Abstract
Several groups have attempted to develop gene therapy strategies to treat cancer via introduction of the wild-type (wt) p53 cDNA into cancer cells. Unfortunately, these approaches do not result in regulated expression of the p53 gene and do not reduce expression of the mutant p53 that is overexpressed in cancerous cells. These shortcomings may greatly limit the utility of this gene replacement approach. We describe an alternative strategy with trans-splicing ribozymes that can simultaneously reduce mutant p53 expression and restore wt p53 activity in various human cancers. The ribozyme accomplished such conversion by repairing defective p53 mRNAs with high fidelity and specificity. The corrected transcripts were translated to produce functional p53 that can transactivate p53-responsive promoters and down-modulate expression of the multidrug resistance (MDR1) gene promoter. The level of wt p53 activity generated was significant, resulting in a 23-fold induction of a p53-responsive promoter and a 3-fold reduction in MDR1 promoter expression in transfected cancer cells. Once efficient delivery systems are developed, this strategy should prove useful for making human cancers more responsive to p53 activity and more sensitive to chemotherapeutic agents.
In many cancers, the p53 tumor suppressor gene is mutated, and the most frequent changes are missense point mutations (1) that cluster in the central region of the gene that encodes the DNA-binding domain of the p53 protein (2). p53 is a transcription factor (3, 4) that binds as a tetramer to specific DNA recognition sequences (5, 6) adjacent to p53-responsive genes that engender cell-cycle arrest (7) and apoptosis (8). The mutant p53s lose sequence-specific DNA-binding properties (2, 5, 6) and cannot regulate expression of those genes (3, 4, 7, 8). Moreover, by binding to and forming inactive tetramers with wild-type (wt) p53, certain mutant versions of the p53 protein can transform cells neoplastically, presumably by inhibiting endogenous wt p53 function in a dominant-negative fashion (9, 10). Finally, certain mutant p53 proteins have acquired a “gain-of-function,” defined as the ability to augment cell proliferation in the absence of endogenous wt p53 (11) and resulting in phenotypic changes such as the overexpression of the multidrug resistance (MDR1) gene (11, 12), which can confer cross-resistance to cytotoxic drugs (13).
Several clinical trials that attempt to introduce expression cassettes for the wt p53 cDNA into cancer cells are underway to evaluate the safety and potential utility of this strategy to combat cancer (14). One significant limitation of this gene transfer approach is the inability to properly regulate gene expression after gene transfer. A second potential complication is that most p53 mutations found in human cancers are not null mutations but rather encode mutant versions of the p53 protein that may have unwanted activities such as a gain-of-function or be dominant negative inhibitors of wt p53 activity (11, 15). Therefore to treat these types of cancers with p53 therapy, one would ideally attempt to restore wt p53 production while simultaneously reducing or eliminating deleterious mutant p53 protein expression.
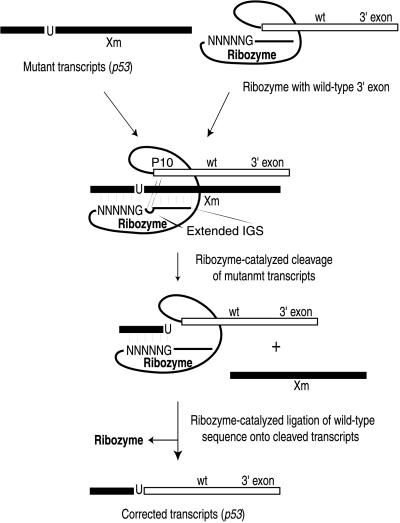
The self-splicing group I intron from Tetrahymena thermophila has been shown previously to mediate trans splicing of an exon attached to its 3′ end onto a targeted 5′ exon RNA that is a separate RNA molecule in vitro (16, 17). More recently, we demonstrated that a slightly shortened version of this ribozyme could repair truncated lacZ transcripts in Escherichia coli (18) and in cultured mammalian cells (19). In these experiments, the ribozyme recognized the lacZ substrate RNA by base pairing to it through the internal guide sequence (IGS) of the ribozyme. Target RNAs can be recognized by the ribozyme at any accessible uridine residue found upstream of the mutation(s) in the target transcript that is to be corrected. The ribozyme then cleaves the target RNA, releases the downstream RNA sequence containing the mutation(s), and replaces the sequence with a 3′ exon that encodes the correct sequence for the wt transcript (Fig. 1). In the process, the regulated expression pattern of the endogenous gene should be maintained.
