Abstract
Heart failure is a leading cause of mortality in South Asians. However, its genetic etiology remains largely unknown1. Cardiomyopathies due to sarcomeric mutations are a major monogenic cause for heart failure (MIM600958). Here, we describe a deletion of 25 bp in the gene encoding cardiac myosin binding protein C (MYBPC3) that is associated with heritable cardiomyopathies and an increased risk of heart failure in Indian populations (initial study OR = 5.3 (95% CI = 2.3–13), P = 2 × 10−6; replication study OR = 8.59 (3.19–25.05), P = 3 × 10−8; combined OR = 6.99 (3.68–13.57), P = 4 × 10−11) and that disrupts cardiomyocyte structure in vitro. Its prevalence was found to be high (~4%) in populations of Indian subcontinental ancestry. The finding of a common risk factor implicated in South Asian subjects with cardiomyopathy will help in identifying and counseling individuals predisposed to cardiac diseases in this region.
Heart failure, an emerging epidemic, is responsible for great loss in the productive years of life in people aged 40 and above. By 2030, in India alone, this loss is expected to rise to 17.9 million years, 10 times more than in the United States1,2. However, the molecular etiologies of such diseases in developing countries such as India remain poorly understood. Here, we focus on cardiomyopathy, a frequent cause of heart failure for which more than 200 rare disease-associated mutations affecting more than 20 different genes have already been identified3.
MYBPC3 encodes cardiac myosin binding protein C (cMyBP-C, Supplementary Fig. 1 online), a key constituent of the thick filaments localized to doublets in the C-zone of the A-band of the sarcomere. By binding to myosin4-6, titin7 and actin8, cMyBP-C contributes to the structural integrity of the sarcomere and regulates cardiac contractility in response to adrenergic stimulation9,10. Because individuals who have heritable cardiomyopathies with cMyBP-C defects have a disorganized sarcomeric structure and late-onset symptoms, MYBPC3 has emerged as a candidate gene for increased risk of heart failure through either hypertrophic or dilated cardiomyopathies (HCM or DCM)11,12.
To identify variant(s) in MYBPC3 associated with increased risk of heart failure, we screened DNA samples from individuals with cardiomyopathies in India. Initially, we observed a 25-bp deletion in intron 32 of individuals with HCM13, but concluded that its relation to disease was not unequivocal. To assess the significance of the 25-bp deletion, we have now done a large-scale case-control study on 800 cases and 699 controls, in two groups (groups 1 and 2).
Group 1 comprised of 354 individuals with cardiomyopathies (including 33 postmortem cases; Supplementary Table 1 online) and 238 healthy controls matched for ancestry, age, sex and geography. Of the 354 cases, 49 (13.8%) carried the 25-bp deletion, 46 as heterozygotes and three as homozygotes. Two out of the three homozygotes died as children younger than 3 years old owing to cardiomyopathies (Fig. 1a-d). These data suggest that homozygous deletion might be associated with a severe and sometimes early-onset form of the disease, although homozygous deletions were also found in normal individuals (see below). In contrast, only 7 (2.9%) of the 238 controls carried the deletion, all as heterozygotes. The allele carrying the 25-bp deletion was in Hardy-Weinberg equilibrium for both cases (P = 0.56) and controls (P = 0.64). However, Fisher’s exact test showed that the overrepresentation of the allele carrying the 25-bp deletion in cardiomyopathy cases was statistically significant, with a P value of 0.000002. The OR of a carrier of the 25-bp deletion (heterozygous or homozygous) showing cardiomyopathy was 5.30 (95% CI = 2.26–13.06, P = 0.000002; Table 1). To replicate these results, we screened for the 25-bp deletion in group 2, comprising 446 cases and 466 controls from six independent cohorts (Table 2). The 25-bp deletion was significantly more common in cases than controls in both groups with a combined study OR of 6.99 (95% CI = 3.68–13.57; P = 4 × 10−11).
