Abstract
The biological role of the mitochondrial DNA (mtDNA) control region in mtDNA replication remains unclear. In a worldwide survey of mtDNA variation in the general population, we have identified a novel large control region deletion spanning positions 16154 to 16307 (m.16154_16307del154). The population prevalence of this deletion is low, since it was only observed in 1 out of over 120,000 mtDNA genomes studied. The deletion is present in a nonheteroplasmic state, and was transmitted by a mother to her two sons with no apparent past or present disease conditions. The identification of this large deletion in healthy individuals challenges the current view of the control region as playing a crucial role in the regulation of mtDNA replication, and supports the existence of a more complex system of multiple or epigenetically-determined replication origins.
Keywords: mitochondrial DNA, mtDNA, control region, replication
INTRODUCTION
The uniparental ancestry and abundance in eukaryotic cells of the α-proteobacterial descended mitochondrion [Lang et al., 1999] has enabled the genome of this “cellular power plant” to serve as a model system for phylogenetic and evolutionary studies [Falkenberg et al., 2007]. Forty years of research on the ~16,569–base pair (bp) circular mitochondrial DNA (mtDNA) genome has fueled an intense debate over the role of the control region, spanning nucleotide positions 16024 to 00576, in mtDNA replication [Bogenhagen and Clayton, 2003; Yasukawa et al., 2005]. Traditionally, replication was believed to involve an asynchronous displacement mechanism of two unidirectional, independent origins [Clayton, 1982; Fernandez-Silva et al., 2003]. This theory has been recently challenged by evidence of a coupled leading- and lagging-strand synthesis of the control region, consistent with strand-coupled replication for mtDNA [Bowmaker et al., 2003; Holt et al., 2000; Yang et al., 2002; Yasukawa et al., 2005]. As a result, the precise role of the control region in mtDNA replication remains unresolved. We now describe a naturally-occurring deletion in a healthy family that removes 154 bp from the control region, showing that this segment, at least, is not required for normal replication.
MATERIAL AND METHODS
This study analyses a novel large mitochondrial control region deletion spanning positions 16154 to 16307 (Fig. 1). The systematic mutation name, m.16154_16307del154, follows the journal's nomenclature recommendations (www.hgvs.org/mutnomen) and is based on the revised Cambridge Reference Sequence (rCRS), reference sequence AC_000021.2 [Andrews et al., 1999]. Initially, this 154-bp deletion was identified in an anonymous Japanese woman tested by the Genographic Project's public participation program, following its reported consent and genotyping protocols [Behar et al., 2007]. Subsequently, and following additional informed consents, the deletion was confirmed in her two sons. No additional maternally-related individual could be identified. All three carriers of the deletion have no known adverse effects on their health or reduced longevity among close family members, as determined by a short questionnaire assessing existing medical conditions such as chronic diseases, severe allergies, major surgeries, birth disabilities, or the long-term use of prescription drugs.
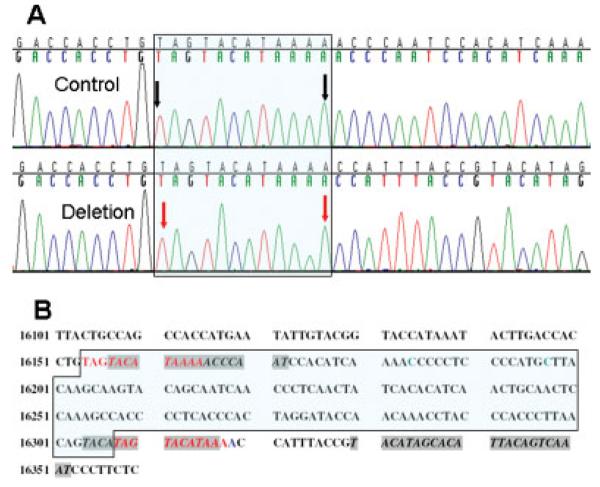
Figure 1. Position of the control region deletion.