Figure 1.
Schema to correct mutant p53 transcripts with targeted trans splicing. Mutant messages can be recognized by a ribozyme at any uridine upstream of the mutation (marked by Xm) by base pairing to the sequence through its IGS. The ribozyme then removes the mutation-containing sequence and replaces it with a 3′ exon that encodes the correct sequence for the wt transcript. The extended IGS and P10 interactions are shown.
Two recent reports demonstrated that trans-splicing ribozymes can be used to repair clinically relevant mutant RNAs in two different human cell types (20, 21). However, neither report demonstrated that the repaired RNAs could be translated to generate functional protein. The question as to whether such trans-spliced transcripts can be translated in mammalian cells is especially important in light of recent work that demonstrated that transcripts do not seem to be translated efficiently after self splicing of a group I intron (22). In the studies described herein, we set out to determine whether trans-splicing group I ribozymes can be used to repair a variety of defective p53 transcripts and in the process restore p53 transcriptional activity to treated cells.
Materials and Methods
Mapping Accessible Sites on p53 RNA.
The mapping library (GN5) was generated as described (20). The BamHI fragment of a plasmid pC53-SN3 (a generous gift from B. Vogelstein, Johns Hopkins University School of Medicine, Baltimore) was inserted into the multiple cloning site of pBluescript SK (+) (Stratagene), and the p53 cDNA was transcribed by T7 RNA polymerase as a run-off transcript that contained the first 432 nt of the p53 mRNA. Transcription of ribozymes and the p53 RNA was performed under the same condition as described (23), except that 5 mM MgCl2 and 25 mM MgCl2 were used for the ribozyme and the substrate, respectively. The GN5 library (100 nM) and either p53 RNA (10 μM) transcribed in vitro or cellular RNA (1–2 μg) were denatured at 95°C for 1 min and then preequilibrated in the reaction buffer (50 mM Hepes, pH 7.0/150 mM NaCl/5 mM MgCl2) at 37°C for 2 min. The substrates were then added to the ribozymes along with guanosine (100 μM) to start the trans-splicing reactions, which proceeded at 37°C for 4 h. The trans-splicing reaction products were reverse transcribed and amplified by PCR as described (19).
Construction of Ribozyme Expression Vectors.
To generate Rib41-3′ p53 and Rib65-3′ p53, PCR was performed with Pfu polymerase (Stratagene) and with pC53-SN3 as a template. The PCR products containing 1,141 nt of p53 3′ exon for Rib41 and 1,117 nt for Rib65 were digested with BamHI and inserted between the NruI and BamHI sites of pL-21 multiple cloning site or pL-21 multiple cloning site dead (a catalytically inactive version of the intron described previously; ref. 18). Each antisense region of the ribozyme was synthesized by Klenow fragment (3′ → 5′ exo−; New England Biolabs) after annealing of the two synthetic oligonucleotides to create a double-stranded DNA sequence that contains the 35- and 36-nt sequence for the antisense region and the 13- and 14-nt helix P1 of Rib41-3′ p53 and Rib65-3′ p53, respectively. After filling in, the products were digested by SacI and SphI and cloned into those sites of a pCS4R61g (provided by N. Lan, Duke University, Durham, NC) that harbor the cytomegalovirus immediate-early (CMV-IE) gene promoter and a polyadenylation signal of simian virus 40 virus to generate pRib41p53 and pRib65p53. The sequences of the PCR primers and the synthetic oligonucleotides used are available on request.
Repair of Truncated p53 Transcripts in Human Osteosarcoma Cells.