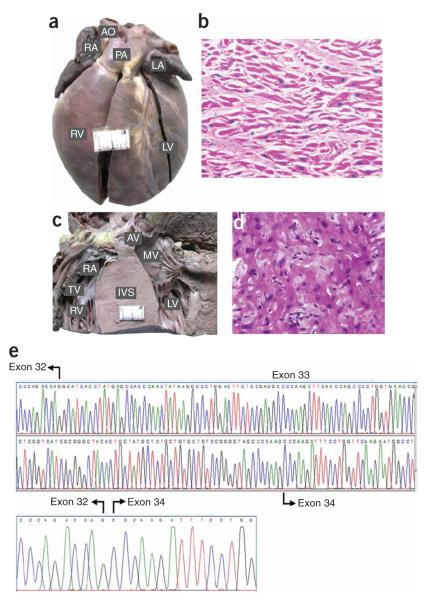
Figure 1.
Morphology and histopathology of heart, and cardiac transcript analysis of individuals with the 25-bp deletion. (a) Morphology of the heart of a subject with DCM. The subject was a child homozygous for the deletion. Note the marked dilation of the left and right ventricles. (b) Histopathological section of the same subject showing hypertrophied myofibers separated from each other by increased connective tissue (hematoxylin and eosin, × 400). (c) Transverse section through both ventricles of a 40-y-old man homozygous for the deletion showing marked asymmetric hypertrophy involving septum (IVS). (d) Histopathological section of the septal myocardium of the same individual showing ‘swirling’ of hypertrophied myofibers amid connective tissue disarray (hematoxylin and eosin, × 400). (e) Characterization of cardiac transcripts. The mRNA from the endomyocardial biopsies of an affected subject heterozygous for the 25-bp deletion was reverse transcribed, amplified, subcloned and sequenced. The sequence shows a normal transcript and a mutant transcript with absence of exon 33. AO, aorta; RA, right atrium; PA, pulmonary artery; RV, right ventricle; LV, left ventricle; TV, tricuspid valve; IVS, intraventricular septum; MV, mitral valve; AV, aortic valve.
Table 1. Frequency of the 25 bp deletion in Indian cardiomyopathy cases and controls.
| Genotype | Cardiomyopathy cases (n = 354)a (%) |
Controls (n = 238) (%) |
Odds ratio (OR) (95% CI)b |
P c |
|---|---|---|---|---|
| D, D | 3 (0.84) | 0 (0.00) | ||
| D, W | 46 (13) | 7 (2.9) | ||
| W, W | 305 (86.1) | 231 (97) | ||
| D, W + D, D | 49 (13.84) | 7 (2.9) | 5.30 (2.26 to 13) | 0.000002 |
D, allele with deletion; W, wild-type allele. Numbers in parentheses indicate the proportion of each genotype as a percentage of total cases or controls.
Total number including 321 cardiomyopathy cases and 33 postmortem cases.
Odds ratio for the likelihood of cardiomyopathy in D carriers (D, W + D, D) versus noncarriers (W, W).
P value for the comparison of carriers (D, W + D, D) and noncarriers (W, W) in cases and controls (Fisher’s exact test.)
Table 2. Replicate association studies in six independent cohorts.
| Cases (%) | Controls (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||
| Cohorts | Locations | n | D, D | D, W | W, W | n | D, D | D, W | W, W | OR (95% CI) | P |
| 1 | North India-2 and Central India |
53 | 0 | 6 (11.3) | 47 (88.68) | 58 | 0 | 1 (1.7) | 57 (98.28) | 7.28 (0.82–166.18) | 0.044 |
| 2 | West India | 46 | 1 (2.17) | 4 (8.6) | 41 (89.1) | 64 | 0 | 1 (1.5) | 63 (98.4) | 7.68 (0.82–180.31) | 0.045 |
| 3 | Southeast India | 163 | 0 | 15 (9.2) | 148 (90.7) | 154 | 0 | 2 (1.3) | 152 (98.7) | 7.70 (1.65–49.66) | 0.001 |
| 4 | South India-2 | 76 | 1 (1.3) | 3 (3.9) | 72 (94.7) | 70 | 0 | 0 | 70 | — | |
| 5 | South India-3 | 38 | 0 | 1 (2.6) | 37 (97.3) | 40 | 0 | 0 | 40 | — | |
| 6 | Southwest India | 70 | 1 (1.4) | 6 (8.5) | 63 (90) | 79 | 0 | 1 (1.2) | 78 (98.7) | 8.67 (1.03–192.44) | 0.021 |
| Overall | 446 | 3 (0.6) | 35 (7.8) | 408 (91.4) | 466 | 0 | 5 (1) | 461 (98.9) | 8.59 (3.19–25.05) | 0.00000003 | |
| Combined association studies | |||||||||||
| Cases | Controls | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| n | D, D | D, W | W, W | n | D, D | D, W | W, W | OR (95% CI) | P | |
| Overall India (n = 1,499) | 800 | 6 (0.7) | 81 (10.1) | 713 (89.1) | 699 | 0 | 12 (1.7) | 687 (98.3) | 6.99 (3.68–13.57) | 4 × 10−14 |
D, allele with deletion; W, wild-type allele. Numbers in parentheses indicate the proportion of each genotype as a percentage of total cases or controls. OR, odds ratio for the likelihood of cardiomyopathy in D carriers (D, W + D, D) versus noncarriers (W, W).
