Abstract
While genetic determinants protecting against development of elevated blood pressure (BP) are well investigated, less is known regarding their impact on longevity. We concomitantly assessed genomic regions of rat chromosomes 3 and 7 (RNO3 and RNO7) carrying genetic determinants of BP without known epistasis, for (1) their independent and combinatorial effects on BP and (2) the presence of genetic determinants of survival using Dahl salt-sensitive (S) strains carrying congenic segments from Dahl salt-resistant (R) rats. While congenic and bicongenic S.R strains carried independent BP QTLs within the RNO3 and RNO7 congenic regions, only the RNO3 allele(s) independently affected survival. The bicongenic S.R strain showed epistasis between R-rat RNO3 and RNO7 alleles for BP under salt-loading conditions, with less-than-additive effects observed on a 2% NaCl diet and greater-than-additive effects observed after prolonged feeding on a 4% NaCl diet. These RNO3 and RNO7 congenic region alleles had more-than-additive effects on survival. Increased survival of bicongenic, compared to RNO3 congenic rats, was attributable, in part, to maintaining lower BP despite chronic exposure to an increased dietary salt (4% NaCl) intake, with both strains showing delays in reaching highest BP. R-rat RNO3 alleles were also associated with superior systolic function, with the S.R bicongenic strain showing epistasis between R-rat RNO3 and RNO7 alleles leading to compensatory hypertrophy. Whether these alleles affect survival by additional actions within other BP-regulating tissues/organs remain unexplored. This is the first report of simultaneous detection of independent and epistatic loci dictating, in part, longevity in a hypertensive rat strain.
Keywords: genetic hypertension, Dahl salt-sensitive rat, Dahl salt-resistant rat, survival, longevity, compensatory hypertrophy, relative wall thickness
Introduction
Most human morbidity and mortality stem from complex diseases and disorders whose phenotypes result from interactions of multiple genes with environmental factors. Hypertension is such a disorder, an independent predisposing factor in the development of several diseases responsible for adult morbidity and mortality, including atherosclerosis, coronary heart disease, peripheral artery disease, heart failure, renal failure, and stroke1. Little is known regarding the relationships between genetic determinants of blood pressure (BP) with genetic determinants of these diseases or overall mortality. We hypothesized that genetic factors contribute to the extended survival of some hypertensive subjects, but not others. The obvious difficulty of using death as an endpoint in studying lifespan in human hypertensive subjects, suggests that hypertension-survival relationships are better-studied using animal models.
Inbred Dahl salt-sensitive (SS/Jr or S) and Dahl salt-resistant (SR/Jr or R) rat strains2 are contrasting models of high and relatively normal BP, respectively, selectively-bred from outbred Sprague-Dawley rats under salt-loading conditions3. Supplemental dietary NaCl increases BP in S rats, with little or no effect on BP in R rats. Segregating populations and congenic strains derived from these inbred strains have been used to screen for and confirm chromosomal locations responsible for heritable BP strain-differences (i.e. BP quantitative trait loci, QTLs)4, 5. S.R congenic strains, and substrains derived from them, were used extensively to identity 11ß-hydroxylase (Cyp11b1) as a genetic determinant of BP on rat chromosome (RNO) 76-8 and to define limits of genomic segments containing BP genetic determinants on other chromosomes4, 9. However, relationships between alleles within BP QTL-containing congenic intervals on different chromosomes have been little studied 10, 11, except when epistatic BP QTL interactions were first identified in genome scans. The effects of epistasis between BP QTLs on mortality have not been addressed in previous substitution mapping studies.
In the present study, we assessed rat genomic regions containing BP genetic determinants lacking known epistasis6-8, 12 for (1) their independent and combinatorial effects on BP and (2) genetic determinants of survival. These BP QTL-containing intervals showed (1) differing epistatic effects on BP, depending on the duration and concentration of the high salt diet, and (2) more-than-additive effects on survival, when chronically fed an even higher salt (4% NaCl) diet. Increased survival of RNO3+RNO7 bicongenic, compared to RNO3 congenic, rats was due, at least in part, to their maintaining lower BP despite prolonged exposure to a higher dietary NaCl intake. R-rat RNO3 congenic region alleles were also associated with measures of superior systolic function, with epistasis between R-rat RNO3 and RNO7 alleles leading to increased compensatory hypertrophy, as evidenced by increased end-diastolic relative wall thickness (RWT). This data is consistent with our hypothesis that interactions between alleles in different BP QTL-containing regions influence both BP and survival under salt-loading conditions and are traceable using S.R congenic strains as genetic tools.
Methods
Inbred and Congenic Rat Strains
Inbred Dahl S and R rat strains were developed2 from outbred stock originally obtained from Dahl3, 13. Development and characterization of rat chromosome 3 and 7 congenic substrains, S.R-(D3Arb14-D3Mco36) and S.R-(D7Mco19-Exon2-Cyp11b1) (Figure S1) were previously described6, 12. Inbred and congenic rat strains were from our colony at the University of Toledo Health Science Campus and will be referred to throughout this manuscript as S, R, RNO3, and RNO7, respectively. Two backcross F1(SxR)xS populations (n = 150 rats) were used to examine epistasis between RNO3 and RNO7 loci. Breeding and phenotyping of these populations were previously described14, 15 and are summarized in the Online Data Supplement available at http://www.hypertensionaha.org.
RNO3 and RNO7 congenic intervals containing R-rat low BP QTL alleles were introgressed into an S-rat genetic background resulting in the S.R-[(D3Arb14-D3Mco36 and D7Mco19-Exon2-Cyp11b1)] rats, hereafter referred to as the RNO3+RNO7 bicongenic strain. Breeding was as follows: F1 rats, bred by crossing RNO3 and RNO7 congenic rats, were backcrossed to RNO3 congenic rats. Progeny heterozygous for the RNO7 and homozygous for the RNO3 congenic intervals were crossed. Resulting progeny homozygous for both congenic intervals were bred to establish the bicongenic strain. All breeding and experimental protocols were approved by the Institutional Animal Care and Use Committee of the University of Toledo Health Science Campus.