Saos-2 human osteosarcoma cells were seeded in six-well plates at a density of 2.0 × 105 cells per well 20 h before transfection. A truncated 5′ p53 expression plasmid, pC53 5′ trunc, was constructed by deleting a PvuII to SmaI fragment of pC53-SN3 (24). The cells were cotransfected with pRib41p53 or pdRib41p53 (5 μg) with or without pC53 5′ trunc (5 μg) and 10 μg of DMRIE-C (GIBCO/BRL). In the control experiments, the empty parental expression vector was substituted for either p53 or ribozyme expression plasmid. Total RNA was isolated from cells 20 h after transfection by TRIzol (GIBCO/BRL). EDTA (45 mM) was added to TRIzol before RNA extraction to chelate magnesium ions and to inhibit ribozyme folding and catalysis. After the cells transfected with pC53 5′ trunc alone and the cells transfected with pRib41p53 alone were solubilized in TRIzol, these two lysates were mixed in the “mix” control sample. RNA was reverse transcribed with a primer specific for the p53 3′ exon sequence at 37°C for 15 min in the presence of 50 mM l-argininamide, a competitive inhibitor of ribozyme activity to squelch trans splicing during reverse transcription (RT). The resulting cDNAs were amplified for 40 cycles by a 5′ primer specific for the trans-splicing junction (5′-GGGGGGATCCGTCGAGCCCCATATCTCC-3′, including two mutations of C to A in a portion of 5′ p53 sequence to avoid amplification of Rib41–3′ p53 itself) and by a 3′ primer specific for the p53 cDNA sequence downstream of the splice junction.
Luciferase Assays on Extracts from Osteosarcoma, Colon Cancer, and Lung Cancer Cells.
Saos-2 and SW480 human colorectal carcinoma (or Calu-6 human lung adenocarcinoma) cells were seeded in 100-mm plates at a density of 7.5 × 105 and 1.5 × 106 cells per plate, respectively, 18–24 h before transfection. The cells were cotransfected with PG13-luc (2 μg; containing 13 copies of a p53 DNA-binding consensus sequence linked to a luciferase reporter gene, a generous gift of B. Vogelstein) and a β-galactosidase expression vector pCMVβgal (1 μg) along with pC53 5′ trunc (5 μg) and pRib41p53 (5 μg) or pRib65p53 (5 μg) with DMRIE-C (26 μg) for Saos-2 cells and pRib41p53 (5 μg) or pRib65p53 (5 μg) along with DMRIE-C (16 μg) for SW480 cells. As a negative control, the empty parental expression vector was substituted for either p53 or ribozyme expression plasmid. In contrast to the active ribozyme constructs, transfection of the inactive ribozyme expression plasmids alone into Saos-2 cells resulted in a modest (2- to 10-fold) increase in luciferase activity, suggesting that the p53-derived 3′ exons after the deleted introns were being translated, thus ruining their utility as controls in these experiments. In previous experiments with a 3′ exon encoding the green fluorescent protein, we observed that several potential translational start sites exist within the sequence of the Tetrahymena group I intron that are activated by deletion of the ribozyme's core in the inactive ribozyme construct (unpublished results). Calu-6 cells were cotransfected with pMDR-luc containing the human MDR1 promoter upstream of the luciferase gene (graciously provided by E. J. Stanbridge, University of California, Irvine; 3 μg) and pCMVβgal (1 μg) along with pRib41p53 (5 μg) or pRib65p53 (5 μg) with DMRIE-C (18 μg). Cell lysates were harvested 24 h after transfection for Saos-2 cells and 48 h after transfection for SW480 and Calu-6 cells with the Reporter lysis buffer provided with the Luciferase Assay System (Promega) and assayed for luciferase activity by using a Lumat LB9507 (Berthold, Nashua, NH). Transfection efficiency was quantified by measuring β-galactosidase activity in the extracts, and luciferase values were normalized to account for differences in transfection efficiency.
Results
To ascertain which uridines in the p53 transcript are accessible, we developed an RNA mapping strategy based on a trans-splicing ribozyme library (25) and RNA tagging (19). A ribozyme library was constructed that contains a randomized IGS, 5′-GNNNNN-3′ (GN5), where G represents guanine and N represents equal amounts of the four nucleotides (Fig. 1). The library was incubated under splicing conditions either with a portion of p53 RNA generated by in vitro transcription (nucleotides 1–432) or with total RNA isolated from MDA-MB-231 human breast cancer cells, which overexpress a mutant version of p53 mRNA (mutation at codon 280, AGA changed to AAA). The trans-splicing reaction products were amplified by RT-PCR with primers specific for the ribozyme's 3′ exon tag (3′ lacZ exon sequence) and the target p53 RNA. Sequence analyses of the splice junctions showed that several uridines appeared accessible (Table 1). To delineate further which uridines are the most accessible, we evaluated a number of the ribozymes for their ability to cleave p53 transcripts in vitro. Only ribozymes Rib41 and Rib65, which recognize the uridines at position 41 (U41) and 65 (U65) of the p53 coding sequence, cleaved the majority of the p53 target RNA to yield products of the expected size (data not shown). Moreover, p53 RNA is most often mutated in human cancers at sites downstream of nucleotide 328 (1). Both reasons encouraged us to focus on further developing ribozymes Rib41 and Rib65.
