Abstract
Classically, recombination between immunoglobulin gene segments uses a pair of recombination signal sequences (RSSs) with dissimilar spacers (the “12/23 rule”). Using a series of different genotyping assays, four different kinds of atypical rearrangements were identified at the murine kappa locus: (1) Vκ to Vκ, (2) Jκ to Jκ, (3) Vκ to iRS, a heptameric sequence found in the JκCκ intron, and (4) a possible by-product of a rearrangement between a Vκ and the hypothetical 12-RSS side of a pre-existing signal joint. The novel Vκ-Vκ structure prompted further characterization. Sequence analysis of 14 different Vκ–Vκ rearrangements cloned from murine splenocytes and hybridomas revealed a Vκ4 family member as one participant in 13 rearrangements, but no rearrangements contained two Vκ4 genes. The Vκ4 partner in the Vκ-Vκ rearrangement exhibited more trimming of nucleotides at the Vκ-Vκ junction. A signal joint derived from the inversional rearrangement of two neighboring Vκs was also recovered. These data suggest that the Vκ-Vκ structures arise via RAG-mediated, intrachromosomal recombination.
Keywords: B cells, antibodies, gene rearrangement, molecular biology, RAG, V(D)J recombination
1. Introduction
The ability of the adaptive immune system to recognize an immense range of antigens stems from the process of V(D)J recombination at the B cell and T cell antigen receptor loci. Each immunoglobulin (Ig) receptor gene segment is flanked by a recombination signal sequence (RSS) consisting of conserved heptamer and nonamer sequences separated by either a 12 or a 23 base pair spacer (12-RSS or 23-RSS, respectively). Classically, recombination requires a pair of RSSs with dissimilar spacers (the “12/23 rule”) (Sakano et al., 1979; Tonegawa, 1983). Previous investigations of the 12/23 rule have focused primarily on in vitro assays using extrachromosomal rearrangement substrates (Hesse et al., 1987; Hiom and Gellert, 1998; Lieber et al., 1988; van Gent et al., 1996). A few 12/23 rule violations have been reported in vivo (Hirama et al., 1991; Langerak et al., 2004; Shimizu et al., 1991), but such rearrangements are generally deemed quite rare, unless the immune system is forced to use incompatible RSSs (Koralov et al., 2005).
After encountering several peculiar κ rearrangements in unrelated experiments, we set out to molecularly characterize the range of 12/23 rule violations seen at the Igκ locus in vivo. The Igκ locus is well suited for this analysis because of its large size and ability to undergo inversional rearrangement, with the retention of signal joints and prior rearrangement coding joints on the chromosome (Feddersen and Van Ness, 1985; Shapiro and Weigert, 1987). Using a degenerate Vκ primer, we characterized 14 independent Vκ-Vκ fusions from spleen and splenic hybridoma DNA, of which 13 contained Vκ4 sequences. We also used a semi-quantitative PCR assay to measure the frequency of Vκ-Vκ rearrangements in wild type mice. The data suggest that these rearrangements are infrequent compared to conventional Vκ-Jκ rearrangements. The biological function of these aberrant rearrangements is unknown.
2. Materials and Methods
2.1 Mice
All mice used for these studies are on the tenth or greater backcross generation onto the C57B6 background. The 56R mouse has a somatically mutated anti-DNA heavy chain that was introduced into the heavy chain J region by homologous recombination in embryonic stem cells (Chen et al., 1995). The bcl-xL mouse, a gift from Tullia Lindsten at the University of Pennsylvania, expresses the anti-apoptotic gene, bcl-xL, in B cells on the C57B6 background (Grillot et al., 1996). Hybridoma panels were generated from 3- to 6-month-old mice. Animals were housed in the University mouse colony and experiments were performed in accordance with a protocol approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
2.2 Hybridomas
Spontaneous hybridomas from 3-month old B6 and B6.56R.BclxL mice were produced by fusion of the murine myeloma cell line Sp2/0 (Kohler, 1980) to freshly harvested splenocytes as described previously (Prak et al., 1994). Hybridomas were cultured at limiting dilution and expanded into duplicate 6-well plates for analysis of culture supernatants and nucleic acid extraction, as described previously (Prak et al., 1994). Hybridomas from B6.56R mice were produced for a separate study, but characterized for atypical κ rearrangements in this study (Sekiguchi et al., 2006).