Experiment 1
Age and weight-matched rats [Group 1: S, n=21; RNO3, n=15; RNO3+RNO7, n=20; and RNO7, n=19] were bred, housed, and studied concomitantly. Rats were weaned at 30 days of age onto a low salt diet (0.3% NaCl, diet 7034; Harlan-Teklad). Two rats of different strains were randomly assigned to each cage. At 40-42 days of age all rats were transferred to a 2% NaCl diet (diet 94217, Harlan-Teklad) for 28 days.
Systolic BP was measured using tail-cuff plethysmography for a subset of Group 1 rats [S, n=15; RNO3, n=15; RNO3+RNO7, n=16; and RNO7, n=14] that were conscious, restrained, and warmed to 28°C16, by operators unaware of their identity. BP of each rat was measured on consecutive days during weeks three (days 17 and 18) and four (days 26 and 27) of the 2% NaCl diet. Daily BP values were the mean of 3-4 consistent readings. Final BP values were the mean of the daily BP values. Following BP measurement (day 28 after initiation of 2% NaCl diet) rats were transferred to a high salt diet (4% NaCl, diet 83033, Harlan-Teklad) until they died or became obviously terminally ill, in which case they were euthanized by carbon dioxide hypoxia. Rats were examined twice daily for signs of distress.
Experiment 2
Two groups of rats were concomitantly bred to further characterize effects of R-rat RNO3 and RNO7 alleles on BP and survival: Group 2A [S, n=8; RNO3, n=8; and RNO3+7, n=8], was used to measure BP by telemetry and Group 2B [S, n=23; RNO3, n=10; RNO3+7, n=10; and RNO7, n=10], was used to assess cardiac function by echocardiography and for terminal experiments. Both groups (2A and 2B) received the dietary NaCl regimen described in Experiment 1, with renal function assessed by 24-hour urinary protein excretion (UPE). Group 2A rats were fed a 4% NaCl diet until they died or became terminally ill. Group 2B rats were fed a 4% NaCl diet for up to 46-48 days, when surviving rats were used in terminal experiments.
Radiotelemetric BP Measurement
Transmitters were surgically implanted into rats 20-24 days after initiating the 2% NaCl diet, as previously described17, 18. Rats recovered for a week before collecting BP data. Five sets of BP measurements (systolic and diastolic) were recorded at 5 min intervals for 24-48 hour periods over 12 weeks. For each rat, a series of six moving averages (each over a 4 h period) were calculated over the first 24 h measured for each time point. An overall mean BP was calculated as the mean of six consecutive, 4-hour moving averages, for each of five time points measured in a rat.
Urinary Protein Excretion
UPE was determined as described previously19. Urine was collected three times: 1) on a 0.3% NaCl diet, just prior to the 2% NaCl diet start, 2) after 27 days on a 2% NaCl diet, and 3) after 38 days on a 4% NaCl diet [a total of 65 days on a higher salt (2% or 4% NaCl) diet].
Echocardiography
Left ventricular function was evaluated by echocardiography as previously described20. Briefly, rats were anesthetized with 1.5–2.0% isoflurane by O2 inhalation, the chest shaved, and situated in the supine position on a warming pad. Two-dimensional guided M-mode studies were performed from parasternal long window using a 15-MHz linear array transducer. Study duration was typically 15–20 min per rat.
Terminal Experiments and Histology
Group 2B rats surviving 46 days on the 4% NaCl diet [S, n=4; RNO3, n=6, RNO3+RNO7, n=6; and RNO7, n=3] were euthanized by pentobarbital overdose, plasma collected, and body weights measured. After blood collection, hearts were removed, blotted, and weighed. Kidneys were removed, de-capsulated, blotted, and weighed separately. Left kidneys and portions of the heart were fixed in 10% neutral-buffered formalin and embedded in paraffin blocks for subsequent sectioning. Plasma glucose, creatinine, and urea nitrogen concentrations were determined by the Department of Pathology, University of Toledo Medical Center.
Additional details regarding the phenotyping of Experiment 2 rats are in Supplemental Methods available online at http://www.hypertensionaha.org.
Statistical Analysis
Normally distributed data was analyzed by one-way ANOVA to determine overall significance followed by Tukey HSD or Games-Howell post-hoc tests. Non-parametric data was analyzed by Kruskal-Wallis tests, followed by Mann-Whitney U pair-wise comparison tests if significant differences were observed. P<0.05 was the criterion for statistical significance. Data is presented as the mean ± the standard error of the mean (SEM).
Equality of the survival functions of the strains was evaluated by Kaplan-Meier and log-rank tests. Survival functions were compared pair-wise, with the statistical significance criterion adjusted for multiple comparisons (Bonferroni correction). The effects of R-rat alleles within the RNO3 (R3) and RNO7 (R7) congenic regions on survival in Experiment 1 and measures of cardiac and renal function in Experiment 2 were examined using general linear models. Additional details are in Supplemental Methods available online at http://www.hypertensionaha.org.
Results
To investigate the relationship between RNO3 and RNO7 BP QTLs, we analyzed the relationship between Cyp11b and Edn3 (near these BP QTL peaks) with BP and body weight-adjusted heart weight (BW-adjusted HW) in a previously studied F1(SxR)xS backcross population6, 7, 12, 14, 15. No interactions were observed between these loci for either trait (Table S1). A bicongenic strain (RNO3+RNO7) was bred to (1) confirm additive actions of R-rat alleles in these congenic regions on BP and (2) examine effects of these alleles on survival in an excessive dietary NaCl intake context.
