Abstract
Since the mTOR pathway is commonly deregulated in human cancer, mTOR inhibitors, rapamycin and its derivatives, are being actively tested in cancer clinical trials. Clinical updates indicate that the anti-cancer effect of these drugs is limited, perhaps due to rapamycin-dependent induction of oncogenic cascades by an, as yet, unclear mechanism. As such, we investigated rapamycin-dependent phosphoproteomics and discovered that 250 phosphosites in 161 cellular proteins were sensitive to rapamycin. Among these, rapamycin regulated 4 kinases and 4 phosphatases. A siRNA-dependent screen of these proteins showed that AKT induction by rapamycin was attenuated by depleting cellular CDC25B phosphatase. Rapamycin induces the phosphorylation of CDC25B at Serine375, and mutating this site to Alanine substantially reduced CDC25B phosphatase activity. Additionally, expression of CDC25B (S375A) inhibited the AKT activation by rapamycin indicating phosphorylation of CDC25B is critical for CDC25B activity and its ability to transduce rapamycin-induced oncogenic AKT activity. Importantly, we also found that CDC25B depletion in various cancer cell lines enhanced the anti-cancer effect of rapamycin. Together, using rapamycin phosphoproteomics, we not only advance the global mechanistic understanding of rapamycin’s action in cancer but also demonstrate that CDC25B may serve as a drug target for improving mTOR-targeted cancer therapies.
Introduction
Mammalian target of rapamycin (mTOR) is a cellular 289 kDa protein mediating signals derived from both growth factors and nutrients and is known to regulate cell growth, proliferation and survival through controlling mRNA translation, metabolism, ribosome biogenesis and autophagy (1,2,3). The mTOR pathway is commonly deregulated in human cancer. For example, in human breast cancer, mTOR is commonly deregulated by loss of PTEN (30% of human breast tumor) (4), by mutation of PI3KCA (18–26%) (4) and by overexpression of Her 2 (15–30%) (5): all of which are associated with a poor prognosis for breast cancer patients (6,7,5). Similarly, in human prostate cancer, mTOR is commonly deregulated by genetic aberrations such as low expression of PTEN, increased PI3K activity and increased expression or activation of AKT in advanced prostate cancer (8,9,10). These aberrations also are indicators of a poor prognosis for prostate cancer patients(11,12). More importantly, long term androgen deprivation treatment for prostate cancer patients that reinforces the PI3K/AKT pathway also upregulates mTOR activation in prostate tumor (9,10). These abovementioned experimental and clinical data lead to the supposition that mTOR inhibitors (rapamycin and its derivatives) should be effective in treating human cancer. Unfortunately, recent clinical data indicates that rapamycin demonstrates therapeutic potential in only few types of human cancer: endometrial carcinoma, renal cell carcinoma and mantle cell lymphoma (13). These results could be explained by recent findings that mTOR inhibition by rapamycin phosphorylates and activates the oncogenic AKT and elF4E pathways while still suppressing the phosphorylation of p70S6K and 4E-BP1 (14) in cancer cells. However, the detailed molecular mechanisms regulating this rapamycin-dependent activation of oncogenic cascades are not clear. Progress toward understanding the underlying mechanisms is hindered by the limited number of known cellular targets for rapamycin. We recently improved the methodology for profiling the cellular phosphoproteome (15) and, using this technology, simultaneously profiled 6,179 phosphosites in cancer cells and identified 161 cellular proteins sensitive to rapamycin. Within these proteins, there are 4 kinases and 4 phosphatases, key mediators for cell signaling. We screened these proteins and found that depletion of cellular CDC25B blocked oncogenic AKT activation by rapamycin and enhanced the anti-cancer effect of rapamycin. Interestingly, we also discovered that a large percentage of the rapamycin-regulated proteins are involved in regulation of cellular transcription. These results demonstrate that rapamycin phosphoproteomics enables us to improve mTOR-targeted therapies, as well as advance the general mechanistic comprehension of rapamycin treatment in cancer.
Materials and Methods
Materials
The human cell lines HeLa, MCF-7 and Du145 were obtained from American Type Culture Collection (Manassas, VA, USA). The human H157 was kindly provided by Dr. Shi-Yong Sun (Winship Cancer Institute, Emory University School of Medicine, Atlanta, Georgia). All cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 5% fetal bovine serum (FBS), 1% penicillin/streptomycin and 1% L-glutamine (Invitrogen, Carlsbad, CA, USA). Anti-phospho-AKT (Ser473) antibody was from Upstate (Lake Placid, NY, USA). Anti-phospho-p38 (T180/Y182), p38, p-S6K1(T389) and p-eIF4E (S209) antibodies were from Cell Signaling (Boston, MA, USA). Anti-GAPDH antibody and rapamycin were from Calbiochem (San Diego, CA, USA). Anti-CDC25B (C-20), anti-rabbit and anti-mouse secondary antibodies were from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA). Topflash reporter cells were kindly provided by Dr. Willert (University of California San Diego). pCRE-Luc reporter plasmid is from Stratagene (La Jolla, CA).