Table 1.
In vitro mapping results of the ribozyme-accessible sites in the p53 RNA
| Reaction sites, nucleotide position* | Number of clones†
|
|
|---|---|---|
| p53 RNA‡ (in vitro transcribed) | Cellular RNA from MDA-MB-231 cells§ | |
| +41 | 4 | 4 |
| +65 | 2 | 2 |
| +307 | — | 1 |
| +332¶ | — | 1 |
| +340¶ | 1 | 1 |
| +384¶ | 6 | 5 |
| +389¶ | — | 1 |
Nucleotide positions are indicated as the nucleotide numbers after the translation initiation site and presented for the accessible uridines identified from the mapping analyses.
The number of individual clones containing a given uridine at the splice.
A portion of p53 target RNA (nucleotides 1–432).
A human breast cancer cell line that overexpresses a mutant p53 mRNA (AGA to AAA at codon 280).
The p53 gene is frequently mutated in human tumors downstream of nucleotide 328 (1).
To create Rib41 and Rib65 derivatives that could convert mutant p53 transcripts into wt p53 RNAs, we attached the wt human p53 sequences to the ribozymes (nucleotides 42 through 1,179 for Rib41 and nucleotides 66 through 1,179 for Rib65) as a 3′ exon. These ribozymes, called Rib41–3′ p53 and Rib65–3′ p53, were expressed by using an RNA polymerase II-based transcription unit containing the CMV-IE gene promoter. Because group I trans-splicing ribozymes that contain 6-nt-long IGS are not very active after expression in bacteria (26), we extended the IGS of Rib41–3′ p53 and Rib65–3′ p53 such that they would contain an extended P1 and a 35-nt-long antisense region as well as a P10 helix. Moreover, we have been unable to detect trans-splicing activity after pol II-based expression of trans-splicing ribozymes that contain only 6-nt-long guide sequences in mammalian cells (N. Lan and B.A.S., unpublished work). To distinguish repaired p53 transcripts from mutant p53 RNA easily, we took advantage of the degeneracy of the genetic code to alter the sequence of the restorative p53 3′ exon without changing the encoded protein. These changes also allowed us to amplify trans-splicing products selectively by using a 5′ primer complementary to the cDNA derived from repaired p53 RNAs and allowed us to reduce potential problems engendered by making the antisense region complementary to the 3′ exon.
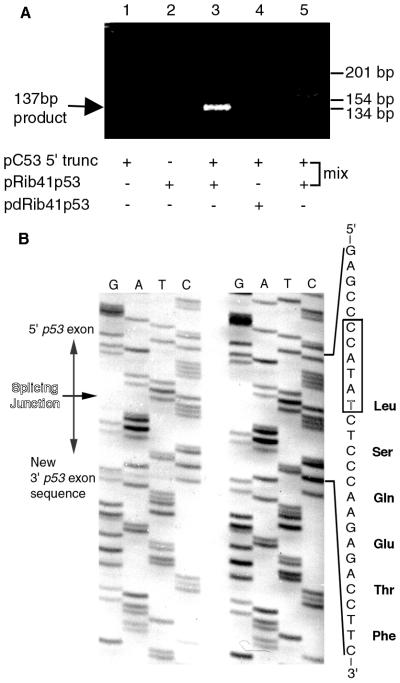
To test whether Rib41–3′ p53 could repair truncated p53 transcripts in cancer cells, we cotransfected a plasmid containing the Rib41–3′ p53 expression cassette (pRib41p53) along with a plasmid containing the CMV promoter and a truncated p53 cDNA (pC53 5′ trunc) into the human osteosarcoma cell line, Saos-2, which does not express endogenous p53 transcripts (27). Total RNA was isolated from transfected cells and analyzed by RT-PCR with primers specific to the repaired p53 transcript. Trans-spliced products were detected only in RNA from cells transfected with pRib41p53 and pC53 5′ trunc (Fig. 2A, lane 3). No such product was detected in cells transfected with pC53 5′ trunc alone or in those cotransfected with pC53 5′ trunc and an inactive ribozyme plasmid (pdRib41p53) that lacks part of the catalytic core of the enzyme (Fig. 2A, lanes 1 and 4). Moreover, no trans-splicing product was detected in a “mix” RNA sample, where lysate from pRib41p53-transfected cells was mixed with lysate from pC53 5′ trunc-transfected cells before RNA extraction (Fig. 2A, lane 5). Sequence analysis demonstrated that the ribozyme had correctly repaired the truncated p53 transcript by splicing its restorative p53 3′ exon onto the U41 of the mutant p53 mRNA (Fig. 2B). Thus, this experiment indicated that the ribozyme was able to repair truncated p53 transcripts in these human osteosarcoma cells and restore the ORF for translation of the p53 protein inside these cells.
