Abstract
The association of DNA copy-number variation (CNV) with specific gene function and human disease has been long known, but the wide scope and prevalence of this form of variation has only recently been fully appreciated. The latest studies using microarray technology have demonstrated that as much as 12% of the human genome and thousands of genes are variable in copy number, and this diversity is likely to be responsible for a significant proportion of normal phenotypic variation. Current challenges involve developing methods not only for detecting and cataloging CNVs in human populations at increasingly higher resolution but also for determining the association of CNVs with biological function, recent human evolution, and common and complex human disease.
From the earliest days of human cytogenetics with the study of chromosomes under the microscope, variation in chromosome copy number, rearrangement and structure was identified and in many cases could be associated with disease. One of the first observations was that an additional copy of chromosome 21 was associated with Down’s syndrome1, and many other syndromes later became associated with visible deletions or duplications of chromosomal material2-4. Seemingly benign chromosome variation in normal individuals was also identified, particularly in the size of regions of heterochromatin on chromosomes 1, 9 and 16 (ref. 5) and in the short arms of the acrocentric chromosomes6. At the other end of the resolution spectrum, the development of methods for analyzing and sequencing short segments of DNA led to the discovery of short tandem repeats7 and single nucleotide polymorphisms (SNPs)8-11. It has thus become clear that human genetic variation ranges from single base-pair changes at the sequence level up to multi-megabase chromosome differences detectable by microscopy.
Recently, our view of human genetic variation has been extended by the observation of abundant and widespread variation in the copy number of submicroscopic DNA segments12-21. This new perspective on human variation has been driven largely by the implementation of whole-genome scanning methods that enable us to interrogate the genome at a resolution intermediate between that of cytogenetic analysis using microscopy (>5–10 Mb) and that of DNA sequencing (1–700 bp). Although many methods have been developed to assay DNA in this intermediate size range, DNA microarrays have probably been most instrumental in advancing our current understanding of copy number variation. In the most comprehensive study to date, Redon et al.17 used two different microarray platforms to analyze copy number variation in 270 normal individuals. They identified almost 1,500 regions of the genome which were variable in copy number, encompassing 360 Mb and thousands of genes. As only a relatively small proportion of the CNV regions overlap with regions found in other studies, the current total of more than 6,000 CNVs detailed in the Database of Genomic Variants (http://projects.tcag.ca/variation/) is most likely an underestimate.
In this article I review the current microarray technologies being used for genome-wide CNV detection and for the association of CNVs with normal and disease phenotypes.
Comparative genomic hybridization to arrays
One of the main methods by which CNVs can be identified using DNA microarrays is comparative genomic hybridization (CGH). CGH was first developed as a method for comparing the copy number of differentially labeled test and normal reference DNAs using fluorescence in situ hybridization (FISH) onto metaphase spreads from a normal individual22. Measuring the fluorescence ratio along the length of each chromosome identified regions of relative loss and gain in the test sample (the reference DNA being assumed to be diploid in copy number). Although this method had a huge impact, particularly on understanding chromosomal rearrangements in tumor biology23, a major drawback was the low resolution, typically 5–10 Mb, afforded by metaphase FISH. Improvements in the resolution of CGH were driven largely by using the resources generated for the public-domain Human Genome Project, where large-insert clone libraries were developed and clones assembled into overlapping contigs for sequencing24,25. With this resource it became possible to replace metaphase chromosomes for CGH with arrays of clones accurately mapped onto the human genome and spotted robotically onto glass slides using split metal pins or glass capillaries. For this method, the resolution of the CGH was determined by the size and number (density) of sequences spotted onto the array. The first hybridizations of this kind were reported in 1997 as matrix-CGH26 and then in 1998 as array-CGH27, and it is this latter term which is now widely used. In array-CGH, as in conventional CGH, test and reference DNAs are differentially fluorescent labeled and hybridized together to the array. The resulting fluorescent ratio is then measured, clone by clone, and plotted relative to each clone’s position in the genome. The main advantage of the cohybridization of the test and reference DNAs to spotted arrays is that the reported ratios are influenced less by spotted probe concentration (signal intensity) and variations in slide production and processing. Array-CGH has revolutionized the study of copy-number changes in tumors and is rapidly becoming a new standard method for clinical cytogenetics.
