Abstract
Sulfation is important in the metabolism and inactivation of steroidal compounds and hormone replacement therapeutic (HRT) agents in human tissues. Although generally inactive, many steroid sulfates are hydrolyzed to their active forms by sulfatase activity. Therefore, the specific sulfotransferase (SULT) isoforms and the levels of steroid sulfatase (STS) activity in tissues are important in regulating the activity of steroidal and HRT compounds. Tibolone (Tib) is metabolized to three active metabolites and all four compounds are readily sulfated. Tib and the Δ4-isomer are sulfated at the 17β-OH group by SULT2A1 and the 17-sulfates are resistant to hydrolysis by human placental STS. 3α-OH and 3β-OH Tib can form both 3- and 17-monosulfates as well as disulfates. Only the 3β-sulfates are susceptible to STS hydrolysis. Raloxifene monosulfation was catalyzed by at least seven SULT isoforms and SULT1E1 also synthesizes raloxifene disulfate. SULT1E1 forms both monosulfates in a ratio of approximately 8:1 with the more abundant monosulfate migrating on HPLC identical to the SULT2A1 synthesized monosulfate. The raloxifene monosulfate formed by both SULT isoforms is sensitive to STS hydrolysis whereas the low abundance monosulfate formed by SULT1E1 is resistant. The benzothiophene sulfates of raloxifene and arzoxifene were hydrolyzed by STS whereas the raloxifene 4′-phenolic sulfate was resistant. These results indicate that tissue specific expression of SULT isoforms and STS could be important in the inactivation and regeneration of the active forms of HRT agents.
Keywords: sulfation, sulfotransferase, tibolone, raloxifene, sulfatase, arzoxifene, sulfate hydrolysis
INTRODUCTION
Tissue-specific modulation of steroid activity is critical in regulating the effects of therapeutic compounds used to alleviate the climacteric symptoms of menopause as well as osteoporosis. Tibolone (Tib) has been used for many years in hormone replacement therapy (HRT), effectively preventing bone loss while relieving climacteric symptoms of menopause without estrogenic stimulation of either the endometrium or breast (1–4). Raloxifene has been used more specifically in the treatment of osteoporosis and, similarly to Tib, antagonizes the effects of estrogens in the breast and endometrium (5). To understand the specific mechanisms of action of these compounds, it is important to analyze the pathways involved in their activation and inactivation. Conjugation of estrogens, therapeutic estrogenic compounds as well as HRT agents with a sulfonate group is important in the regulation of their activity and disposition. Sulfation is essentially an inactivation reaction, inhibiting the biological activity of steroids by preventing binding to specific hormone receptors (6–8). In humans, Tib is rapidly metabolized into its main active metabolites, 3α-OH Tib and 3β-OH Tib, as well as a Δ4-isomer (1, 9). Sulfation is the major Phase II conjugation pathway involved in the metabolism and inactivation of Tib and its metabolites and is also involved in the metabolism of raloxifene in human tissues (10–12). In conjunction with sulfation, desulfation of Tib and raloxifene sulfates by sulfatase (STS) activity may have a role in regulation of in situ activity via regeneration of the active compounds (13). Therefore, the interactions between sulfation and desulfation in a particular tissue may be critical in regulation of the availability and activity of these therapeutic compounds, especially if the different sulfated forms of Tib and raloxifene are differentially sensitive to STS activity. The combination of sulfation and desulfation could result in either an increased or decreased availability of biologically active forms of these drugs in different tissues.
In human tissues, several members of the cytosolic sulfotransferase (SULT) family are responsible for the sulfation of estrogens, therapeutic estrogenic compounds and HRT agents (14–16). As reported previously, SULT1E1 and SULT 2A1 are the major isoforms involved in the sulfation of Tib and its metabolites (10). Raloxifene is sulfated in vitro by at least seven expressed human SULT isoforms including SULTs 1E1 and 2A1 (12, 17). SULT1E1 is responsible for the high affinity sulfation of estrogens (14, 18, 19) while SULT2A1 sulfates hydroxysteroids, bile acids and several therapeutic drugs (10, 15, 20, 21). SULT1E1 and SULT2A1 are also of interest since SULT1E1 can form raloxifene disulfate and SULT2A1 can generate the 3-OH Tib disulfates (10, 12). SULT1E1 is expressed in hormonally responsive tissues including breast, endometrium and prostate as well as the liver and GI tract (8, 14, 22–24). SULT2A1 is present in relatively large amounts in the liver and adrenals but is not detectable in the breast, endometrium and prostate (15, 24, 25).
In contrast to sulfation, desulfation hydrolyzes sulfated inactive compounds to their unconjugated active forms. The enzyme responsible for the majority of steroid desulfation in human tissues is human STS or aryl sulfatase C, and is known to hydrolyze both 17β-estradiol (E2) and dehydroepiandrosterone (DHEA) sulfates to generate biologically active hormones from their sulfated precursors (26). STS is a membrane-bound, hydrophobic enzyme that is difficult to solubilize, and thus attempts to clone and express the full-length active protein have been relatively unsuccessful (26, 27). Therefore, the potential contribution of STS in the metabolism of sulfated therapeutic drugs to render them biologically active has been inadequately examined. In this report purified human placental STS was utilized to characterize the desulfation of sulfated Tib and its metabolites as well as the different raloxifene sulfates. Both the Tib 3-OH metabolites and raloxifene possess two hydroxyl groups and can form two monosulfates as well as disulfates (10, 12). The ability of purified placental STS to desulfate the different raloxifene and Tib monosulfates and disulfates was analyzed to understand the possible role of desulfation in the regulation of the tissue specific activity of these therapeutic reagents. Preliminary studies of the sulfation/desulfation of arzoxifene, a recently described SERM related to raloxifene, were also carried out to clarify the results of the raloxifene studies.
MATERIALS AND METHODS
Materials
Tib, 3β-OH Tib, 3α-OH Tib, Δ4-Tib and arzoxifene were provided by N.V. Organon (the Netherlands). Raloxifene hydrochloride was purchased from Sigma Chemical Co. (St. Louis, MO). 3′-Phosphoadenosine 5′-phosphosulfate (PAPS) was purchased from Dr. Sanford Singer (University of Dayton, Dayton, OH). [35S]-PAPS (2.2 Ci/mmol) and [1,2,6,7-3H(N)] DHEA-sulfate (60 Ci/mmol) were purchased from New England Nuclear (Boston, MA). LK6DF 60 Å silica gel thin layer chromatography (TLC) plates with a layer thickness of 250 μm were obtained from Whatman Inc. (Clifton, NJ). Con-A Sepharose was purchased from GE Healthcare (Pistcataway, NJ). All other chemicals used were of reagent grade from Fisher Scientific (Norcross, GA).
Methods
Preparation of human SULT1E1, SULT2A1 and STS
SULT1E1 and SULT2A1 were expressed in E. coli using the pKK233-2 vector to generate the native form of the enzymes and purified by DEAE-Sepharose chromatography to obtain a preparation suitable for enzymatic characterization as described previously (14, 28, 29).