Figure 2.
(A) Repaired p53 RNAs generated in Saos-2 cells. Cells were transfected with a truncated p53 expression plasmid (pC53 5′ trunc; lanes 1 and 3–5) alone (lane 1) or with the active (pRib41p53, lanes 2 and 3; lane 5 mix sample) or with the inactive (pdRib41p53, lane 4) ribozyme expression plasmid. Corrected p53 RNAs were amplified by RT-PCR, yielding a DNA fragment of 137 bp. The migration of size markers of 134, 154, and 201 bp is indicated. (B) Sequences of amended p53 transcripts produced in Saos-2 cells. Sequences of two representative clones are shown. The expected sequence for a corrected transcript around the splicing junction is shown along with the predicted translation product. The ribozyme recognition sequence is boxed, and the nucleotide at the position 41 is outlined.
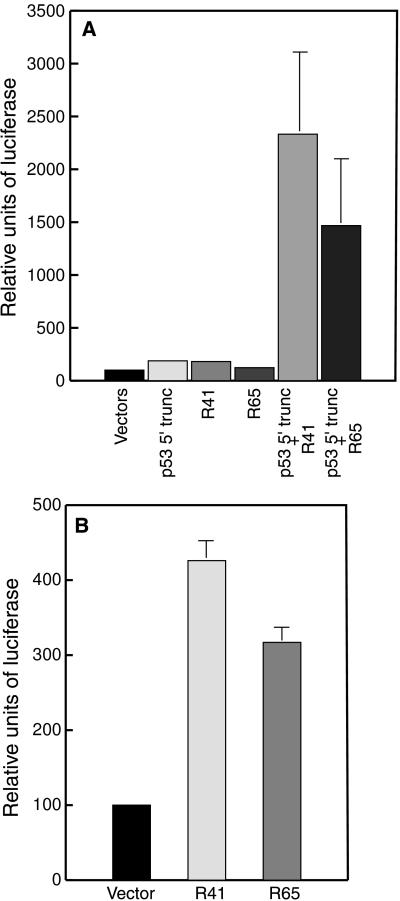
To determine whether the repaired p53 transcripts were being translated to produce functional p53 protein inside transfected cells, we cotransfected a reporter plasmid containing the luciferase gene under the control of a p53-dependent promoter (PG13-luc) into the Saos-2 cells that had been transfected with the ribozyme and p53 substrate expression plasmids. Production of functional p53 was then monitored by measuring the induction of luciferase activity. In Saos-2 cells transfected with PG13-luc and the empty parental expression vectors, for both truncated p53 and ribozyme, only very low levels of luciferase activity were induced, as would be expected because Saos-2 cells do not produce endogenous p53 protein (27). Transfection of pRib41p53 or pRib65p53 along with pC53 5′ trunc and PG13-luc in Saos-2 cells resulted in a 23-fold and a 15-fold increase in p53-induced luciferase activity, respectively (Fig. 3A). No significant increase in luciferase activity was noted in cells transfected either with ribozyme or with the substrate plasmid alone (Fig. 3A). These results demonstrate that trans-splicing ribozymes can repair mutant p53 transcripts to generate mRNAs that are translated to produce functional p53 protein. These newly generated transcription factors can then transactivate p53 responsive promoters in human cancer cells.
Figure 3.
p53-mediated transcriptional transactivation after RNA repair. (A) Saos-2 cells were transfected with the PG13-luc reporter construct and the empty parental expression vectors (Vectors), the truncated 5′ p53 expression plasmid alone (pC53 5′ trunc), each ribozyme expression plasmid alone [pRib41p53 (R41) or pRib65p53 (R65)], or pC53 5′ trunc along with pRib41p53 or pRib65p53. (B) SW480 cells were transfected with the PG13-luc and each ribozyme expression vector (pRib41p53 and pRib65p53) or an empty vector. Relative luciferase activity was quantitated as a percentage of the vector control sample, and average values ± SD for luciferase activity from three independent experiments are shown.