P value for the comparison of carriers (D, W + D, D) and noncarriers (W, W) in cases and controls (Fisher’s exact test).
A potential concern with these results is that they may reflect population stratification: systematic differences in allele frequency between cases and controls due to difference in their ancestry rather than to any true disease association. In India, such stratification is most likely to take the form of genetic proximity to Europeans, due to a history of migrations between India and ‘West Eurasia’, a term that we use to refer to Central Asia, the Middle East and Europe14. To maximize our chances of detecting such stratification, we used a panel of 50 ancestry-informative markers for inferring ancestry on a West Eurasian–related axis and genotyped 456 cases and 338 control samples (Tables 1 and 2) from the South, Central and North Indian subpopulations (Supplementary Methods and Supplementary Table 2 online). We then carried out a principal-components analysis (Supplementary Fig. 2 online) together with CEU samples from HapMap15 (Supplementary Methods). For each of the South, Central and North groups, no significant difference in ancestry along the West Eurasian–related axis was observed between cases and controls (each ANOVA P > 0.05; Supplementary Fig. 3 online). Thus, population stratification along the West Eurasian–related axis can be ruled out as the cause of the disease association.
To investigate genotype–phenotype correlations, we more carefully assessed 28 unrelated families containing a total of 120 members. Although many young and middle-aged individuals carrying the deletion were asymptomatic or had only mild hypertrophy, the majority (~90%) of the oldest members of each family were symptomatic. In most carriers the effects remained dormant until the third decade and then manifested themselves as mild hypertrophy (Supplementary Fig. 4a,b and Supplementary Table 3 online). However, seven probands with the deletion showed severe symptoms of cardiomyopathies (mean age 20), illustrating the variability of the phenotype. Notably, in the middle-aged men >40, when symptoms develop the phenotype is not mild; at least 20 of the probands who carried the deletion had family members who had died of sudden cardiac death (SCD). In three cases, hypertension coexisted with the deletion in the family members and these individuals showed severe phenotypes. Also, the observation of recurrent ventricular tachyarrhythmias in one homozygous subject, a 40-y-old man with dilated cardiomyopathy, is of interest, as mice with homozygous mutant Mybpc3 and DCM are vulnerable to ventricular tachyarrhythmias. Thus, a homozygous deletion might be a pathogenic substrate for ventricular tachyarrhythmias and subsequently SCD risk16.
Variability in the onset of the disease and prognosis have been recorded in several longitudinal studies of the progression of cardiac phenotypes associated with MYBPC3 mutations in different world populations, but MYBPC3 mutations have usually been associated with late-onset, mild hypertrophy, incomplete penetrance and a better prognosis17,18. This is also shown by our study of 28 families (Supplementary Fig. 4b) and thus suggests that the deletion results in genetic predisposition for heart failure and lifelong threat to carriers, with risk increased further by secondary factors including compound heterozygosity, ventricular arrthymia, hypertension, age and other environmental factors. (Supplementary Fig. 4c).
To further understand the risk induced by the deletion, we analyzed RNA and protein from the endomyocardial biopsy specimens of two young index subjects, who were 25 and 32 y old. Cardiac MyBP-C cDNA of two heterozygous individuals revealed two transcript structures: a normal transcript and a mutated allele with skipping of the associated exon (Fig. 1e and Supplementary Fig. 1). However, the altered protein could not be detected in tissue samples obtained from affected individuals (Supplementary Fig. 5 online), as in three previous reports of MYBPC3 mutations19-21.
