Summary
This chapter describes a method of sample preparation called “the rock and roll method,” which is basically a solvent evaporation technique with controlled manual sample movement during evaporation of solvent from lipid/solvent mixtures that produces well-oriented thick stacks of about 2000 lipid bilayers. Many lipid types have been oriented using different solvent mixtures that balance solubilization of the lipid with uniform deposition of the lipid solution onto solid substrates. These well-oriented thick stacks are then ideal samples for collection of both X-ray diffraction data in the gel phase and X-ray diffuse scattering data in the fluid phase of lipids. The degree of orientation is determined using visual inspection, polarizing microscopy, and a mosaic spread X-ray experiment. Atomic force microscopy is used to compare samples prepared using the rock and roll method with those prepared by spin-coating, which produces well-oriented but less homogeneous lipid stacks. These samples can be fully hydrated through the vapor provided that the hydration chamber has excellent temperature and humidity control.
Keywords: AFM, hydration, mosaic spread, oriented lipid bilayers, polarizing microscopy, rock and roll method, X-ray scattering
1. Introduction
Lipid bilayers form the underlying structure of every plant and animal cell membrane wherein, in general, they occur in a fluid, fully hydrated state. Whereas X-rayed, multilamellar arrays of lipid bilayers produce sharp Bragg diffraction peaks only in the lower-temperature crystalline phases due to their well-ordered structure (1), fluid phase lipid bilayers produce diffuse scattering in the fully hydrated state due to thermal fluctuations in the water space between fluctuating bilayers (for a review, see ref. 2). The Nagle laboratory pioneered a diffuse scattering technique that collects and analyzes two-dimensional (2D) diffuse X-ray scattering from fully hydrated, fluid-phase lipid stacks oriented onto a solid substrate (3–5). These data yield structural and material parameters about specific lipids, but require thick samples (~4–10 µm) since diffuse scattering is weaker than sharp, Bragg diffraction peaks. This chapter will present the step-by-step procedures of how to prepare oriented, thick bilayer sample films and how to hydrate them.
The name “rock and roll method” derives from the popular culture; it was borrowed since it describes well the procedure for making oriented stacks of lipid bilayers first published by the lab in 1993 (6). The appropriate amount of dry lipid to form a 10-µm thick film (see Note 1 and Subheading 3.1., step 1) is dissolved in the appropriate solvent(s). The choice of solvent(s) depends on the lipid chain length, unsaturation, backbone, and headgroup type, and is crucial for the success of the films (see Subheading 3.1., step 3). The lipid solution is poured onto one of the substrates (see Subheading 2., step 3), which is hand-rocked in four directions (N, S, E, and W), forcing the lipid solution to roll out to the edges of the substrate. These actions subject the lipid bilayers to shear force, which helps them orient during evaporation of the solvent. The rock and roll procedure requires a large glove box to produce defect-free films, because a solvent-rich vapor slows down the evaporation of solvent from the films (see Subheading 3.3. and Note 2).
The rock and roll procedure is compared herein with two other methods that were previously used to form thin films (~20 bilayers):
A simple stationary solvent evaporation technique described in detail by Seul and Sammon (7) (see Subheading 3.4.)
A “high-tech” method called spin-coating (8) (see Subheading 3.5.).
A third method published recently (9) that codissolves solid naphthalene with the solvents and lipid, renders well-aligned and easily hydrated thick lipid films. However, it was deemed unsuitable for the analysis because the resulting films contain many small holes, which preclude an accurate estimation of the sample thickness.
2. Materials
Lipids: most lipids are purchased from Avanti Polar Lipids (Alabaster, AL) due to their high purity and, more recently, low acyl-chain migration in mixed chain lipids. Occasionally, novel lipids are synthesized for the Nagle lab by collaborators; these are checked by electrospray ionization mass spectrometry to verify their purity. Lyophilized lipids are stored at −20°C in their original vials in dessicators before use. The original vials are equilibrated at room temperature for at least 15 min before opening to avoid condensation of water into the lipid, which can lead to hydrolysis of chains. When stored and opened this way, the saturated chain lipids do not degrade for many years, as verified using thin-layer chromatography (46:15:3, chloroform:methanol:7 N NH4OH [v:v:v]), stained with molybdic acid stain (10).
Solvents: solvents are of high-pressure liquid chromatography grade; purity of the solvents and cleanliness in forming the sample films are essential. Solvents are poured from the original solvent container into disposable 20-mL glass scintillation vials, which are discarded after 1 d of sample preparation.
Substrates: usually a 1-mm thick silicon wafer (100 face) cut to 1.5× 3 cm2 is used. The size of the wafer is cut to be the same as the size of the Peltier cooler (Melcor, Trenton, NJ) that supports the silicon wafer during the hydration experiment, so that the sample is cooled evenly (see Subheading 3.7.). The silicon wafers can be used with or without dopant. The first substrate that was used with some success for gel phase samples was a glass microscope slide. Microscope slides have the advantage that orientation can be checked easily using a polarizing microscope (see Subheading 3.4.); however, the diffuse background from this substrate is large and not easily subtracted from the fluid phase diffuse data. Alternatively, the substrate is a 3× 3 cm2 piece of freshly cleaved 35-µm thick mica (Mica New York Corp, NY), which can be curved and glued onto a cut, glass beaker. Another type of substrate is a thin (70 µm) 3× 3 cm2 00 glass cover slip (Precision Glass and Optics, Santa Ana, CA).