The current interest in CNV was stimulated by the publication of two papers in 2004 that used array-CGH to compare the genomes of normal individuals13,18. Using standard array-CGH on commercial arrays with one BAC clone every 1 Mb across the human genome, Iafrate et al.13 identified 255 variable regions in 55 normal individuals, and using representational oligonucleotide microarray analysis (ROMA), Sebat et al.18 found 76 variable regions in 20 individuals. This initial demonstration of the importance of CNV in normal human variation has been confirmed and extended in several subsequent studies using array-CGH15,17,21,28.
There have been many developments in array technology over the past 7 or 8 years, and in particular there has been a significant trend toward increased numbers of features (spots) and toward shorter DNA sequences as hybridization targets, both of which have an impact on the resolution at which CNVs can be detected. The DNA sequences that have been used to construct arrays have included large-insert clones (40–200 kb in size), small insert clones (1.5–4.5 kb), cDNA clones (0.5–2 kb), genomic PCR products (100 bp–1.5 kb) and oligonucleotides (25–80 bp). While the resolution of an array covering the whole genome depends on the number, distribution and the length of the probes, the ability to detect copy-number changes depends on signal-to-noise ratio and probe response characteristics29.
Clone and PCR-product arrays
Of the sequences used to produce DNA microarrays, BAC clones provide the most comprehensive coverage of the genome and robust low-noise hybridizations. As long as signal from repeat sequences in the clones is well suppressed by competitive hybridization with Cot 1 DNA (DNA enriched for common repetitive sequences), probe responses to ratio changes are also very close to theoretical values. However, although BAC arrays consisting of over 30,000 overlapping clones tiling the human genome are now available within the research community30,31, BACs are typically 80–200 kb in length, so that even in good hybridizations with high signal-to-noise ratios it is difficult to identify with confidence single-copy-number differences smaller than 50 kb. Fosmid and cosmid clones of approximately 40 kb in length also provide high signal-to-noise targets for array-CGH, and the smaller size of the insert DNA improves CNV detection resolution to about 20 kb. In particular, fosmid clones are available mapped onto the human reference sequence at high density with considerable sequence overlap. By constructing arrays using this fosmid sequence redundancy, array resolution can be improved further, but ultimately it becomes difficult to identify confidently CNVs smaller than 15 kb.
Many groups have used cDNA clones for array construction32-36. cDNA clones are able to increase array-CGH resolution down to single genes or parts of genes. However, for CNV detection, the uneven distribution of genes produces variable resolution across the genome, and the mismatch between cDNA and genomic DNA during the hybridization reduces signal and probe response to ratio changes. Higher resolution and more complete coverage can be achieved with genomic PCR products, but signal-to-noise ratios can be poor and costs of probe generation, particularly for genome-wide arrays, can be high37,38. A major limitation for clone and PCR-product arrays is that they are generally constructed using mechanical spotting of DNA solutions onto glass microscope slides. Unfortunately, it is difficult to spot more than 60,000 DNAs onto a glass slide using current spotting devices, and so this limits the resolution of a single array covering the complete genome. It should also be noted that probes containing segmental duplications or low-copy-number repeats should be analyzed with care, as these probes will show, with a reduced response, copy-number changes that have occurred at any (and all) of the locations having homology.
Oligonucleotide arrays
Oligonucleotides provide the highest potential resolution for array-CGH, and early arrays were constructed by mechanically spotting previously synthesized oligonucleotides39. As is true for clone-based arrays, a maximum of approximately 60,000 oligonucleotides can be spotted, typically onto glass microscope slides, so the coverage of such arrays is restricted to one probe every 50 kb. More recently, commercial oligonucleotide arrays in which the oligonucleotides are synthesized directly onto the glass slide have become widely available. Agilent and OGT use ink-jet technology to apply the reagents necessary for oligonucleotide synthesis on a spot-by-spot basis. At present this technology can achieve 244,000 oligonucleotides per array. NimbleGen, however, uses a programmable mirror array to generate a software-defined digital mask for oligonucleotide synthesis by photolithography. This technology currently achieves 385,000 oligonucleotides per array, but during 2007, arrays (HD2) comprising 2.1 million and potentially 4.2 million oligonucleotides will be available. A disadvantage of oligonucleotide arrays, however, is the poor signal-to-noise ratio of hybridizations leading to considerable variation in reported CGH ratio. Typically, the s.d. of log2 ratios in an oligonucleotide array is on the order of 0.25, whereas the s.d. for a BAC clone–based array is typically 0.05. Furthermore, probe responses to ratio changes tend to be suppressed. Because of these limitations, data from several oligonucleotides need to be averaged to reduce measurement variance to acceptable limits, reducing the overall resolution of the array.