Less Than Additive Effects of RNO3 and RNO7 QTLs on BP
Systolic BP was measured by tail-cuff plethysmography in concomitantly raised male S, RNO3 and RNO7 congenic, and RNO3+RNO7 bicongenic rats during the third and fourth weeks of a 2% NaCl diet (Table 1 and Figure 1, Panels A and B). These BP measurement sets were strongly correlated (r = 0.660, P<0.0001; Figure S2). Compared to the parental S strain, lower BP was observed for all three congenic strains at both time points (Table 1; Figure 1, Panels A and B). Lower BP was observed in RNO3, compared to RNO7, congenic rats (160.5 ± 2.3 mm Hg vs. 170.8 ± 3.3 mm Hg, respectively, P=0.018) and in RNO3+RNO7 bicongenic, compared to RNO7 congenic rats in week 4 (157.8 ± 3.2 mm Hg vs. 170.8 ± 3.3 mm Hg, respectively, P=0.013) but not between RNO3+RNO7 bicongenic and RNO3 congenic rats (Table 1). Indeed, the differential BP [ΔBP; i.e., mean BP of a congenic strain — mean BP of S] for RNO3+RNO7 bicongenic rats in both weeks 3 and 4 were lower than expected if the RNO3 and RNO7 congenic region low BP QTL alleles had additive effects (Figure 1, A and B).
Table 1.
Strain-Differences in Blood Pressure, Body Weight and Survival among S and Congenic Rat Strains.
| Strain | BP, week 3, mm Hg |
BP, week 4, mm Hg |
Body weight, initial, g |
Days Survived on 4% NaCl diet |
|---|---|---|---|---|
| S | 173.5 ± 4.0 [15] | 181.9 ± 4.1 [15] | 161.0 ± 2.1 [20] | 64.8 ± 6.4 [21] |
| RNO3 | 154.7 ± 1.7 [15] | 160.5 ± 2.3 [15] | 145.1 ± 3.6 [15] | 96.9 ± 7.1 [15] |
| RNO3 + RNO7 | 152.7 ± 2.6 [16] | 157.8 ± 3.2 [16] | 150.2 ± 3.4 [20] | 126.4 ± 8.5 [20] |
| RNO7 | 160.5 ± 3.0 [14] | 170.8 ± 3.3 [14] | 150.2 ± 2.5 [19] | 72.5 ± 6.8 [19] |
| P (overall) | <0.0001 | <0 .0001 | <0.0001 | 0.002 |
| P (S vs. RNO3) | 0.002 | 0.001 | 0.002 | 0.002 |
| P (S vs. RNO3 + RNO7) | 0.001 | 0.0004 | 0.015 | <0.0001 |
| P (S vs. RNO7) | NS, 0.067 | 0.052 | 0.004 | NS |
| P (RNO3 vs. RNO3 + RNO7) |
NS | NS | NS | 0.016 |
| P (RNO3 vs. RNO7) | NS | 0.018 | NS | 0.009 |
| P (RNO7 vs. RNO3 + RNO7) | NS | 0.013 | NS | <0.0001 |
BW was measured on 40-42 day-old Experiment 1 rats. Tail-cuff BP was measured during the third (days 17 and 18) and fourth weeks (days 26 and 27) on a 2% NaCl diet. After BP measurement, rats were maintained on a 4% NaCl diet until they died or became moribund. Numbers of rats per strain measured for each trait are in brackets. NS, not significant (P>0.05).
Figure 1. Epistasis between Genetic Determinants of Blood Pressure and Survival in RNO3 and RNO7 Congenic Regions.
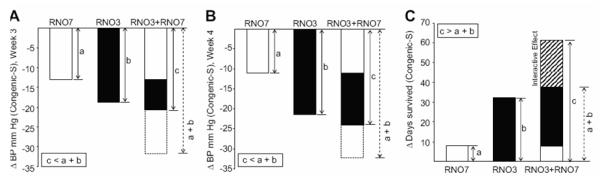
Bars depict decreased BP (Δ BP) in congenic strains compared to S rats after 4 weeks on a 2% NaCl diet (Panels A) and increased survival (Δ survival) in congenic strains compared to S rats subsequently fed a 4% NaCl diet (Panel B). Increments labeled a and b depict effects of R-rat RNO3 and RNO7 congenic interval alleles, respectively, on Δ BP and Δ survival. The increment labeled a+b depicts the expected combined effects of R-rat alleles in both congenic intervals on Δ BP and Δ survival, if additive effects were exerted. The increment labeled c depicts the observed effects of R-rat alleles in both congenic intervals on Δ BP and Δ survival. Rats used were those described in Experiment 1.
Greater Than Additive Effects of RNO3 and RNO7 QTLs on Survival, Under Salt-Loading Conditions
As non-additive BP effects were observed between R-rat alleles within these congenic intervals after 4 weeks on a 2% NaCl diet, our primary phenotypic measurement, we examined their effects on longevity following chronic salt-loading. All rats (including some whose BP were not measured) were fed a higher (4% NaCl) salt diet. RNO3 and RNO3+RNO7 strains survived significantly longer (96.9 ± 7.1 and 126.4 ± 8.5 days, P=0.002 and P<0.0001, respectively) compared to the parental S strain (64.8 ± 6.4 days; Table 1). Survival differences were not observed between RNO7 and S rats, though RNO7, compared to RNO3, rats survived significantly fewer days (Table 1). Surprisingly, differential survival of RNO3+RNO7 [Δ survival; i.e., mean survival of a congenic strain — mean survival of S rats, days on 4% NaCl diet] was much greater compared to the sum of the Δ survivals for RNO3 and RNO7 congenic rats, indicating a strong interactive effect (Figure 1C). Survival functions of these four rat strains were significantly different (P<0.0001, Figure 2), with all pair-wise survival function comparisons significantly different (following Bonferroni correction), excepting those of RNO7 with RNO3 or S rats.
Figure 2. Cumulative Survival Curves for S, Congenic, and Bicongenic Rats.

Cumulative survival curves, with days survived on the 4% NaCl diet as the time variable, are shown for male S, RNO3, RNO3+RNO7, and RNO7 rats (Experiment 1).
Effects of RNO3 and RNO7 congenic interval alleles on survival were examined using a general linear model, with BP (measured during week 4 of the 2% NaCl diet) a covariate. Both RNO3 congenic interval alleles (R3, P=0.037) and main effects interactions (R3×R7, P=0.030; Figure S2), but not RNO7 congenic interval alleles (R7, P=0.11), were associated with BP-adjusted survival. Increased BP-adjusted survival was observed for RNO3+RNO7 rats (115.3 days), compared with S (84.5 days, P=0.020), RNO3 (86.9 days, P=0.009), or RNO7 (78.9 days, P=0.002) rats (Figure S3).