Water: nanopure water is used to hydrate the lipids from the vapor phase in a freshly cleaned hydration chamber (described in Subheading 3.7.). Water purity and cleanliness of the chamber are essential to reach full hydration.
3. Methods
3.1. Preparation of Samples
To calculate the mass of lipid required to make a 10-µm film, consider a 10-µm thick column of many copies of a single lipid pair in a bilayer stack. Convert to number of molecular bilayers by dividing by the thickness of a dried lipid bilayer (compare with ~60 Å for dried dipalmitoylphosphatidylcholine [DPPC]). Determine the total number of lipids in the film by first multiplying the aforementioned number of bilayers by two, then dividing by the surface area of a dried lipid (~50 Å2), and then multiplying by the total surface area of the substrate. Then divide by Avogradro’s number to get moles, and multiply by the gram molecular weight of the lipid to get grams of lipid. The actual thickness of these films measured by atomic force microscopy (AFM) is slightly smaller than the calculated thickness, presumably because of some collection of lipid near the edges of the substrate (11).
Weigh lyophilized lipid using an analytical balance and place the lipid (~4–10 mg, calculated as aforementioned) into a disposable, small glass test tube. Disposable test tubes are used to assure cleanliness. Add 100–200 μL solvent to the weighed lipid in the test tube in the glove box. Agitate the tube to solubilize the lipid. The solvents required for each lipid were determined by the method of trial and error, within the general guidelines that a hydrophobic solvent is needed to dissolve the lipid, whereas an amphipathic solvent is needed to decrease the contact angle of the lipid solution with the hydrophilic substrate, thus causing the solution to spread. Often one solvent is not sufficient to both solubilize the lipid and cause the solution to spread on the silicon, mica, or glass surface. Listed below are the solvents or solvent mixtures (shown as volume ratios), which were successful for each lipid in the lab. Results may vary across laboratories due to relative humidity, temperature, and size (or lack) of the glove box (see Note 2). If the solvents below do not successfully form a smooth-looking film to the naked eye on the first attempt, then a different ratio may be used to redissolve the film.
- Suggested solvents for specific lipids:
- Dioleoylphosphatidylcholine (DOPC)—easiest lipid to orient. 2:1, 2.5:1, or 3:1 chloroform: methanol, or 1:1 chloroform: trifluoroethanol (TFE).
- Dilauroylphosphatidylcholine (DLPC)—difficult to orient. 1:1:1 chloroform:methanol:TFE.
- Dimyristoylphosphatidylcholine (DMPC)—chloroform:TFE 1:1 or 2:1, chloroform:methanol 1:1 or 1.5:1, and neat isopropanol.
- DiC15phosphatidylcholine—chloroform:methanol 2.5:1.
- Dipalmitoylphosphatidylcholine (DPPC)—chloroform:methanol 2–3.5:1.
- D,L-DPPC—chloroform:methanol 2:1.
- DiC17phosphatidylcholine—chloroform:methanol 3:1.
- Distearoylphosphatidylcholine (DSPC)—chloroform:methanol 2:1 or 3:1.
- DiC19phosphatidylcholine—chloroform:methanol 2.5:1, methanol:carbon tetrachloride 5:1.
- DiC20phosphatidylcholine—methanol:carbon tetrachloride 4:1.
- DiC22phosphatidylcholine—methanol:carbon tetrachloride 4:1.
- DiC24phosphatidylcholine—difficult to orient. Methanol:carbon tetrachloride 3:1 and chloroform: methanol 2:1.
- DSPC-Br4—chloroform:TFE, 1:1.
- Dihexadecylphosphatidylcholine (DHPC)—chloroform:methanol 2:1.
- Dierucoylphosphatidylcholine DiC22:1PC—chloroform:TFE, 1:1.
- Palmitoyloleoylphosphatidylcholine (POPC)—chloroform:TFE, 1:1.
- Dimyristoylphosphatidylserine (DMPS)—difficult to orient. TFE:toluene, 4:1, or neat toluene.
- Dioleolyphosphatidylserine (DOPS)—chloroform:TFE, 2:1.
- Stearoyldocosahexaenoylphosphatidylcholine—methylene chloride:methanol, 3:1.
- Stearoyldocosapentaenoylphosphatidylcholine—methylene chloride:methanol, 3:1.
- Dilaurylphosphatidylethanolamine (DLPE)—neat hexafluoroisopropanol in the hood (see Note 3)
- Myristoylpalmitoylphosphatidylcholine (MPPC)—chloroform:methanol, 2:1.
- Myristoylstearoylphosphatidylcholine (MSPC)—chloroform:methanol, 2.5:1.
- 1,2-Di-O-myristoyl-3-N,N,N-trimethylaminopropane (DM-TAP)—chloroform:methanol, 10:1.