2.3 PCR primers and conditions
All PCRs were performed with 100–250 ng of genomic DNA from spleen or individual spontaneous B6 hybridomas, in 1× PCR Buffer I (Applied Biosystems, Foster City, CA) with 1.5 U AmpliTaq Gold (Applied Biosystems) and 250 μM dNTPs. The Vs PCR was performed as described above in a 20 μL reaction volume, with 40 pmol of a degenerate primer in Vκ (Schlissel and Baltimore, 1989). Thermal cycling conditions were: primary denaturation at 94°C for 10 minutes; 40 cycles of 94°C for 30 sec, 67°C for 30 sec, and 72°C for 30 sec; and final extension at 72°C for 10 minutes. Assays to characterize rearrangements in individual hybridomas to Vκ20 and Vκ21 were performed as described previously (Li et al., 2001). Assays to detect signal joints remaining on the chromosome after Jκ to Jκ inversion were performed as described above, with 20 pmol of each primer:
Jκ1for: 5′-AATCAGCAGTTCTCTGTCAGAGAAGCC-3′
Jκ4for: 5′-CACGTTCGGCTCGGGGACAAAGTTGGAA-3′
Thermal cycling conditions were: primary denaturation at 94°C for 10 minutes; 40 cycles of 94°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec; and final extension at 72°C for 5 minutes. PCR assays to detect signal joints remaining on the chromosome after Vκ to Vκ inversion were performed using primers situated in genomic DNA sequences flanking individual Vκ RSSs. The primers used for this analysis are:
Vκ4-86 SJP: 5′-TCCTGCCAGTGTGAAGACAG-3′
Vκ1-88 SJP: 5′-TGATGAAGGCTGTCATGCTCA-3′
The signal joint amplification was performed in a 50 μL volume using 50 pmol of each primer and the same concentrations of all of the other mix components as the Jκ-Jκ PCR described above. Cycling conditions were: primary denaturation at 94°C for 10 minutes; 40 cycles of 94°C for 30 sec, 65°C for 30 sec, and 72°C for 30 sec; and final extension at 72°C for 10 minutes.
2.4 Cloning and sequence analysis
PCR products were band purified using a Qiaquick gel extraction kit, per the manufacturer's instructions (Qiagen, Valencia, CA) and either sequenced directly or cloned into pCR4 TOPO per the manufacturer's instructions (Invitrogen, Carlsbad, CA). Sequencing was performed on an ABI 3730 using BigDye Taq FS terminator V 3.1 in the University of Pennsylvania DNA Sequencing facility (http://www.med.upenn.edu/genetics/core-facs/dna-seq/). Sequences (in both directions) were aligned and compared to germline Vκ sequences using IgBLAST (http://ncbi.nih.gov/igblast/). Nomenclature used for Vκ gene segments follows the system described in reference (Brekke and Garrard, 2004).
2.5 Statistical analysis
As described in Results, we encountered a predominance of Vκ4–non-Vκ4 rearrangements, without any Vκ4–Vκ4 rearrangements. To calculate the likelihood these results could be due to chance, we considered a model wherein different Vκ genes have independent probabilities of undergoing Vκ–Vκ rearrangement. This model assumes that the assay, which relies upon the use of a degenerate Vκ primer, does not result in the biased amplification of particular Vκ gene families. Based on our previous experience, we know that the Vs primer can amplify approximately 80% of all Vκ gene family members, including Vκ4 and non-Vκ4 genes (Prak et al., 1994). Applying this model, there is some unknown probability p that any given gene we recover is from the Vκ4 family. Assuming that the 14 Vκ-Vκ sequences shown in table 1 are derived from independent clones of B cells (based on sequence differences), p, the frequency of Vκ4, is estimated to be 13/28. The chance that both Vκs in a given pairing are Vκ4 is (0.464)2 = 0.21, assuming that Vκ4 and non-Vκ4 genes rearrange independently. The chance of not seeing Vκ4-Vκ4 in 14 Vκ-Vκ pairings is (1-0.21)14 = 0.037. A Student's t-test (one-tailed, equal variance) was used to compare the 3′ trim length of Vκ4 to non-Vκ4 partners in the 14 Vκ-Vκ rearrangements.
Table 1. Vκ usage and DNA source of cloned Vκ-Vκ rearrangements.
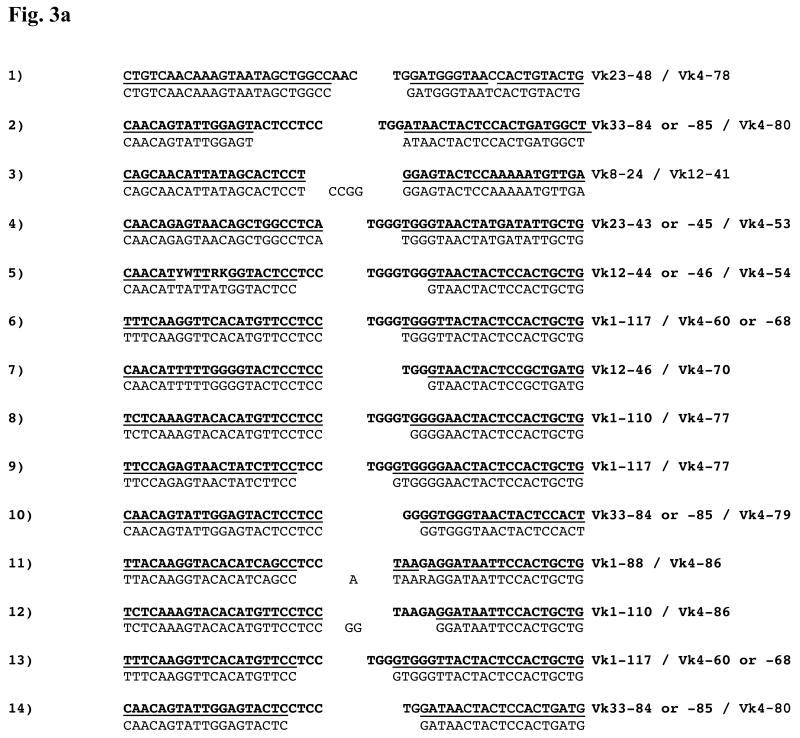
Four different mice provided splenocytes. Spleen refers to spleen DNA. Hybridoma refers to spontaneous hybridomas produced from the spleen (see Methods). The Vκ gene assignments are based on DNA sequence analysis (see Methods). Vκ-Vκ rearrangements using the same Vκ gene segments are shaded. The junction of each Vκ-Vκ rearrangement is shown in fig. 3a.