A: L-strand chromatograms reading 5′ to 3′ of a control mtDNA sample that matches the rCRS [Andrews et al.,1999] (top) and of the sample containing the 154-bp deletion (bottom). The repeated sequences flanking the deletion are marked in the box. The black arrows point to mtDNA positions 16154 and 16165. The red arrows point to positions 16308 and 16319 of the mtDNA. Position 16308 also marks the 3′ end of the deletion breakpoint. B: L-strand stretch of haplogroup A1a sequence reading 5′ to 3′. The deleted segment between positions 16154 and 16307 is marked in the box and proposed to have arisen by recombination between the two flanking repeats in red fonts. The adenine in position 16319 is marked in blue as it differs from the rCRS in the same position [Andrews et al.,1999]. The TAS are marked in italics and gray background [Roberti et al., 1998]. Positions 16197 and 16184, thought to have important roles in replication [Yasukawa et al., 2005], are marked in green.
To confirm that the reported deletion does not represent an undetected heteroplasmic state [Torroni et al., 1994] or a nuclear insertion resulting from horizontal gene transfer, we repeated the amplification and sequencing of the control region using a number of primer pairs both internal and external to the deleted region (Fig. 2A and B). The amplification and sequencing of the control region was run using three pairs of primers lying outside the deleted region and using as positive controls samples from various human phylogenetic branches. Two pairs of primers have been previously reported, namely, 15876F and 639R [Behar et al., 2007], and D1F and D1R [Taylor et al., 2001]. The third pair of primers was designed as the M13-tailed primers 11CF and 11CR:
● 11CF 5′-TGTAAAACGACGGCCAGTGAAAAAGTCTTTAACTCCAC-3′ and
● 11CR 5′-CAGGAAACAGCTATGACCATACCAAATGCATGGAGAGC-3′.
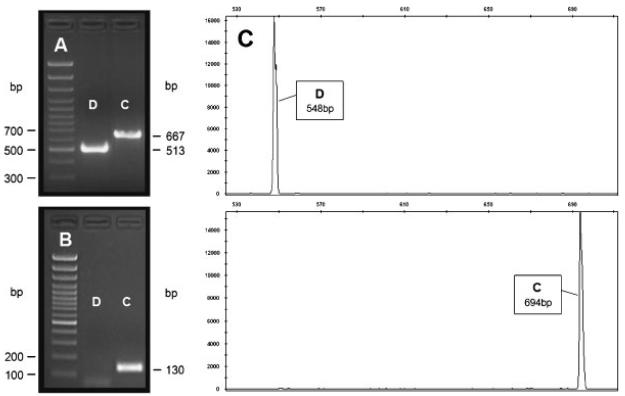
Figure 2. Molecular analysis of the control region deletion.
A: Detection of the deletion by 2% agarose gel electrophoresis of amplified fragments using the external to the deletion primers D1F and D1R [Taylor et al., 2001]. Molecular size marker (100-bp ladder, lowest band 300 bp) is shown on the left. The amplification product of the control (C) mtDNA sequence is seen at the 667-bp position and the deleted (D) mtDNA sequence at the 513-bp position. B: Detection of the deletion by 2% agarose gel electrophoresis of amplified fragments using the internal to the deletion primers 16158F and 16287R. Molecular size marker (100-bp ladder, lowest band 100 bp) is shown on the left. The amplification product of the control mtDNA sequence is seen at the 130-bp position while the deleted mtDNA showed no amplification. C: Electropherogram. The PCR was performed using the M13-tailed primers 11CF and 11CR, followed by a secondary PCR using aTAMRA-labeled M13 primer (Applied Biosystems). The secondary PCR products were then run on an ABI3730 using standard protocols for fragment analysis. Base pair insertions or deletions can be seen by an increase or decrease in fragment length when compared to a reference sequence.
In addition, we verified the deletion length by means of a fragment length analysis technique, which accurately measured the size of the PCR products on an ABI3730 (Applied Biosystems, Foster City, CA) using standard allele-size ladders and the same tailed primers (Fig. 2C). To confirm that the deleted segment was not transferred to the nuclear genome (i.e., a NUMT) or had been missed by our PCR amplification primers, we designed a pair of primers internal to the deleted segment:
● 16158F 5′-ACATAAAAACCCAATCCACA-3′ and
● 16287R 5′-GGTTTGTTGGTATCCTAGTGG-3′.
Finally, to confirm that the deleted segment was not transferred to a different location in the mitochondrial genome, we sequenced the entire mtDNA genome of the mother [Taylor et al., 2001]. The complete sequence of the index case (GenBank accession number EU558518) was colinear with the rCRS and revealed no sign of the deleted segment.