- Most mixtures combining DOPC, DPPC, DLPC, brain sphingomyelin, and cholesterol (raft and super lattice mixtures)—chloroform:TFE, 1:1 (see Note 4).
- Mixtures with alamethicin and DOPC or DOPC-Br4—chloroform, then redissolved in chloroform: TFE, 1:1.
- Mixtures with human immunodeficiency virus fusion peptide FP-23 and DOPC or diC22:1PC—neat hexafluoroisopropanol, in the hood (see Note 3).
Salts can be added to any of the aforementioned lipid–solvent mixtures. Salts crystallize on drying, yet they diffuse readily between bilayers on subsequent hydration (see Subheading 3.7.) (see also ref. 12). In the glove box, close the test tube prepared with above-containing solvent and lipid, with a silicone or neoprene stopper during the following preparation of the substrate, so that the ratio of solvents does not change due to unequal evaporation. Rubber stoppers should not be used as they are degraded by the solvents.
3.2. Preparation of the Substrates
The normal procedure is to simply bathe the Si wafers in chloroform in a glass Petri dish inside the glove box; then, the wafers are rubbed with a cotton swab (see Note 5). This procedure not only cleans the wafer, but also fills the glove box with a chloroform-saturated atmosphere. The Si wafers are sometimes rubbed with a Kimwipe in the final step to remove any remaining residue, and any lint is carefully blown away with a small rubber bulb. When mica is used, the 3× 3 cm2 piece is freshly cleaved using a technique shown to the author by Jacob Israelachvilli; this consists of wedging a fine gauge needle or pin into the edge of a square of grade 2 mica to separate a thin layer of mica from the original slab, which is then slowly teased away using a tweezer. The thickness should be about 35 µm (i.e., flexible enough to become slightly curved, yet thick enough to contain lipid and solvent), with no cracks or holes. The mica should be used within 1 or 2 h after cleaving to avoid dust collecting on its surface.
The third substrate, i.e., 3×3 cm2 thin glass cover slips, should be cleaned with chloroform swabbing as for the silicon wafer. At just 70-µm thick, these cover slips break easily, especially during cleaning. The thin glass cover slip is useful for X-raying lipid samples through the back of the glass at a 45° angle of incidence; the scattering from the sample is then totally unobstructed by the substrate, which is not the case for a grazing angle of incidence on the silicon wafer or curved mica. These substrates are then attached to the cap of a 20-mL disposable glass scintillation vial using either sticky tack or clay (see Fig. 1). This method of attachment leaves the sides of the substrate completely free, which is essential to allow surface tension and hold the solvent solution on the top of the substrate. If the substrates are placed directly onto a larger substrate, such as a microscope slide, the lipid–solvent mixture tends to run off at the edges, due to a small contact angle of the lipid solution with the substrate.
Fig. 1.
A silicon substrate (15×30 mm2) is attached through sticky tack (synthetic clay) to a 20-mL scintillation vial. (Left) Side view, (Right) top view. The highly polished silicon surface appears black in this photograph.
3.3. Rock and Roll Method
When the author first began making films in the early 1990s, she tried to evaporate the solvents in the hood, but rapid evaporation of the solvent caused many surface defects (a frosted appearance) that were poorly oriented. Even working on the open lab bench close to an open window or air conditioner caused the lipid surface to become “frosty” during evaporation of the solvent. Therefore, investment in a suitable glove box is key for the success of this method. The Nagle lab glove box is 8 ft3 made from 3/16 in. thick Plexiglas, with rubber, gloveless ports for entry of hands. Humidity is monitored with a probe (Rotronics, Dallas, TX), but not controlled. The humidity generally increases from about 40 to 70% relative humidity (RH) during the film formation, because of perspiration from human hands in the glove box (see also Note 2).
In the glove box, the lipid in solvent is poured onto the substrate attached as described in Subheading 3.2., with rocking and rolling (see Subheading 1.). As the sample dries, a higher angle of rocking (up to 90°) may be required in order to induce the now-viscous solution to move down the wafer. The rocking continues until the rolling of lipid in solvent over the surface stops; at this point the substrate is removed from the sticky tack and placed onto a flat surface in the glove box, which is why flat substrates work best (although the author has also used a hanging drop rock and roll method on an inverted curved beaker). The entire process takes about 5 min. Rotating the substrate on the sticky tack before detaching it avoids breaking the thinner substrates. The film is left to dry for 1 d in the glove box and an additional day in air on the lab bench or in a fume hood to assure complete removal of solvent (see Note 6).