For use in activity assays, STS was purified from human full-term placenta based upon the method of Hernandez-Guzman et al. (26). Fresh placental tissue was obtained from the Tissue Procurement Service of the UAB Comprehensive Cancer Center with Institutional Review Board approval. The placental tissue was homogenized in 67 mM phosphate buffer, pH 7.4 containing 0.5 mM DTT and 0.24 M sucrose in a volume of approximately 1.5 liters and stored at −80° C. STS activity was determined using reactions (125 μl) containing 20 μM [3H]-DHEA-sulfate as substrate in 0.1 M Tris-maleate buffer, pH 7.4, and 25 μl enzyme. Reactions were stopped by the addition of 200 μl 0.25 M Tris-HCl, pH 8.7 to alkalinize the reaction and extracted with 800 μl ethyl acetate. Radioactivity was determined in 500 μl of the ethyl acetate phase to quantify desulfated [3H]-DHEA. For STS purification, 100 ml of placental homogenate was thawed on ice and diluted with 200 ml of buffer containing 10 mM potassium phosphate, pH 7.4, 20% glycerol, 0.5 μM androstenedione and 0.1 mM EDTA, then centrifuged at 105,000 × g for 60 min. The pellet was homogenized in the same buffer, adjusted to 0.3% Na cholate and 0.3% Emulgen 911, and stirred at 4° C for 30 min then centrifuged at 105,000 × g for 1 hr. The supernatant fraction was diluted with an equal volume of 10 mM potassium phosphate, pH 7.4, 20% glycerol, 0.5 μM androstenedione and 0.1 mM EDTA then loaded on a 60 ml DE52 column equilibrated in the same buffer. The column was washed with buffer containing 0.15% Emulgen 911 then with 20 mM Tris-HCl, pH 7.4 containing 0.1% Triton X-100. STS activity was eluted with 0.15 M NaCl in the same buffer in 3 ml fractions. Fractions containing STS activity were pooled and adjusted to contain 5 mM CaCl2, 5 mM MgCl2, 5 mM MnCl2 and 1 M NaCl. The pool was stirred for 30 min at 4° C then centrifuged at 105,000 × g for 30 min. STS was further purified from the supernatant fraction on a 5 ml Con-A Sepharose column equilibrated in 20 mM Tris-HCl, pH 7.4, containing 0.1% Triton X-100, 5 mM CaCl2, 5 mM MgCl2, 5 mM MnCl2 and 1 M NaCl. Subsequent to the wash step, STS was eluted with 10% α-p-mannopyranoside and fractions containing STS activity were pooled. The α-p-mannopyranoside was removed by repeated concentration and dilution using a Centricon 30,000 MWCO membrane. The purified STS was aliquotted and stored at −80° C.
Sulfation/sulfatase assays
Sulfation activity was determined using the appropriate substrate concentrations and [35S]-PAPS as the sulfate donor with the subsequent resolution of [35S]-sulfates by TLC as described previously (10, 12, 16). Briefly, reactions contained the appropriate substrate dissolved in ethanol (Tib compounds) or DMSO (raloxifene, arzoxifene), 50 mM Tris-HCl, pH 7.4, 10 mM MgCl2 and 25 μM [35S]-PAPS in a volume of 100 μl. Control reactions were run with no substrate but contained the appropriate volume of the appropriate ethanol or DMSO vehicle. Reactions were incubated for the appropriate time interval at 37° C then terminated by extraction with 100 μl CHCl3. Aliquots of the aqueous phase were spotted onto the loading zones of channeled silica gel TLC plates and the plates were developed in methylene chloride:MeOH:ammonium hydroxide (85:15:5 by volume) and the radiolabeled sulfated products were localized by autoradiography. The sulfated products were scraped into scintillation fluid and radioactivity was determined by scintillation spectroscopy.
For the STS assays, the aqueous phase of the appropriate SULT assay containing the sulfated reaction products was removed and duplicate 40 μl aliquots were removed to fresh tubes. The concentrations of raloxifene and tibolone as well as the SULT isoforms chosen to generate the mono- and disulfated compounds was based on the reactivity of the human SULT isoforms with these compounds as described previously (10, 12). The concentration of sulfated substrates was determined from the incorporation of the 35S-label. Human STS was added to one tube while the other tube received the STS buffer only as a control. STS reactions were incubated at 37° C for the appropriate time (15 to 60 min) and terminated by spotting the reaction on a silica gel TLC plate. The plates were developed in methylene chloride:MeOH:ammonium hydroxide (85:15:5 by volume) and the radiolabeled sulfated products were localized by autoradiography. The sulfated products were scraped into scintillation fluid and radioactivity was determined by scintillation spectroscopy (16, 21, 30). Significant differences between reactions with and without STS was determined using the Students T-test.
HPLC analysis of raloxifene sulfates
Raloxiene sulfation and STS reactions were run as described previously. Subsequent to CHCl3 extraction, the aqueous phase was analyzed by HPLC on a Luna 5 μm C18 column (250 × 4.6 mm) (Phenomenex). Reaction products were eluted with a 25 min linear gradient of 80% 5 mM ammonium acetate/20% acetonitrile to 50% 5 mM ammonium acetate/50% acetonitrile with detection at 290 nm. Fractions were collected at 1 min intervals and quantified by scintillation spectroscopy.
Mass spectroscopy
To confirm the identity of the sulfated arzoxifene reaction product, non-radioactive reactions were run in parallel with reactions containing 35S-PAPS that were monitored by TLC. Identification of arzoxifene monosulfate was carried out by HPLC mass spectroscopy using a Sciex 3000 mass spectrometer with two Perkin-Elmer series 200 micropumps and a Keystone Scientific Aquasil (100 × 2 mm ID) analytical column with a C-18 guard column. The mobile phases were (A) 5 mM ammonium acetate, and (B) acetonitrile. The gradient profile was 0 to 1 min, 80% (A); 1 to 5 min, 10% (A) (linear); 5 to 7.5 min 10% (A); and 7.5 to 10 min 80% A (step). Mass spectroscopy results were analyzed with the Analyst 1.4 software.
