montreal.
Abbreviations
- pDC
plasmacytoid dendritic cells
- IKDC
interferon-producing killer dendritic cells
References
- 1.Josien R, Heslan M, Soulillou JP, Cuturi MC. Rat spleen dendritic cells express natural killer cell receptor protein 1 (NKR-P1) and have cytotoxic activity to select targets via a Ca2+-dependent mechanism. J Exp Med. 1997;186:467–472. doi: 10.1084/jem.186.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan CW, Crafton E, Fan HN, Flook J, Yoshimura K, Skarica M, Brockstedt D, Dubensky TW, Stins MF, Lanier LL, Pardoll DM, Housseau F. Interferon-producing killer dendritic cells provide a link between innate and adaptive immunity. Nat Med. 2006;12:207–213. doi: 10.1038/nm1352. [DOI] [PubMed] [Google Scholar]
- 3.Taieb J, Chaput N, Menard C, Apetoh L, Ullrich E, Bonmort M, Pequignot M, Casares N, Terme M, Flament C, Opolon P, Lecluse Y, Metivier D, Tomasello E, Vivier E, Ghiringhelli F, Martin F, Klatzmann D, Poynard T, Tursz T, Raposo G, Yagita H, Ryffel B, Kroemer G, Zitvogel L. A novel dendritic cell subset involved in tumor immunosurveillance. Nat Med. 2006;12:214–219. doi: 10.1038/nm1356. [DOI] [PubMed] [Google Scholar]
- 4.Vremec D, O’Keeffe M, Hochrein H, Fuchsberger M, Caminschi I, Lahoud M, Shortman K. Production of interferons by dendritic cells, plasmacytoid cells, natural killer cells, and interferon-producing killer dendritic cells. Blood. 2007;109:1165–1173. doi: 10.1182/blood-2006-05-015354. [DOI] [PubMed] [Google Scholar]
- 5.Blasius AL, Barchet W, Cella M, Colonna M. Development and function of murine B220+CD11c+NK1.1+ cells identify them as a subset of NK cells. J Exp Med. 2007 doi: 10.1084/jem.20070991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caminschi I, Ahmet F, Heger K, Brady J, Nutt SL, Vremec D, Pietersz S, Lahoud MH, Schofield L, Hansen DS, O’Keeffe M, Smyth MJ, Bedoui S, Davey GM, Villadangos JA, Heath WR, Shortman K. Putative IKDCs are functionally and developmentally similar to natural killer cells, but not to dendritic cells. J Exp Med. 2007 doi: 10.1084/jem.20071351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vosshenrich CA, Lesjean-Pottier S, Hasan M, Goff OR, Corcuff E, Mandelboim O, Di Santo JP. CD11cloB220+ interferon-producing killer dendritic cells are activated natural killer cells. J Exp Med. 2007 doi: 10.1084/jem.20071451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welner RS, Pelayo R, Garrett KP, Chen X, Perry SS, Sun XH, Kee BL, Kincade PW. Interferon-producing killer dendritic cells (IKDC) arise via a unique differentiation pathway from primitive c-kitHiCD62L+lymphoid progenitors. Blood. 2007 doi: 10.1182/blood-2006-08-043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ullrich E, Bonmort M, Mignot G, Jacobs B, Bosisio D, Sozzani S, Jalil A, Louache F, Bulanova E, Geissman F, Ryffel B, Chaput N, Bulfone-Paus S, Zitvogel L. Trans-presentation of IL-15 dictates IFN-producing killer dendritic cells effector functions. J Immunol. 2008;180:7887–7897. doi: 10.4049/jimmunol.180.12.7887. [DOI] [PubMed] [Google Scholar]
- 10.Robbins SH, Walzer T, Dembele D, Thibault C, Defays A, Bessou G, Xu H, Vivier E, Sellars M, Pierre P, Sharp FR, Chan S, Kastner P, Dalod M. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 2008;9:R17. doi: 10.1186/gb-2008-9-1-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Himoudi N, Nabarro S, Buddle J, Eddaoudi A, Thrasher AJ, Anderson J. Bone marrow-derived IFN-producing killer dendritic cells account for the tumoricidal activity of unpulsed dendritic cells. J Immunol. 2008;181:6654–6663. doi: 10.4049/jimmunol.181.9.6654. [DOI] [PubMed] [Google Scholar]
- 12.McDuffie M. Derivation of diabetes-resistant congenic lines from the nonobese diabetic mouse. Clin Immunol. 2000;96:119–130. doi: 10.1006/clim.2000.4893. [DOI] [PubMed] [Google Scholar]
- 13.Spits H, Lanier LL. Natural killer or dendritic: what’s in a name? Immunity. 2007;26:11–16. doi: 10.1016/j.immuni.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Podolin PL, Pressey A, DeLarato NH, Fischer PA, Peterson LB, Wicker LS. I-E+ nonobese diabetic mice develop insulitis and diabetes. J Exp Med. 1993;178:793–803. doi: 10.1084/jem.178.3.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damotte D, Colomb E, Cailleau C, Brousse N, Charreire J, Carnaud C. Analysis of susceptibility of NOD mice to spontaneous and experimentally induced thyroiditis. Eur J Immunol. 1997;27:2854–2862. doi: 10.1002/eji.1830271117. [DOI] [PubMed] [Google Scholar]
- 16.Serreze DV, Prochazka M, Reifsnyder PC, Bridgett MM, Leiter EH. Use of recombinant congenic and congenic strains of NOD mice to identify a new insulin-dependent diabetes resistance gene. J Exp Med. 1994;180:1553–1558. doi: 10.1084/jem.180.4.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jevremovic D, Billadeau DD, Schoon RA, Dick CJ, Irvin BJ, Zhang W, Samelson LE, Abraham RT, Leibson PJ. Cutting edge: a role for the adaptor protein LAT in human NK cell-mediated cytotoxicity. J Immunol. 1999;162:2453–2456. [PubMed] [Google Scholar]
- 18.Monteleone G, Pallone F, MacDonald TT. Interleukin-21: a critical regulator of the balance between effector and regulatory T-cell responses. Trends Immunol. 2008;29:290–294. doi: 10.1016/j.it.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 19.Choi P, Xanthaki D, Rose SJ, Haywood M, Reiser H, Morley BJ. Linkage analysis of the genetic determinants of T-cell IL-4 secretion, and identification of Flj20274 as a putative candidate gene. Genes Immun. 2005;6:290–297. doi: 10.1038/sj.gene.6364192. [DOI] [PubMed] [Google Scholar]
- 20.Haskins K, Kench J, Powers K, Bradley B, Pugazhenthi S, Reusch J, McDuffie M. Role for oxidative stress in the regeneration of islet beta cells? J Investig Med. 2004;52:45–49. doi: 10.1136/jim-52-01-25. [DOI] [PubMed] [Google Scholar]
- 21.Chen J, Reifsnyder PC, Scheuplein F, Schott WH, Mileikovsky M, Soodeen-Karamath S, Nagy A, Dosch MH, Ellis J, Koch-Nolte F, Leiter EH. “Agouti NOD”: identification of a CBA-derived Idd locus on Chromosome 7 and its use for chimera production with NOD embryonic stem cells. Mamm Genome. 2005;16:775–783. doi: 10.1007/s00335-005-0007-1. [DOI] [PubMed] [Google Scholar]
- 22.Chiu PP, Ivakine E, Mortin-Toth S, Danska JS. Susceptibility to lymphoid neoplasia in immunodeficient strains of nonobese diabetic mice. Cancer Res. 2002;62:5828–5834. [PubMed] [Google Scholar]