| B6 spleen | ||
|---|---|---|
| 1. | Vκ23-48 | Vκ4-78 |
| 2. | Vκ33-84 or Vκ33-85 | Vκ4-80 |
| 3. | Vκ12-41 | Vκ8-24 |
| B6.Bcl-xL spleen | ||
| 4. | Vκ23-43 or Vκ23-45 | Vκ4-53 |
| 5. | Vκ12-44 or Vκ12-46 | Vκ4-54 |
| 6. | Vκ1-117 | Vκ4-60 or Vκ4-68 |
| 7. | Vκ12-46 | Vκ4-70 |
| 8. | Vκ1-110 | Vκ4-77 |
| 9. | Vκ1-117 | Vκ4-77 |
| 10. | Vκ33-84 or Vκ33-85 | Vκ4-79 |
| 11. | Vκ1-88 | V4-86 |
| 12. | Vκ1-110 | Vκ4-86 |
| B6.56R.Bcl-xL hybridoma | ||
| 13. | Vκ1-117 | Vκ4-60 or Vκ4-68 |
| B6 hybridoma | ||
| 14. | Vκ33-84 or Vκ33-85 | Vκ4-80 |
3. Results
3.1 Atypical Vκ–Vκ gene rearrangements occur in vivo
During routine hybridoma genotyping, we noted a PCR product of unexpected size that, on sequence analysis, appeared to be a Vκ–Vκ rearrangement. We first confirmed that the unexpected product could be amplified with Vs (a degenerate Vκ primer, see Methods) alone in the reaction mix. We then used Vs PCR to identify additional examples from spleen DNA of mice. Table 1 illustrates the range of Vκ–Vκ rearrangements that were recovered.
3.2 Vκ–Vκ rearrangements likely invert and may also delete
To better understand the mechanism of Vκ–Vκ rearrangement, we examined the germline positions and orientations of the participating gene segments. The gene pairs involved have a variety of relative configurations in the germline (fig. 1). Assuming that these rearrangements arise by recombining Vκ segments that are on the same chromosome and are in the germline configuration, these data suggest that Vκ-Vκ rearrangements can occur by inversion or deletion (fig. 2b; the conventional Vκ-Jκ rearrangement is shown in fig. 2a for general orientation). Consistent with this possibility, we recovered a reciprocal product using primers that faced towards the recombination signal sequences of two neighboring Vκ1-88 and Vκ4-86 (fig. 2c, the annotated sequence is given in fig. S1 of the electronic supplement). In the germline configuration, these primers do not efficiently amplify genomic DNA because they are facing in the same direction. Vκ1-88 and Vκ4-86 genes are adjacent in the germline Igκ locus, thus a single rearrangement can produce their Vκ-Vκ fusion and the corresponding signal joint. However, primary rearrangement is not the only possible pathway for Vκs that are not immediately adjacent to one another. Some of the Vκ–Vκ rearrangements could represent secondary rearrangements, on alleles already modified by deletions and/or inversions from preceding rearrangements.
Figure 1. Chromosomal locations of Vκ gene segments that were found in Vκ-Vκ rearrangements.

The Vκ gene segments that have been identified in 14 independent Vκ-Vκ rearrangements are shown, based on the positions of their Vκ gene segments in the germline locus. The color of the triangles is used to identify partners of a Vκ-Vκ fusion (partners share the same color and are listed in table 1). The direction of the triangles is used to denote the Vκ gene segment orientation in the germline κ locus, as described previously (Brekke and Garrard, 2004; Thiebe et al., 1999).
Figure 2.


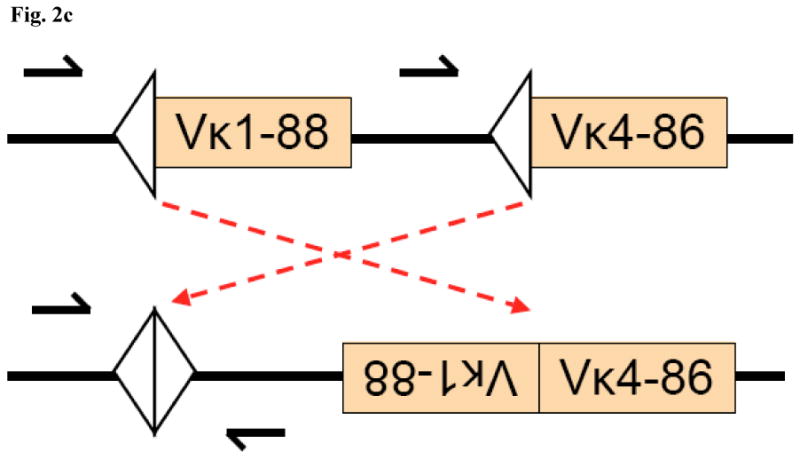
Figure 2a: Conventional Vκ-Jκ rearrangement. A deletional rearrangement of Vκ to Jκ is depicted. The Vκ gene segment is flanked by a 3′ recombination signal sequence with a 12 base pair spacer (12-RSS, white triangle). The Jκ gene segment is flanked by a 5′ recombination signal sequence with a 23 base pair spacer (23-RSS, black triangle). The recombination results in the generation of a coding joint (Vκ-Jκ, on the left) and a signal joint (an episome containing the two fused RSSs, on the right).