RESULTS
STS and Tib
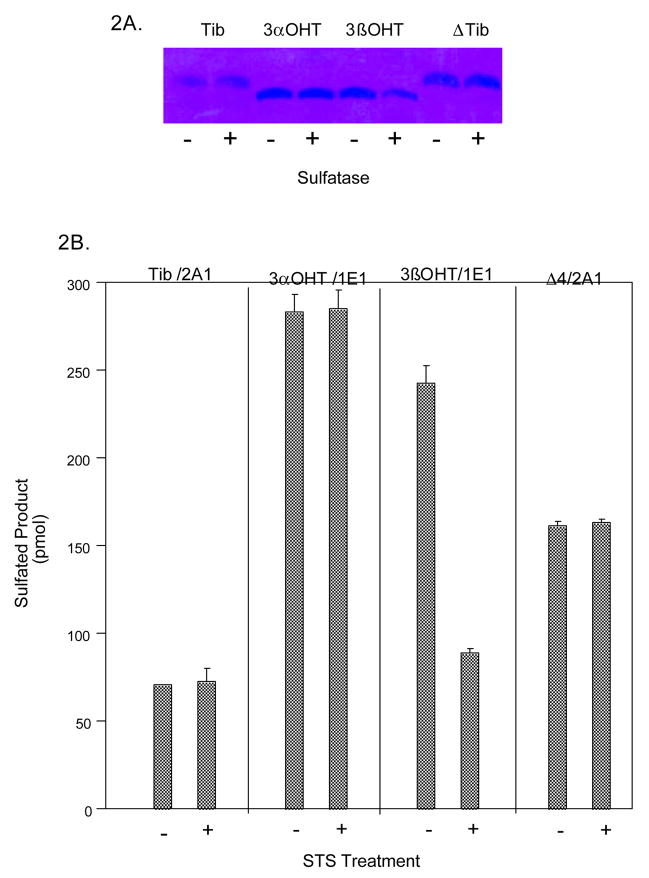
To evaluate the ability of STS to hydrolyze the sulfates of Tib and its metabolites, sulfation reactions were performed at substrate concentrations of 10 μM with SULT2A1, which sulfates all four Tib compounds or SULT1E1, which selectively conjugates the 3α-OH and 3β-OH Tib metabolites (10). Since Tib and the Δ4-isomer possess only a 17β-OH group these compounds can form only 17β-monosulfates (Figure 1)(10). Sulfated Tib and metabolites were then incubated with STS, and all STS reactions were analyzed by TLC to visualize and quantify reaction products. As shown in Figure 2A, the 17β-monosulfates of Tib and the Δ4-isomer were generated with SULT2A1 and the 3α- and 3β-Tib monosulfates synthesized with SULT1E1 (10). However, after incubation with STS, only the 3β-Tib monosulfate showed significant desulfation (Figure 2B). The 17β-monosulfates of Tib and the Δ4-isomer as well as 3α-Tib monosulfate were resistant to STS hydrolysis. Quantification of the monosulfated Tib-derived compounds by scintillation spectroscopy confirms that only 3β-Tib monosulfate was significantly hydrolyzed by STS.
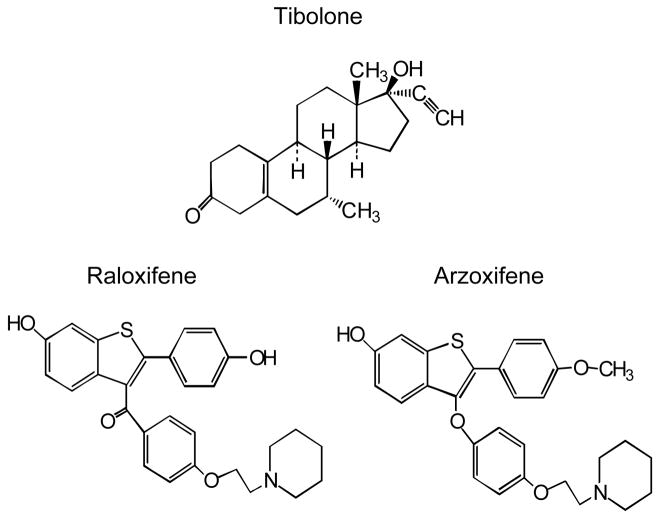
Figure 1.
Structures of tibolone, raloxifene and arzoxifene.
Figure 2.
Sulfation and desulfation of Tib and metabolites. The formation of Tib and the Δ4 monosulfates was carried using SULT2A1 and the 3α- and 3β-monosulfates were generated using SULT1E1 in the presence of 35S-PAPS. The four monosulfates were then tested for sensitivity for hydrolysis by STS. Panel A is the autoradiograph of the formation and hydrolysis of the four monosulfates. Panel B shows the selective hydrolysis of Tib 3β-monosulfate by STS. Each reaction was run in quadruplicate and the mean ± s.d. of the 35S-Tib metabolites generated or remaining after STS incubation is shown.
SULT2A1 is the only human SULT isoform reported to form disulfates with the 3-OH metabolites of Tib (10). Sulfation of both 3α-OH Tib and 3β-OH Tib by SULT2A1 at a substrate concentration of 5 μM also yields low levels of Tib disulfate in addition to two monosulfated products (10). Upon incubation with STS, the levels of 3α-Tib monosulfate and 3α-Tib disulfate are both unaltered, demonstrating a resistance to STS hydrolysis (Figure 3). In contrast, disulfated 3β-Tib was significantly hydrolyzed by STS to a monosulfate. After STS treatment, the level of the 3β-Tib monosulfate in the reaction was decreased as well. Approximately 30% of the SULT2A1 generated 3β-Tib monosulfate was hydrolyzed. The SULT2A1 generated 3β-Tib monosulfate levels were not decreased to the same extent as was the 3β-Tib monosulfate produced by SULT1E1 (Fig. 2B). This may be due to the fact that all or part of the 3β-Tib disulfate generated by SULT2A1 was desulfated to generate the STS-resistant 3β-OH Tib 17-monosulfate that contributes to the 3β-OH Tib 17-monosulfate levels produced via sulfation by SULT2A1.
Figure 3.
Hydrolysis of the 3α- and 3β-Tib mono- and disulfates by STS. The 3α- and 3β-Tib monosulfates and disulfates were synthesized using SULT1E1 and 35S-PAPS. The levels of the synthesized sulfates were quantified by resolution of an aliquot of the reaction by TLC, autoradiography and scintillation spectroscopy. Aliquots of the reactions were then extracted with chloroform and the Tib sulfates used as substrates for STS. After 60 min incubations the reactions were resolved by TLC and the Tib sulfates quantified by scintillation spectroscopy. The mean ± s.d. of four reactions is shown. An asterisk (*) indicates a significant difference between the assays with and without STS (p<0.01).
Desulfation time course
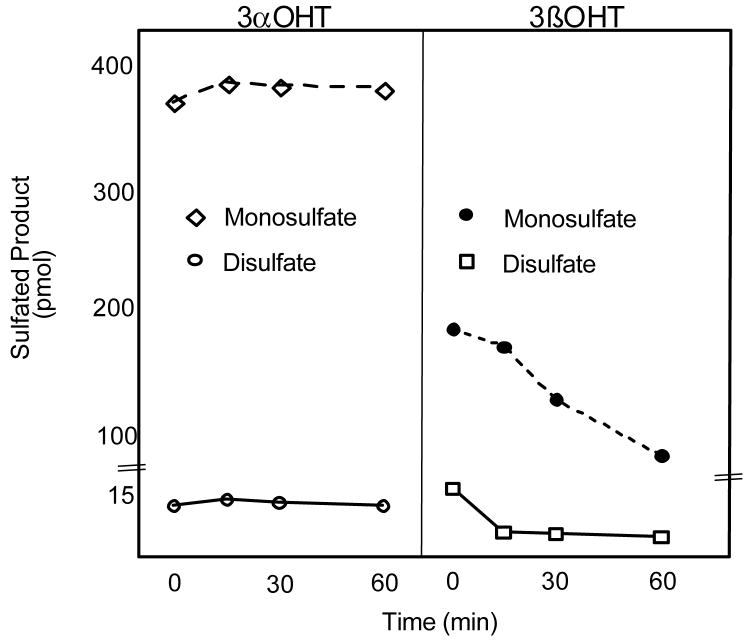
Figures 2 & 3 demonstrate that 3α-Tib monosulfates as well as 3α-Tib disulfate were unaffected by STS, whereas the levels of 3β-Tib monosulfates and disulfate were decreased. To further understand Tib desulfation, STS reactions were monitored at 0, 15, 30 and 60 min after the addition of STS. Figure 4 shows that even after 60 min incubation with STS, neither the 3α-Tib disulfate or 3β-Tib monosulfate were completely desulfated. However, after 15 min with STS the 3β-Tib disulfate was reduced to 37% of its initial level, but was decreased only slightly more after 30 and 60 min incubation. Approximately 55% of the corresponding 3β-Tib monosulfate was hydrolyzed over the course of the 60 min reaction (Figure 4). The 3β-Tib monosulfate was desulfated more slowly for the first 15 min, most likely due to the addition of monosulfate generated by desulfation of the 3β-Tib disulfate, which was more rapid in this initial 15 min interval.
