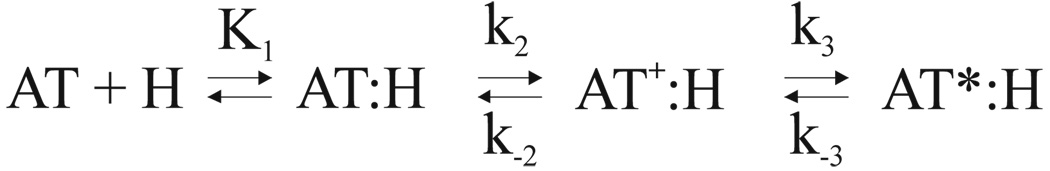Summary
Antithrombin (AT) is the most important inhibitor of the coagulation proteases. Its activity is stimulated by glycosaminoglycans such as heparin, through allosteric and template mechanisms. AT utilises an induced-fit mechanism to bind with high affinity to a pentasaccharide sequence found in about one-third of heparin chains. The conformational changes behind this mechanism have been characterised by several crystal structures of AT in the absence and presence of the pentasaccharide. Pentasaccharide binding ultimately results in a conformational change that improves affinity by about 1000-fold. Crystal structures show several differences, including the expulsion of the hinge region of the reactive centre loop from β-sheet A, known to be critical for the allosteric activation of AT. Here we present data that reveals an energetically distinct intermediate on the path to full activation, where the majority of conformational changes have already occurred. A crystal structure of this intermediate shows that the hinge region is in a native like state, in spite of having the pentasaccharide bound in the normal fashion. We engineered a disulphide bond to lock the hinge in its native position to determine the energetic contributions of the initial and final conformational events. Approximately 60% of the free energy contribution of conformational change is provided by the final step of hinge region expulsion and subsequent closure of the main β-sheet A. A new analysis of the individual structural changes provides a plausible mechanism for propagation of conformational change from the heparin binding site to the remote hinge region in β-sheet A.
Keywords: allosteric, antithrombin, heparin, pentasaccharide, serpin
Introduction
Antithrombin (AT) is the principal inhibitor of the coagulation proteases (for recent review see1). Its deficiency results in venous thrombosis2, and the homozygous mouse knock-out is embryonic lethal3. AT circulates in the blood plasma at the relatively high concentration of 2.3µM in a low activity state. A fraction of the circulating AT associates to the blood vessel wall by binding to glycosaminoglycans (GAGs), such as heparan sulphate, resulting in the activation of AT towards thrombin and factors IXa and Xa. The therapeutic effect of heparin is similarly due to the activation of the inhibitory activity of AT. A fraction of the heparin and heparan sulphate chains contain a special sequence and sulphation pattern that promotes a high affinity interaction by inducing a large-scale conformational change in AT. The minimal pentasaccharide sequence binds to AT by a well characterised induced-fit mechanism involving at least two steps: an initial weak interaction with a dissociation constant K1 in the µM range, followed by a conformational change in AT (AT*) that improves the affinity by 1000-fold characterised by a rapid forward rate constant k2 and slow reverse rate constant k−24 (Scheme 1).
Scheme 1.
AT is thus unique amongst the known heparin binding proteins in its specificity of interaction and for utilizing conformational change to improve affinity. Defining the conformational events which lie behind the specific binding of AT to its pentasaccharide is the starting point for understanding the determinants of GAG-protein specificity and the action of the widely-used drug heparin.
AT was one of the first identified members of the serpin family of serine protease inhibitors5. The serpins share a common fold consisting of an upper β-barrel domain and a lower helical domain bridged by a large five-stranded β-sheet (the A sheet), supporting a long (20 residue) exposed reactive centre loop (RCL) that serves as the protease bait. The serpin mechanism of protease inhibition has been well characterised and is shared by all inhibitory members of the family6. Proteases engage the RCL of the serpin within the active site cleft and begin hydrolysis of the scissile P1-P1′ bond (nomenclature of Schechter and Berger7). At the acyl-enzyme intermediate step, where an ester bond exists between the catalytic Ser195 of the protease and the P1 residue of the serpin, the RCL rapidly incorporates into β-sheet A carrying the protease 70Å to the opposite pole of the serpin. This event results in a hyperstable serpin, and the pulling force exerted on the active site loop of the protease distorts the catalytic architecture and partially unfolds the protease, thereby preventing hydrolysis of the ester linkage.