Cell Culture, SILAC-labeling and Sample preparation for MS
GIBCO SILAC DMEM basal cell culture medium (Invitrogen, Carlsbad, CA) containing 2 mM L-Glutamine, 10% dialyzed fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA) and 100 U/ml penicillin and streptomycin was supplemented with 100 mg/L L-Lysine and 20 mg/L L-Arginine or 100 mg/L [U-13C6]-L-Lysine and 20 mg/L [U-13C6, 15N4]-L-Arginine (Invitrogen, Carlsbad, CA) to make the “Light” SILAC or “Heavy” SILAC culture media, respectively. HeLa cells were obtained from ATCC and were propagated in SILAC medium for more than nine generations to ensure nearly 100% incorporation of labeled amino acids before the experiment was performed. Before cell treatment, both heavy and light labeled cells were cultured overnight in the corresponding SILAC media without FBS. “Heavy” cells were treated for 15 minutes with EGF (R&D Systems, Minneapolis, MN). “Light” cells were pretreated with rapamycin for one hour followed by addition of EGF. After being washed with cold PBS buffer, HeLa cells were harvested and lysed in an Extraction/Loading buffer from the TALON PMAC Phosphoprotein Enrichment Kit (Clontech, Mountain View, CA) supplemented with 10 mM sodium fluoride and a cocktail of protease inhibitors (Roche, Indianapolis, IN). “Light” cell lysate and “Heavy” lysates were mixed at 1:1 ratio and 16 mg of the mixed lysate was loaded equally onto two phosphoprotein affinity columns (Talon PMAC, Clontech) following the manual. The eluate was reduced with DTT, and then alkylated with iodoacetamide. The resulting solution was dialyzed against 1M urea/100 mM NH4HCO3 at 4°C overnight. The samples were digested with trypsin (Promega, Madison, WI). The digestion was subjected to solid phase extraction by Extract-clean SPE C18 column (Alltech Associates, Deerfield, IL) and later lyophilized. The lyophilized peptide sample was loaded onto four IMAC spin columns (Pierce Biotechnology, Rockford, IL). The IMAC-retained peptides were then washed and eluted as recommended by the manufacturer’s manual. Formic acid was added to the phosphopeptide-enriched eluate to a final concentration of 4%, and the eluate was subsequently centrifuged to remove any particulate matter prior to analysis by MudPIT(16).
Purified phosphopeptide samples were centrifuged to remove any particulate matter before being analyzed by LC/LC-MS/MS using an online HPLC system (Nano LC-2D, Eksigent) coupled with a hybrid LTQ Orbitrap mass spectrometer (Thermo Electron). The LC/LC and MS/MS were performed as previously described (15).
Acquired MS/MS (MS2) and MS/MS/MS (MS3) spectra were subjected to SEQUEST searches against the EBI-IPI human database (version 3.17 released 05-09-2006) attached with common contaminants (e.g. proteases and keratins) with its reverse decoy for the assessment of false positive rates. Phosphorylation searches were performed where serine, threonine, and tyrosine were allowed to be differentially modified as previous described (15). The SEQUEST identifications were additionally filtered using a false positive cut-off by the DTASelect program based on XCorr and DeltaCN scores using a reversed database approach (17). All identified phosphopeptides were subjected to relative quantification analysis using the program Census (18). The Census program quantifies relative abundances of light and heavy versions of precursor peptides identified by MS2 and/or MS3 spectra. The R-square statistics were calculated by regression analysis of the profile of the heavy-peptide and the profile of the light-peptide using the Census program. A p-value for each phosphopeptide ratio was calculated according to the distribution of peptide ratio measurements of the 1:1 heavy:light mixed lysates.
siRNA-dependent function screen
Eighteen rapamycin-sensitive kinases and phosphatases were screened for their ability to mediate AKT activation by rapamycin. HeLa cells were individually transfected with siRNAs (Dharmacon, Lafayette, CO, USA) of these kinases and phosphatases. After 72 hours, these cells were treated with 100 nM rapamycin for 3 hr. Western blot was used to detect phosphorylation changes in AKT at Ser-473.
Phosphatase assays
Expression plasmid encoding Myc-tagged CDC25B or CDC25B (S375A) (a gift from Dr. Brian Gabrielli, NCI) were transfected into HEK293 cells. After 48 h, CDC25B protein was immunoprecipitated using anti-Myc antibody. These immunoprecipitates were incubated with 300 µM OMFP (3-O-methyl fluorescein phosphate; Sigma, St. Louis, MO) in 50 mM Tris-HCl, pH 8.2, 50 mM NaCl, 1 mM DTT, 20% glycerol for 15 minutes at 30 °C. Hydrolysis of OMFP to OMF was monitored at 477 nm.
Dual luciferase assay
HeLa cells were plated in 24-well plates in DMEM medium containing 2 mM L-Glutamine and 10% fetal bovine serum (FBS). Cells were cotransfected with various promoter-firefly luciferase reporters (200 ng DNA/well) and pRL-TK reporter (20 ng DNA/well) using GeneJet DNA In Vitro Transfection reagent (SignaGen Laboratories, Gaithersburg, MD) following the manufacturer’s protocol. After 24 hr, cells were changed to serum-free medium and treated with indicated reagents. The resultant cells were lysed and analyzed for luciferase activity using the dual luciferase assay (Promega, Madison, WI) according to the manufacturer’s instruction. All experiments were repeated at least three times with similar results.