Figure 4.
Time course of tibolone monosulfate and disulfate hydrolysis by STS. The tibolone monosulfates and disulfate were generated using SULT2A1 and 10 μM 3α-OH or 3β-OH tibolone and 35S-PAPS. The 3-OH tibolone sulfates were extracted with chloroform and hydrolyzed with STS as described in Methods. Panel A shows the results of the hydrolysis of the mono- and disulfates formed with 3α-OH tibolone by STS. Panel B shows the results of the hydrolysis of the 3β-OH tibolone mono- and disulfates by STS. Aliqouts were taken from the STS reactions at different times, resolved by TLC and quantified. The mean ± s.d. of four reactions is shown.
STS and raloxifene
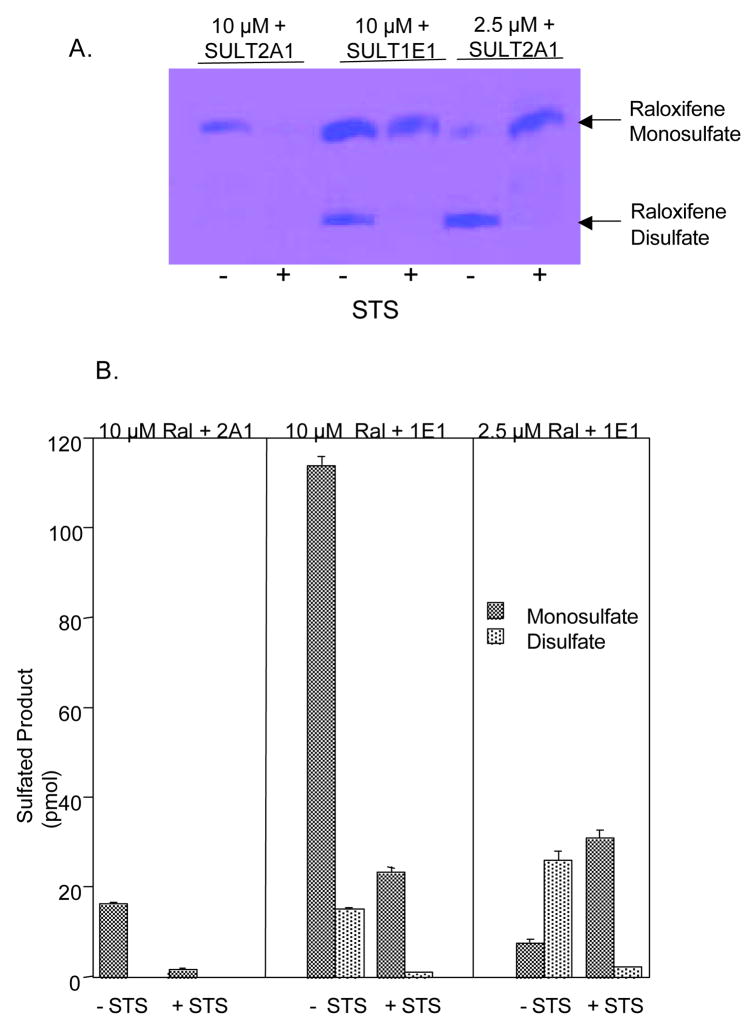
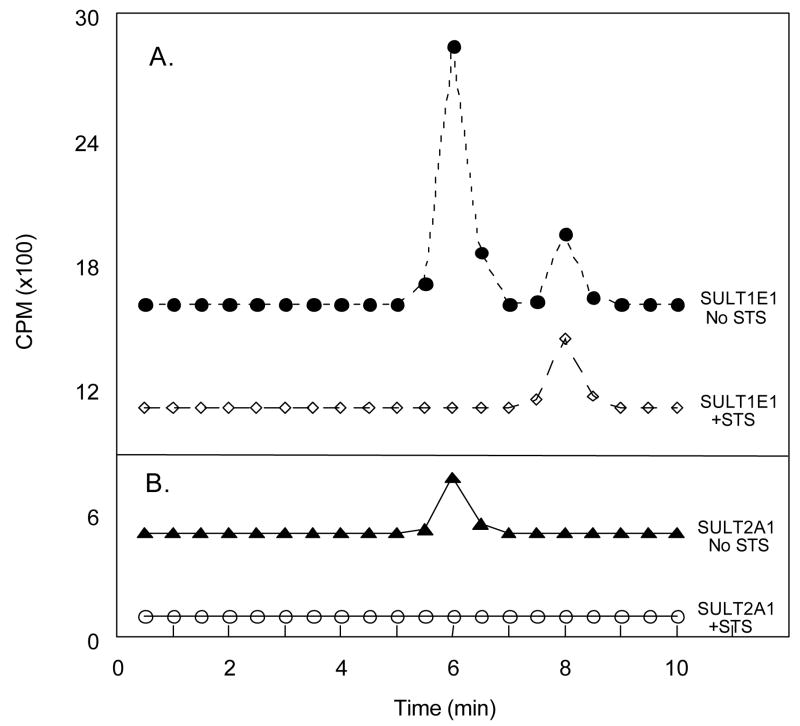
Our laboratory has previously reported that raloxifene, which has two hydroxyl groups (Figure 1), is sulfated at only one position by SULT2A1 (12). In contrast, at a concentration of 10 μM raloxifene, SULT1E1 generates both raloxifene monosulfates in a ratio of approximately 8 to 1 as well as relatively small amounts of the disulfate. However, at a concentration of 2.5 μM raloxifene, the disulfate is the primary reaction product formed by SULT1E1 (12). Figure 5A shows an autoradiograph of a TLC plate demonstrating the formation of raloxifene monosulfate by SULT2A1, and both raloxifene monosulfates and the disulfate by SULT1E1. The generation of only one monosulfate by SULT2A1, but two different monsosulfates as well as the disulfate by SULT1E1, has been confirmed by MS/MS (12). As shown in Figure 5A, after treatment with STS most of the raloxifene monosulfate formed by SULT2A1 is desulfated indicating that this monosulfate is readily hydrolyzed by STS. In contrast, when the raloxifene mono- and disulfate formed by SULT1E1 at both the 10 μM and 2.5 μM raloxifene concentrations are treated with STS, almost all of the disulfate and a significant fraction of the monosulfate were hydrolyzed (Fig. 5B), but in both cases there remained a significant amount of monsosulfate that was resistant to desulfation.