To determine whether the altered protein could distort the normal architecture of the sarcomere, we expressed the myc-tagged altered protein along with wild type (WT) protein in neonatal rat cardiomyocytes. Staining with antibodies to the myc tag showed a highly disorganized and diffused pattern of sarcomeric architecture as a result of aberrant incorporation of altered proteins (Fig. 2). The lack of sarcomeric organization in mutant individuals is one mechanism by which pathogenesis is triggered in the heart4,22. It also promotes the degradation of truncated proteins by the ubiquitin-proteasome system (UPS), perhaps competitively inhibiting breakdown of other UPS substrates and leading to an impairment of UPS activity23. Also, considering the decline in function of the UPS with age and oxidative stress24,25, which ultimately may result in failure to eliminate the altered protein, it seems likely that the accumulation of the altered protein might disrupt the cellular homeostasis and initiate late-onset cardiomyopathy. This might explain why most individuals with the 25-bp deletion remain healthy until the third decade of their life26. This illustrates the need for long-term clinical follow-up for deletion carriers, as the deletion poses a lifelong risk. Although we cannot exclude the possibility that the deletion is in linkage disequilibrium with unknown variants at the MYBPC3 locus that cause or contribute to the disease, possibly by modifying the risk associated with the deletion itself, the data presented above support a role for the deletion itself in disease pathogenesis.
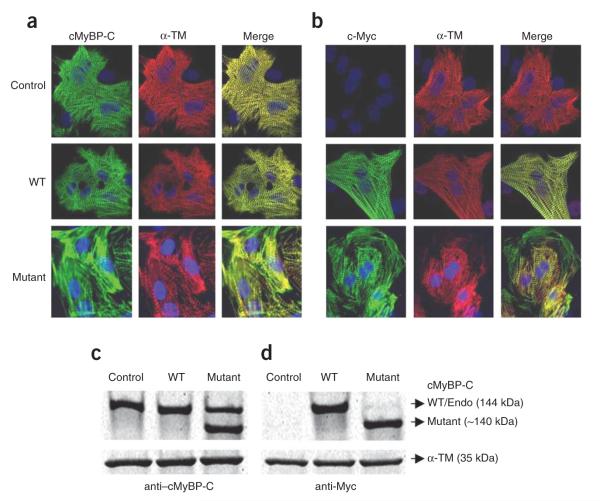
Figure 2.
Expression and localization of cMyBP-CWT and cMyBP-CΔEx33 proteins in neonatal rat cardiomyocytes. Recombinant adenoviruses that expressed mouse cMyBP-CWT (WT) or cMyBP-CΔEx33 (mutant) were introduced into rat neonatal cardiomyocytes. Noninfected cardiomyocytes were used as control. (a,b) Three days after infection, cardiomyocytes were fixed and immunostained with either cMyBP-C antibodies (green) (a) raised against the C0-C1 domain carried by native, WT and mutant proteins or myc-tag antibodies carried only by the WT and mutant proteins (green) (Roche) (b) and costained with α-tropomyosin (red) (Sigma). (c,d) Representative protein blots obtained by using cMyBP-C (c) and Myc (d) antibodies in control and WT- and mutant-infected neonatal rat cardiomyocytes. Twenty μg of total cardiomyocytes lysates was separated on a 4–15% linear gradient SDS-PAGE, transferred to nitrocellulose membrane, and immunoblotted as described in Supplementary Methods. In lysates from noninfected neonatal cardiomyocytes and WT-infected cells, the cMyBP-C antibody detected a major band of the expected molecular weight of 144 kDa and overexpression of mutant cMyBP-C showed the predicated truncated protein of approximately 140 kDa. α-TM was used as a load control.
As the epidemiology of cardiomyopathies in India may be influenced by the frequency of the allele carrying the 25-bp deletion in different populations that are genetically isolated, we also screened 6,273 individuals belonging to 107 ethnic populations across 35 states representing all of India for the presence of this deletion. The deletion was found in all major Indian populations at frequencies ranging from 2% to 8% (Supplementary Fig. 6 online). Of the 6,273 individuals, 287 possessed the 25-bp deletion (4.6%), and 8 were homozygous, although their heart failure and cardiomyopathy status is unknown. The mutation was absent from Northeast Indians (who show affinities with East Asians), Siddis (recent migrants from Africa) and Onges (Andaman Islands)27. The results also show that the presence of the deletion is not influenced by climatic or geographical features (Supplementary Table 4 online). The frequency of the deletion is significantly higher in the southern and western states compared with the northern states (P < 4 × 10−8, Fisher 2-by-5 test). This frequency difference might contribute to the wide disparity in cardiac mortality at old age between Indian states: heart failure is more prevalent in the south than in the north and northeast (386–422 cardiac deaths per 100,000 persons compared to 76–99 per 100,000 persons, respectively28). Overall, the population attributable risk of the deletion is ~4.5%.