Results
Rapamycin phosphoproteome generation and results
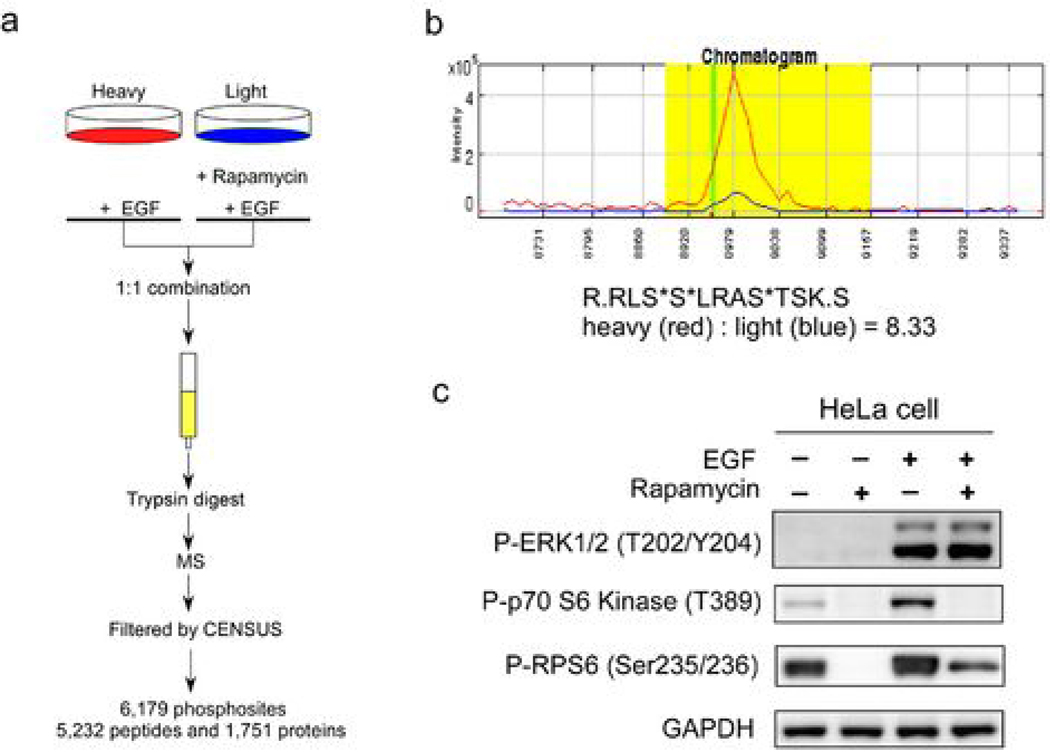
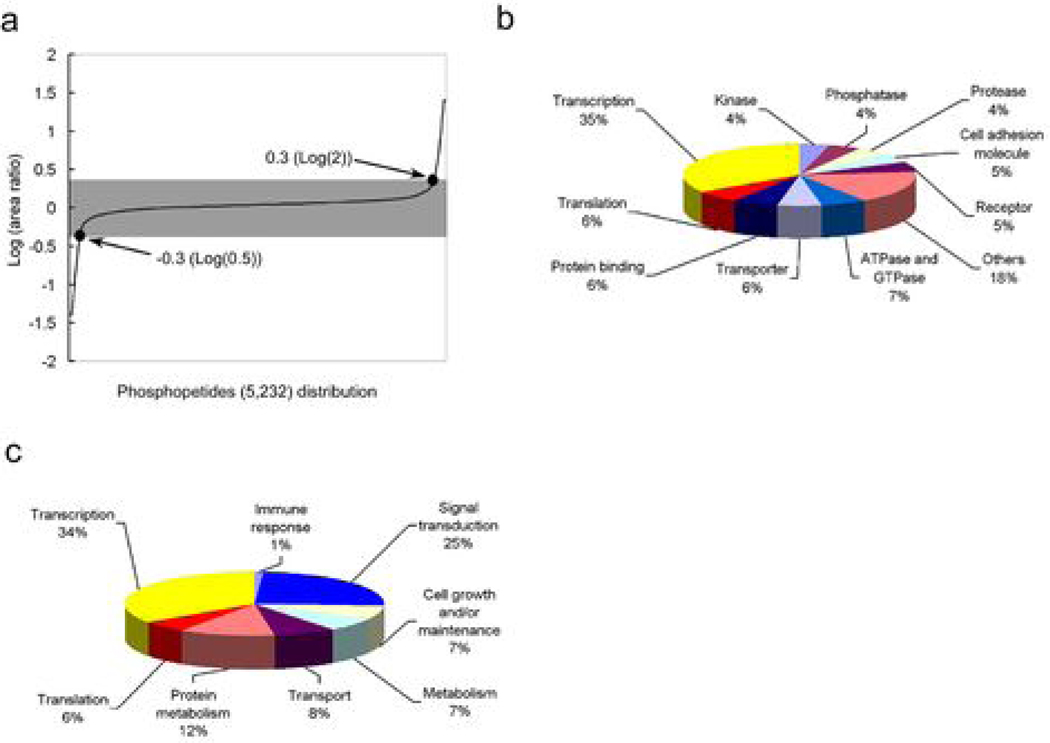
Rapamycin inhibits the ability of mTOR protein kinase to phosphorylate cellular proteins thereby blocking the cancerous signals derived from mTOR. Therefore, to profile rapamycin-modulated cellular proteins, we compare the phosphoproteome of two growth factor induced-cell cultures pretreated with or without rapamycin. Briefly, cells in normal media (light culture) were treated with rapamycin and cells grown in media containing stable-isotopes (heavy culture) were treated with vehicle. These two populations of cells were stimulated with growth factor for 15 min, after which the two populations of cells were harvested, mixed at a 1:1 ratio, and subjected to double IMAC purifications followed by MudPIT LC/MS/MS analysis (15). Peptide sequences were obtained from the MS2 and MS3 spectra by SEQUEST software. Phosphopeptides were obtained by filtering total peptides by DTASelect 2.0 using the default settings, followed by quantifying the abundance (peak area) of the light and heavy phosphopeptides by CENSUS 1.05 using the default filter setting. We detected 6,179 phosphosites derived from 1,751 phosphoproteins (Figure 1a). As an example that this phosphoproteomic profiling succeeded, the phosphorylation of ribosomal protein S6 (RPS6) at S235 and S236 is known to be controlled by mTOR and inhibited by rapamycin (19). Our rapamycin phosphoproteome data showed that a phosphopeptide from RPS6 was phosphorylated at S235, S236 and S240 and the phosphorylation was inhibited 8.33 fold by rapamycin (Figure 1b). This was verified by Western blotting using an anti-phospho RPS6 antibody (Figure 1c). To quantify the phosphorylation change for each phosphopeptide, we used the Census software to calculate area ratio (AR), defined as the ratio of the “light” peak area over the “heavy” peak area in the chromatogram (Fig 1b). An R-square statistic was provided to indicate the accuracy of each area ratio measurement (Supplemental Table 1). The 1:1 mixed un-enriched lysates were analyzed by LC/LC-MS/MS to measure ratios of heavy and light peptides. The distribution of the log ratios of base 2 of this 1:1 mixture was studied. The mean of the log 2 ratios is −0.0892 and standard deviation is 0.2546. Based on this distribution, the 2-sigma cutoffs are log 2 ratio > 0.4200 or log 2 ratio < −0.5984, corresponding to ratios of >1.34 and < 0.66, respectively. Because we believe phosphopeptides more differentially expressed would also be more biologically interesting, we chose to use a more stringent 2-fold cutoff. Using >2 or <0.5 as the threshold for area ratio, we found that the phosphorylation of 250 sites (161 proteins) out of 6,179 sites (1,751 proteins) was significantly modulated by rapamycin (Figure 2a & 2b). In other words, only about 4% of the total phosphosites detected showed significant alteration after rapamycin treatment. Among these 250 rapamycin-sensitive phosphosites, only 3 have been reported previously as downstream targets of mTOR (19) (Figure 2b).