3.4. Assessment of Orientation
After the sample on mica or glass cover slip has dried for at least 1 d on the lab bench, it is first examined by eye; gross misorientation appears as large holes, crusted lipid, and a frosted appearance. If there is no evidence of gross misorientation, the sample is examined using an optical microscope under crossed polarizers as described by Asher and Pershan (13). If the film is completely smooth, well oriented, and homogeneous, there will be a uniform black field under crossed polars. Any defects retard the light along the defect, which results in loss of total cancellation of crossed polarized light, giving a bright spot or streak. Many curious and astonishing structures have been observed by the author and her students (see also Note 7). The sample in Fig. 2 was prepared by pouring DPPC solubilized in 3:1 chloroform:methanol onto a cleaned glass microscope slide; this was rocked briefly to evenly distribute the lipid solution over the substrate. It was then left to dry on the lab bench without any further movement, similar to the procedure of Seul and Sammon (7), except that the amount of lipid in this sample was for a 10-µm thick film. The visual appearance of the film after drying on the lab bench is shown in Fig. 2A. The various parts of the same film visualized under crossed polars in the optical microscope are shown in Figs. 2B–D. A photograph of a sample prepared using the rock and roll procedure is shown in Fig. 6.
Fig. 2.
(A) Visual appearance of a sample of DPPC dried from chloroform:methanol (3:1) onto a cleaned, stationary glass microscope slide. (B–D) Defects observed using crossed polars in the optical microscope on the far left side of the sample shown in A. The lipid in the center of the slide appeared dark gray under crossed polars, i.e., fairly well oriented but not uniform. Lipid was not deposited on the far right side of the slide. Results like these were the motivation for the rock and roll procedure.
Fig. 6.
DPPC dissolved in chloroform:methanol (3.5:1) was oriented onto a silicon substrate using the rock and roll procedure and trimmed as described in Subheading 3.6.
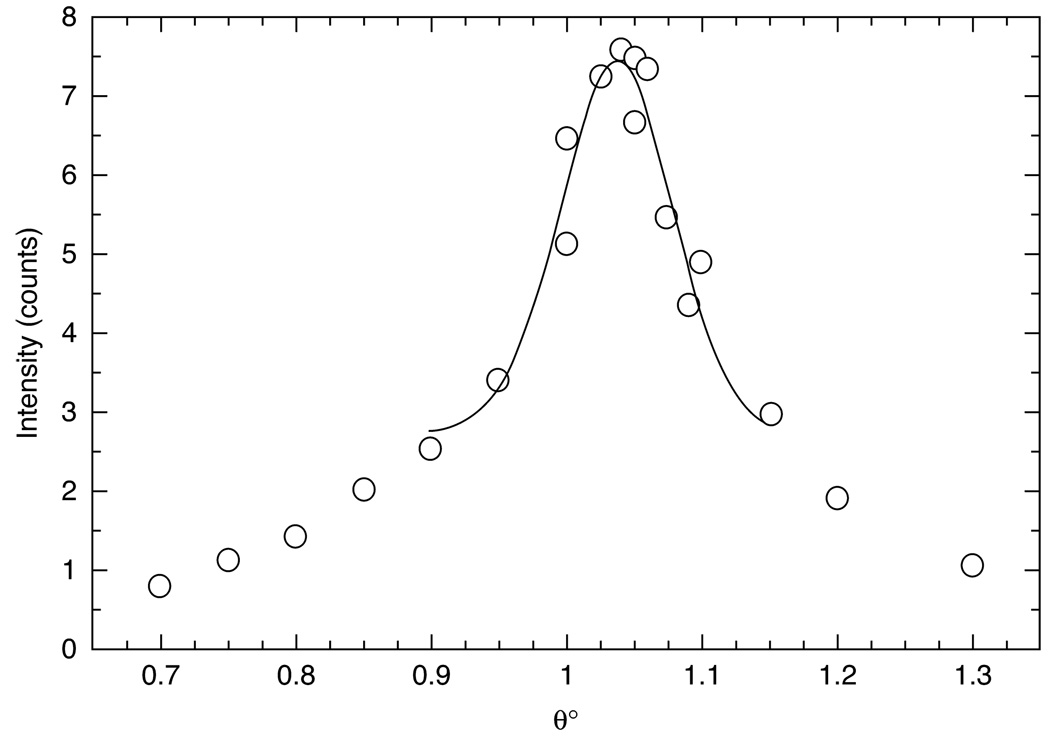
Once the sample appears uniform under crossed polars, it can be X-rayed either in the dried or hydrated state using the lab-rotating anode or during the synchrotron experiment. The measurement of mosaic spread is obtained by rotating the flat sample in 0.005° steps through a chosen Bragg θ angle. The Bragg peak intensity is measured and plotted vs angle of rotation. This is fit with a Gaussian function, and the sigma of the Gaussian function is reported in degrees as the mosaic spread. An example of a rocking scan to determine mosaic spread is shown in Fig. 3.
Fig. 3.
Mosaic spread of a 10-µm thick DMPC sample oriented onto a silicon wafer. This rocking scan was centered on the second order Bragg reflection from a lamellar stack of hydrated bilayers. Each data point was collected at a separate angle of incidence. The half-width of the Gaussian fit (solid line) is 0.08°.