AT shares the overall serpin fold and mechanism, but the first structures of native AT published in 19948,9 surprisingly revealed a subtle but important difference with other native serpins; the N-terminal portion of the RCL (the so-called ‘hinge-region’) was inserted into β-sheet A (Fig. 1a, left panel). The result was a conformationally constrained RCL, less flexible than for other serpins, and it was proposed that the loss in RCL accessibility to proteolytic attack accounted for the poor inhibitory activity of native AT10. This hypothesis was supported by biochemical data that showed mutations in the hinge (at the P14 position) affected heparin affinity and the rate of factor Xa inhibition11,12. The structure of AT bound to a synthetic high-affinity pentasaccharide (Idraparinux) followed in 199713, and revealed the expected expulsion of the hinge region and a closure of β-sheet A to the normal five-stranded form (Fig. 1b). Other conformational changes were associated, including the apparent N-terminal extension of helix A, the formation of a new helix P at the N-terminus of helix D, and a two turn extension at the C-terminus of helix D. In addition, helix D rotated by ∼17° and the lower helical domain was observed to rotate relative to the β-barrel domain. These secondary and tertiary structural changes were accompanied by multiple subtle alterations in hydrogen bonding and the packing of hydrophobic side chains on the interior of AT, all of which might contribute to the overall free-energy of heparin binding.
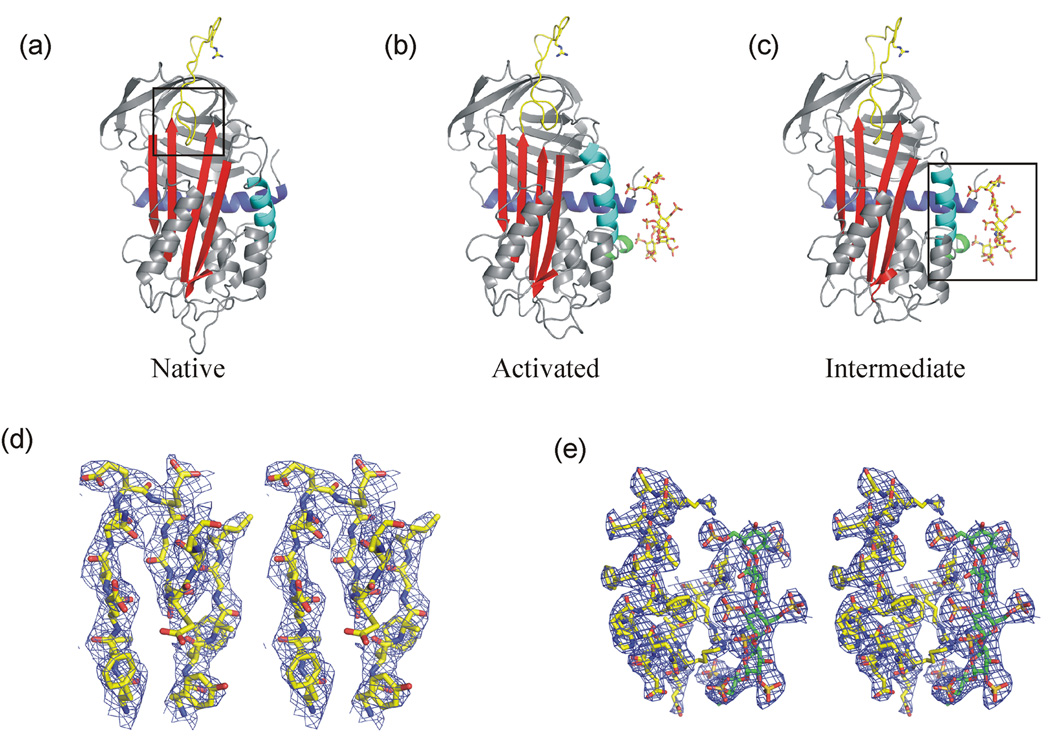
Figure 1.
Structures of native and pentasaccharide bound forms of AT. Ribbon diagrams of antithrombin in its native (a), pentasaccharide-bound activated (b) and pentasaccharide-bound intermediate states (c) are shown in the classic orientation, with β-sheet A (red) facing and the reactive centre loop (yellow) on top. The P1 Arg residue and the heparin pentasaccharide are shown as sticks. Helix A is coloured blue and the major heparin binding helix (helix D) is cyan. Heparin binding results in the formation of the P-helix (green) and the 2-turn C-terminal extension of the D-helix (the heparin binding site is indicated by a box in (c)). In the native and intermediate states, the N-terminal portion of the RCL (the hinge region, indicated by a box in (a)) is partially inserted into the middle of β-sheet A. Expulsion of the hinge and closure of the gap between strands 3 and 5A allows the extension of helix D in the activated conformation. (d) Stereo representation of electron density (blue, contoured at 1σ) surrounding residues in the hinge region and the tops of strands 3 and 5A reveal a native like state in the new intermediate structure described in this manuscript. (e) Similar high-quality density is seen in the heparin binding region (pentasaccharide shown in green and AT in yellow).