Figure 1. Experimental design for rapamycin-dependent phosphoproteomic profiling.
(a) “Heavy” cells were pretreated with 25 nM rapamycin for one hour followed by 15 min treatment with 10 ng/ml EGF. “Light” cells were only treated with 10 ng/ml EGF for 15 min. Heavy and light cell lysates were combined in a 1:1 ratio. Phosphorylation of each peptide was quantified by measuring the peak area of light and heavy peptides in the MS spectra with the CENSUS program. A total of 6,179 phosphosites were detected. (b) A chromatogram of heavy and light phosphopeptides of RPS6 (R.RLS*S*LRAS*TSK.S), showing a rapamycin-dependent 8.33 fold inhibition by area ratio. Red line: heavy phosphopeptide treated with EGF; Blue line: light phosphopeptide treated with EGF and rapamycin. (c) Western blot analysis of cell lysates showing modification of the phosphorylation of ERK1/2, p70S6K and RPS6 proteins by the indicated treatment, using anti-phospho-p70S6K (S389), anti-phospho-ERK1/2 (T202/Y204) and anti-phospho-RPS6 S235/S236 antibodies. GAPDH was used as loading control.
Figure 2. Results of rapamycin-dependent phosphoproteomic analysis and categorization of rapamycin-regulated proteins by their activities and functions.
(a) Distribution of the area ratio (AR) of phosphopeptides detected in rapamycin phosphoproteomics. (b) Summary of phosphosites, phosphopeptides and phosphoproteins identified in rapamycin phosphoproteomics. (c) Categorizing rapamycin-regulated proteins by their biological activities and (d) by their cellular functions. (94 and 99 rapamycin-modulated proteins without known activity or function, respectively, were excluded from these two charts).
Categorizing rapamycin-regulated proteins by their activities and functions
To perceive the scope of rapamycin action in cancer cells, we used the human protein reference database (HPRD) to classify the 161 rapamycin-sensitive proteins by their cellular activities and functions (Figure 2c and 2d). Previous studies indicated that rapamycin exerts its anti-cancer function by inhibiting protein translation in cancer cells (20). Interestingly, in our screen the largest percentage (35%) of the rapamycin-regulated proteins were involved in regulation of transcription and not translation (6%) (Figure 2c). Additional examination showed that 32 out of 345 (9.3%) transcription-related proteins and 6 out of 112 (5.4%) translation-related proteins detected in phosphoproteomic analysis with their phosphorylation significantly altered by rapamycin (Supplemental Table 1). Thus, rapamcyin has significant effect on 9.3% of transcription regulators detected versus 5.4% of translational regulators detected. This data suggested that the cancer-inhibitory effect of rapamycin may also rely on its regulatory role in mRNA transcription.