3.5. AFM to Characterize Uniformity and Smoothness of Sample Films
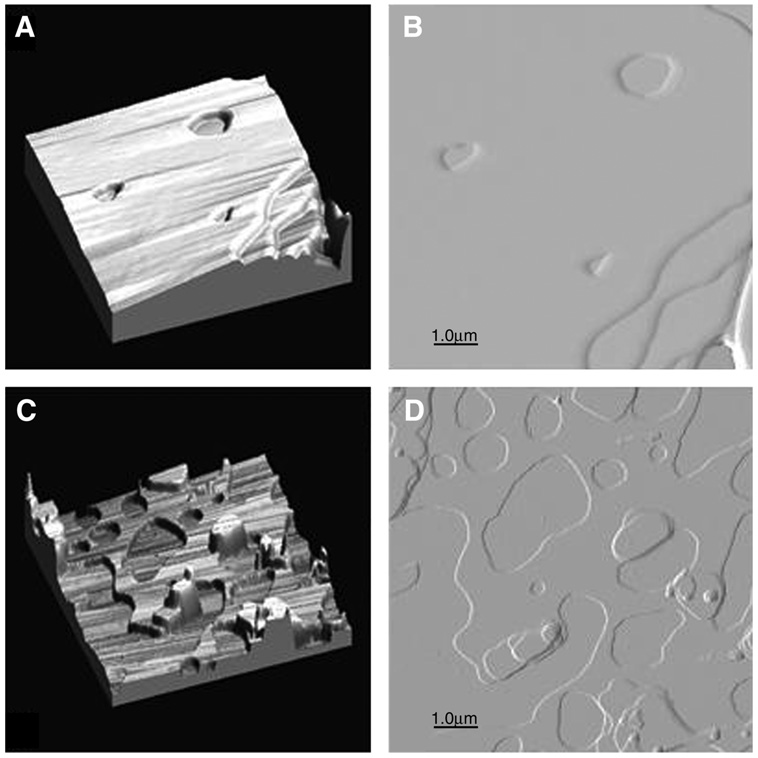
With the help of Justin Legleiter, a talented graduate student in Chemistry at Carnegie Mellon University, the author used AFM to probe microscopic differences between lipid films prepared using the rock and roll procedure, with those using spin-coating. Details of the tapping mode AFM are presented in ref. 11. Many variables such as spinning speed, method of application of the lipid in solvent (dropwise or stationary), concentration of lipid in solvent, and choice of solvent were attempted. The samples shown below in this comparison study were deposited on 3×3 cm2 pieces of freshly cleaved mica and the thicknesses were approx 200 bilayers. The AFM 3D and the phase images of a 1.1 mg DMPC sample dissolved in 100 µL TFE:chloroform (2:1), and prepared using the rock and roll method are shown in Fig. 4A,B, respectively. Three steps of one bilayer thickness are shown in the lower right-hand corner of both Fig. 4A,B. These are sometimes called edge dislocations. The corresponding images for a sample prepared by spin-coating are shown in Fig. 4C,D. The sample was 0.94 mg of DMPC in 100 µL TFE:chloroform (2:1), added drop-by-drop as the sample was spinning at approx 20g. The main difference between the images in Fig. 4 is that in the spin-coated sample there are many large holes, and several protrusions of one or more bilayers. Although the lipid appears to be flat and well oriented in Fig. 4C,D, the film is less homogeneous and extensive than that in Fig. 4A,B.
Fig. 4.
Tapping mode AFM of DMPC deposited from TFE:chloroform spin-oating (2:1) onto freshly cleaved mica either by the rock and roll method (A and B) or by spin-coating (C and D). The 3D images have a 25-nm height range.
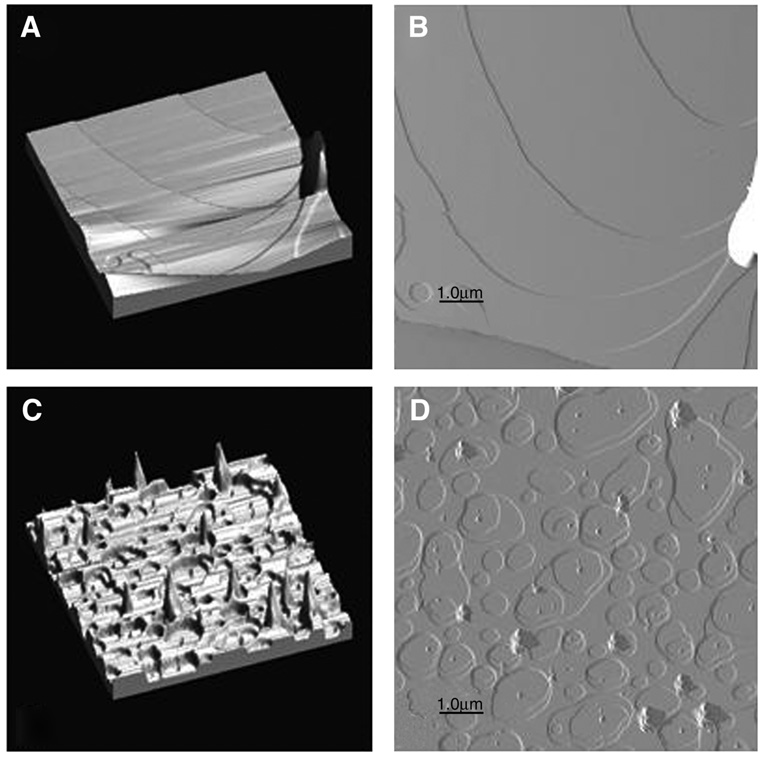
Another comparison of these two methods was carried out using the neat solvent, isopropanol, first reported by Seul and Sammon (7). Figure 5A,B show the 3D and phase images of a DMPC sample (1.2 mg DMPC dissolved in100 µL isopropanol) prepared by the rock and roll method. The figure shows that the surface is quite smooth, with four extensive steps of one bilayer thickness each. For Fig. 5C,D, the DMPC was dissolved in 200 µL isopropanol, placed onto the freshly cleaved mica and then spun at approx 20g until no more solvent remained (about 2 min). As in the case of TFE:chloroform, the film prepared by the rock and roll method is shown to be smoother, more homogenous, and more uniform. Another comparison using neat TFE gave the same result. A combined technique of rock and roll followed by spin-coating during the final drying step resulted in less loss of material, but did not produce films of significantly better orientation than the rock and roll procedure alone, as judged by polarizing microscopy.