To improve the signal-to-noise ratio, some groups have used a method to reduce the complexity of the genomic DNA used for hybridization. This method is called representational oligonucleotide microarray analysis (ROMA)40. With ROMA, the genomic DNA is digested using a restriction enzyme and the fragments ligated to adapters and then amplified using universal primers. During the PCR amplification only the smaller restriction fragments (up to about 1.2 kb in size) become amplified, reducing the complexity (and the representation) of the DNA. This is then hybridized to an oligonucleotide array consisting of probes that have been selected to match the reduced set of amplified restriction fragments. However, signal-to-noise ratios still remain well below those of BAC arrays, and typically at least three probes are averaged to reduce data variance. And ROMA technology does present further potential problems for CNV detection. First, the complexity reduction may lead to differential representation of parts of the genome, which may be interpreted erroneously as copy number changes, Second, different individuals will have different restriction digestion patterns, and it is possible that some individual probe ratios may be related to restriction fragment size differences rather than to true copy-number changes.
A further consideration for oligonucleotide arrays is the design of the sequences themselves. Little useful data can be generated by array-CGH from highly repetitive regions of the genome, and so it is common practice to consider only the repeat-masked fraction (approximately 55%) of the human genome for sequence design. As is true for clone and PCR product arrays, data from low-copy-number repeats and segmental duplications can be difficult to interpret, as the genomic location of detected copy-number changes could be the result of copy-number changes at any combination of the shared sequence locations. However, low-copy-number repeats and segmental duplications are associated with CNVs, and it is important that array designs interrogate these regions of the genome. In designs we have specified for custom NimbleGen arrays, we allow oligonucleotides ranging in size from 45 to 75 bp to have up to five short matches elsewhere in the genome, enabling such arrays to assay, at least to some extent, these regions of the genome. Precise determination of which sequence location(s) are variable in these cases requires further, more detailed investigation using other molecular techniques.
Because the design of oligonucleotides can be specified, it is possible to achieve almost nucleotide-level resolution in array-CGH on oligonucleotide arrays by synthesizing overlapping oligonucleotides with as little as a single base-pair shift across the sequence of interest. Clearly, covering the whole repeat-masked genome at this resolution would require more than 2 billion probes and would be prohibitively expensive, but for custom interrogation of specific regions of the genome this approach can be particularly useful. Indeed, we have been able to map chromosome translocation breakpoints to within 4 bp using this approach41.
Genotyping arrays
SNPs have received considerable attention as a source of human variation and are particularly amenable to high-throughput genotyping and disease association studies. A particularly important study was the HapMap project (http://www.hapmap.org/), which catalogued SNPs in four of the major ethnic human populations42. Disease association studies are now being extended to large case-control studies such as the Wellcome Trust Case Control Consortium (involving 19,000 samples), which is investigating SNP association in tuberculosis, coronary heart disease, type 1 diabetes, type 2 diabetes, rheumatoid arthritis, Crohn’s disease and ulcerative colitis, bipolar disorder, and hypertension (http://www.wtccc.org.uk/). Such studies have been made possible largely by the development of high-throughput array technologies for SNP genotyping from commercial companies such as Affymetrix and Illumina. Although they were originally developed simply for determining the base(s) present at SNPs, these arrays are increasingly being mined for intensity information that can be used to determine genomic copy number. With the Affymetrix SNP chips, 20 matched and mismatched probe pairs 25 bases long are designed to each SNP allele. Hybridizations are not performed using cohybridization of two DNA as in array-CGH, but by hybridization of a single DNA. To improve the signal-to-noise ratio, the DNA is first digested with a restriction enzyme and ligated with adapters and then the smaller fragments amplified using universal primers to reduce the complexity of the hybridization. As with ROMA, the reduced complexity of the hybridization brings with it the possibility of amplification bias of different regions of the genome and detection of changes reflecting differences in restriction digestion patterns between individuals rather than in true copy number. For CNV detection, the signal intensities of the match and mismatch probes are compared with values from another individual (or group of individuals) and the relative copy number per locus is determined. Highly standardized slide handling and processing procedures and precision in array fabrication help to reduce the variance of ratios calculated from independent hybridizations. Further noise reductions can be achieved by taking length and GC content of the probes into account43, and various algorithms for copy-number detection have been developed to aid CNV detection, for example that in Komura et al.44. Again, for robust detection, significant ratio shifts in several sequential probes are required. For the current Affymetrix GeneChip 500k, SNPs have a median spacing of 2.5 kb. However, the probes are not uniformly distributed across the genome and are particularly sparse in regions of segmental duplication and complex CNV that create problems for the design of robust genotyping SNP assays in these regions. As a result, the resolution of the array is variable across the genome and CNV detection has a lower limit of 10–40 kb. To overcome this limitation, Affymetrix and Illumina are proposing to include additional nonpolymorphic probes on their next-generation arrays. As an example, the Affymetrix Genome-Wide Human SNP Array 5.0 will contain approximately 500,000 genome-wide SNPs and an additional 500,000 nonpolymorphic probes that can be used to assess chromosomal copy-number change in areas of the genome not covered well by SNPs.