RESULTS AND DISCUSSION
In this study we describe a large mtDNA control region deletion of 154 bp (m.16154_16307del154) inherited in a nonheteroplasmic state in a healthy trio of one mother and two sons. Our molecular analysis indicated that the three samples with the deletion produced a PCR amplification product only when primers external to the deletion were used, and product sizes were as expected from the deletion length (Fig. 2A). When primers internal to the deletion were used, control samples yielded a PCR product concordant with the expected sizes, while the studied samples containing the 154-bp deletion showed no amplification (Fig. 2B). In all attempts, only one specific PCR product was observed, demonstrating the existence of a single DNA template in both the control and studied samples at this level of sensitivity (Fig. 2). The mutation, classified as a class I deletion [Samuels et al., 2004], is flanked by two short homologous direct repeat sequences of 12 nucleotides, TAGTACATAAAA (Fig. 1), and contains several positions previously reported by two models as involved in mtDNA replication [Fernandez-Silva et al., 2003; Yasukawa et al., 2005]. It is important to note that even if our molecular analysis missed a low level of heteroplasmy and a subset of the mtDNA genomes carried by the studied individuals does not contain the deletion, the identification of the deletion in two different generations is a clear-cut proof of their capability to replicate. While the prevalence of the deletion in the general population remains unknown, it should be noted that it was identified in only 1 out of over 120,000 mtDNA genomes tested so far by the Genographic Project's public participation program [Behar et al., 2007]. An update of the Genographic mtDNA database is also being made available as part of this report, and can be accessed at www3.nationalgeographic.com/genographic/resources.html.
Interestingly, the phylogenetic branch of the index case, haplogroup A1a according to standard nomenclature [Tanaka et al., 2004], is characterized by an adenine at position 16319 as compared to a guanine found at the same position in the rCRS. This increases the length of the repeated sequences to 12 nucleotides (TAGTACATAAAA) rather than 11, as found in mitochondrial genomes devoid of an adenine in position 16319 (Fig. 1) and may thus predispose to the deletion. Preliminary analyses of potential DNA folding in the region where the deletion is located using the mfold program [Zuker, 2003] have revealed two folding structures depending on whether a G or an A was at position 16319 (results not shown). Even though the mutation at position 16319 seems to affect the mtDNA folding structure, this mutation is found in many other phylogenetic branches [Behar et al., 2007], suggesting that the formation of the deletion is still a rare event.
Associations of large deletions in the mitochondrial genome with human pathologies have been used to elucidate the function of specific genomic regions [Copeland, 2008]. A comprehensive review of 263 large mtDNA deletions noted only four deletions with a 3′ end extending beyond position 16085, while deletions removing the region beyond 16268 have never been reported [Samuels et al., 2004]. One deletion extended to position 16191 and was associated with Wolfram syndrome [Barrientos et al., 1996]. The other three were observed only as sublimons present in very low levels in human cells and extending to positions 16085, 16116, and 16268 [Kajander et al., 2000]. This absence of large deletions within the control region has actually been suggested to circumstantially enforce the importance of this region as containing potential elements important for the origin or termination of mtDNA replication [Samuels et al., 2004; Yasukawa et al., 2005]. Our data show that that the deletion reported herein is not overtly deleterious. Its rarity among humans might suggest a subtle reduction in fitness, but we have no way to refute or confirm this possibility.
The novel deletion event, from positions 16154 to 16307, can however contribute significantly to our understanding of the functionality of this region in mtDNA replication. In particular, it is pertinent to assessing the two competing models of mtDNA replication, as it encompasses a region labeled as significant to the regulation of mtDNA replication by the asynchronous displacement model, as well as positions that have been highlighted as potential replication origins by the strand-coupled replication model. Under the classic asynchronous displacement model, unidirectional replication begins at the origin of the heavy strand (OH), around mtDNA position 191, and proceeds to displace the parental heavy strand [Andrews et al., 1999; Brown et al., 2005; Crews et al., 1979] until it induces the second replication origin at the light strand (OL) located outside of the control region [Fernandez-Silva et al., 2003]. While the asynchronous displacement mechanism is not fully resolved, this model has emphasized the control region as important for the termination of replication [Fernandez-Silva et al., 2003]. The regulation of replication termination and elongation is reportedly controlled by a series of cis-acting termination associated sequences (TAS) located at the 5′ end of the termination sites in a subregion of the first hypervariable segment (HVS-I) [Brown and Clayton, 2002; Doda et al., 1981; Kai et al., 1999]. More specifically, in vivo footprints of protein-binding sites were detected in the TAS region, particularly around mtDNA positions 16147 to 16176 on the H-strand and positions 16309 to 16336 on the L-strand, suggesting the existence of five possible TAS elements, A–E [Roberti et al., 1998]. The reported deletion completely eliminates the first identified TAS [Doda et al., 1981], later labeled as TAS-D (H strand), and 4 out of the 14 nucleotides comprising TAS-C (L strand) [Roberti et al., 1998], and therefore calls into question the presumed role of the TAS in the regulation of mtDNA replication.