Longer Exposure to Elevated Dietary NaCl Significantly Reduced BP in Bicongenic, Compared to RNO3 Congenic Rats
We next sought to identify factors responsible for increased survival of the RNO3+RNO7 bicongenic rats. Despite BP additivity not being observed in congenic rats fed a 2% NaCl diet (Figure 1 and Table 1), we hypothesized that RNO3 and RNO7 BP QTL allelic products might interact in rats maintained longer on a higher salt (4% NaCl) diet, causing lower BP in bicongenic, compared to RNO3 congenic rats. Experiment 2 was conducted to test this hypothesis. Similar to our earlier results (Table 1), S rats had higher BP (systolic and diastolic) compared to both RNO3 and RNO3+RNO7 rats after 27 days on a 2% NaCl diet, with no significant difference observed between RNO3 and RNO3+RNO7 rats (Table S2). However, BP strain-differences were observed between RNO3 and RNO3+RNO7 after additional time on a diet with an even higher salt (4% NaCl) content. Lower systolic BP was observed for bicongenic rats compared to RNO3 congenic rats after 38 days (157.7 ± 2.3 vs. 169.6 ± 2.1 mm Hg, P=0.0004), 68 days (196.6 ± 3.6 vs. 215.6 ± 4.1 mm Hg, P=0.039), and 75 days (203.8 ± 3.7 vs. 223.7 ± 4.6 mm Hg, P=0.0003) of salt loading (Figure 3A and Table S2). Similarly, lower diastolic BP was also observed for RNO3+RNO7, compared to RNO3, rats after 38 days of salt loading (110.2 ± 2.0 vs. 115.7 ± 1.8 mm Hg, P=0.039; Figure 3B and Table S2). Interestingly, the BP of S and RNO3 congenic (but not bicongenic) rats plateaued, with S rats reaching this level first (Figure 3 and Table S2).
Figure 3. Longitudinal Study of Blood Pressure Strain-Differences among S and Congenic Rats Under an Increasing Dietary NaCl Intake.
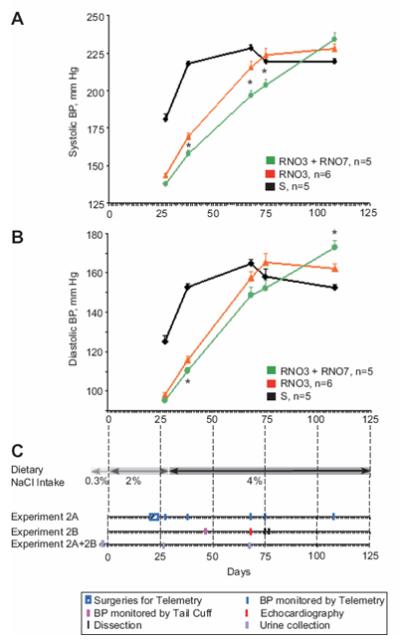
Panels A and B depict systolic and diastolic BP measurements by radio-telemetry, in male S, RNO3 congenic, and RNO3+RNO7 bicongenic rats (Group 2A) chronically exposed to an elevated dietary NaCl intake. *, significant BP differences between RNO3 congenic and RNO3+RNO7 bicongenic rats. Numbers of rats per strain measured and statistical analyses of systolic and diastolic BP are in Table S2. Panel C depicts the Experiment 2 design. Experimental manipulations and measurements are shown separately for each rat group above a timeline showing days after 2% NaCl diet initiation.
In addition to the above telemetry experiment, we assessed the BP of Group 2B rats surviving for 17-18 days on a 4% NaCl diet (69-70 days on a higher dietary NaCl intake) by tail-cuff plethysmography. The time-line of all experiments is given in Figure 3C. RNO3+RNO7 rats had lower BP compared to S and RNO7 rats (P=0.001 and P=0.015, respectively; Table S3), but not RNO3 rats, which approached significance (P=0.08). Both RNO3 (R3, P=0.002) and RNO7 (R7, P=0.026) congenic interval alleles, but not main effects interactions (R3 × R7, NS), were associated with significant differences in tail-cuff BP.
R-rat RNO3 Alleles Are Associated with Superior Cardiac Function After Prolonged Exposure to Excessive Dietary NaCl
The cardiac function of Group 2B rats surviving 40-41 days on a 4% NaCl diet was assessed by echocardiography, with representative M-mode images shown in Figure S4. Overall, inbred and congenic rat strains in this study could be ranked for echocardiographic parameters, from best to worst, as follows: RNO3+RNO7 ≥ RNO3 ≥ S ≥ RNO7 (Table 2). Measures of systolic function [left ventricular (LV) fractional shortening, FS and mean velocity of circumferential fiber shortening, Vcf] were similarly improved in RNO3 and RNO3+RNO7 rats compared to S and RNO7 rats (Table 2). Bicongenic rats showed the most cardiac hypertrophy (as determined by RWT, left ventricular relative wall thickness), compared to the other tested strains (Table 2). RNO3 congenic interval alleles were associated with significant differences in the following parameters: FS (P=0.0005), Vcf (P=0.0002), RWT (P=0.026), LV end-diastolic diameter (LVDd, P=0.019), and LV end-systolic diameter (LVSd, P=0.001; Table 2). Interestingly, RNO3 and RNO7 congenic interval alleles were epistatic for LVDd, (P=0.042) and RWT (P=0.025, Table 2).
Table 2.