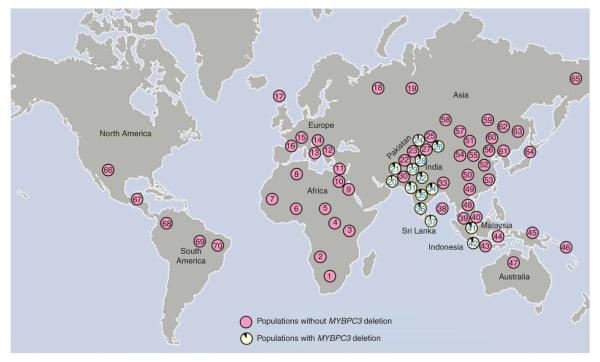
The presence of this deletion in many Indian populations with varied geographical and ancestral backgrounds raises the question of how geographically widespread it is outside India. We therefore also analyzed 63 world population samples, comprising 2,085 indigenous individuals from 26 countries including all five continents. The 25-bp deletion was observed in Pakistan, Sri Lanka, Indonesia and Malaysia, (all heterozygotes) but was absent from other samples. Thus, the deletion is a common variant in individuals from South Asia, present in Southeast Asia, but undetectable elsewhere (Fig. 3 and Supplementary Table 5 online).
Figure 3.
Global distribution of MYBPC3 deletion in indigenous populations. For populations with the MYBPC3 deletion, the frequency of the deletion-containing allele is shown. See Supplementary Table 5 for details of the populations studied.
The presence of a disease-associated variant at substantial frequency raises an evolutionary question: if it is disadvantageous, how did it become so common? In principle, it could be evolutionarily neutral, manifesting its disadvantages only late in life; alternatively, its disadvantages could be outweighed by advantages early in life, or in a different environment, so that it could have been positively selected. To address this question, we examined the haplotype structure surrounding the deletion. Using five short tandem repeat (STR) markers, spanning ~3.4 Mb surrounding the deletion in 287 heterozygous individuals, we found similar high degrees of variation in the inferred haplotypes from chromosomes with and without the deletion (Supplementary Fig. 7 and Supplementary Table 6 online). We then used allele-specific amplification to resequence ~10-kb haplotypes centered on the 25-bp deletion from nine heterozygous individuals (Supplementary Tables 7 and 8 online). The chromosomes carrying the 25-bp deletion showed five closely related haplotypes (Supplementary Fig. 8 online). After excluding variants likely to have arisen by recombination, we estimated a time to most recent common ancestry (TMRCA) of ~33 ± 23 thousand years for the deletion haplotypes (Supplementary Methods). This time slightly postdates the initial peopling of the subcontinent 30,000–50,000 years ago and together with its restricted geographical distribution suggests that the deletion did not arrive with the first modern human settlers from Africa ≥50,000 years ago, but arose subsequently within the subcontinent. Its occurrence in two populations from Southeast Asia can be explained by recent gene flow from India (Supplementary Note online). Collectively, these observations provide no evidence for rapid spread of a recent founder haplotype or any departure from neutral evolution.
We conclude that the 25-bp deletion, a common MYBPC3 variant in South Asians, is associated with chronic risk of heart failure. The delayed symptoms, mild hypertrophy and influence of secondary risk factors pose a lifelong threat to carriers. Notably, such gene-based insights into pathophysiology may allow more subtle clinical manifestations to be identified and additional phenotypes associated with the variant to be investigated. Furthermore, genotyping could be used for the identification of persons at risk of heart failure among South Asians and could be accompanied by advice for a lower-risk lifestyle.
METHODS
Ethical approval
Samples were collected from cases and controls with their informed consent. Ethical committees of all institutions involved gave approval to conduct this study. A full description of methods is provided in the Supplementary Methods and Supplementary Tables 6-9 online.