Fig. 5.
Tapping mode AFM of DMPC from isopropanol deposited onto freshly cleaved mica either by the rock and roll method (A and B) or by spin-coating (C and D). Height ranges were 25 and 50 nm for 5A,C, respectively.
An additional benefit of the rock and roll procedure, when compared with spin-coating, is that no material is lost because of spinning; this result is particularly important when expensive peptides are added to the films. Thus, these AFM studies show that (at least for films of thickness ~200 bilayers) the rock and roll method produces better-oriented, smoother, and more uniform films.
3.6. Packing Samples
The sample on the 1.5×3 cm2 silicon wafer is then trimmed to a thin strip of 4 or 5×30 mm2 long, by removing sample along both long edges of the silicon wafer using a fresh single-edge razor blade, followed by wiping off any remaining lipid with a dry cotton swab. One reason for trimming the samples is to remove lipid that may be less well oriented near the edges of the wafer. The other reason concerns the X-ray experiment (5). The trimmed samples are stored in multicompartment plastic boxes within a large glass dessicator at 4°C. The samples are usually prepared 1–3 wk before a synchrotron trip, but samples on all three substrates are quite stable for several months when stored in a dessicator at 4°C. They are carried to the Cornell high-energy synchrotron source (CHESS) in the dessicator in an ice-filled container. A picture of a sample prepared in this way is shown in Fig. 6.
3.7. Sample Hydration Through the Vapor
Hydration of lipid bilayers through the vapor phase has been difficult (14). To summarize briefly, although it was possible to fully hydrate gel phase bilayers through the vapor (6,15,16), it was at first impossible to hydrate through the vapor in the more biologically relevant fluid phase (17,18). The seeming paradox (i.e., that the vapor above pure water is not at 100% relative humidity even though it is in equilibrium with pure water) generated controversy in several labs (19). The trick that worked in the gel phase was to cool the sample relative to the surrounding vapor using a Peltier cooler under the sample substrate (6); however, this solution was not sufficient to condense the considerably higher water content into a fluid phase lipid. The key to successfully hydrating a sample through the vapor was excellent temperature control; when this is done, the vapor pressure paradox does not exist (19,20). The researchers have been fortunate to be able to use two X-ray chambers for hydrating lipids in the fluid phase. The details and pictures of these chambers are described in refs. 5 and 21 and will not be repeated in this chapter. One important design feature is a rapid flow to the chamber of the coolant that is temperature-controlled by an external water bath. In addition, thick or double walls (double 6-µm thick mylar windows for entry and exit of X-rays) and additional foam insulation on the outside of the chamber for extreme temperatures (>50°C and <10°C) are needed. A final important feature is to add a water-filled piece of filter paper (suggestion of Peter Rand) to the inside top of the chamber with fingers that extend into the hydrating water reservoir. This increases the water evaporation surface and helps to decrease hydration equilibration times. When the Peltier current under the sample is set to cool the sample by 0.1°C relative to the hydration chamber temperature (5), hydration at 30°C occurs in about 30 min. If a slower hydration is desired (as when collecting data at many D-spacings within 10 Å of full hydration), a smaller current is used. The current can also be reversed to slow down the hydration or slightly dry out the sample by heating the surface of the Peltier in contact with the sample. Controlling the hydration speed and extent this way was a huge achievement, allowing data collection of many samples during a single trip to CHESS.
The chamber uses flat samples, which are rotated during the X-ray data collection. Alternatively, mica can be used as substrate (described in Subheading 2., step 3). The mica with lipid sample is curved and glued onto a cut, 50-mL glass beaker. Devcon 5 min epoxy and clamps are used to hold the mica in place during the drying of the epoxy. Since the sample is curved, all the angles of incidence that are obtained by rotating a flat sample can be obtained in one scan at a grazing angle of incidence. The cut, glass beaker sits on a semicylinder of solid aluminum, which is in contact with the Peltier cooler (see Fig. 7). One small drawback to using this sample is that there are several sharp and intense mica reflections that are difficult to subtract out with a background scan, resulting in their inclusion in the final data. However, as the diffuse background scattering from mica is very low, these sharp peaks were found to be acceptable.