Strain-Differences in Echocardiographic Parameters among S and Congenic Rat Strains Maintained on Elevated Dietary NaCl.
| Strain, (n) | LVDd, cm | LVSd, cm | FS, % | Vcf, 1/sec | RWT |
|---|---|---|---|---|---|
| S (6) | 0.74 ± 0.04 | 0.42 ± 0.04 | 44 ± 4 | 5.48 ± 0.59 | 0.67 ± 0.01* |
| RNO3 (7) | 0.73 ± 0.04 | 0.34 ± 0.04 | 54 ± 3 | 7.94 ± 0.60 | 0.67 ± 0.09 |
| RNO3 + RNO7 (7) | 0.69 ± 0.02 | 0.30 ± 0.02 | 57 ± 3 | 8.48 ± 0.50 | 0.85 ± 0.03† |
| RNO7 (4) | 0.86 ± 0.04 | 0.52 ± 0.05 | 40 ± 3 | 5.30 ± 0.78 | 0.50 ± 0.05 |
| P (overall) | 0.036 | 0.004 | 0.005 | 0.002 | 0.023 |
| P (S vs. RNO3) | NS | NS | NS | 0.031 | NS |
| P (S vs.RNO3 + RNO7) | NS | NS | 0.039 | 0.007 | 0.010 |
| P (S vs. RNO7) | NS | NS | NS | NS | 0.041 |
| P (RNO3 vs. RNO3 + RNO7) |
NS | NS | NS | NS | NS, 0.08 |
| P (RNO3 vs. RNO7) | NS | 0.017 | 0.035 | 0.043 | NS |
| P (RNO7 vs. RNO3 + RNO7) |
0.024 | 0.004 | 0.013 | 0.012 | 0.010 |
| P (R3) | 0.019 | 0.001 | 0.0005 | 0.0002 | 0.026 |
| P (R7) | NS | NS | NS | NS | NS |
| P (R3 × R7) | 0.042 | NS, 0.08 | NS | NS | 0.025 |
Echocardiography was performed on surviving Group 2B rats exposed to a 4% NaCl diet for 40-41 days (i.e., 69-70 days on higher NaCl diets). Numbers of rats per strain studied for each trait are in parentheses, with the following exceptions
four S rats
six RNO3+RNO7 rats.
NS, not significant (P>0.05). R3, allelic content within the RNO3 congenic interval; R7, allelic content within the RNO7 congenic interval. LVDd: LV end-diastolic diameter, LVSd: LV end-systolic diameter, FS: left ventricular (LV) fractional shortening, Vcf: mean velocity of circumferential fiber shortening, RWT: left ventricular relative wall thickness.
Effects of RNO3 and RNO7 Congenic Interval Alleles on Renal Function were Highly-Dependent upon Dietary NaCl intake
Male S, RNO3 and RNO7 congenic, and RNO3+RNO7 bicongenic (Groups 2A and 2B) rats were assessed for renal function on three dietary NaCl regimens by measuring 24-hour UPE. As significant strain-differences in body weight (BW) were observed at each urine collection, 24-hour UPE/BW was analyzed. 24-hour UPE/BW was first measured in rats maintained on a low (0.3% NaCl) salt diet, with higher 24-hour UPE/BW observed for S and RNO3 rats compared to RNO3+RNO7 and RNO7 rats (P≤0.001, Table 3), with RNO7 congenic interval alleles associated with differences in 24-hour UPE/BW (<0.0001, Table 3). However, after 28 days on a higher (2% NaCl) salt diet, S and RNO7 rats had higher 24-hour UPE/BW compared to RNO3 and RNO3+RNO7 rats (P<0.01, Table 3), with RNO3 congenic interval alleles associated with 24-hour UPE/BW differences (P<0.0001). After 38 days on a 4% NaCl diet, RNO3 rats had lower 24-hour UPE/BW compared to RNO3+RNO7 rats (P=0.012, Table 3) with RNO7 congenic interval alleles associated with 24-hour UPE/BW differences (P=0.042).
Table 3.
Longitudinal Study of Urinary Protein Excretion among S and Congenic Rat Strains Under an Increasing Dietary NaCl Intake.
| Dietary Conditions | Collection 1 | Collection 2 | Collection 3 |
|---|---|---|---|
| Dietary salt content | 0.3% NaCl | 2% NaCl | 4% NaCl |
| Days on higher salt diet (2% and/or 4% NaCl) |
0 | 28 | 67 (38 days on 4% NaCl diet) |
| Strain | 24 hour UPE/BW, g/kg | ||
| S | 83.4 ± 5.8 [18] | 907 ± 59 [18] | 1621 ± 169 [9] |
| RNO3 | 81.9 ± 7.2 [18] | 467 ± 26 [18] | 1042 ± 153 [13] |
| RNO3 + RNO7 | 33.1 ± 4.8 [18] | 565 ± 41 [17] | 1672 ± 135 [15] |
| RNO7 | 37.1 ± 11.0 [10] | 814 ± 57 [10] | 1716 ± 143 [5] |
| P (overall) | <0.0001 | <0.0001 | 0.009 |
| P (S vs. RNO3) | NS | <0.0001 | NS, 0.06 |
| P (S vs. RNO3 + RNO7) | <0.0001 | <0.0001 | NS |
| P (S vs. RNO7) | 0.0003 | NS | NS |
| P (RNO3 vs. RNO3 + RNO7) | <0.0001 | NS | 0.012 |
| P (RNO3 vs. RNO7) | 0.001 | <0.0001 | NS, 0.08 |
| P (RNO7 vs. RNO3 + RNO7) | NS | 0.007 | NS |
| P (R3) | NS | <0.0001 | NS, 0.08 |
| P (R7) | <0.0001 | NS | 0.042 |
| P (R3 × R7) | NS | 0.052 | NS |
Numbers of rats per strain measured for each trait are in brackets. R3, allelic content within the RNO3 congenic interval; R7, allelic content within the RNO7 congenic interval. NS, not significant (P>0.05).
Terminal Morphometric and Biochemical Assessment of Inbred and Congenic Rats
No significant strain-differences in body, kidney, or heart weights were observed (Table S4). There were also no significant strain-differences in circulating creatinine, glucose, or urea nitrogen values (Table S4). However, mean circulating creatinine values for all four strains were higher than the rat reference range 21, as were mean circulating urea nitrogen values for all but bicongenic rats.
S, RNO3 and RNO7 congenic, and RNO3+RNO7 bicongenic rat kidney sections showed similar, extensive renal vascular changes, consistent with the presence of malignant hypertension (data not shown). Similarly, heart sections from these inbred and congenic strains were evaluated for arterial stenosis, hypertrophic myocytes, and interstitial fibrosis. No strain differences were observed among these four strains for these three phenotypes (Table S5).