Figure 5.
Sulfation and desulfation of raloxifene. Raloxifene monosulfate was formed using SULT2A1 and both raloxifene monosulfates and disulfate were synthesized using SULT1E1 and 35S-PAPS. As reported previously (12), panel A shows that using SULT1E1, raloxifene disulfate formation at 2.5 μM is favored over monosulfate synthesis although monosulfate formation is greater at the 10 μM concentration. Panel B shows the sensitivity of the raloxifene monosulfates and disulfate for sensitivity to hydrolysis by STS. The products of the STS reactions were resolved by TLC and the products quantified by scintillation spectroscopy Each reaction was run in quadruplicate and the mean ± s.d. of the 35S-raloxifene sulfates generated or remaining after STS incubation is shown.
Time course of raloxifene desulfation
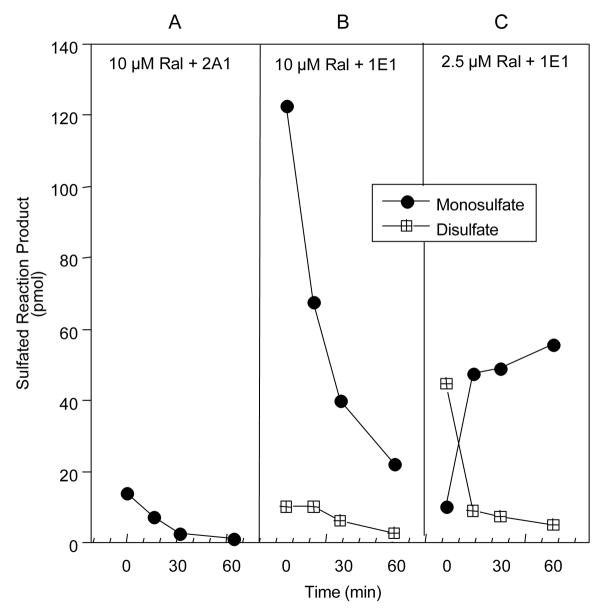
To further characterize raloxifene desulfation, time courses for desulfation by STS were carried out with the products of a reaction containing 10 μM raloxifene and SULT2A1, as well as reactions using both 10 μM and 2.5 μM raloxifene and SULT1E1. Figure 6 demonstrates that after 60 min incubation with STS, all of the raloxifene monosulfate produced by SULT2A1 was hydrolyzed. With the high levels of raloxifene monosulfates and relatively low levels of disulfate formed using 10 μm raloxifene and SULT1E1, almost all of the disulfate was metabolized by 60 min but approximately 15% of the monosulfate remained. In the case of the high level of raloxifene disulfate formed with 2.5 μM raloxifene and SULT1E1, the disulfate level drops rapidly for the initial 15 min of STS incubation, then the hydrolysis rate levels off, with only a small amount of disulfate remaining after 60 min. In contrast, the level of raloxifene monosulfate increases from 0 to 15 min STS incubation, suggesting that the disulfate undergoes a single desulfation event to produce a raloxifene monosulfate that is resistant to further STS hydrolysis.
Figure 6.
Time courses of the hydrolysis of raloxifene mono- and disulfate by human placental sulfatase (STS). Expressed human SULT2A1 and SULT1E1 were used to generate the two raloxifene monosulfates and the disulfate radiolabeled with 35S-PAPS. The 35S-labeled raloxifene products were then used as substrates for hydrolysis by STS. The formation and hydrolysis of the raloxifene monosulfates and disulfates were analyzed by HPLC. Panel A; STS hydrolysis of the single raloxifene monosulfate formed by SULT2A1. Panel B; STS hydrolysis of the monosulfates and disulfate of raloxifene formed by SULT1E1 at a raloxifene concentration of 10 μM. SULT1E1 with 10 μM raloxifene forms mostly monosulfates with a ratio of approx. 8:1, and only a relatively low level of the disulfate; Panel C. STS hydrolysis of the monosulfates and disulfate of raloxifene formed by SULT1E1 at a raloxifene concentration of 2.5 μM. SULT1E1 with 2.5 μM raloxifene forms mostly the disulfate.
HPLC analysis of raloxifene sulfates
HPLC analysis of the raloxifene reaction products generated with SULT2A1 or SULT1E1 and their subsequent hydrolysis by STS was performed. Of the two raloxifene monsosulfates generated by SULT1E1, one monosulfate appears unaffected by STS (Fig. 7A). HPLC analysis indicates that the STS-resistant raloxifene monosulfate generated by SULT1E1 was different from the STS-sensitive monosulfate produced by SULT2A1 (Fig. 7B). Previous results from our laboratory established that at a concentration of 10 μM raloxifene, SULT1E1 produces two raloxifene monosulfates in a ratio of approximately 8 to 1 (12). Upon HPLC on a C-18 column, the two monosulfates migrate with slightly different retention times. The predominant monosulfate has a shorter retention time, corresponding to the single raloxifene monosulfate produced by SULT2A1. As shown in Figure 7, the predominant raloxifene monosulfate formed by SULT1E1 corresponds to the SULT2A1 generated raloxifene monosulfate hydrolyzed by STS. The smaller raloxifene monosulfate peak with the longer retention time during HPLC was resistant to STS hydrolysis. However, the identity of each peak as either the benzothiophene or 4′-phenolic sulfates could not be confirmed by the available HPLC MS/MS procedure.
Figure 7.
HPLC analysis of the hydrolysis of the raloxifene monosulfates and disulfate by human placental STS. Raloxifene monosulfates and disulfate were synthesized using SULT2A1 and 1E1 as described in Figure 6. Panel A shows the elution of the monosulfates formed by SULT1E1 before and after STS hydrolysis. Panel B shows the elution of the monosulfate formed by SULT2A1 before and after STS hydrolysis.
STS and arzoxifene
Arzoxifene was utilized to gain insight into the sulfation and desulfation of raloxifene. Arzoxifene and raloxifene are similar in structure (Figure 1); however, the carbonyl function of raloxifene has been replaced with an ether linkage in arzoxifene, and the equivalent 4′-phenolic hydroxyl group of raloxifene is methylated in arzoxifene. Preliminary evaluation of arzoxifene sulfation demonstrated that it was sulfated by both SULT2A1 and SULT1E1 (data not shown). Because arzoxifene has only one hydroxyl group, there is only one potential site for sulfation at the benzothiophene hydroxyl. This site is analogous to the benzothiophene hydroxyl group of raloxifene (Figure 1). Thus, the arzoxifene monosulfates generated with SULT2A1 and SULT1E1 are presumed to be similar. Upon treatment with STS, the raloxifene and arzoxifene sulfates produced by SULT2A1 were completely desulfated (Fig. 8). By correlation, this provides a strong indication that the raloxifene monosulfate resistant to STS treatment is the 4′-phenolic monosulfate rather than the benzothiophene sulfate. Additionally, this indicates that the raloxifene monosulfate produced by SULT1E1 that was susceptible to STS treatment is the benzothiophene sulfate.