Supplementary Material
ACKNOWLEDGMENTS
We thank all participants for making this study possible. We acknowledge the help of G.S. Selvam, C. Rajamanickam, R. Srinivasan, V. Madhavan, S. Madhavan, A.G. Reddy, A. Vanniarajan, P. Govindaraj, B.D. Gelb and Q. Long. P.S.D. was supported by Council of Scientific and Industrial Research – Senior Research Fellowship (CSIR-SRF) and P.N. by Department of Biotechnology (DBT). K.T. was supported by Council of Scientific and Industrial Research (CSIR) - Raman Research Fellowship and Y.X., G.T.P., Q.A., S.Q.M. and C.T.-S. were supported by The Wellcome Trust. T.S.R. received an Senior Research Fellowship (SRF) from Indian Council of Medical Research (ICMR), M.K. and A.B. were supported by Post Graduate Institute of Medical Education and Research (PGIMER), Chandigarh. S.S. received a post-doctoral training grant from the American Heart Association, Ohio Valley Affiliate, USA. D.R. is supported by a Burroughs Wellcome Career Development Award in the Biomedical Sciences.
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
References
- 1.Reddy KS, Shah B, Varghese C, Ramadoss A. Responding to the threat of chronic diseases in India. Lancet. 2005;366:1744–1749. doi: 10.1016/S0140-6736(05)67343-6. [DOI] [PubMed] [Google Scholar]
- 2.Joshi P, et al. Risk factors for early myocardial infarction in South Asians compared with individuals in other countries. J. Am. Med. Assoc. 2007;297:286–294. doi: 10.1001/jama.297.3.286. [DOI] [PubMed] [Google Scholar]
- 3.Liew CC, Dzau VJ. Molecular genetics and genomics of heart failure. Nat. Rev. Genet. 2004;5:811–825. doi: 10.1038/nrg1470. [DOI] [PubMed] [Google Scholar]
- 4.Flavigny J, et al. Biomolecular interactions between human recombinant h-MyHC and cMyBP-Cs implicated in familial hypertrophic cardiomyopathy. Cardiovasc. Res. 2003;60:388–396. doi: 10.1016/j.cardiores.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Flavigny J, et al. COOH-terminal truncated cardiac myosin-binding protein C mutants resulting from familial hypertrophic cardiomyopathy mutations exhibit altered expression and/or incorporation in fetal rat cardiomyocytes. J. Mol. Biol. 1999;294:443–456. doi: 10.1006/jmbi.1999.3276. [DOI] [PubMed] [Google Scholar]
- 6.Gruen M, Gautel M. Mutations in beta-myosin S2 that cause familial hypertrophic cardiomyopathy (FHC) abolish the interaction with the regulatory domain of myosin binding protein-C. J. Mol. Biol. 1999;286:933–949. doi: 10.1006/jmbi.1998.2522. [DOI] [PubMed] [Google Scholar]
- 7.Freiburg A, Gautel M. A molecular map of the interactions between titin and myosin-binding protein C. Implications for sarcomeric assembly in familial hypertrophic cardiomyopathy. Eur. J. Biochem. 1996;235:317–326. doi: 10.1111/j.1432-1033.1996.00317.x. [DOI] [PubMed] [Google Scholar]
- 8.Squire JM, Luther PK, Knupp C. Structural evidence for the interaction of C-protein (MyBP-C) with actin and sequence identification of a possible actin-binding domain. J. Mol. Biol. 2003;331:713–724. doi: 10.1016/s0022-2836(03)00781-2. [DOI] [PubMed] [Google Scholar]
- 9.McClellan G, Kulikovskaya I, Winegrad S. Changes in cardiac contractility related to calcium-mediated changes in phosphorylation of myosin-binding protein C. Biophys. J. 2001;81:1083–1092. doi: 10.1016/S0006-3495(01)75765-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadayappan S, et al. Cardiac myosin binding protein c phosphorylation is cardio-protective. Proc. Natl. Acad. Sci. USA. 2006;103:16918–16923. doi: 10.1073/pnas.0607069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niimura H, et al. Sarcomere protein gene mutations in hypertrophic cardiomyopathy of the elderly. Circulation. 2002;105:446–451. doi: 10.1161/hc0402.102990. [DOI] [PubMed] [Google Scholar]