Discussion
Over the past two decades, hundreds of QTLs for BP and related traits have been identified in rodent models and humans4, 5, 22-24, though few were characterized with respect to either interaction with other BP QTLs or effects on mortality. Two QTLs for survival in the context of an excessive dietary NaCl intake were identified in the present study. RNO3 congenic rats carried a newly-identified survival QTL, while R-rat RNO7 congenic interval alleles did not independently affect survival. The latter contrasts with our previous results, where in males, R-rat RNO7 alleles within a much larger congenic region (Figure S1) significantly increased survival, compared to S rats14. However, in the present study, R-rat RNO7 alleles were associated with increased survival under salt-loading conditions in RNO3+RNO7 bicongenic rats, where their products could interact with those of R-rat RNO3 alleles.
Dietary NaCl, Epistasis, and Blood Pressure
Surprisingly, low BP QTL alleles within the RNO3 and RNO7 congenic intervals of bicongenic rats showed BP epistasis highly dependent upon the content and/or duration of exposure to a high dietary NaCl intake (Tables 1 and S2; Figures 1 and 3). In these RNO3+RNO7 bicongenic rats, less-than-additive effects were observed after a 2% NaCl diet, compared with the greater-than-additive effects observed with additional, prolonged exposure to a higher (4% NaCl) salt diet. Also, when BP was measured under our standard conditions (i.e., after 4 weeks on a 2% NaCl diet) low BP QTL alleles in RNO3+RNO7 rats showed less-than-additive effects in contrast with the greater-than-additive effects previously observed in another bicongenic rat strain10, 25. The differing interactive effects of low BP QTL alleles on different chromosomes observed in these two bicongenic strains further reflects the intricate gene-environment relationships in complex traits like BP.
R-rat RNO7 congenic interval alleles demonstrated modest BP effects in this study compared to the much larger BP and survival affects of the R-rat RNO7 alleles in S.R-Cyp11b14, from which it was derived. This suggests S.R-Cyp11b rats carried additional R-rat RNO7 BP QTL and survival QTL allele(s). Substitution mapping8 suggests the additional RNO7 low BP QTL allele(s) in S.R-Cyp11b congenic rats14 do not act independently of those of the RNO7 congenic substrain6 used in this study.
Overall, it is clear that BP alone does not completely explain the observed extended survival of the bicongenic rats. There may be factors within and/or outside the cardiovascular and renal systems that dictate the extended survival of the bicongenic rats. In this report, we chose to test the hypothesis that differential functionality of the heart and/or kidney may contribute to differences in survival.
Cardiac Function and Survival
Echocardiographic evaluation of cardiac function suggested that RNO3 congenic region alleles were associated with preservation of systolic function under salt-loading conditions (Table 2). RNO3+RNO7 rats displayed superior systolic function (significantly higher FS and Vcf), compared to S rats. However, no FS and Vcf differences were observed between RNO3+RNO7 and RNO3 rats, suggesting these strains exhibited similar increases in systolic function compared with S rats (Table 2). Furthermore, no epistasis between RNO3 and RNO7 congenic interval alleles was observed for either measure of systolic function (Table 2). In contrast, RNO7 congenic rats did not demonstrate improved systolic function compared with S rats. Together, this data indicates that under salt-loading conditions, RNO3 congenic region alleles are primarily responsible for the observed increased systolic function of RNO3+RNO7 bicongenic and RNO3 congenic rats, compared to S rats.
However, the above systolic function differences do not explain the longer survival of RNO3+RNO7, compared to RNO3, rats. Echocardiographic evaluation found bicongenic rats to have the highest cardiac hypertrophy (as determined by RWT) among the tested strains. This observation, combined with epistasis between RNO3 and RNO7 congenic interval alleles for RWT (Table 2) suggested that greater RWT contributes to the increased longevity of bicongenic rats (Table 2), consistent with previous studies of pressure overload-induced heart failure where increased survival was observed for rats with greater RWT26.
Renal Function and Survival
Clearly, the inbred and congenic strains used in this study showed heritable differences in renal function. R-rat RNO7 alleles (in bicongenic and RNO7 congenic rats) were associated with decreased UPE/BW (Table 3), compared to strains (S and RNO3 congenic) lacking these alleles, on a low salt (0.3% NaCl) diet. After exposure to the 2% NaCl diet this effect disappeared and instead, R-rat RNO3 alleles were associated with decreased UPE/BW (Table 3), possibly reflecting the lower BP of RNO3 and bicongenic rats, compared to S and RNO7 rats (Figure 3, Table 1).
While these differences in measures of renal function may be related to initial BP strain-differences, they are unlikely responsible for increased survival of RNO3+RNO7 rats. Treatment with an even higher, 4% NaCl, diet paradoxically led to lower UPE/BW for RNO3 rats compared to RNO3+RNO7 (as well as S and RNO7) rats. Indeed, histologic examination of their renal sections after such treatment found similar, substantial renal vascular changes, consistent with malignant hypertension that were also reflected in high circulating creatinine and urea nitrogen levels in these four rat strains (Table S5).
Mortality as a BP QTL Study Criterion
While genomic regions containing alleles protecting from morbidity and simultaneously increasing longevity are clearly advantageous, few BP QTLs have been tested for effects on survival14. In this context, while transgenic Sprague-Dawley rats over-expressing Npy27 and transgenic Dahl S rats expressing the R-rat Atp1a1 allele28 showed decreased BP and increased survival, other studies did not associate decreased BP with increased survival 29-32 33. The dual beneficial effects of decreasing morbidity (by lowering BP) and increasing survival, suggest that the RNO3 and RNO7 BP QTLs can be viewed as priorities for further genetic dissection.