Figure 2b: Vκ-Vκ rearrangements may occur by inversion or deletion. Shown are two schematic pairs of Vκ gene segments undergoing either inversional rearrangement (VκA, VκB) or deletional rearrangement (VκC, VκD). Whether Vκ-Vκ rearrangement results in inversion or deletion is dictated by the orientation of the Vκ gene segments. As shown in fig. 1, both deletional and inversional rearrangements are possible, based on the orientations of the Vκ-Vκ pairs in the germline locus. Boxes denote exons, lines introns and white triangles represent 12-RSS.
Figure 2c: Inversional Vκ-Vκ Rearrangement and Signal Joint. Shown is a proposed inversional rearrangement that involves two neighboring Vκ gene segments, Vκ1-88 and Vκ4-86. The signal joint, consisting of two facing 12-RSSs, is retained on the chromosome. The Vκ1-88/Vκ4-86 rearrangement and the corresponding signal joint were amplified and cloned from independent PCR amplifications of spleen DNA from two different mice (see Materials and Methods). The nucleotide sequence of the signal joint is excerpted in fig. 3b (#1) and the full sequence is provided in fig. S1 of the electronic supplement.
3.3 Vκ–Vκ rearrangement commonly involves the Vκ4 gene family
Almost every Vκ–Vκ rearrangement we recovered (13/14) contains exactly one Vκ4 gene (table 1). The large size of the Vκ4 family and the possibility that the degenerate Vs primer may not recognize all Vκ genes equally well could contribute to an increased likelihood of recovering Vκ4 rearrangements. However, such causes of bias would, as described in Methods, predict that Vκ4–Vκ4 rearrangements should also be present. Using the assumptions described in the Methods, we calculate a probability of 4% of encountering no Vκ4-Vκ4 rearrangements due to chance.
3.4 Vκ–Vκ rearrangements demonstrate junctional modifications suggesting RAG involvement
Vκ-Vκ rearrangements resemble canonical Vκ-Jκ rearrangements in that they appear to use the 3′ RSS. Examination of the 14 Vκ–Vκ junctions reveals frequent “nibbling” (nucleotide deletion at junction ends) of up to 8 nt per end and 3 instances of probable “P addition” (insertion of palindromic nucleotides complementary to a non-nibbled end, fig. 3a). These modifications resemble those seen at normal VJ coding joints (Martin et al., 1992; Meier and Lewis, 1993; Victor et al., 1994). Vκ4 gene segments appear to harbor fewer 3′ nucleotides than their non-Vκ4 partners; on average, 3.3 residues were missing from the 3′ end of the Vκ4 gene compared to 2.1 residues from the non-Vκ4 gene (p=0.07, 1-tailed Student's t-test).
Figure 3.
Figure 3a: Nucleotide Sequences of Vκ–Vκ junctions. Each Vκ gene contributes nucleotides from its 3′ end to the Vκ–Vκ junction. Here, the corresponding germline sequences appear in bold font above each junction to permit analysis of junctional modification. A bar between the germline and experimental sequences indicates regions of identity. Nucleotides that cannot be attributed to a particular germline sequence are shown centered in the junction. Displayed sequences are from the CDR3 of each gene, aligned against each other, and the Vκ families contributing to each junction are shown. The sequence numbering corresponds to the numbering in table 1, which provides the mouse/tissue/hybridoma origin of each sequence. For consistency, the top strands of the non-Vκ4 genes are shown on the left and the bottom strands of the Vκ4 genes (when present) are shown on the right. An example of one of these rearrangements (involving Vκ1-88 and Vκ4-86) and its corresponding signal joint is shown in fig. 2c.
Figure 3b: Sequences of other atypical κ rearrangements. Sequence data from the atypical junctions shown in fig. 4a-4c appear with the corresponding portions of the germline Igκ locus. The notation is as described for fig. 3a. In the hybrid joint involving the Jκ4 RSS, “<23bp>” indicates the spacer between the heptamer (shown, with junctional modification) and the nonamer (shown, preserved). Similarly, in the Vκ-Vκ signal joint, the 12-RSS spacer is denoted “<12bp>”. The DNA sources of the sequences are: 1. B6 spleen; 2. B6.bcl-xL spleen; 3. B6.bcl-xL spleen; 4. B6.56R.bcl-xL hybridoma; 5. B6.56R hybridoma; 6. B6.56R hybridoma.
3.5 Vκ-Vκ rearrangements are infrequent in splenocytes
To determine the frequency of Vκ-Vκ rearrangements, a semi-quantitative PCR assay was performed on different quantities of wild type spleen DNA (fig. S2). Vκ-Vκ amplification was present with ∼100 ng of input DNA from a C57B6 mouse. Assuming that half of the DNA mass in the spleen is due to B cells, that each cell contains approximately 6.7 pg genomic DNA, that the Vκ-Vκ PCR efficiently recovers all Vκ-Vκ rearrangements and that each cell harbors at most one Vκ-Vκ rearrangement, this corresponds to a Vκ-Vκ rearrangement frequency of approximately one in 7500 B cells.