To determine whether trans-splicing ribozymes can repair endogenous transcripts expressed from mutant p53 genes present in cancer cells and restore p53 function therein, we evaluated the ribozymes in the human colorectal carcinoma cell line SW480 that expresses a p53 gene harboring two point mutations (G818 to A818 and C925 to U925). Transfection of these cells with PG13-luc and a control vector resulted in the generation of only low levels of luciferase activity, because the cells express only mutant p53 protein that poorly transactivates luciferase gene expression (Fig. 3B). However, cotransfection of SW480 cells with PG13-luc and either pRib41p53 or pRib65p53 resulted in a 4-fold or 3-fold induction, respectively, of luciferase expression in these cells (Fig. 3B). By comparison, cotransfection of these SW480 cells with PG13-luc and pC53-SN3, which harbors the CMV-IE promoter and the wt p53 cDNA, stimulated luciferase production by approximately 180-fold (data not shown). Thus, the ribozymes are able to repair endogenous mutant p53 transcripts in human colorectal carcinoma cells, and the amended RNAs can be translated to produce functional p53 protein. Moreover, the amount of p53-mediated transcriptional transactivation generated after such repair can reach levels that are at least 1–2% of the level of p53 activity generated after high-level expression of the protein in these same cells with a strong, virally derived promoter.
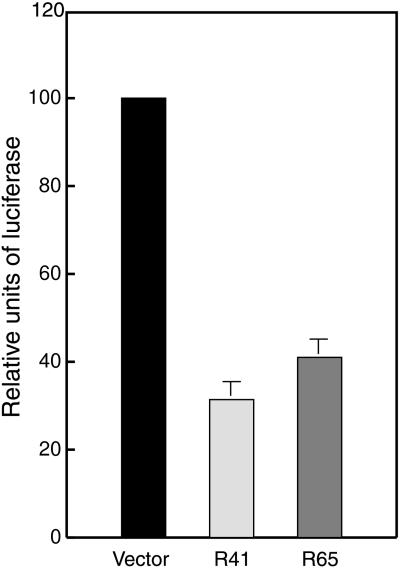
Because it has been observed that certain mutant p53s activate transcription of the human MDR1 gene, whereas wt p53 represses such expression (12), we sought to determine whether the wt p53 generated after RNA repair was able to repress mutant p53-mediated transactivation of the MDR1 promoter in cancer cells. A construct containing the human MDR1 promoter upstream of the luciferase gene (pMDR-luc) was cotransfected with pRib41p53 or pRib65p53 into the Calu-6 lung carcinoma cells that harbor a homozygous nonsense p53 mutation at codon 196 (CGA to TGA). Cotransfection of either pRib41p53 or pRib65p53 along with pMDR-luc significantly repressed transcription from the MDR1 promoter (67 ± 5% and 60 ± 6%, respectively; Fig. 4) as compared with the level of expression observed when the cells were cotransfected with pMDR-luc and a control vector (Fig. 4). By comparison, transfection of Calu-6 cells with pC53-SN3 suppressed activation of the MDR1 promoter by approximately 95% (data not shown). Thus, the functional p53 protein generated after RNA repair can significantly repress the activation of the MDR1 gene promoter by mutant p53 protein present in the lung adenocarcinoma cells. Moreover, this level of repression is approximately 65–75% the level obtained when high levels of p53 protein are expressed in the Calu-6 cells.
Figure 4.
Repression of transcription from the MDR1 gene promoter after p53 repair. Calu-6 cells were cotransfected with the MDR-luc reporter construct and an empty vector or each ribozyme expression vector (pRib41p53 or pRib65p53). Relative luciferase activity was quantitated as a percentage of the vector control sample, and average values ± SD for luciferase activity from three independent experiments are shown.
Discussion
We have used a trans-splicing group I ribozyme to amend mutant p53 transcripts in human cancer cells (Fig. 1). To determine which regions of the p53 transcript are accessible for ribozyme binding, we mapped the p53 RNA with a trans-splicing ribozyme library (Table 1). Such analysis suggested that the uridines at positions 41 and 65 in the p53 coding sequence are particularly accessible for ribozyme binding and activity. Using ribozymes designed to target these particular residues, we demonstrated that a single trans-splicing ribozyme can be used to revise a variety of different mutant p53 transcripts in cells from different types of human cancers (Figs. 2–4). The revision of p53 transcripts seems to proceed with high fidelity in these settings (Fig. 2). Moreover, we showed that the RNAs repaired by trans splicing go on to be translated to produce functional protein inside human cancer cells, which can lead to transactivation of a p53 responsive promoter (Fig. 3 A and B) and down-modulate expression from the MDR1 gene promoter (Fig. 4).