Illumina has developed an alternative platform using 50-bp oligonucleotides attached to indexed beads randomly deposited onto glass slides. Their current highest-resolution array has 650,000 different oligonucleotides with a median SNP spacing of 2 kb. On this platform, after whole genome amplification, the test DNA is hybridized to the slide; this is followed by primer extension and immunohistochemical fluorescence detection. This technology is also now being used for the identification of CNVs. As this is a SNP-based platform, the same constraints with regard to probe distribution apply, although the incorporation of nonpolymorphic probes on the bead arrays will similarly overcome this problem.
CNVs from SNP genotyping errors
CNVs, and in particular deletions, can also be identified directly from genotyping determinations in a variety of ways. Using parent-offspring trios from the phase 1 HapMap Project genotyping calls, Conrad et al.45 used apparent errors in Mendelian inheritance to identify 586 potential deletions ranging in size from 300 bp to 1.2 kb. Using the same data, McCarroll et al.46 identified clusters of SNPs that were not in Hardy-Weinberg equilibrium, or that showed other evidence of genotyping errors, to discover 541 potential deletions ranging in size from 1 to 745 kb. However, these approaches are only capable of identifying deletions and are subject to the same limitations of SNP distribution as discussed above and so should be considered as a useful additional method to identify CNVs from genotyping data rather than a direct discovery tool.
Calling CNVs
A major concern for the detection of CNVs using array technology is how a putative CNV is defined. There is a plethora of different methods being used to call significant changes in relative ratio changes from arrays. These vary from simple defined thresholds to complex statistical modeling. For example, the use of threshold values is discussed in Vermeesch et al.47, and a more complex multithreshold approach has been adopted by Fiegler et al.30. Examples of widely implemented segmentation algorithms used for calling copy number changes are SW-Array48 and circular binary segmentation49. However, a discussion of the pros and cons of all of the methods used so far is of little utility, as rarely have different approaches been compared on the same datasets, and different platforms introduce their own specific problems to data analysis. It is inevitable that in any hybridization, measurement variance will lead to false positive and false negative results however the data are analyzed. It is particularly important that these two rates are assessed in studies using array-CGH or SNP arrays for CNV detection, as high false positive rates will lead to the databases becoming populated with regions incorrectly called as CNVs. Indeed, many of the regions in the databases today identified as CNVs will prove to be false discoveries, particularly where loci have not been validated independently or are not replicated between studies. Interestingly, we have identified reproducible local small variations in reported ratios that in regions of equal copy number in the two genomes compared can be identified as ‘waves’ of increasing or decreasing ratios. These waves, which we have observed in copy-number data from several array platforms (including BAC, Affymetrix and NimbleGen arrays), not only affect the setting of calling thresholds or algorithms by their contribution to the overall variance of the data, but also contribute to false positives where the peaks of the waves may pass a calling threshold or condition. However, as the waves in the data are largely reproducible and indeed seem to be correlated in some way with genomic GC content, it is possible to normalize datasets to remove the majority of this effect and so improve the accuracy and sensitivity of CNV calling (J.C. Marioni et al., University of Cambridge, unpublished data). Nevertheless, what is urgently required is a ‘gold standard’ for platform performance testing. In our recent studies, we have selected two DNA samples generally available from Coriell Cell Depositories from which we have validated independently the CNV status of several hundred regions to allow the estimation of false positive and false negative rates for the two microarray platforms that we have used17,30. To test array performance, we now run as routine these same DNAs on any new array design or platform we use. The DNAs we chose were NA15510, the source of the fosmid library used to confirm genome assembly during the finishing of the human genome, which was analyzed for structural variants against the human genome by Tuzun et al.20; and NA10851, a well characterized trio-offspring DNA from the HapMap collection of DNAs45,46. We will continue to release increasingly higher resolution CNV data from these two DNAs from our ongoing studies. Elsewhere in this issue50, we recommend that at least one of these DNAs be included for comparison as a standard control sample in all studies.