Perspectives
The genetic contribution to human lifespan is estimated to be ∼20%34. However, study designs to identify such genes in humans and determine whether they remain operational in a morbid human condition such as hypertension are limited35. These newly-identified survival QTLs, one acting independently (on RNO3) and the other epistatically (on RNO7), illustrate how available congenic strains can be exploited for dissecting genes underlying life-span differences among hypertensive subjects, and facilitate further positional cloning of causative genes. Further, our study in rat models demonstrates how heritable elements dictating small BP changes in hypertensive subjects can lead to differential mortality through epistatic mechanisms. As our analysis of the effects of these alleles on cardiac and renal function was limited, it remains to be investigated whether these RNO3 and RNO7 alleles might also exert their effects through actions occurring in other tissues/organ systems involved in the maintenance of BP homeostasis.
Acknowledgements
Authors thank the Physiology and Pharmacology Departmental Phenotyping Core for facilitating the Echocardiography experiments.
Sources of Funding: NHLBI/NIH (RO1-HL020176 and RO1-HL075414) to BJ and NHLBI/NIH (RO1-68994) to GTC and SJL.
Footnotes
Disclosures: None
Literature Cited
- 1.Kannel WB, Wolf WFB. Cardiovascular risk factors and hypertension. In: Izzo JI Jr., Black HR, editors. Hypertension Primer. 3rd ed Lippincott Williams & Wilkens; Philadelphia: 2003. pp. 235–238. [Google Scholar]
- 2.Rapp JP, Dene H. Development and characteristics of inbred strains of Dahl salt-sensitive and salt-resistant rats. Hypertension. 1985;7:340–349. [PubMed] [Google Scholar]
- 3.Dahl LK, Heine M, Tassinari L. Role of genetic factors in susceptibility to experimental hypertension due to chronic excess salt ingestion. Nature. 1962;194:480–482. doi: 10.1038/194480b0. [DOI] [PubMed] [Google Scholar]
- 4.Rapp JP. Genetic analysis of inherited hypertension in the rat. Physiol Rev. 2000;80:135–172. doi: 10.1152/physrev.2000.80.1.135. [DOI] [PubMed] [Google Scholar]
- 5.Joe B, Garrett MR. Genetic analysis of inherited hypertension in the rat. In: Dominiczak A, Connell JM, editors. Genetics of Hypertension. Vol 24. Elsevier Science; Amsterdam: 2006. pp. 177–200. [Google Scholar]
- 6.Garrett MR, Rapp JP. Defining the blood pressure QTL on chromosome 7 in Dahl rats by a 177-kb congenic segment containing Cyp11b1. Mamm Genome. 2003;14:268–273. doi: 10.1007/s00335-002-2245-9. [DOI] [PubMed] [Google Scholar]
- 7.Cicila GT, Rapp JP, Wang J-M, Lezin E, Ng SC, Kurtz TW. Linkage of 11®-hydroxylase mutations with altered steroid biosynthesis and blood pressure in the Dahl rat. Nat Genet. 1993;3:346–353. doi: 10.1038/ng0493-346. [DOI] [PubMed] [Google Scholar]
- 8.Cicila GT, Garrett MR, Lee SJ, Liu J, Dene H, Rapp JP. High resolution mapping of the blood pressure QTL on chromosome 7 using Dahl rat congenic strains. Genomics. 2001;72:51–60. doi: 10.1006/geno.2000.6442. [DOI] [PubMed] [Google Scholar]
- 9.Rapp JP, Deng AY. Detection and positional cloning of blood pressure quantitative trait loci: Is it possible? Identifying the genes for genetic hypertension. Hypertension. 1995;25:1121–1128. doi: 10.1161/01.hyp.25.6.1121. [DOI] [PubMed] [Google Scholar]
- 10.Rapp JP, Garrett MR, Deng AY. Construction of a double congenic strain to prove an epistatic interaction on blood pressure. J Clin Invest. 1998;101:1591–1595. doi: 10.1172/JCI2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monti J, Plehm R, Schulz H, Ganten D, Kreutz R, Hübner N. Interaction between blood pressure quantitative trait loci in rats in which trait variation at chromosome 1 is conditional upon a specific allele at chromosome 10. Hum Mol Genet. 2003;12:435–439. doi: 10.1093/hmg/ddg041. [DOI] [PubMed] [Google Scholar]
- 12.Lee SJ, Liu J, Westcott AM, Vieth JA, DeRaedt SJ, Yang S, Joe B, Cicila GT. Substitution mapping in Dahl rats identifies two distinct blood pressure quantitative trait loci within 1.12 Mb and 1.25 Mb intervals on chromosome 3. Genetics. 2006;174:2203–2213. doi: 10.1534/genetics.106.061747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahl LK, Heine M, Tassinari L. Effects of chronic excess salt ingestion: evidence that genetic factors play an important role in the susceptibility to experimental hypertension. J Exp Med. 1962;115:1173–1190. doi: 10.1084/jem.115.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cicila GT, Dukhanina OI, Kurtz TW, Walder R, Garrett MR, Dene H, Rapp JP. Blood pressure and survival of a chromosome 7 congenic strain derived from Dahl rats. Mamm Genome. 1997;8:896–902. doi: 10.1007/s003359900607. [DOI] [PubMed] [Google Scholar]
- 15.Cicila GT, Choi C, Dene H, Lee SJ, Rapp JP. Two blood pressure/cardiac mass quantitative trait loci are present on chromosome 3 of Dahl salt-sensitive and salt-resistant rats. Mamm Genome. 1999;10:112–116. doi: 10.1007/s003359900954. [DOI] [PubMed] [Google Scholar]
- 16.Buñag R, Butterfield J. Tail cuff blood pressure measurement without external preheating in awake rats. Hypertension. 1982;4:898–903. doi: 10.1161/01.hyp.4.6.898. [DOI] [PubMed] [Google Scholar]