CDC25B mediates the activation by rapamycin of the oncogenic Akt pathway
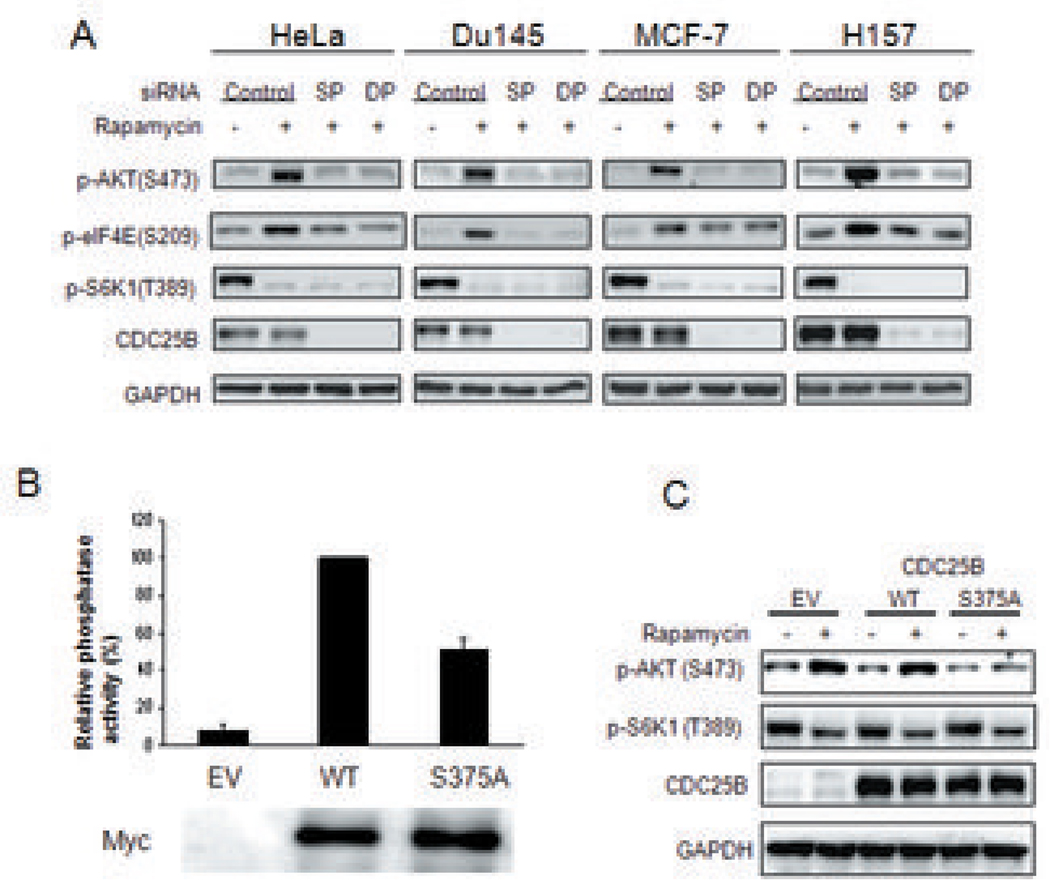
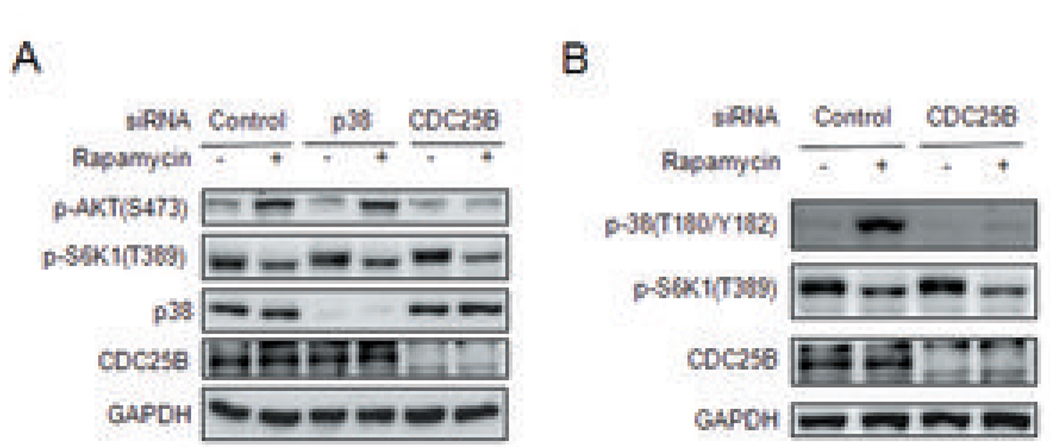
Kinases and phosphatases are critical signal mediators, and thus excellent cancer drug targets. We found that rapamycin modulates 4 kinases and 4 phosphatases (Figure 2c) and suspected that these kinases/phosphatases may transduce the oncogenic signal induced by rapamycin. We screened these kinases/phosphatases for their ability to mediate rapamycin-induced AKT and elF4E activation. We found that silencing CDC25B phosphatase blocked the activation of AKT and eIF4E by rapamycin in various human tumor cell lines (Figure 3a). Rapamycin induced phosphorylation of CDC25B at Serine 375 (supplemental table 1). To examine the effect of this phosphorylation on CDC25B’s phosphatase activity, we expressed either wild type (WT) or a mutant form of CDC25B [CDC25B (S375A)] in cells and subsequently analyzed their phosphatase activity. We found that mutation of Serine 375 to Alanine in CDC25B significantly attenuate its phosphatase activity (Figure 3b). We next expressed this CDC25B mutant in cells followed by rapamycin treatment and found that CDC25B (S375A) effectively blocked AKT activation by rapamycin while WT CDC25B did not (Figure 3c). In UV-induced DNA damage, p38 is the upstream regulatory kinase for CDC25B (21). However, silencing of p38 did not affect the activation of AKT by rapamycin (Figure 4a). Instead, surprisingly, CDC25B knockdown blocked p38 activation by rapamycin (Figure 4b) indicating that p38 is downstream of CDC25B in the activation of AKT. As blocking CDC25B inhibits the activation of the oncogenic pathways, AKT and elF4E, by rapamycin, we hypothesized that CDC25B knockdown in tumor cells should enhance the anti-cancer effects of rapamycin. To test this, in the presence or absence of rapamycin, we knocked down CDC25B in various types of cancer cells containing genetic mutations (supplemental table 2) that augment or deregulate the mTOR pathway. Moreover, as anti-mTOR treatment is only effective in few types of human cancer (e.g. kidney cancer and mantle B cell) (13), we want to test whether our finding may help enhancing the anti-mTOR treatment of some other kinds of cancers such as prostate (Du145), breast (MCF7), Non-small cell lung (H157) and cervical (HeLa) cancers. We found that CDC25B knockdown enhanced the anti-proliferative effect of rapamycin on cancer cells by 0.5 to 2.5 fold (Figure 