How important is resolution?
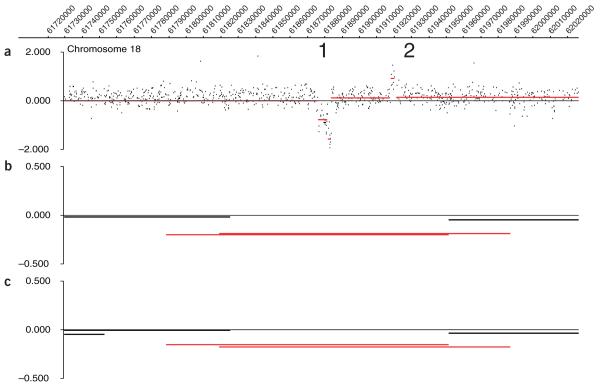
Current array-CGH methods for identifying CNVs typically assay the genome in the 40-kb to several megabase range. However, specifically for deletions, CNVs as small as a few kilobases can be detected from genotyping data by identifying errors in Mendelian inheritance. Conrad et al.45 modeled the size distribution of deletions in their study of the 30 trios of Americans of European ancestry (the CEU cohort; http://www.hapmap.org) and 30 Yoruban trios in the HapMap phase 1 study using observed data with size-related detection-power estimates. They concluded that the frequency of deletions in the genome increased with decreasing size (down to their smallest class interval of 5 kb), implying that deletions smaller than this may be even more numerous. It is therefore an important challenge to develop array-based CNV assays which access copy number changes in this smaller size range. For these reasons, it is clear that large-insert clone arrays will no longer be optimal for CNV discovery and that the higher-density oligonucleotide or SNP arrays offer the most likely platforms for development in the near future. This is illustrated in Figure 1, which shows a CNV region detected by array-CGH to a whole genome tiling-path large-insert clone array and to a custom high-resolution NimbleGen array. The CNV is detected on the BAC array within the overlap of two BACs by a small but significant ratio drop. In contrast, two CNVs in the BAC overlap region can be seen using the NimbleGen array. CNV 1 has a complex structure involving different levels of loss of a 40-kb region, while CNV 2 is a small gain of approximately 10 kb. On the BAC array, the ratio of these two CNVs is averaged, masking the complexity of the region and resulting in the reported small copy number loss.
Figure 1.
Array-CGH profiles of a small region of chromosome 18 for two normal DNAs. Copy-number ratio is graphed on the y axis. (a) High-resolution NimbleGen oligonucleotide array. The red horizontal lines indicate regions of similar copy number. Numbers 1 and 2 indicate clusters of probes (CNVs) with low and high ratios, respectively. (b,c) Replicate whole-genome large-insert clone tiling path array hybridizations. Clones called as identifying CNVs are indicated by the red horizontal lines and other clones by black horizontal lines; the lengths represent the sizes of the clone inserts. Modified from a display generated in SignalMap (NimbleGen Systems).