Figure 8.
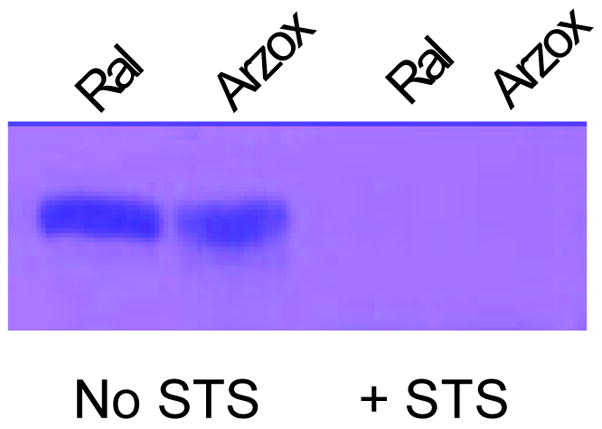
Hydrolysis of arzoxifene sulfate by human placental STS. Arzoxifene possesses a single benzothiophene hydroxyl group capable of being sulfated. The figure shows the monosulfates formed by SULT2A1 with raloxifene (Ral) and arzoxifene (Arz), respectively, with and without subsequent incubation with placental STS (+ STS).
DISCUSSION
The conjugation of estrogens, HRTs and therapeutic estrogenic compounds with a sulfonate group is an important mechanism in their metabolism and disposition. Therefore, since both Tib and raloxifene were readily sulfated by human tissues the regulation of the levels of these sulfates may be critical in regulating their physiological functions especially in target tissues (12, 13). Although conjugation with a sulfonate group is generally an inactivation process, sulfation is a reversible process since many sulfate esters can be hydrolyzed by STS activity (31, 32). Desulfation regenerates the active form of the compound and renders it capable of exerting a biological effect. Both the SULTs and STS are widely distributed in human tissues (6, 7, 31) although several of the SULT isoforms demonstrate important tissue specific activity in steroid metabolism (8, 13, 22, 33). The sulfation of Tib and its active metabolites in human endometrial cells by SULT1E1 is important for preventing the estrogenic activity of Tib in human endometrial tissues by selecting the generation of the progestagenic Δ4-isomer (13). Also, STS activity is increased in both breast and endometrial tumors (34, 35) suggesting increased generation of free steroids from STS-sensitive steroid sulfates present in the circulation in these tissues. Thus, the levels of bioavailable therapeutic drugs that are sulfated may be differentially regulated in peripheral target tissues depending upon the interactions of different SULT isoforms and expression levels of STS.
The hormonal effects of Tib and raloxifene depend upon metabolism in peripheral tissues (9). Tib is readily metabolized to its two estrogenic metabolites, 3αOH-Tib and 3βOH-Tib, which then circulate predominantly in their mono- or disulfated inactive forms. The 3α- and 3β-Tib sulfates can become estrogenically active when desulfated in target tissues (4). However, the differential ability of STS to hydrolyze the 3α- and 3β-Tib sulfates indicates a selectivity of the enzyme that is important in the regeneration of the active Tib metabolites. The 3β-Tib sulfate and 3β-Tib disulfate were efficiently desulfated by STS although the disulfate gave rise to a monosulfate that was resistant to hydrolysis. The resistant monosulfate is apparently the 3β-OH Tib 17-sulfate. The inability of STS to hydrolyze the 17-sulfate of tibolone is consistent with previous reports that the 17-sulfate of estrogens are also resistant (36, 37). Thus, 3β-Tib monosulfate can be desulfated to generate the hormonally active 3β-OH Tib in tissues with STS activity. The 3β-Tib disulfate is metabolized to the 17-sulfate that is not efficiently hydrolyzed by STS. In contrast, neither of the 3α-Tib monosulfates or the 3α-Tib disulfate were hydrolyzed by STS. This implies that the high levels of 3α-Tib sulfates observed in the plasma may be due to their lack of desulfation by STS. In contrast, the Tib 3β-sulfate is capable of being desulfated and spontaneously converting into the Δ4-isomer. Since the Δ4-isomer is progestagenic rather than estrogenic, the hormonal effect of Tib in a given tissue will be greatly dependent on the SULT isoforms expressed as well as the level of STS activity. The Δ4-Tib isomer is only inactivated by SULT2A1 that is not expressd in tissues such as the breast and endometrium, but is highly expressed in liver (15, 38). This suggests that sulfation of 3α-OH Tib is a permanent inactivation reaction and since 3β-Tib sulfate can be desulfated it represents a more important therapeutic metabolite.
The sulfation and desulfation of raloxifene was investigated in a similar manner to that of Tib. Our laboratory has previously reported that raloxifene is sulfated at only one position by SULT2A1 with no disulfate formation (12). In contrast, at a concentration of 10 μM raloxifene, SULT1E1 generates both raloxifene monosulfates as well as the disulfate whereas at lower concentrations the formation of raloxifene disulfate is increased (12). The single raloxifene monosulfate formed by SULT2A1 is hydrolyzed by STS. Upon treatment with STS the raloxifene monosulfate produced by SULT2A1 is essentially desulfated (Figure 4). In particular, the reaction at the 2.5 μM raloxifene concentration demonstrates that there is a higher level of monosulfated reaction product present after exposure to STS than in the initial reaction (Fig. 5). These results indicate that although almost all of the raloxifene disulfate is hydrolyzed by STS, some of the monosulfate generated is resistant to further metabolism by STS. This suggests that the sulfate of either the 4′-phenolic or benzothiophene hydroxyl group is not extensively sulfohydrolyzed by STS, resulting in a significant amount of raloxifene monosulfate that is not susceptible to STS activity. It is not known whether the different raloxifene monosulfates are receptor active. Nowell et al. (39) have suggested that in breast cancer tamoxifen 4-sulfate is associated with a better disease prognosis following tamoxifen therapy. The function or biological activity of selected STS resistant sulfates of the therapeutic HRT agents has not been evaluated.