We and others have shown by mutagenesis and structural studies that the expulsion of the hinge region is crucial for accelerating AT inhibition of factors IXa and Xa, but that thrombin is insensitive to the RCL conformation14–17. Heparin thus allosterically activates AT toward factors IXa and Xa resulting in a two order-of-magnitude increase in rate of inhibition, whereas thrombin inhibition is accelerated by ∼1000-fold solely through a bridging mechanism. Thus, we know that binding of the specific heparin pentasaccharide ultimately and necessarily results in the expulsion of the hinge region as seen in the original crystal structures.
However, we recently solved several crystal structures of AT bound to pentasaccharides that surprisingly revealed a native-like hinge region conformation18 (Fig. 1c)19. These structures call into question the contribution of hinge region expulsion and closure of β-sheet A to the overall energetics of heparin binding, and suggest a three step induced-fit binding mechanism (as opposed to the two step mechanism of Scheme 1) where a native-like state is in equilibrium with a hinge region expelled state.
To test this hypothesis we engineered an internally oriented disulphide bond between the P14 (S380C) and an adjacent residue on strand 5A (V375C) to prevent hinge region expulsion. We determined that ∼60% of the free energy provided by the AT conformational change to pentasaccharide binding affinity is supplied by hinge region expulsion, and that this step is the final conformational change in a three step induced-fit binding mechanism.
Results
Crystal structure of AT bound to the heparin pentasaccharide
Two previous structures of AT bound to synthetic heparins have shown a native-like hinge conformation. The first was an enzymatically deglycosylated recombinant S137A (β-glycoform) variant bound to the high-affinity pentasaccharide Idraparinux, grown in the presence of latent AT (to make the active/latent heterodimer), the hydrophobic compound ANS and 20% glycerol18. This surprising structure revealed the normal heparin-induced conformational changes in the lower portion of the molecule, except for the C-terminal extension of helix D. Videos depicting the changes from native to this intermediate to the activated form are given as supplemental information. The second structure was of a stabilised variant of AT (S137A/V317C/T401C) bound to a pentasaccharide-containing hexadecasaccharide19, and although it was not crystallised with latent AT as for previous structures of AT, it also revealed a native-like hinge region conformation. These structures suggested that hinge-region expulsion did not contribute significantly to the energetics of pentasaccharide binding to AT. However, both structures involved recombinant variants that were chemically different from plasma AT, and one was grown in the presence of additives (ANS and glycerol). For this reason we decided to determine the structure of plasma AT bound to the natural pentasaccharide, in the context of the active/latent heterodimer, as for the original structure that showed hinge region expulsion.
Multiple unsuccessful attempts were made to obtain diffraction quality crystals using the conditions from the original structure of the AT-pentasaccharide complex (batch method with large drops of protein in 20mM Tris, pH 8 and precipitant containing 30% PEG 4000 in 100mM sodium cacodylate, pH 7)13. We therefore used hanging drop vapour diffusion with a commercial precipitant (Peg/Ion screen from Hampton Research) to grow suitable crystals. The structure was solved to 3.0Å resolution (Table 1), and gave a normal latent monomer and an intermediate, native-like monomer, both of which were bound to the pentasaccharide in the normal fashion. The structure of the active component is shown in Fig. 1c, and is essentially identical to the previous intermediate solved in the context of the latent/active heterodimer18 (RMSD 0.66Å for 401 equivalent Cα atoms). The quality of the density was very good, allowing unequivocal tracing of the hinge region (Fig. 1d) and the heparin binding site (Fig. 1e). This new intermediate structure adds weight to the hypothesis that hinge region expulsion does not contribute significantly to the energetics of heparin binding to AT.
Table 1.