CNV discovery
CNV discovery requires screening of the whole genome for copy-number changes. In practical terms, the resolution required for the next few years in CNV discovery needs to include CNVs larger than those detected by sequence analysis and smaller than those now assayed by array technology (that is, between 500 bp and 40 kb). So far, the highest density oligonucleotide array is the HD2 array from NimbleGen, comprising over 2 million probes. Assuming that five probes need to be averaged to produce acceptable quality data, the effective resolution of this array to the repeat-masked fraction of the genome will be approximately 5 kb (that is, one probe every 1 kb). Thus, at least an order of magnitude increase in probe density will be required for a resolution of 500 bp using a single array. Alternatively, this higher resolution can be achieved by using ten arrays to cover the genome, albeit at a ten-fold increase in cost.
CNV association
Array design parameters for CNV association are different from those for CNV discovery. For association studies, probes need only be developed to assay known CNVs, so the whole genome need not be covered. Furthermore, it may also be sufficient for most studies simply to detect the presence of the CNV and its copy number using a minimal number of probes, sacrificing information about CNV breakpoints. Thus an array design using oligonucleotides might require only ten probes per CNV, so that for the study of several thousand CNVs the number of probes required could easily be accommodated using current array technologies. Multisample arrays are particularly cost-effective for this purpose. Agilent currently produces a slide comprising four subarrays of 44,000 probes, and NimbleGen similarly divides their 385,000-probe format in four to provide approximately 75,000 probes per subarray. Even further economies of scale will be achieved using NimbleGen’s HD2 platform, allowing the assay of thousands of CNVs on up to 12 samples per slide. These multiplex approaches substantially reduce array costs and, owing to the smaller areas being hybridized, also reduce labeling costs to some extent. Although it is inevitable that there will be a degree of platform convergence, particularly with the rapid development of SNP genotyping platforms containing nonpolymorphic probes, cost will remain a major driver of platform choice, particularly where specific CNVs are being studied.
CNV detection now and in the future
Array technology is clearly not the only technology that can be used to identify CNVs and associate them with disease. Methods such as quantitative PCR, multiplex amplifiable probe hybridization (MAPH), multiplex ligation-dependent probe amplification (MLPA) and dynamic allele-specific hybridization (DASH) all are capable of assaying CNVs, albeit with restricted throughput and scale. For association studies with CNVs, mass spectrometry holds some promises for population screening. Sequencing approaches, such as the mapping of fosmid ends onto the reference sequence, are particularly powerful at identifying, besides copy number changes, structural rearrangements such as inversions and translocations. Unfortunately, using current capillary-based technology, the cost of sequencing the fosmid library ends per sample remains very high (in the order of $1 million). However, the new generation of sequencing technologies, such as those produced by 454 Life Sciences, Solexa and ABI, are predicted to reduce substantially this cost in the future. New assay methods for paired end reads will need to be developed to allow this technology to identify larger structural rearrangements. Ultimately, all forms of genomic variation will be accessible by personalized sequencing, and new technologies such as microelectrophoretic and nanopore sequencing seem very promising in this regard51,52. Nevertheless, it is most probable that most CNV discovery and studies of phenotype association over the next few years will be achieved using DNA microarray technology of continually increasing resolution and cost effectiveness.
ACKNOWLEDGMENTS
I would like to thank H. Fiegler, R. Redon and M. Hurles for critical reading of the manuscript. N.P.C. is supported by the Wellcome Trust.
Footnotes
COMPETING INTERESTS STATEMENT The author declares no competing financial interests.