HPLC analysis of the SULT2A1 and SULT1E1 sulfated raloxifene products indicates that the common raloxifene monosulfate formed by both SULTs was sensitive to STS hydrolysis. This monosulfate corresponds to the first peak to elute during HPLC/MS of the monosulfates formed by both SUL1E1 and 2A1. In contrast, the second slower eluting monosulfate peak is resistant to STS hydrolysis indicating that it cannot be desulfated to generate active raloxifene. Since the identity of the raloxifene monosulfates was not known, the newer SERM arzoxifene was utilized to probe the sensitivity of the benzothiophene sulfate ester to STS hydrolysis. Arzoxifene is structually related to raloxifene, however the 4-hydroxyl group is methylated allowing sulfation only at the benzothiophene hydroxyl group. Arzoxifene is sulfated by both SULT2A1 and 1E1, and Liu et al. (11) report the formation of arzoxifene sulfates with human Caco-2 cells. The identity of the reaction product as arzoxifene sulfate was confirmed by HPLC MS/MS (data not shown). Treatment of the arzoxifene monosulfate with STS resulted in complete hydrolysis of the sulfated product. By correlation, the raloxifene monosulfate that is sensitive to STS hydrolysis is the benzothiophene sulfate rather than the 4′-phenolic sulfate. Thus the raloxifene monosulfate produced by SULT2A1 is most likely the benzothiophene sulfate because it is completely eliminated by STS treatment. This would also correspond the predominant monosulfate generated by SULT1E1 because they possess the same retention time during HPLC.
For both Tib and raloxifene, net biologic activity in a target tissue depends in part upon the levels of the SULT isoforms and STS activity expressed in the tissue. For example, SULT1E1 is relatively highly expressed in normal breast epithelial tissues, whereas the expression of SULT1E1 is significantly lower in breast cancer tissues (40). In contrast, SULT1A1 is more highly expressed in breast cancers than in normal breast tissues (41). For raloxifene, a buildup of the STS-resistant phenolic sulfate would be greater in normal breast tissues than breast cancer tissues since this reaction is primarily catalyzed by SULT1E1 (12). Generally, the form of the sulfated compounds in the circulation that are susceptible to STS usually depends on SULT activity in the GI tract and liver (10). The conversion from the conjugated biologically inactive forms generally found in plasma to the unconjugated forms capable of initiating biological effects is dependent upon the availability of STS in the specific tissue. Additionally, the potential exists to resulfate the desulfated compound and return it to its inactive state if the appropriate SULT is present. Complicating this balance is the fact that STS is a microsomal enzyme, whereas the SULTs are generally cytosolic; thus, cellular compartmentalization of these reactions may also be significant in establishing net sulfation levels. Evaluation of the sulfation and desulfation of both Tib and raloxifene may yield important information as to the ways in which the selectivity of the SULT isoforms and STS interact to regulate the activity of these compounds in different tissues.
Acknowledgments
This research was supported in part by research grants from Organon and NIH grant GM38953 to CNF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kloosterboer HJ. Tibolone: a steroid with a tissue-specific mode of action. J Steroid Biochem Mol Biol. 2001;76:231–8. doi: 10.1016/s0960-0760(01)00044-9. [DOI] [PubMed] [Google Scholar]
- 2.Kenemans P, Speroff L. Tibolone: clinical recommendations and practical guidelines. A report of the International Tibolone Consensus Group. Maturitas. 2005;51:21–8. doi: 10.1016/j.maturitas.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Davis SR. The effects of tibolone on mood and libido. Menopause. 2002;9:162–70. doi: 10.1097/00042192-200205000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Reed MJ, Kloosterboer HJ. Tibolone: a selective tissue estrogenic activity regulator (STEAR) Maturitas. 2004;48(Suppl 1):S4–6. doi: 10.1016/j.maturitas.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Jordan V, Morrow M. Raloxifene as a multifunctional medicine? Brit Med J. 1999;319:331–332. doi: 10.1136/bmj.319.7206.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falany CN. Enzymology of human cytosolic sulfotransferases. FASEB J. 1997;11(4):206–16. doi: 10.1096/fasebj.11.4.9068609. [DOI] [PubMed] [Google Scholar]
- 7.Glatt H, Engelke CE, Pabel U, Teubner W, Jones AL, Coughtrie MW, Andrae U, Falany CN, Meinl W. Sulfotransferases: genetics and role in toxicology. Toxicol Lett. 2000;112–113:341–8. doi: 10.1016/s0378-4274(99)00214-3. [DOI] [PubMed] [Google Scholar]
- 8.Raftogianis R, Creveling C, Weinshilboum R, Weisz J. Estrogen metabolism by conjugation. J Natl Cancer Inst Monogr. 2000;(27):113–24. doi: 10.1093/oxfordjournals.jncimonographs.a024234. [DOI] [PubMed] [Google Scholar]
- 9.Kloosterboer HJ. Tissue-selectivity: the mechanism of action of tibolone. Maturitas. 2004;48(Suppl 1):S30–40. doi: 10.1016/j.maturitas.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Falany JL, Macrina N, Falany CN. Sulfation of tibolone and tibolone metabolites by expressed human cytosolic sulfotransferases. J Steroid Biochem Mol Biol. 2004;88:383–91. doi: 10.1016/j.jsbmb.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Bolton JL, Thatcher GR. Chemical modification modulates estrogenic activity, oxidative reactivity, and metabolic stability in 4′F-DMA, a new benzothiophene selective estrogen receptor modulator. Chem Res Toxicol. 2006;19:779–87. doi: 10.1021/tx050326r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falany JL, Pilloff DE, Leyh TS, Falany CN. Sulfation of raloxifene and 4-hydroxytamoxifen by human cytosolic sulfotransferases. Drug Metab Dispos. 2006;34:361–8. doi: 10.1124/dmd.105.006551. [DOI] [PubMed] [Google Scholar]
- 13.Falany JL, Falany CN. Regulation of SULT1E1 expression in Ishikawa adenocarcinoma cells by tibolone. Steroids. 2006;71(10):880–5. doi: 10.1016/j.steroids.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 14.Falany CN, Krasnykh V, Falany JL. Bacterial expression and characterization of a cDNA for human liver estrogen sulfotransferase. J Steroid Biochem Mol Biol. 1995;52:529–39. doi: 10.1016/0960-0760(95)00015-r. [DOI] [PubMed] [Google Scholar]
- 15.Falany CN, Comer KA, Dooley TP, Glatt H. Human dehydroepiandrosterone sulfotransferase. Purification, molecular cloning, and characterization. Ann N Y Acad Sci. 1995;774:59–72. doi: 10.1111/j.1749-6632.1995.tb17372.x. [DOI] [PubMed] [Google Scholar]