These results suggest that repair of defective p53 RNA may become a useful strategy to engender production of functional p53 activity in neoplastic cells to combat cancer. Aberrant expression of p53 by conventional gene replacement strategies may lead to unintended phenotypic changes in both tumorigenic and normal cells (28, 29). For example, it has been demonstrated that overexpression of the wt p53 gene aberrantly decreases growth rate and alters morphological differentiation of normal human keratinocytes (28). Thus, coordinated expression of the p53 gene is apparently critical for proper growth, development, and differentiation of certain primary human cells, and incorrect expression can lead to dramatic phenotypic aberrations. By contrast, mRNA repair should result in the production of correct gene products at the appropriate times in human development and cellular differentiation, because the ribozyme can amend the mutant mRNA only when it is expressed. Our observation that p53 activity is engendered only in Saos-2 cells when such cells are cotransfected with a ribozyme plasmid and a plasmid encoding a truncated p53 transcription unit highlights the fact that mRNA repair can occur only when substrate RNAs are present in the cell with the ribozyme (Fig. 3A). In addition, trans-splicing ribozymes can simultaneously reduce the production of a deleterious protein and induce the production of a functional gene product to lead to desired phenotypic changes in treated cells, as is the case when functional p53 protein suppressed mutant p53 protein-mediated activation of the MDR1 promoter in Calu-6 cells after RNA repair (Fig. 4). Thus, mRNA repair represents a potentially very powerful approach to genetic therapy that offers gene therapists some unique advantages over currently used techniques.
Our findings that significant levels of p53 activity can be generated from repaired transcripts are quite encouraging with regard to long-term development of this technology, especially because the efficiency of mutant p53 RNA repair will almost assuredly not have to be 100% to benefit patients. The wt p53 protein has been shown to be able to suppress cell growth totally and decrease colony formation when present at quantities 10-fold lower than mutant p53 in neoplastic cells (30). Thus, repair of as little as 10% of the mutant p53 transcripts may be efficient enough to revert neoplastic transformation, induce apoptosis, or render cancerous cells sensitive to lower doses of chemotherapeutic agents. To determine whether trans-splicing ribozymes can repair this level of p53 RNA in neoplastic cells and impact the development and progression of cancer in animals, more efficient ribozyme delivery systems must be developed. Of the currently available gene delivery systems, adenoviral vectors may prove the most useful for such studies because of their ability to transfer genes fairly efficiently into a variety of human cancer cells (14). However, in the long term, new, less immunogenic, and even more efficient gene transfer systems will likely be required to deliver trans-splicing ribozyme in a clinically relevant fraction of premalignant or transformed cells. The demonstration that trans-splicing ribozymes can repair a variety of mutant p53 transcripts and induce p53 activity in a number of different human cancer cell lines should continue to encourage the development of more effective gene transfer systems.
Acknowledgments
We thank T. Burke, P. Zarrinkar, C. Rusconi, L. Milich, J. P. Jones, N. Lan, J. T. Jones, and M. Long for helpful discussions and technical assistance. This project was supported by Department of the Army Grant DAMD17-97-1-7278 and National Institutes of Health Grant GM53525 (to B.A.S.).