5). Furthermore, CDC25B silencing only additively but not synergistically enhanced cell cycle effect by rapamycin (Supplemental figure 1). These data suggest that CDC25B mediates the rapamycin-induced activation of AKT, elF4E and p38 cascades, and CDC25B knockdown significantly promotes the inhibitory effect of rapamycin on the growth of cancer cells not only through blocking cell cycle progression but also by inhibiting oncogenic pathways activation by rapamycin.
Figure 3. CDC25B mediates activation of the oncogenic AKT pathway by rapamycin.
(a) Effect of CDC25B depletion on AKT activation by rapamycin in cancer cell lines. Cells were transfected with CDC25B-specific smartpool siRNA (SP), duplex siRNAs (DP) or control siRNAs for 60 hours. Cells were then treated with 100 nM rapamycin for 3 hours. The phosphorylation of AKT(S473), eIF4E (S209), S6K1(T389), and expression of CDC25B and GAPDH proteins were detected by immunoblotting (b) HEK293 cells were transfected with empty vector (EV) or expression plasmids encoding Myc-tagged wild type (WT) or mutant CDC25B with its Serine 375 mutated to Ala (S375A). 48 h post transfection, cells were collected and phosphatase activities of EV, WT, and S375A were assessed as described in Materials and Methods. Relative phosphatase activity in these cells was normalized using WT transfected cells whose value was taken as 100%. (c) EV or expression plasmids encoding WT or mutant CDC25B, CDC25B (S375A) were transfected into cells, followed by 100 nM rapamycin stimulation. The phosphorylation of AKT(S473), S6K1(T389), and expression of CDC25B and GAPDH proteins were detected by immunoblotting.
Figure 4. CDC25B mediates activation of the p38 MAP kinase by rapamycin.
(a) Depletion of cellular p38 has no effect in AKT activation by rapamycin. Cells were transfected for 60 hours with p38-specific, CDC25B-specific or control siRNAs as indicated, before being treated with 100 nM rapamycin for 15 min. Phosphorylation of AKT, S6K1, p38, CDC25B was detected by immunoblotting. GAPDH protein was used as loading control. (b) CDC25B depletion attenuated p38 activation by rapamycin. Cells were transfected with CDC25B-specific siRNA or control siRNA for 60 hours before treatment with 100 nM rapamycin for 15 min. Phosphorylation of p38 (T180/Y182), S6K1 (T389) and expression of CDC25B were detected by immunoblotting.
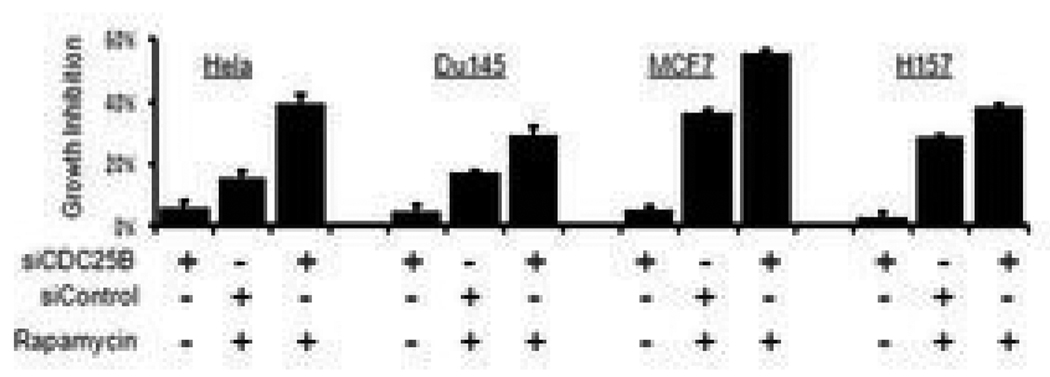
Figure 5. Cellular depletion of CDC25B enhances the anti-cancer effect of rapamycin.