- 16.Meloche CA, Falany CN. Expression and characterization of the human 3 beta-hydroxysteroid sulfotransferases (SULT2B1a and SULT2B1b) J Steroid Biochem Mol Biol. 2001;77(4–5):261–9. doi: 10.1016/s0960-0760(01)00064-4. [DOI] [PubMed] [Google Scholar]
- 17.Jeong EJ, Lin H, Hu M. Disposition Mechanisms of Raloxifene in the Human Intestinal Caco-2 Model. J Pharmacol Exp Ther. 2004;310:376–385. doi: 10.1124/jpet.103.063925. [DOI] [PubMed] [Google Scholar]
- 18.Schrag ML, Cui D, Rushmore TH, Shou M, Ma B, Rodrigues AD. Sulfotransferase 1E1 is a low km isoform mediating the 3-O-sulfation of ethinyl estradiol. Drug Metab Dispos. 2004;32:1299–303. doi: 10.1124/dmd.32.11.. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Varlamova O, Vargas FM, Falany CN, Leyh TS, Varmalova O. Sulfuryl transfer: the catalytic mechanism of human estrogen sulfotransferase. J Biol Chem. 1998;273:10888–92. doi: 10.1074/jbc.273.18.10888. [DOI] [PubMed] [Google Scholar]
- 20.Radominska A, Comer KA, Zimniak P, Falany J, Iscan M, Falany CN. Human liver steroid sulphotransferase sulphates bile acids. Biochem J. 1990;272:597–604. doi: 10.1042/bj2720597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meloche CA, Sharma V, Swedmark S, Andersson P, Falany CN. Sulfation of budesonide by human cytosolic sulfotransferase, dehydroepiandrosterone-sulfotransferase (DHEA-ST) Drug Metab Dispos. 2002;30:582–5. doi: 10.1124/dmd.30.5.582. [DOI] [PubMed] [Google Scholar]
- 22.Falany JL, Azziz R, Falany CN. Identification and characterization of cytosolic sulfotransferases in normal human endometrium. Chem Biol Interact. 1998;109:329–39. doi: 10.1016/s0009-2797(97)00143-9. [DOI] [PubMed] [Google Scholar]
- 23.Duanmu Z, Weckle A, Koukouritaki SB, Hines RN, Falany JL, Falany CN, Kocarek T, Runge-Moris M. Developmental expression of aryl, estrogen, and hydroxysteroid sulfotransferases in pre- and postnatal human liver. J Pharmacol Exp Ther. 2006;316:1310–7. doi: 10.1124/jpet.105.093633. [DOI] [PubMed] [Google Scholar]
- 24.Tashiro A, Sasano H, Nishikawa T, Yabuki N, Muramatsu Y, Coughtrie MW, Nagura H, Hongo M. Expression and activity of dehydroepiandrosterone sulfotransferase in human gastric mucosa. J Steroid Biochem Mol Biol. 2000;72:149–54. doi: 10.1016/s0960-0760(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 25.Chen G, Zhang D, Jing N, Yin S, Falany CN, Radominska-Pandya A. Human gastrointestinal sulfotransferases: identification and distribution. Toxicol Appl Pharmacol. 2003;187:186–97. doi: 10.1016/s0041-008x(02)00073-x. [DOI] [PubMed] [Google Scholar]
- 26.Hernandez-Guzman FG, Higashiyama T, Osawa Y, Ghosh D. Purification, characterization and crystallization of human placental estrone/dehydroepiandrosterone sulfatase, a membrane-bound enzyme of the endoplasmic reticulum. J Steroid Biochem Mol Biol. 2001;78:441–50. doi: 10.1016/s0960-0760(01)00119-4. [DOI] [PubMed] [Google Scholar]
- 27.Hernandez-Guzman FG, Higashiyama T, Pangborn W, Osawa Y, Ghosh D. Structure of human estrone sulfatase suggests functional roles of membrane association. J Biol Chem. 2003;278:22989–97. doi: 10.1074/jbc.M211497200. [DOI] [PubMed] [Google Scholar]
- 28.Ganguly TC, Krasnykh V, Falany CN. Bacterial expression and kinetic characterization of the human monoamine-sulfating form of phenol sulfotransferase. Drug Metab Dispos. 1995;23(9):945–50. [PubMed] [Google Scholar]
- 29.Wang J, Falany JL, Falany CN. Expression and characterization of a novel thyroid hormone-sulfating form of cytosolic sulfotransferase from human liver. Mol Pharmacol. 1998;53:274–82. doi: 10.1124/mol.53.2.274. [DOI] [PubMed] [Google Scholar]
- 30.Falany CN, Wheeler J, Oh TS, Falany JL. Steroid sulfation by expressed human cytosolic sulfotransferases. J Steroid Biochem Mol Biol. 1994;48:369–75. doi: 10.1016/0960-0760(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 31.Pasqualini JR, Chetrite GS. Estrone sulfatase versus estrone sulfotransferase in human breast cancer: potential clinical applications. J Steroid Biochem Mol Biol. 1999;69:287–92. doi: 10.1016/s0960-0760(99)00082-5. [DOI] [PubMed] [Google Scholar]
- 32.Pasqualini JR, Chetrite GS. Recent insight on the control of enzymes involved in estrogen formation and transformation in human breast cancer. J Steroid Biochem Mol Biol. 2005;93:221–36. doi: 10.1016/j.jsbmb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 33.Kotov A, Falany JL, Wang J, Falany CN. Regulation of estrogen activity by sulfation in human Ishikawa endometrial adenocarcinoma cells. J Steroid Biochem Mol Biol. 1999;68:137–44. doi: 10.1016/s0960-0760(99)00022-9. [DOI] [PubMed] [Google Scholar]
- 34.Reed MJ, Purohit A, Woo LW, Newman SP, Potter BV. Steroid sulfatase: molecular biology, regulation, and inhibition. Endocr Rev. 2005;26:171–202. doi: 10.1210/er.2004-0003. [DOI] [PubMed] [Google Scholar]
- 35.Ahmed S, Owen CP, James K, Sampson L, Patel CK. Review of estrone sulfatase and its inhibitors--an important new target against hormone dependent breast cancer. Current Med Chem. 2002;9:263–73. doi: 10.2174/0929867023371210. [DOI] [PubMed] [Google Scholar]
- 36.Pasqualini JR, Gelly C, Lecerf F. Estrogen sulfates: Biological and ultrastructural responses and metabolism in MCF-7 human breast cancer cells. Breast Cancer Res Treat. 1986;8:233–240. doi: 10.1007/BF01807336. [DOI] [PubMed] [Google Scholar]
- 37.Pasqualini JR, Gelly C, Lecerf F. Biological effects and morphological responses to estriol, estriol-3-sulfate, estriol-17-sulfate and tamoxifen in a tamoxifen-resistant cell line (R-27) derived from MCF-7 human breast cancer cells. Eur J Cancer Clin Oncol. 1986;22:1495–1501. doi: 10.1016/0277-5379(86)90086-6. [DOI] [PubMed] [Google Scholar]
- 38.Falany CN, Vazquez ME, Kalb JM. Purification and characterization of human liver dehydroepiandrosterone sulphotransferase. Biochem J. 1989;260:641–6. doi: 10.1042/bj2600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nowell S, Sweeney C, Winters M, Stone A, Lang NP, Hutchins LF, Kadlubar FF, Ambrosone CB. Association between sulfotransferase 1A1 genotype and survival of breast cancer patients receiving tamoxifen therapy. J Natl Cancer Inst. 2002;94:1635–40. doi: 10.1093/jnci/94.21.1635. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki T, Nakata T, Miki Y, Kaneko C, Moriya T, Ishida T, Akinaga S, Hirakawa H, Kimura M, Sasano H. Estrogen sulfotransferase and steroid sulfatase in human breast carcinoma. Cancer Res. 2003;86:2762–2770. [PubMed] [Google Scholar]
- 41.Falany JL, Falany CN. Expression of cytosolic sulfotransferases in normal mammary epithelial cells and breast cancer cell lines. Cancer Res. 1996;56:1551–1555. [PubMed] [Google Scholar]