3.6 A variety of atypical rearrangements can occur in vivo
In addition to Vκ–Vκ rearrangements, we have recovered evidence of several other atypical rearrangements. In a splenic hybridoma from an anti-DNA heavy chain knock-in mouse (B6.56R (Chen et al., 1995; Li et al., 2001; Sekiguchi et al., 2006)), we recovered a Jκ1–Jκ5 rearrangement (fig. 4a) as well as a hybrid joint involving a Vκ20 and the Jκ4 RSS (fig. 4b). The two junctions were in close proximity and oriented to permit inadvertant amplification on a routine genotyping PCR. In a hybridoma from a B6.56R.bcl-xL mouse, we encountered a rearrangement involving a Vκ12 and the JκCκ intron upstream of the intronic RS (fig. 4c). The existence of Vκ to JκCκintron rearrangements has been demonstrated previously in the B cell line MPC-11 (Seidman and Leder, 1980) and further substantiated by the analysis Abelson murine leukemia virus transformant subclones (Feddersen et al., 1990). Atypical rearrangements involving the JκCκ intron RSS also include Jκ1-iRS fused signal join in the plasmacytoma PC 8701 (Kelley et al., 1985) as well as a reciprocal product (Shimizu et al., 1991).
Figure 4. Schematics of Different Atypical Kappa Rearrangements.
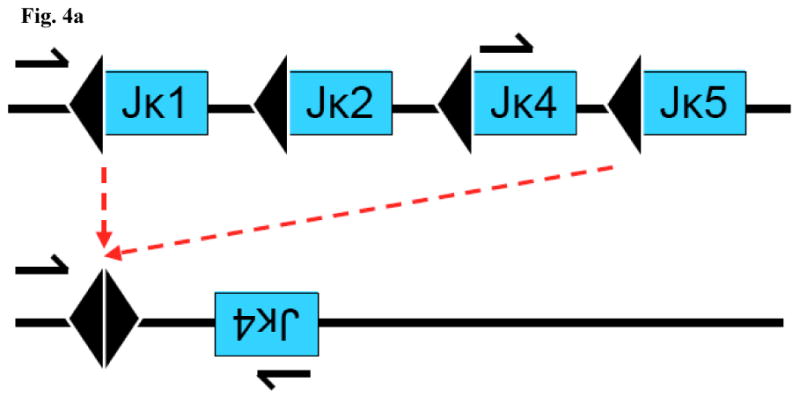


Figure 4a: Probable inversional rearrangement between Jκ5 and Jκ1. The signal joint contains 23-RSS sequences derived from Jκ1 and Jκ5 (because they are flanked by intronic sequences upstream of Jκ1 and Jκ5). Only the Jκ4 sequence is shown to the right of the signal joint because only the Jκ4 segment was recovered in the PCR due to the use of primers upstream of Jκ1 and within Jκ4 (arrows). Two distinct rearrangements of this type were recovered; the sequences are excerpted in fig. 3b (#2 and #3) and provided in detail in fig. S3.
Figure 4b: Complex Aberrant κ Rearrangements on one chromosome of a hybridoma. Rearrangements involving Vκ20, probably Jκ4 (RSS-23), Jκ2 and Jκ5 were recovered in a single PCR amplification using primers in Jκ5 and Vκ20 (arrows) in a splenic hybridoma derived from a 56R anti-DNA heavy chain knock in mouse (Sekiguchi et al., 2006). Two possible rearrangement scenarios are illustrated, both of which begin with an inversional rearrangement between Jκ1 and Jκ4. After the presumed inversion, Vκ20 is postulated to invade the proposed Jκ1/Jκ4 signal joint and Jκ2 is postulated to rearrange to Jκ5, deleting the intervening Jκs. The open triangle with wavy edging indicates an incomplete 23-RSS with bases missing from the heptamer. Based on the flanking sequence, this heptamer most likely derives from the Jκ4 gene segment. Both junctions from these complex rearrangements were recovered; the sequences are excerpted in fig. 3b (#5 and #6) and provided in detail in fig. S4.
Figure 4c: Probable Inversional Rearrangement of Vκ12 to the JCintron Heptamer. A PCR product containing a conventional Vκ4-Jκ2 rearrangement, the Jκ4 and Jκ5 gene segments, part of the JC-intron and an inverted Vκ12 gene segment was obtained, demonstrating loss of the JCintron heptamer (fig. 3b). The simplest explanation is an inversional rearrangement of Vκ12 to the cryptic heptamer in the JCintron on an allele that has already undergone conventional Vκ4-Jκ2 rearrangement. The dashed triangle represents the cryptic heptamer of the intronic RSS. The sequence of the atypical rearrangement is excerpted in fig. 3b (#4) and provided in detail in fig. S5.
All three of these atypical rearrangements exhibit junctional modifications on one or both ends (fig. 3b). Each Jκ has a 23-RSS, so the Jκ–Jκ rearrangement violates the 12/23 rule. The Vκ–JκCκintron rearrangement “bends” the 12/23 rule, in that the intronic RS is degenerate, but does classically recombine with the 23-RSS of the downstream RS element. Finally, the Vκ–JκRSS rearrangement, involving secondary rearrangement into a signal joint, appears to require a 12/23 rule violation; however, if we postulate a Jκ–iRS signal joint (Langerak et al., 2004) as an intermediate, the Vκ would then recombine with the iRS heptamer, also only “bending” the 12/23 rule.