According to the strand-coupled replication mechanism, different control region positions are highlighted as potential replication origins within and between different organs which attests for redundancy and plasticity of mtDNA replication origin. In solid tissues a broad origin of replication was suggested, which extend into the coding region, including the genes for cytochrome b and reduced nicotinamide adenine dinucleotide (NADH) dehydrogenase subunits five and six [Bowmaker et al., 2003]. The same group reported that in cultured human cells, bidirectional replication most frequently initiated at positions 16197 and 16184 of the H- and L-strands, respectively [Yasukawa et al., 2005]. It is clear, however, that these positions and the cluster of adjacent possible replication initiation site are eliminated by the reported deletion arguing against an essential role of positions 16197 and 16184 as replication origins.
The discovery of the novel deletion reported here can neither directly support nor refute these competing models of mtDNA replication, but it does have important biological implications. First, the deletion may indicate a more complex system of multiple replication origins, suggesting that mtDNA replication in humans is more dispersed than previously thought. The classic asynchronous displacement mechanism has emphasized the deleted region as important for the termination of replication [Fernandez-Silva et al., 2003]. However, the deletion did not abolish all previously suggested TAS A–E elements [Roberti et al., 1998] and in theory the deletion could have yielded a new functional TAS. Furthermore, a comprehensive survey of mammalian mtDNA regions has shown that these TAS are part of two main conserved blocks, named the extended TAS (ETAS-1 and ETAS-2) [Sbisa et al., 1997]. While the former is present in all mammals, the latter can be missing. It is also worth noting that the deleted region largely overlaps with a deletion previously reported in gorilla and orangutan [Sbisa et al., 1997], suggesting an evolutionary precedent for this event. Indeed, ETAS-1 (16081–16138) is retained in the reported samples, which might be consistent with the idea of its key role, rather than that of TAS-D, in mtDNA replication termination. In agreement with this idea is the observation that deletion of ETAS-1 has been associated with Wolfram syndrome [Barrientos et al., 1996]. It can also be argued that the existence of a few TAS elements in a concentrated area is actually indicative for the biological importance of that region as the repeated elements provide redundancy. The strand-coupled replication mechanism has emphasized the deleted region as important for the origin of replication [Yasukawa et al., 2005]. However, it is possible that other replication origins previously believed to be of minor importance, such as those observed around the H strand positions 16370 or 16411, might assume a dominant role in the mtDNA genomes reported herein [Yasukawa et al., 2005]. Alternatively, mtDNA replication origins might show little sequence specificity and be determined by a largely epigenetic mechanism, like nuclear replication origins [Aladjem and Fanning, 2004]. Second, we anticipate this finding will lead to further research on the reported samples in an attempt to further elucidate our understanding of the role of specific sequences within the control region for mtDNA replication. Finally, this work illustrates the importance of creating large databases of human genetic variation (i.e., in the general healthy population) to discover rare genetic variants that would otherwise remain unidentified. The discovery of such rare mtDNA haplotypes will be important to identify the relative power of adaptive and nonadaptive forces acting on the evolution of the mtDNA genome.