- 12.Van Driest SL, et al. Sarcomeric genotyping in hypertrophic cardiomyopathy. Mayo Clin. Proc. 2005;80:463. doi: 10.1016/S0025-6196(11)63196-0. [DOI] [PubMed] [Google Scholar]
- 13.Waldmuller S, et al. Novel deletions in MYH7 and MYBPC3 identified in Indian families with familial hypertrophic cardiomyopathy. J. Mol. Cell. Cardiol. 2003;35:623–636. doi: 10.1016/s0022-2828(03)00050-6. [DOI] [PubMed] [Google Scholar]
- 14.Thanseem I, et al. Genetic affinities among the lower castes and tribal groups of India: inference from Y chromosome and mitochondrial DNA. BMC Genet. 2006;7:42. doi: 10.1186/1471-2156-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berul CI, et al. Ventricular arrhythmia vulnerability in cardiomyopathic mice with homozygous mutant myosin-binding protein C gene. Circulation. 2001;104:2734–2739. doi: 10.1161/hc4701.099582. [DOI] [PubMed] [Google Scholar]
- 17.Niimura H, et al. Mutations in the gene for cardiac myosin-binding protein C and late-onset familial hypertrophic cardiomyopathy. N. Engl. J. Med. 1998;338:1248–1257. doi: 10.1056/NEJM199804303381802. [DOI] [PubMed] [Google Scholar]
- 18.Kubo T, et al. Lifelong left ventricular remodeling of hypertrophic cardiomyopathy caused by a founder frameshift deletion mutation in the cardiac myosin-binding protein C gene among Japanese. J. Am. Coll. Cardiol. 2005;46:1737–1743. doi: 10.1016/j.jacc.2005.05.087. [DOI] [PubMed] [Google Scholar]
- 19.Rottbauer W, et al. Novel splice donor site mutation in the cardiac myosin-binding protein-C gene in familial hypertrophic cardiomyopathy. Characterization of cardiac transcript and protein. J. Clin. Invest. 1997;100:475–482. doi: 10.1172/JCI119555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moolman JA, et al. A newly created splice donor site in exon 25 of the MyBP-C gene is responsible for inherited hypertrophic cardiomyopathy with incomplete disease penetrance. Circulation. 2000;101:1396–1402. doi: 10.1161/01.cir.101.12.1396. [DOI] [PubMed] [Google Scholar]
- 21.Vignier N, et al. Cardiac myosin-binding protein C and familial hypertrophic cardiomyopathy: from mutations identification to human endomyocardial proteins analysis. Circulation. 2001;104(Suppl.):II–1. [Google Scholar]
- 22.Yang Q, Osinska H, Klevitsky R, Robbins J. Phenotypic deficits in mice expressing a MyBP-C lacking the titin and myosin binding domains. J. Mol. Cell. Cardiol. 2001;33:1649–1658. doi: 10.1006/jmcc.2001.1417. [DOI] [PubMed] [Google Scholar]
- 23.Sarikas A, et al. Impairment of the ubiquitin-proteasome system by truncated cardiac myosin binding protein C mutants. Cardiovasc. Res. 2005;66:33–44. doi: 10.1016/j.cardiores.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Okada K, et al. 4-Hydroxy-2-nonenal-mediated impairment of intracellular proteolysis during oxidative stress. Identification of proteasomes as target molecules. J. Biol. Chem. 1999;274:23787–23793. doi: 10.1074/jbc.274.34.23787. [DOI] [PubMed] [Google Scholar]
- 25.Bulteau AL, Szweda LI, Friguet B. Age-dependent declines in proteasome activity in the heart. Arch. Biochem. Biophys. 2002;397:298–304. doi: 10.1006/abbi.2001.2663. [DOI] [PubMed] [Google Scholar]
- 26.Sato N, et al. A novel variant of cardiac myosin-binding protein-C that is unable to assemble into sarcomeres is expressed in the aged mouse atrium. Mol. Biol. Cell. 2003;14:3180–3191. doi: 10.1091/mbc.E02-10-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thangaraj K, et al. Reconstructing the origin of Andaman Islanders. Science. 2005;308:996. doi: 10.1126/science.1109987. [DOI] [PubMed] [Google Scholar]
- 28.Gupta R, Misra A, Pais P, Rastogi P, Gupta VP. Correlation of regional cardiovascular disease mortality in India with lifestyle and nutritional factors. Int. J. Cardiol. 2006;108:291–300. doi: 10.1016/j.ijcard.2005.05.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.