CDC25B depletion enhanced the growth-inhibitory effect of rapamycin in different cancer cell lines. Cells were transfected with CDC25B-specific siRNAs or control siRNAs for 24 h followed by treatment with 100 nM rapamycin for another 48 h as indicated. Viable cell count was estimated by the MTT assay. % Growth inhibition was calculated relative to the MTT value from cells transfected with control siRNA only.
Discussion
The CDC25 family of proteins is comprised of dual specificity phosphatases that regulate cell cycle transitions, and are key targets of the checkpoint machinery to maintain genome stability (22) during DNA damage. Three isoforms of CDC25 have been identified in mammalian cells: CDC25A, CDC25B and CDC25C. CDC25A and CDC25B overexpression has been reported in many types of human cancers (22,23) but is insufficient to cause cancer (22) and the mechanism responsible for CDC25 overexpression is unclear. Nevertheless, CDC25A and CDC25B overexpression correlates with aggressive, high grade and late-stage tumors (22), as well as with a poor prognosis for cancer patients (24,22,25), possibly resulting from genome instability caused by the checkpoint-abrogating effect of their overexpression. However, CDC25 may contribute to tumorigenesis by other mechanism(s). Herein, we discovered that the anti-cancer drug, rapamycin, induced the phosphorylation of CDC25B at Serine 375 which is critical for its phosphatase activity, and depletion of cellular CDC25B inhibited the rapamycin-dependent activation of the survival/oncogenic AKT, elF4E and p38 pathways suggesting that CDC25B phosphatase activity is required for the activation of Akt and/or p38. It is possible that CDC25B might dephosphorylate and subsequently activate the upstream regulator(s) of AKT and/or p38, but the exact mechanism for this needs further investigation. Moreover, the depletion of cellular CDC25B resulted in the augmentation of anti-cancer effect of rapamycin on various types of tumor cells. This result indicates that CDC25B is the key mediator for supplying survival/oncogenic signals to tumor cells during mTOR-targeted cancer treatment and provide us with a potential drug target to improve the efficacy of anti-cancer drugs rapamycin and its derivatives.
It is well known that the mTOR/Raptor (rapamycin-sensitive) pathway is critical in modulating cellular translational machineries thereby regulating physiological and pathological responses in cell. However, the role of this pathway in altering cellular transcription apparatuses has been emphasized much less. Interestingly, 35% of the mTOR/Raptor-regulated cellular proteins identified herein are transcriptional factors/transcription regulators and only 6% are proteins controlling cellular translation processes (Figure 2C). Further analysis showed that 32 out of 345 (9.3%) transcription-related proteins and 6 out of 112 (5.4%) translation-related proteins detected in phosphoproteomic profiling have their phosphorylation levels significantly altered by rapamycin (Supplemental Table 1). It seems that the impact of the mTOR/Rapamycin pathway in altering mRNA transcription and the consequent effect on gene expression, biological function and disease progression may has been underestimated. Further studies to clarify molecular mechanisms and identify key cellular mediator(s) for these rapamycin-induced phosphorylation alterations of transcription regulators should facilitate the development of approaches to improve the efficacy or reduce unwanted side effects during the mTOR-targeted cancer therapies.
In conclusion, using rapamycin phosphoproteomics, we identified hundreds of novel rapamycin-targeted cellular proteins and their phosphorylation sites. This information enabled us to identify CDC25B as the key enzyme in mediating rapamycin induced oncogenic AKT activation. Importantly, we demonstrate that phosphoproteomic profiling of a certain therapeutic agent can not only identify potential drug target(s) to improve the efficacy of that therapeutic approach in disease treatment but also provide cellular information of possible beneficial and adverse side effects of a certain disease therapy when treating patients.
Supplementary Material
Acknowledgements
This work was supported by grants from the NIH, CA079871 and CA114059, (to J.-D.L.). This research was supported by funds from the Tobacco-Related Disease, Research Program of the University of California, 15RT-0104, (to J.-D.L.).