ACKNOWLEDGMENTS
We thank the three individuals that voluntarily donated their DNA sample to the study. C.T.S. is supported by The Wellcome Trust. The Genographic Consortium includes: Theodore G. Schurr, Department of Anthropology, University of Pennsylvania, Philadelphia, PA 19104-6398, USA; Fabricio R. Santos, Departamento de Biologia Geral, Universidade Federal de Minas Gerais, Belo Horizonte, Minas Gerais 31270-010, Brazil; Lluis Quintana-Murci, Unit of Human Evolutionary Genetics, CNRS URA3012, Institut Pasteur, Paris 75724, France; Jaume Bertranpetit, Evolutionary Biology Unit, Department of Experimental and Health Sciences, Universitat Pompeu Fabra, Barcelona 08003, Catalonia, Spain; David Comas, Evolutionary Biology Unit, Department of Experimental and Health Sciences, Universitat Pompeu Fabra, Barcelona 08003, Catalonia, Spain; Chris Tyler-Smith, The Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridgeshire CB10 1SA, UK; Elena Balanovska, Research Centre for Medical Genetics, Russian Academy of Medical Sciences, Moscow 115478, Russia; Oleg Balanovsky, Research Centre for Medical Genetics, Russian Academy of Medical Sciences, Moscow 115478, Russia; Doron M. Behar, Molecular Medicine Laboratory, Rambam Health Care Campus, Haifa 31096, Israel and Genomics Research Center, Family Tree DNA, Houston, TX 77008, USA; R. John Mitchell, Department of Genetics, La Trobe University, Melbourne, Victoria, 3086, Australia; Li Jin, Fudan University, Shanghai, China; Himla Soodyall, Division of Human Genetics, National Health Laboratory Service, Johannesburg, 2000, South Africa; Ramasamy Pitchappan, Department of Immunology, Madurai Kamaraj University, Madurai 625021, Tamil Nadu, India; Alan Cooper, Division of Earth and Environmental Sciences, University of Adelaide, South Australia 5005, Australia; Ajay K. Royyuru, Computational Biology Center, IBM T.J. Watson Research Center, Yorktown Heights, NY 10598, USA; Saharon Rosset, Department of Statistics and Operations Research, School of Mathematical Sciences, Tel Aviv University, Tel Aviv 69978, Israel and Data Analytics Research Group, IBM T.J. Watson Research Center, Yorktown Heights, NY 10598, USA; Laxmi Parida, Computational Biology Center, IBM T.J. Watson Research Center, Yorktown Heights, NY 10598, USA; Jason Blue-Smith, Mission Programs, National Geographic Society, Washington, DC 20036, USA; David Soria Hernanz, Mission Programs, National Geographic Society, Washington, DC 20036, USA; R. Spencer Wells, Mission Programs, National Geographic Society, Washington, DC 20036, USA.
Grant sponsors: National Geographic Society; IBM; Waitt Family Foundation; Seaver Family Foundation; Family Tree DNA; Wellcome Trust.
REFERENCES
- Aladjem MI, Fanning E. The replicon revisited: an old model learns new tricks in metazoan chromosomes. EMBO Rep. 2004;5:686–691. doi: 10.1038/sj.embor.7400185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- Barrientos A, Volpini V, Casademont J, Genis D, Manzanares JM, Ferrer I, Corral J, Cardellach F, Urbano-Marquez A, Estivill X, Nunes V. A nuclear defect in the 4p16 region predisposes to multiple mitochondrial DNA deletions in families with Wolfram syndrome. J Clin Invest. 1996;97:1570–1576. doi: 10.1172/JCI118581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar DM, Rosset S, Blue-Smith J, Balanovsky O, Tzur S, Comas D, Mitchell RJ, Quintana-Murci L, Tyler-Smith C, Wells RS. The Genographic Project public participation mitochondrial DNA database. PLoS Genet. 2007;3:e104. doi: 10.1371/journal.pgen.0030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen DF, Clayton DA. The mitochondrial DNA replication bubble has not burst. Trends Biochem Sci. 2003;28:357–360. doi: 10.1016/S0968-0004(03)00132-4. [DOI] [PubMed] [Google Scholar]
- Bowmaker M, Yang MY, Yasukawa T, Reyes A, Jacobs HT, Huberman JA, Holt IJ. Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J Biol Chem. 2003;278:50961–50969. doi: 10.1074/jbc.M308028200. [DOI] [PubMed] [Google Scholar]