Fig. 7.
DPPC dissolved in 2:1 chloroform:methanol is deposited onto mica, which is glued to a cut, glass, 50-mL beaker. This is in contact with a curved aluminum holder; Dow heat sink compound is normally spread under the beaker for better thermal contact during the hydration experiment. There is a Peltier cooler under the sample (not visible); a second Peltier cooler is shown to the right of the holder as a demonstration. An alternate sample holder for flat silicon substrates does not have the semicylindrical aluminum piece.
Acknowledgments
I would like to thank my husband and colleague, Prof. John Nagle, for continued collaboration and support, and Dr. Norbert Kučerka for supplying the mosaic spread graph. I would also like to thank Dr. Horia Petrache for help with construction of the hydration chamber described in ref. 5. Supported by National Institutes of Health grant no. GM44976 (JFN).
Footnotes
Thicker films up to 40 µm were attempted, but the mosaic spread, or degree of misorientation, increased. Thinner films were tried (2 µm), but the intensity of diffuse scattering was reduced substantially when compared with the reflectivity from the substrate and general background. Films between 4 and 10 µm are a compromise between satisfactory X-ray signal-to-noise and low mosaic spread. However, films less than 8 µm are sometimes not successful because there is not enough lipid to cover the surface homogeneously.
When teaching the rock and roll method to colleagues, the author reached success only when a suitable glove box was available. The use of a small, plastic disposable glove box was not successful because the atmosphere in the glove box rapidly became too humid due to perspiration from the hands. The use of thin neoprene gloves is possible, yet somewhat awkward, given the necessary manipulations. Even in a large glove box, the humidity sometimes rises above 80% RH when gloves are not worn. If this occurs, then the ratio of the hydrophobic/hydrophilic solvent should be increased: for example, chloroform:TFE, from 1:1 to 2:1.
Hexafluoroisopropanol’s label warns that it is dangerous to all organs. Wear neoprene gloves and work with this solvent in the hood only. This procedure is different from use of the glove box described in Subheading 3.3., but this solvent is unique in that it can produce quite uniform films while drying quickly. This solvent is only successful for some lipids and should be avoided if possible.
Films formed from cholesterol mixtures are usually not as well oriented as phospholipids, but the orientation of mixtures of lipids and cholesterol is improved by annealing at 10–20°C above the main phase-transition temperature of the highest melting lipid. This is carried out in a humid environment in an annealing chamber with a water-filled sponge attached above and below the lipid sample for 6–12 h.
For cleaning the silicon wafers, the author has tried a chromic acid wash, followed by copious rinsing with Barnstead/Thermolyne (Dubuque, IA) nanopure water, then an HCl wash, followed by copious rinsing with Barnstead nanopure water, with or without a final step of swabbing with high-pressure liquid chromatography chloroform using a cotton swab. The results are similar with or without the two acid washes before the chloroform swabbing. The final step of chloroform swabbing imparts a slightly hydrophobic surface to the wafers, which is beneficial. On one occasion the author tried to save time by using a solution of hot, concentrated Contrad (Decon Labs, King of Prussia, PA). This is a basic surfactant solution that is used for washing glassware, which was recommended by a colleague. This was nearly disastrous: the Contrad etched the silicon wafer surface so that it lost its shine; this was not restored with a hydrofluoric acid dip, as used for the removal of a silicon oxide layer. The mosaic spread from lipid samples prepared on these wafers was worse than that with the normal chloroform swabbing technique, and there was a large diffuse scatter from these substrates, similar to that from glass substrates. Therefore, it is best not to aggressively clean the polished silicon wafer surface.
Some care is needed to master the rock and roll technique. The rocking should be slow and shallow at first, when there is still much of the solvent on the substrate. Excessive rocking at this time can cause the lipid solution to fall off the edges of the substrate. The angle and speed of rocking can be increased as the sample dries. Rocking too slowly at this time, or not forcing the lipid solution to roll out to the edges of the substrate, produces a poorly oriented sample.
Defects can be described as volcanoes, feathers, tubules, squigglies, simple crosses, and fatty streaks. If there is water present, a familiar “Maltese cross” appears because of the circular shape of the multilamellar vesicles that retard the light in a radial fashion (22). If Maltese crosses are observed, the sample is not well oriented and should be discarded. Simple crosses are more prominent if the methanol content of the solvent mixture is too high, and squigglies and feathers are more prominent when the chloroform content of the solvent mixture is too high. Squigglies and feathers are more disruptive than a surface layer of methanol-induced simple crosses. Another feature, termed “Grandjean terraces” and described nearly 100 yr ago (23), looks like steps or plateaus (see Fig. 4A,B and Fig 5A,B). These edge dislocations, unlike in the thin films of Seul and Sammon (7), do not degrade the orientation of the sample, probably because they are largely confined just to the outer surface of thick films.