References
- 1.Bjornsti MA, Houghton PJ. The TOR pathway: a target for cancer therapy. Nat Rev Cancer. 2004;4:335–348. doi: 10.1038/nrc1362. [DOI] [PubMed] [Google Scholar]
- 2.Sawyers CL. Will mTOR inhibitors make it as cancer drugs? Cancer Cell. 2003;4:343–348. doi: 10.1016/s1535-6108(03)00275-7. [DOI] [PubMed] [Google Scholar]
- 3.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 4.Easton JB, Houghton PJ. mTOR and cancer therapy. Oncogene. 2006;25:6436–6446. doi: 10.1038/sj.onc.1209886. [DOI] [PubMed] [Google Scholar]
- 5.Rampaul RS, Pinder SE, Gullick WJ, Robertson JF, Ellis IO. HER-2 in breast cancer--methods of detection, clinical significance and future prospects for treatment. Crit Rev Oncol Hematol. 2002;43:231–244. doi: 10.1016/s1040-8428(01)00207-4. [DOI] [PubMed] [Google Scholar]
- 6.Li SY, Rong M, Grieu F, Iacopetta B. PIK3CA mutations in breast cancer are associated with poor outcome. Breast Cancer Res Treat. 2006;96:91–95. doi: 10.1007/s10549-005-9048-0. [DOI] [PubMed] [Google Scholar]
- 7.Depowski PL, Rosenthal SI, Ross JS. Loss of expression of the PTEN gene protein product is associated with poor outcome in breast cancer. Mod Pathol. 2001;14:672–676. doi: 10.1038/modpathol.3880371. [DOI] [PubMed] [Google Scholar]
- 8.Koksal IT, Dirice E, Yasar D, et al. The assessment of PTEN tumor suppressor gene in combination with Gleason scoring and serum PSA to evaluate progression of prostate carcinoma. Urol Oncol. 2004;22:307–312. doi: 10.1016/j.urolonc.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 9.Pfeil K, Eder IE, Putz T, et al. Long-term androgen-ablation causes increased resistance to PI3K/Akt pathway inhibition in prostate cancer cells. Prostate. 2004;58:259–268. doi: 10.1002/pros.10332. [DOI] [PubMed] [Google Scholar]
- 10.Edwards J, Krishna NS, Witton CJ, Bartlett JM. Gene amplifications associated with the development of hormone-resistant prostate cancer. Clin Cancer Res. 2003;9:5271–5281. [PubMed] [Google Scholar]
- 11.McMenamin ME, Soung P, Perera S, Kaplan I, Loda M, Sellers WR. Loss of PTEN expression in paraffin-embedded primary prostate cancer correlates with high Gleason score and advanced stage. Cancer Res. 1999;59:4291–4296. [PubMed] [Google Scholar]
- 12.Graff JR, Konicek BW, McNulty AM, et al. Increased AKT activity contributes to prostate cancer progression by dramatically accelerating prostate tumor growth and diminishing p27Kip1 expression. J B C. 2000;275:24500–24505. doi: 10.1074/jbc.M003145200. [DOI] [PubMed] [Google Scholar]
- 13.Faivre S, Kroemer G, Raymond E. Current development of mTOR inhibitors as anticancer agents. Nat Rev Drug Discov. 2006;5:671–688. doi: 10.1038/nrd2062. [DOI] [PubMed] [Google Scholar]
- 14.Sun SY, Rosenberg LM, Wang X, et al. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 2005;65:7052–7058. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]
- 15.Cantin GT, Yi W, Lu B, et al. Combining protein-based IMAC, peptide-based IMAC, and MudPIT for efficient phosphoproteomic analysis. J Proteome Res. 2008;7:1346–1351. doi: 10.1021/pr0705441. [DOI] [PubMed] [Google Scholar]
- 16.Washburn MP, Wolters D, Yates JR., III Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 17.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 18.Park SK, Venable JD, Xu T, Yates JR., III A quantitative analysis software tool for mass spectrometry-based proteomics. Nat Methods. 2008;5:319–322. doi: 10.1038/nmeth.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiu T, Santiskulvong C, Rozengurt E. EGF receptor transactivation mediates ANG II-stimulated mitogenesis in intestinal epithelial cells through the PI3-kinase/Akt/mTOR/p70S6K1 signaling pathway. Am J Physiol Gastrointest Liver Physiol. 2005;288:G182–G194. doi: 10.1152/ajpgi.00200.2004. [DOI] [PubMed] [Google Scholar]
- 20.Hidalgo M, Rowinsky EK. The rapamycin-sensitive signal transduction pathway as a target for cancer therapy. Oncogene. 2000;19:6680–6686. doi: 10.1038/sj.onc.1204091. [DOI] [PubMed] [Google Scholar]
- 21.Bulavin DV, Higashimoto Y, Popoff IJ, et al. Initiation of a G2/M checkpoint after ultraviolet radiation requires p38 kinase. Nature. 2001;411:102–107. doi: 10.1038/35075107. [DOI] [PubMed] [Google Scholar]
- 22.Boutros R, Lobjois V, Ducommun B. CDC25 phosphatases in cancer cells: key players? Good targets? Nat Rev Cancer. 2007;7:495–507. doi: 10.1038/nrc2169. [DOI] [PubMed] [Google Scholar]
- 23.Galaktionov K, Lee AK, Eckstein J, et al. CDC25 phosphatases as potential human oncogenes. Science. 1995;269:1575–1577. doi: 10.1126/science.7667636. [DOI] [PubMed] [Google Scholar]
- 24.Ito Y, Yoshida H, Tomoda C, et al. A. Expression of cdc25B and cdc25A in medullary thyroid carcinoma: cdc25B expression level predicts a poor prognosis. Cancer Lett. 2005;229:291–297. doi: 10.1016/j.canlet.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 25.Takemasa I, Yamamoto H, Sekimoto M, et al. Overexpression of CDC25B phosphatase as a novel marker of poor prognosis of human colorectal carcinoma. Cancer Res. 2000;60:3043–3050. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.