4. Discussion
Diversity is both important and dangerous for the immune system. As such, mechanisms that influence diversity, such as the 12/23 rule, are complex in their biological effects. On one hand, efficient recombination between dissimilar RSS spacers promotes diversification. For example, at the heavy chain locus, the 12/23 rule enforces the incorporation of DH segments, increasing CDR3 length and repertoire complexity (Ippolito et al., 2003; Sakano et al., 1981). On the other hand, given the fact that all gene segments of a given type (V, D, or J) at each antigen receptor locus use the same size spacer, the 12/23 rule discourages recombinations that are unlikely to yield a meaningful antigen receptor.
In this investigation of V(D)J recombination at the mouse Igκ locus, we describe a variety of rearrangements that apparently violate the 12/23 rule, including Vκ–Vκ rearrangement, Jκ–Jκ rearrangement, and others. Most of the rearrangements analyzed in this study harbor junctional modifications (nucleotide deletion and occasionally P addition). All 14 Vκ-Vκ sequences that were recovered were unique (based on the Vκ-Vκ junction). However, there does appear to be a preference for particular Vκ gene segment combinations (fig. 1). Two Vκ-Vκ rearrangements were each observed twice: Vκ80 to Vκ33-84/85 and Vκ60 to Vκ1-117.
In addition to the seemingly non-random usage of particular Vκ-Vκ pairs, there is an intriguing tendency for Vκ–Vκ rearrangements to involve gene segments from the Vκ4 family. 13 out of the 14 Vκ-Vκ rearrangements use gene segments from the Vκ4 family. The high frequency of Vκ4 usage is not unique to a particular mouse, as these rearrangements were independently cloned from 2 different mouse spleens and recovered from hybridomas from two other mice. While Vκ4 is not absolutely required, its usage is favored amongst Vκ-Vκ rearrangements. Vκ4 is the largest Vκ gene family in the mouse, consisting of 27 members and comprising 28% of functional murine Vκ gene segments (Brekke and Garrard, 2004). If rearrangements to different Vκ gene segments are uniformly distributed, then Vκ4 should be present in a sizable fraction of Vκ-Vκ rearrrangements. However, only one Vκ4 is found in all of the Vκ4-containing rearrangements. Attempts to amplify Vκ4-Vκ4 rearrangements with a Vκ4-specific primer failed (data not shown). Failure to amplify Vκ4-Vκ4 rearrangements is likely to reflect the rarity of Vκ4-Vκ4 rearrangement, but could also be due to difficulty in cloning and/or sequencing rearrangements with highly homologous Vκs.
We wondered if there could be a structural feature of Vκ4 family members that would make them more likely to participate in aberrant rearrangement. We noticed that the 3′ ends of the Vκ4 partner in the Vκ-Vκ rearrangement were shorter (being recessed an average of 3.3 nt compared to the germline sequence), than the non-Vκ4 partner (which was recessed 2.1 nt, compared to the germline sequence). Most murine kappa light chains have a highly conserved proline residue at position 95 (Pro95) that is important for CDR3 folding (Chothia and Lesk, 1987; Kabat, 1983). Most of the Vκ4 genes in our Vκ-Vκ collection have four nucleotides between Pro95 and the RSS heptamer, whereas most Vκ genes, including the non-Vκ4 genes in our Vκ-Vκ collection, only have two bases (Milstein et al., 1992). This asymmetric trimming was first noted in conventional Vκ4 to Jκ2 or Jκ5 rearrangements cloned from BALB/c spleen DNA (Milstein et al., 1992). Thus, on average, Vκ4 genes exhibit more “trimming” (or RAG is permitted to cut more sloppily), but there is usually more DNA “to spare” between Pro95 and the heptamer (Milstein et al., 1992). Our data, as well as the out of frame rearrangements recovered in the earlier analysis of Vκ4-Jκ2/5 rearrangements, suggest that this 3′ length asymmetry is intrinsic to the rearrangement mechanism, rather than being due to selection for Vκ4 rearrangements of a particular CDR3 length.
We also noticed that Vκ4 genes tend to have nucleotide sequences that are rich in Gs and Ts on the non-coding strand (the Vκ4 sequences in fig. 3a are aligned to illustrate this) and include stretches of 2-4 Gs and GTGs. It is possible, as suggested by Gellert, that these sequences result in an unusual DNA structure that may be recognized by the recombination machinery (Gellert, 1992). Because Vκ-Vκ rearrangements involve the apposition of two RSS-12 sequences, having two altered DNA structures in close apposition (such as two Vκ4 family members) could be prohibitive. It is interesting that coding sequences can influence the efficiency of recombination over 250-fold, although this has not been directly tested for two RSS-12 containing recombination substrates (Gerstein and Lieber, 1993).
Our agnostic approach to recovering atypical κ rearrangement products provides insights into the stringency of V(D)J recombination in a physiologic in vivo system. Presumably, these rearrangements are mediated by the RAG enzymes, given the pattern of cleavage: the recombination signal sequence at the 3′ end of the Vκs is missing from all of the Vκ-Vκ rearrangements that were recovered. The recovery of a reciprocal product is consistent with intrachromosomal RAG-mediated inversional recombination to generate at least one of the Vκ-Vκ rearrangements. The signal joint in this reciprocal product was perfectly intact, which is different from a mechanism proposed for re-entry of damaged signal joints into the genome (Neiditch et al., 2002). In the latter case, a damaged signal joint is postulated to re-invade an RSS or cryptic RSS.