References
- 1.Luzzati V. X-Ray Diffraction Studies of Lipid-Water Systems. In: Chapman D, editor. Biological Membranes. London: Academic Press; 1968. pp. 71–123. [Google Scholar]
- 2.Tristram-Nagle S, Nagle JF. Lipid Bilayers: Thermodynamics, structure, fluctuations, and interactions. Chem. Phys. Lipids. 2004;127:3–14. doi: 10.1016/j.chemphyslip.2003.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lyatskaya Y, Liu Y, Tristram-Nagle S, Katsaras J, Nagle JF. Method for obtaining structure and interactions from oriented lipid bilayers. Phys. Rev. E. 2001;63 doi: 10.1103/PhysRevE.63.011907. 011907 (1–9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y, Nagle JF. Diffuse scattering provides material parameters and electron density profiles of biomembranes. Phys. Rev. E. 2004;69 doi: 10.1103/PhysRevE.69.040901. 040901(R). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kučerka N, Liu Y, Chu N, Petrache HI, Tristram-Nagle S, Nagle JF. Structure of fully hydrated fluid phase DMPC and DLPC bilayers using X-ray scattering from oriented multilamellar arrays and from unilamellar vesicles. Biophys. J. 2005;88:2626–2637. doi: 10.1529/biophysj.104.056606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tristram-Nagle S, Zhang R, Suter RM, Worthington CR, Sun W-J, Nagle JF. Measurement of chain tilt angle in fully hydrated bilayers of gel phase lecithins. Biophys. J. 1993;64:1097–1109. doi: 10.1016/S0006-3495(93)81475-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seul M, Sammon MJ. Preparation of surfactant multilayer films on solid substrates by deposition from organic solution. Thin Solid Films. 1990;185:287–305. [Google Scholar]
- 8.Mennicke U, Salditt T. Preparation of solid-supported lipid bilayers by spin-coating. Langmuir. 2002;18:8172–8177. [Google Scholar]
- 9.Hallock KJ, Wildman KH, Lee D-K, Ramamoorthy A. An innovative procedure using a sublimable solid to align lipid bilayers for solid-sate NMR studies. Biophys. J. 2002;82:2499–2503. doi: 10.1016/S0006-3495(02)75592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dittmer JC, Lester RL. A simple, specific spray for the detection of phospholipids on thin-layer chromatograms. J. Lipid Res. 1964;5:126–127. [PubMed] [Google Scholar]
- 11.Tristram-Nagle S, Liu Y, Legleiter J, Nagle JF. Structure of gel phase DMPC determined by X-ray diffraction. Biophys. J. 2002;83:3324–3335. doi: 10.1016/S0006-3495(02)75333-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petrache HI, Tristram-Nagle S, Harries D, Kučerka N, Nagle JF, Parsegian VA. Swelling of Phospholipids by Monovalent Salt. J. Lipid Res. 2006;47:302–309. doi: 10.1194/jlr.M500401-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asher SA, Pershan PS. Alignment and defect structures in oriented phosphatidylcholine multilayers. Biophys. J. 1979;27:393–421. doi: 10.1016/S0006-3495(79)85225-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rand RP, Parsegian VA. Hydration forces between phospholipid bilayers. Biochim. Biophys. Acta. 1989;988:351–376. [Google Scholar]
- 15.Levine YK. X-Ray Diffraction Studies of Membranes. Prog. Surf. Sci. 1973;3:279–352. [Google Scholar]
- 16.Katsaras J, Yang DSC, Epand RM. Fatty-acid chain tilt angles and directions in dipalmitoylphosphatidylcholine bilayers. Biophys. J. 1992;63:1170–1175. doi: 10.1016/S0006-3495(92)81680-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jendrasiak GL, Hasty JH. Hydration of phospholipids. Biochim. Biophys. Acta. 1974;337:79–91. doi: 10.1016/0005-2760(74)90042-3. [DOI] [PubMed] [Google Scholar]
- 18.Smith GS, Safinya CR, Roux D, Clark NA. X-ray study of freely suspended films of a multilamellar lipid system. Mol. Cryst. Liq. Cryst. 1987;144:235–255. [Google Scholar]
- 19.Nagle JF, Katsaras J. Absence of a vestigial vapor pressure paradox. Phys. Rev. E. 1999;59:7018–7024. doi: 10.1103/physreve.59.7018. [DOI] [PubMed] [Google Scholar]
- 20.Katsaras J. Adsorbed to a rigid substrate, dimyristoyl-phosphatidylcholine multibilayers attain full hydration in all mesophases. Biophys. J. 1998;75:2157–2162. doi: 10.1016/S0006-3495(98)77658-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katsaras J, Watson MJ. Sample cell capable of 100% relative humidity suitable for x-ray diffraction of aligned lipid multibilayers. Rev. Sci. Inst. 2000;71:1737–1739. [Google Scholar]
- 22.Tristram-Nagle S, Wingert LM. A thermotropic study of 1-deoxy-1-(N-methyloctanamido)-D-glucitol (MEGA-8) using microscopy, calorimetry and x-ray diffraction. Mol. Cryst. Liq. Cryst. 1990;188:41–56. [Google Scholar]
- 23.Grandjean F. The orientation of anisotropic liquids on the surface of crystals. Bull. Soc. Franc. Min. 1916;39:164–213. [Google Scholar]