References
- 1.Jacobs PA, Baikie AG, Brown W.M. Court, Strong JA. The somatic chromosomes in mongolism. Lancet. 1959;1:710. doi: 10.1016/s0140-6736(59)91892-6. [DOI] [PubMed] [Google Scholar]
- 2.Kunze J. Neurological disorders in patients with chromosomal anomalies. Neuropediatrics. 1980;11:203–249. doi: 10.1055/s-2008-1071392. [DOI] [PubMed] [Google Scholar]
- 3.Lejeune J, Lafourcade J, Berger R, Rethore MA. [The crying cat syndrome and its reciprocal] [in French] Ann. Genet. 1965;8:11–15. [PubMed] [Google Scholar]
- 4.Sedano HO, Look RA, Carter C, Cohen MM., Jr. B group short-arm deletion syndrome. Birth Defects Orig. Artic. Ser. 1971;7:89–97. [PubMed] [Google Scholar]
- 5.Morton CC, Corey LA, Nance WE, Brown JA. Quinacrine mustard and nucleolar organizer region heteromorphisms in twins. Acta Genet. Med. Gemellol. (Roma) 1981;30:39–49. doi: 10.1017/s0001566000006607. [DOI] [PubMed] [Google Scholar]
- 6.Verma RS, Dosik H, Lubs HA. Size variation polymorphisms of the short arm of human acrocentric chromosomes determined by R-banding by fluorescence using acridine orange (RFA) Hum. Genet. 1977;38:231–234. doi: 10.1007/BF00527408. [DOI] [PubMed] [Google Scholar]
- 7.Edwards A, Civitello A, Hammond HA, Caskey CT. DNA typing and genetic mapping with trimeric and tetrameric tandem repeats. Am. J. Hum. Genet. 1991;49:746–756. [PMC free article] [PubMed] [Google Scholar]
- 8.Kwok PY, Deng Q, Zakeri H, Taylor SL, Nickerson DA. Increasing the information content of STS-based genome maps: identifying polymorphisms in mapped STSs. Genomics. 1996;31:123–126. doi: 10.1006/geno.1996.0019. [DOI] [PubMed] [Google Scholar]
- 9.Mir KU, Southern EM. Sequence variation in genes and genomic DNA: methods for large-scale analysis. Annu. Rev. Genomics Hum. Genet. 2000;1:329–360. doi: 10.1146/annurev.genom.1.1.329. [DOI] [PubMed] [Google Scholar]
- 10.Taillon-Miller P, Gu Z, Li Q, Hillier L, Kwok PY. Overlapping genomic sequences: a treasure trove of single-nucleotide polymorphisms. Genome Res. 1998;8:748–754. doi: 10.1101/gr.8.7.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang DG, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 12.Freeman JL, et al. Copy number variation: new insights in genome diversity. Genome Res. 2006;16:949–961. doi: 10.1101/gr.3677206. [DOI] [PubMed] [Google Scholar]
- 13.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 14.Khaja R, et al. Genome assembly comparison identifies structural variants in the human genome. Nat. Genet. 2006;38:1413–1418. doi: 10.1038/ng1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Locke DP, et al. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am. J. Hum. Genet. 2006;79:275–290. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perry GH, et al. Hotspots for copy number variation in chimpanzees and humans. Proc. Natl. Acad. Sci. USA. 2006;103:8006–8011. doi: 10.1073/pnas.0602318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 19.Sharp AJ, Cheng Z, Eichler EE. Structural variation of the human genome. Annu. Rev. Genomics Hum. Genet. 2006;7:407–442. doi: 10.1146/annurev.genom.7.080505.115618. [DOI] [PubMed] [Google Scholar]
- 20.Tuzun E, et al. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 21.Wong KK, et al. A comprehensive analysis of common copy-number variations in the human genome. Am. J. Hum. Genet. 2007;80:91–104. doi: 10.1086/510560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kallioniemi OP, et al. Comparative genomic hybridization: a rapid new method for detecting and mapping DNA amplification in tumors. Semin. Cancer Biol. 1993;4:41–46. [PubMed] [Google Scholar]
- 23.Kallioniemi A, Visakorpi T, Karhu R, Pinkel D, Kallioniemi OP. Gene copy number analysis by fluorescence in situ hybridization and comparative genomic hybridization. Methods. 1996;9:113–121. doi: 10.1006/meth.1996.0015. [DOI] [PubMed] [Google Scholar]
- 24.Bentley DR, et al. The physical maps for sequencing human chromosomes 1, 6, 9, 10, 13, 20 and X. Nature. 2001;409:942–943. doi: 10.1038/35057165. [DOI] [PubMed] [Google Scholar]
- 25.Cheung VG, et al. Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature. 2001;409:953–958. doi: 10.1038/35057192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solinas-Toldo S, et al. Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosom. Cancer. 1997;20:399–407. [PubMed] [Google Scholar]