RAG-mediated recombination beyond the traditional boundaries of V(D)J recombination is inherently dangerous (Hiom et al., 1998) and many previously characterized translocation breakpoints involve the immunoglobulin or TCR loci. It is possible that the frequency of Vκ-Vκ rearrangement in mature splenocytes (which have survived negative selection) underestimates the frequency of these aberrant rearrangements during lymphocyte maturation. In addition to the potential dangers of generating Vκ-Vκ rearrangements, the rearrangement product, if transcribed, has the potential to form a hairpin, due to oppositely facing Vκs. Vκ hairpin RNAs, if they exist, could silence κ.
Supplementary Material
Acknowledgments
We thank members of the Luning Prak laboratory, Martin Weigert and Craig Bassing for helpful discussions. We thank the University of Pennsylvania DNA Sequencing facility for their expertise and technical contributions to this study. E.L.P. is supported by grants from the NIH, Alliance for Lupus Research and Southern New Jersey Lupus Society. J.M.V. was supported by a T32 training grant from the NIDDK and D.C. was supported by the Goldie Simon Award from the Southeastern Pennsylvania Lupus Society (re-named the Philadelphia Tri-State Chapter of the Lupus Foundation of America).
Abbreviations
- RSS
(recombination signal sequence)
- nt
(nucleotide)
- 12-RSS and 23-RSS
(RSS with 12 or 23 nt spacer)
- iRS
recombination sequence located in the Jκ-Cκ intron
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brekke KM, Garrard WT. Assembly and analysis of the mouse immunoglobulin kappa gene sequence. Immunogenetics. 2004;56:490–505. doi: 10.1007/s00251-004-0659-0. [DOI] [PubMed] [Google Scholar]
- Chen C, Nagy Z, Prak EL, Weigert M. Immunoglobulin heavy chain gene replacement: a mechanism of receptor editing. Immunity. 1995;3:747–55. doi: 10.1016/1074-7613(95)90064-0. [DOI] [PubMed] [Google Scholar]
- Chothia C, Lesk AM. Canonical structures for the hypervariable regions of immunoglobulins. J Mol Biol. 1987;196:901–17. doi: 10.1016/0022-2836(87)90412-8. [DOI] [PubMed] [Google Scholar]
- Feddersen RM, Martin DJ, Van Ness BG. Novel recombinations of the IG kappa-locus that result in allelic exclusion. J Immunol. 1990;145:745–50. [PubMed] [Google Scholar]
- Feddersen RM, Van Ness BG. Double recombination of a single immunoglobulin kappa-chain allele: implications for the mechanism of rearrangement. Proc Natl Acad Sci U S A. 1985;82:4793–7. doi: 10.1073/pnas.82.14.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M. Molecular analysis of V(D)J recombination. Annu Rev Genet. 1992;26:425–46. doi: 10.1146/annurev.ge.26.120192.002233. [DOI] [PubMed] [Google Scholar]
- Gerstein RM, Lieber MR. Coding end sequence can markedly affect the initiation of V(D)J recombination. Genes Dev. 1993;7:1459–69. doi: 10.1101/gad.7.7b.1459. [DOI] [PubMed] [Google Scholar]
- Grillot DA, Merino R, Pena JC, Fanslow WC, Finkelman FD, Thompson CB, Nunez G. bcl-x exhibits regulated expression during B cell development and activation and modulates lymphocyte survival in transgenic mice. J Exp Med. 1996;183:381–91. doi: 10.1084/jem.183.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse JE, Lieber MR, Gellert M, Mizuuchi K. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V-(D)-J joining signals. Cell. 1987;49:775–83. doi: 10.1016/0092-8674(87)90615-5. [DOI] [PubMed] [Google Scholar]
- Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–9. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–70. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- Hirama T, Takeshita S, Yoshida Y, Yamagishi H. Structure of extrachromosomal circular DNAs generated by immunoglobulin light chain gene rearrangements. Immunol Lett. 1991;27:19–23. doi: 10.1016/0165-2478(91)90238-6. [DOI] [PubMed] [Google Scholar]
- Ippolito GC, Pelkonen J, Nitschke L, Rajewsky K, Schroeder HW., Jr Antibody repertoire in a mouse with a simplified D(H) locus: the D-limited mouse. Ann N Y Acad Sci. 2003;987:262–5. doi: 10.1111/j.1749-6632.2003.tb06058.x. [DOI] [PubMed] [Google Scholar]
- Kabat EA, Wu TT, Bilofsky H, Reid-Milner M, Perry H. Sequences of Proteins of Immunological Interest. 3rd. Public Heath Service; Washington, D.C.: 1983. [Google Scholar]