Abbreviations
- wt
wild-type
- CMV
cytomegalovirus
- IGS
internal guide sequence
- RT
reverse transcription
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.150104097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.150104097
References
- 1.Hollstein M, Sidransky D, Vogelstein B, Harris C C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 2.Cho Y, Gorina S, Jeffery P D, Pavletich N P. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 3.Farmer G, Bargonetti J, Zhu H, Friedman P, Prywes R, Prives C. Nature (London) 1992;358:83–86. doi: 10.1038/358083a0. [DOI] [PubMed] [Google Scholar]
- 4.Kern S E, Pietenpol J A, Thiagalingam S, Seymour A, Kinzler K W, Vogelstein B. Science. 1992;256:827–832. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- 5.Kern S E, Kinzler K W, Bruskin A, Jarosz D, Friedman P, Prives C, Vogelstein B. Science. 1991;252:1708–1711. doi: 10.1126/science.2047879. [DOI] [PubMed] [Google Scholar]
- 6.el-Deiry W S, Kern E S, Pietenpol J A, Kinzler K W, Vogelstein B. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 7.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 8.Miyashita T, Reed J C. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 9.Milner J, Medcalf E A. Cell. 1991;65:765–774. doi: 10.1016/0092-8674(91)90384-b. [DOI] [PubMed] [Google Scholar]
- 10.Harvey M, Vogel H, Morris D, Bradley A, Bernstein A, Donehower L A. Nat Genet. 1995;9:305–311. doi: 10.1038/ng0395-305. [DOI] [PubMed] [Google Scholar]
- 11.Dittmer D, Pati S, Zambetti G, Chu S, Teresky A K, Morre M, Finlay C, Levine A J. Nat Genet. 1993;4:42–46. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- 12.Chin K-V, Ueda K, Pastan I, Gottesman M M. Science. 1992;255:459–462. doi: 10.1126/science.1346476. [DOI] [PubMed] [Google Scholar]
- 13.Endicott J A, Ling V. Annu Rev Biochem. 1989;58:137–171. doi: 10.1146/annurev.bi.58.070189.001033. [DOI] [PubMed] [Google Scholar]
- 14.Roth J A, Swisher S G, Meyn R E. Oncology. 1999;13:148–154. [PubMed] [Google Scholar]
- 15.Hann B C, Lane D P. Nat Genet. 1995;9:221–222. doi: 10.1038/ng0395-221. [DOI] [PubMed] [Google Scholar]
- 16.Inoue T, Sullivan F X, Cech T R. Cell. 1985;43:431–437. doi: 10.1016/0092-8674(85)90173-4. [DOI] [PubMed] [Google Scholar]
- 17.Been M D, Cech T R. Cell. 1986;47:207–216. doi: 10.1016/0092-8674(86)90443-5. [DOI] [PubMed] [Google Scholar]
- 18.Sullenger B A, Cech T R. Nature (London) 1994;371:619–622. doi: 10.1038/371619a0. [DOI] [PubMed] [Google Scholar]
- 19.Jones J T, Lee S-W, Sullenger B A. Nat Med. 1996;2:643–648. doi: 10.1038/nm0696-643. [DOI] [PubMed] [Google Scholar]
- 20.Lan N, Howrey R P, Lee S-W, Smith C A, Sullenger B A. Science. 1998;280:1592–1596. doi: 10.1126/science.280.5369.1593. [DOI] [PubMed] [Google Scholar]
- 21.Phylactou L A, Darrah C, Wood M J A. Nat Genet. 1998;18:378–381. doi: 10.1038/ng0498-378. [DOI] [PubMed] [Google Scholar]
- 22.Hagen M, Cech T R. EMBO J. 1999;18:6491–6500. doi: 10.1093/emboj/18.22.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zarrinkar P P, Sullenger B A. Biochemistry. 1998;37:18056–18063. doi: 10.1021/bi982193x. [DOI] [PubMed] [Google Scholar]
- 24.Baker S J, Markowitz E R, Fearon J K, Willson K V, Vogelstein B. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 25.Campbell T B, Cech T R. RNA. 1995;1:598–609. [PMC free article] [PubMed] [Google Scholar]
- 26.Köhler U, Ayre B G, Goodman H M, Haseloff J. J Mol Biol. 1998;285:1935–1950. doi: 10.1006/jmbi.1998.2447. [DOI] [PubMed] [Google Scholar]
- 27.Masuda H, Miller C, Koeffler H P, Battifora H, Cline M J. Proc Natl Acad Sci USA. 1987;84:7716–7719. doi: 10.1073/pnas.84.21.7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woodworth C D, Wang H, Simpson S, Alvarez-Salas L M, Notario V. Cell Growth Differ. 1993;4:357–376. [PubMed] [Google Scholar]
- 29.Slack R S, Belliveau D J, Rosenberg M, Atwal J, Lochmuller H, Aloyz R, Haghighi A, Lach B, Seth P, Cooper E, et al. J Cell Biol. 1996;135:1085–1096. doi: 10.1083/jcb.135.4.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen P-L, Chen Y, Bookstein R, Lee W-H. Science. 1990;250:1576–1580. doi: 10.1126/science.2274789. [DOI] [PubMed] [Google Scholar]