- 27.Pinkel D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 28.Sharp AJ, et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ylstra B, van den Ijssel P, Carvalho B, Brakenhoff RH, Meijer GA. BAC to the future! or oligonucleotides: a perspective for micro array comparative genomic hybridization (array CGH) Nucleic Acids Res. 2006;34:445–450. doi: 10.1093/nar/gkj456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fiegler H, et al. Accurate and reliable high-throughput detection of copy number variation in the human genome. Genome Res. 2006;16:1566–1574. doi: 10.1101/gr.5630906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishkanian AS, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat. Genet. 2004;36:299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- 32.Kauraniemi P, Barlund M, Monni O, Kallioniemi A. New amplified and highly expressed genes discovered in the ERBB2 amplicon in breast cancer by cDNA microarrays. Cancer Res. 2001;61:8235–8240. [PubMed] [Google Scholar]
- 33.Monni O, et al. Comprehensive copy number and gene expression profiling of the 17q23 amplicon in human breast cancer. Proc. Natl. Acad. Sci. USA. 2001;98:5711–5716. doi: 10.1073/pnas.091582298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollack JR, et al. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat. Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 35.Porkka K, Saramaki O, Tanner M, Visakorpi T. Amplification and overexpression of Elongin C gene discovered in prostate cancer by cDNA microarrays. Lab. Invest. 2002;82:629–637. doi: 10.1038/labinvest.3780457. [DOI] [PubMed] [Google Scholar]
- 36.Squire JA, et al. High-resolution mapping of amplifications and deletions in pediatric osteosarcoma by use of CGH analysis of cDNA microarrays. Genes Chromosom. Cancer. 2003;38:215–225. doi: 10.1002/gcc.10273. [DOI] [PubMed] [Google Scholar]
- 37.Dhami P, et al. Exon array CGH: detection of copy-number changes at the resolution of individual exons in the human genome. Am. J. Hum. Genet. 2005;76:750–762. doi: 10.1086/429588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mantripragada KK, Buckley PG, Jarbo C, Menzel U, Dumanski JP. Development of NF2 gene specific, strictly sequence defined diagnostic microarray for deletion detection. J. Mol. Med. 2003;81:443–451. doi: 10.1007/s00109-003-0458-3. [DOI] [PubMed] [Google Scholar]
- 39.Carvalho B, Ouwerkerk E, Meijer GA, Ylstra B. High resolution microarray comparative genomic hybridisation analysis using spotted oligonucleotides. J. Clin. Pathol. 2004;57:644–646. doi: 10.1136/jcp.2003.013029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lucito R, et al. Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gribble SM, et al. Ultra-high resolution array painting facilitates breakpoint sequencing. J. Med. Genet. 2007;44:51–58. doi: 10.1136/jmg.2006.044909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nannya Y, et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65:6071–6079. doi: 10.1158/0008-5472.CAN-05-0465. [DOI] [PubMed] [Google Scholar]
- 44.Komura D, et al. Genome-wide detection of human copy number variations using high-density DNA oligonucleotide arrays. Genome Res. 2006;16:1575–1584. doi: 10.1101/gr.5629106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat. Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 46.McCarroll SA, et al. Common deletion polymorphisms in the human genome. Nat. Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 47.Vermeesch JR, et al. Molecular karyotyping: array CGH quality criteria for constitutional genetic diagnosis. J. Histochem. Cytochem. 2005;53:413–422. doi: 10.1369/jhc.4A6436.2005. [DOI] [PubMed] [Google Scholar]
- 48.Price TS, et al. SW-ARRAY: a dynamic programming solution for the identification of copy-number changes in genomic DNA using array comparative genome hybridization data. Nucleic Acids Res. 2005;33:3455–3464. doi: 10.1093/nar/gki643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 50.Scherer SW, et al. Challenges and standards in integrating surveys of structural variation. Nat. Genet. 2007;39:S7–S15. doi: 10.1038/ng2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Service RF. Gene sequencing: the race for the $1000 genome. Science. 2006;311:1544–1546. doi: 10.1126/science.311.5767.1544. [DOI] [PubMed] [Google Scholar]
- 52.Shendure J, Mitra RD, Varma C, Church GM. Advanced sequencing technologies: methods and goals. Nat. Rev. Genet. 2004;5:335–344. doi: 10.1038/nrg1325. [DOI] [PubMed] [Google Scholar]