- Kelley DE, Wiedemann LM, Pittet AC, Strauss S, Nelson KJ, Davis J, Van Ness B, Perry RP. Nonproductive kappa immunoglobulin genes: recombinational abnormalities and other lesions affecting transcription, RNA processing, turnover, and translation. Mol Cell Biol. 1985;5:1660–75. doi: 10.1128/mcb.5.7.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler G. Immunoglobulin chain loss in hybridoma lines. Proc Natl Acad Sci U S A. 1980;77:2197–9. doi: 10.1073/pnas.77.4.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koralov SB, Novobrantseva TI, Hochedlinger K, Jaenisch R, Rajewsky K. Direct in vivo VH to JH rearrangement violating the 12/23 rule. J Exp Med. 2005;201:341–8. doi: 10.1084/jem.20041577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerak AW, Nadel B, De Torbal A, Wolvers-Tettero IL, van Gastel-Mol EJ, Verhaaf B, Jager U, van Dongen JJ. Unraveling the consecutive recombination events in the human IGK locus. J Immunol. 2004;173:3878–88. doi: 10.4049/jimmunol.173.6.3878. [DOI] [PubMed] [Google Scholar]
- Li H, Jiang Y, Prak EL, Radic M, Weigert M. Editors and editing of anti-DNA receptors. Immunity. 2001;15:947–57. doi: 10.1016/s1074-7613(01)00251-5. [DOI] [PubMed] [Google Scholar]
- Lieber MR, Hesse JE, Mizuuchi K, Gellert M. Studies of V(D)J recombination with extrachromosomal substrates. Curr Top Microbiol Immunol. 1988;137:94–9. doi: 10.1007/978-3-642-50059-6_15. [DOI] [PubMed] [Google Scholar]
- Martin T, Blaison G, Levallois H, Pasquali JL. Molecular analysis of the V kappa III-J kappa junctional diversity of polyclonal rheumatoid factors during rheumatoid arthritis frequently reveals N addition. Eur J Immunol. 1992;22:1773–9. doi: 10.1002/eji.1830220716. [DOI] [PubMed] [Google Scholar]
- Meier JT, Lewis SM. P nucleotides in V(D)J recombination: a fine-structure analysis. Mol Cell Biol. 1993;13:1078–92. doi: 10.1128/mcb.13.2.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein C, Even J, Jarvis JM, Gonzalez-Fernandez A, Gherardi E. Non-random features of the repertoire expressed by the members of one V kappa gene family and of the V-J recombination. Eur J Immunol. 1992;22:1627–34. doi: 10.1002/eji.1830220642. [DOI] [PubMed] [Google Scholar]
- Neiditch MB, Lee GS, Huye LE, Brandt VL, Roth DB. The V(D)J recombinase efficiently cleaves and transposes signal joints. Mol Cell. 2002;9:871–8. doi: 10.1016/s1097-2765(02)00494-x. [DOI] [PubMed] [Google Scholar]
- Prak EL, Trounstine M, Huszar D, Weigert M. Light chain editing in kappa-deficient animals: a potential mechanism of B cell tolerance. J Exp Med. 1994;180:1805–15. doi: 10.1084/jem.180.5.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano H, Huppi K, Heinrich G, Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 1979;280:288–94. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- Sakano H, Kurosawa Y, Weigert M, Tonegawa S. Identification and nucleotide sequence of a diversity DNA segment (D) of immunoglobulin heavy-chain genes. Nature. 1981;290:562–5. doi: 10.1038/290562a0. [DOI] [PubMed] [Google Scholar]
- Schlissel MS, Baltimore D. Activation of immunoglobulin kappa gene rearrangement correlates with induction of germline kappa gene transcription. Cell. 1989;58:1001–7. doi: 10.1016/0092-8674(89)90951-3. [DOI] [PubMed] [Google Scholar]
- Seidman JG, Leder P. A mutant immunoglobulin light chain is formed by aberrant DNA- and RNA-splicing events. Nature. 1980;286:779–83. doi: 10.1038/286779a0. [DOI] [PubMed] [Google Scholar]
- Sekiguchi DR, Yunk L, Gary D, Charan D, Srivastava B, Allman D, Weigert MG, Prak ET. Development and selection of edited B cells in B6.56R mice. J Immunol. 2006;176:6879–87. doi: 10.4049/jimmunol.176.11.6879. [DOI] [PubMed] [Google Scholar]
- Shapiro MA, Weigert M. How immunoglobulin V kappa genes rearrange. J Immunol. 1987;139:3834–9. [PubMed] [Google Scholar]
- Shimizu T, Iwasato T, Yamagishi H. Deletions of immunoglobulin C kappa region characterized by the circular excision products in mouse splenocytes. J Exp Med. 1991;173:1065–72. doi: 10.1084/jem.173.5.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebe R, Schable KF, Bensch A, Brensing-Kuppers J, Heim V, Kirschbaum T, Mitlohner H, Ohnrich M, Pourrajabi S, Roschenthaler F, Schwendinger J, Wichelhaus D, Zocher I, Zachau HG. The variable genes and gene families of the mouse immunoglobulin kappa locus. Eur J Immunol. 1999;29:2072–81. doi: 10.1002/(SICI)1521-4141(199907)29:07<2072::AID-IMMU2072>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–81. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- van Gent DC, Ramsden DA, Gellert M. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 1996;85:107–13. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- Victor KD, Vu K, Feeney AJ. Limited junctional diversity in kappa light chains. Junctional sequences from CD43+B220+ early B cell progenitors resemble those from peripheral B cells. J Immunol. 1994;152:3467–75. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.