- Brown TA, Clayton DA. Release of replication termination controls mitochondrial DNA copy number after depletion with 2′,3′-dideoxycytidine. Nucleic Acids Res. 2002;30:2004–2010. doi: 10.1093/nar/30.9.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TA, Cecconi C, Tkachuk AN, Bustamante C, Clayton DA. Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism. Genes Dev. 2005;19:2466–2476. doi: 10.1101/gad.1352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28:693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- Copeland WC. Inherited mitochondrial diseases of DNA replication. Annu Rev Med. 2008;59:131–146. doi: 10.1146/annurev.med.59.053006.104646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews S, Ojala D, Posakony J, Nishiguchi J, Attardi G. Nucleotide sequence of a region of human mitochondrial DNA containing the precisely identified origin of replication. Nature. 1979;277:192–198. doi: 10.1038/277192a0. [DOI] [PubMed] [Google Scholar]
- Doda JN, Wright CT, Clayton DA. Elongation of displacement-loop strands in human and mouse mitochondrial DNA is arrested near specific template sequences. Proc Natl Acad Sci USA. 1981;78:6116–6120. doi: 10.1073/pnas.78.10.6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- Fernandez-Silva P, Enriquez JA, Montoya J. Replication and transcription of mammalian mitochondrial DNA. Exp Physiol. 2003;88:41–56. doi: 10.1113/eph8802514. [DOI] [PubMed] [Google Scholar]
- Holt IJ, Lorimer HE, Jacobs HT. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell. 2000;100:515–524. doi: 10.1016/s0092-8674(00)80688-1. [DOI] [PubMed] [Google Scholar]
- Kai Y, Miyako K, Muta T, Umeda S, Irie T, Hamasaki N, Takeshige K, Kang D. Mitochondrial DNA replication in human T lymphocytes is regulated primarily at the H-strand termination site. Biochim Biophys Acta. 1999;1446:126–134. doi: 10.1016/s0167-4781(99)00015-9. [DOI] [PubMed] [Google Scholar]
- Kajander OA, Rovio AT, Majamaa K, Poulton J, Spelbrink JN, Holt IJ, Karhunen PJ, Jacobs HT. Human mtDNA sublimons resemble rearranged mitochondrial genoms found in pathological states. Hum Mol Genet. 2000;9:2821–2835. doi: 10.1093/hmg/9.19.2821. [DOI] [PubMed] [Google Scholar]
- Lang BF, Gray MW, Burger G. Mitochondrial genome evolution and the origin of eukaryotes. Annu Rev Genet. 1999;33:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]
- Roberti M, Musicco C, Polosa PL, Milella F, Gadaleta MN, Cantatore P. Multiple protein-binding sites in the TAS-region of human and rat mitochondrial DNA. Biochem Biophys Res Commun. 1998;243:36–40. doi: 10.1006/bbrc.1997.8052. [DOI] [PubMed] [Google Scholar]
- Samuels DC, Schon EA, Chinnery PF. Two direct repeats cause most human mtDNA deletions. Trends Genet. 2004;20:393–398. doi: 10.1016/j.tig.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Sbisa E, Tanzariello F, Reyes A, Pesole G, Saccone C. Mammalian mitochondrial D-loop region structural analysis: identification of new conserved sequences and their functional and evolutionary implications. Gene. 1997;205:125–140. doi: 10.1016/s0378-1119(97)00404-6. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Cabrera VM, González AM, Larruga JM, Takeyasu T, Fuku N, Guo LJ, Hirose R, Fujita Y, Kurata M, Shinoda K, Umetsu K, Yamada Y, Oshida Y, Sato Y, Hattori N, Mizuno Y, Arai Y, Hirose N, Ohta S, Ogawa O, Tanaka Y, Kawamori R, Shamoto-Nagai M, Maruyama W, Shimokata H, Suzuki R, Shimodaira H. Mitochondrial genome variation in eastern Asia and the peopling of Japan. Genome Res. 2004;14:1832–1850. doi: 10.1101/gr.2286304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RW, Taylor GA, Durham SE, Turnbull DM. The determination of complete human mitochondrial DNA sequences in single cells: implications for the study of somatic mitochondrial DNA point mutations. Nucleic Acids Res. 2001;29:E74–E74. doi: 10.1093/nar/29.15.e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Lott MT, Cabell MF, Chen YS, Lavergne L, Wallace DC. mtDNA and the origin of Caucasians: identification of ancient Caucasian- specific haplogroups, one of which is prone to a recurrent somatic duplication in the D-loop region. Am J Hum Genet. 1994;55:760–776. [PMC free article] [PubMed] [Google Scholar]
- Yang MY, Bowmaker M, Reyes A, Vergani L, Angeli P, Gringeri E, Jacobs HT, Holt IJ. Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell. 2002;111:495–505. doi: 10.1016/s0092-8674(02)01075-9. [DOI] [PubMed] [Google Scholar]
- Yasukawa T, Yang MY, Jacobs HT, Holt IJ. A bidirectional origin of replication maps to the major noncoding region of human mitochondrial DNA. Mol Cell. 2005;18:651–662. doi: 10.1016/j.molcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]