- 17.Joe B, Garrett MR, Dene H, Rapp JP. Substitution mapping of a blood pressure quantitative trait locus to a 2.73 Mb region on rat chromosome 1. J Hypertens. 2003;21:2077–2084. doi: 10.1097/00004872-200311000-00017. [DOI] [PubMed] [Google Scholar]
- 18.Saad Y, Garrett MR, Manickavasagam E, Yerga-Woolwine S, Farms P, Radecki T, Joe B. Fine-mapping and comprehensive transcript analysis reveals nonsynonymous variants within a novel 1.17 Mb blood pressure QTL region on rat chromosome 10. Genomics. 2007;89:343–353. doi: 10.1016/j.ygeno.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toland EJ, Yerga-Woolwine S, Farms P, G.T. C, Saad Y, Joe B. Blood pressure and proteinuria effects of multiple quantitative trait loci on rat chromosome 9 that differentiate the spontaneously hypertensive rat from the Dahl salt-sensitive rat. J Hypertens. 2008;26:2134–2141. doi: 10.1097/HJH.0b013e32830ef95c. [DOI] [PubMed] [Google Scholar]
- 20.Morgan EE, Faulx MD, McElfresh TA, Kung TA, Zawaneh MS, Stanley WC, Chandler MP, Hoit BD. Validation of echocardiographic methods for assessing left ventricular dysfunction in rats with myocardial infarction. Am J Physiol Heart Circ Physiol. 2004;287:H2049–H2053. doi: 10.1152/ajpheart.00393.2004. [DOI] [PubMed] [Google Scholar]
- 21.Boehm O, Zur B, Kioch A, Tran N, Freyenhagen R, Hartmann M, Zacharowski K. Clinical chemistry reference database for Wistar rats and C57/BL6 mice. Biol Chem. 2007;388:547–554. doi: 10.1515/BC.2007.061. [DOI] [PubMed] [Google Scholar]
- 22.Joe B, Garrett MR. Substitution mapping: using congenic strains to detect genes controling blood pressure. In: Raizada MK, Paton JFR, Kasparov S, Katovich MJ, editors. Cardiovascular Genomics. Humana Press Inc.; Totowa, N.J.: 2005. pp. 41–58. [Google Scholar]
- 23.Deng AY. Positional cloning of quantitative trait Loci for blood pressure: how close are we? a critical perspective. Hypertension. 2007;49:740–747. doi: 10.1161/01.HYP.0000259105.09235.56. [DOI] [PubMed] [Google Scholar]
- 24.Cowley AW., Jr. The genetic dissection of essential hypertension. Nat Rev Genet. 2006;7:829–840. doi: 10.1038/nrg1967. [DOI] [PubMed] [Google Scholar]
- 25.Deng Y, Rapp JP. Cosegregation of blood pressure with angiotensin converting enzyme and atrial natriuretic peptide receptor genes using Dahl salt-sensitive rats. Nat Genet. 1992;1:267–272. doi: 10.1038/ng0792-267. [DOI] [PubMed] [Google Scholar]
- 26.Norton GR, Woodiwiss AJ, Gaasch WH, Mela T, Chung ES, Aurigemma GP, Meyer TE. Heart failure in pressure overload hypertrophy: The roles of ventricular remodeling and myocardial dysfunction. J Am Coll Cardiol. 2002;39:664–671. doi: 10.1016/s0735-1097(01)01792-2. [DOI] [PubMed] [Google Scholar]
- 27.Michalkiewicz M, Knestaut KM, Bytchkova EY, Michalkiewicz T. Hypotension and reduced catecholamines in neuropeptide Y transgenic rats. Hypertension. 2003;41:1056–1062. doi: 10.1161/01.HYP.0000066623.64368.4E. [DOI] [PubMed] [Google Scholar]
- 28.Herrera VL, Xie HX, Lopez LV, Schork NJ, Ruiz-Opazo N. The ⟨ 1 Na,K-ATPase gene is a susceptibility hypertension gene in the Dahl salt-sensitiveHSD rat. J Clin Invest. 1998;102:1102–1111. doi: 10.1172/JCI3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim-Mitsuyama S, Izumi Y, Izumiya Y, Yoshida K, Yoshiyama M, Iwao H. Additive beneficial effects of the combination of a calcium channel blocker and an angiotensin blocker on a hypertensive rat-heart failure model. Hypertens Res. 2004;27:771–779. doi: 10.1291/hypres.27.771. [DOI] [PubMed] [Google Scholar]
- 30.Pinto YM, Pinto-Sietsma SJ, Philipp T, Engler S, Kossamehl P, Hocher B, Marquardt H, Sethmann S, Lauster R, Merker HJ, Paul M. Reduction in left ventricular messenger RNA for transforming growth factor beta(1) attenuates left ventricular fibrosis and improves survival without lowering blood pressure in the hypertensive TGR(mRen2)27 Rat. Hypertension. 2000;36:747–754. doi: 10.1161/01.hyp.36.5.747. [DOI] [PubMed] [Google Scholar]
- 31.Sironi L, Gelosa P, Guerrini U, Banfi C, Crippa V, Brioschi M, Gianazza E, Nobili E, Gianella A, de Gasparo M, Tremoli E. Anti-inflammatory effects of AT1 receptor blockade provide endorgan protection in stroke-prone rats independently from blood pressure fall. J Pharmacol Exper Therapeut. 2004;311:989–995. doi: 10.1124/jpet.104.072066. [DOI] [PubMed] [Google Scholar]
- 32.Takenaka H, Kihara Y, Iwanaga Y, Onozawa Y, Toyokuni S, Kita T. Angiotensin II, oxidative stress, and extracellular matrix degradation during transition to LV failure in rats with hypertension. J Mol Cell Cardiol. 2006;41:989–997. doi: 10.1016/j.yjmcc.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 33.Libonati JR, Gaughan JP. Low-intensity exercise training improves survival in Dahl salt hypertension. Med Sci Sports Exercise. 2006;38:856–858. doi: 10.1249/01.mss.0000218129.03008.ab. [DOI] [PubMed] [Google Scholar]
- 34.Cournil A, Kirkwood TB. If you would live long, choose your parents well. Trends Genet. 2001;17:233–235. doi: 10.1016/s0168-9525(01)02306-x. [DOI] [PubMed] [Google Scholar]
- 35.Atzmon G, Rincon M, Schechter CB, Shuldiner AR, Lipton RB, Bergman A, Barzilai N. Lipoprotein genotype and conserved pathway for exceptional longevity in humans. PLoS Biol. 2006;4:e113. doi: 10.1371/journal.pbio.0040113. [DOI] [PMC free article] [PubMed] [Google Scholar]