Data processing, refinement and final model (3EVJ)
| Crystal | ||
| Space Group | P21 | |
| Cell dimensions (Å) | a = 65.85 b = 87.05 c = 92.38 | |
| (°) | β = 106.18 | |
| Solvent content (%) | 46.1 | |
| Data Processing Statistics | ||
| Wavelength (Å) | 1.488 (SRS, beam line 14.1) | |
| Resolution (Å) | 63.25-3.00 | 3.16-3.00 |
| Total reflections | 62258 | 9162 |
| Unique reflections | 20068 | 2905 |
| Multiplicity | 3.1 | 3.2 |
| <I/σ(I)> | 10.9 | 2.1 |
| Completeness (%) | 99.2 | 99.4 |
| Rmerge | 0.116 | 0.523 |
| Model | ||
| Number of atoms modeled: | ||
| Protein | 6164 | |
| Water | 45 | |
| Carbohydrate | 183 | |
| Pentasaccharide | 182 | |
| Average B-factor (Å2) | 64.2 | |
| Refinement statistics | 63.24-3.00Å | 3.19-3.00Å |
| Reflections in working/free set | 19039/1016 | 3163/153 |
| R-factor/R-free (%) | 23.1 / 28.9 | 32.9 / 35.8 |
| r.m.s. deviation of bonds(Å)/angles (°) from ideality | 0.008 / 1.5 | |
| Ramachandran plot; residues in most favored region (%) | 74.2 | |
| additionally allowed region (%) | 23.8 | |
| generously allowed region (%) | 2.0 | |
| disallowed region (%) | 0 | |
Design of hinge-region disulphide bond
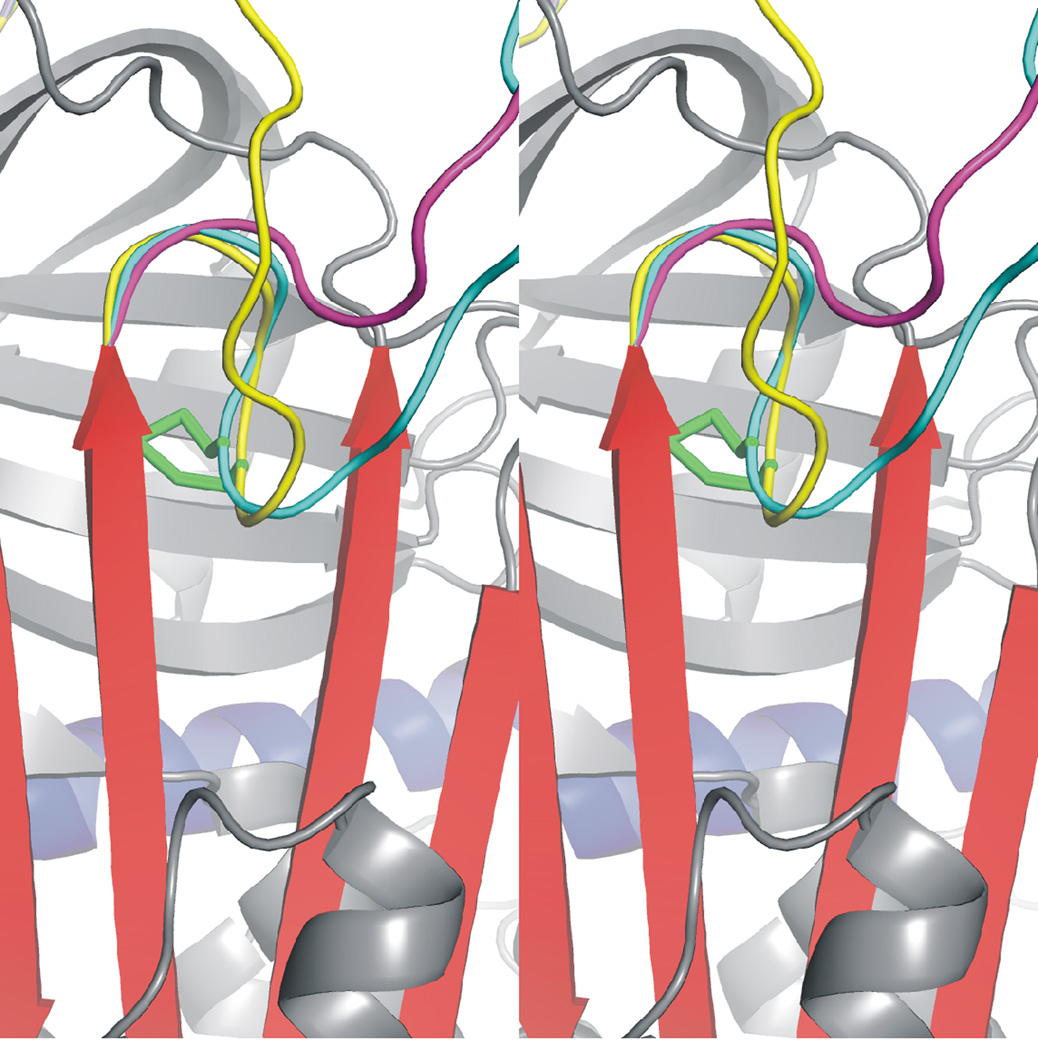
We previously made and characterised an externally oriented disulphide bond that was intended to allow hinge region expulsion but prevent its extension, in order to determine the requirement of hinge region extension in protease recognition15. We found that hinge region extension was absolutely required for pentasaccharide acceleration of factor IXa and Xa inhibition by AT, but did not affect thrombin inhibition. Surprisingly, the new disulphide bond itself did not affect pentasaccharide affinity nor the intrinsic fluorescence enhancement associated with pentasaccharide binding. Here, in order to prevent hinge region expulsion and the closure of sheet A, we engineered an internally oriented disulphide bond between the P14 residue (S380C) and the P19 residue on strand 5A (V375C). Energy minimization of residues 376–400 within the RCL indicated that formation of the disulphide bond renders the hinge incapable of expulsion from the gap between the tops of strands 3 and 5A, in contrast to wild-type which readily expels the hinge under the same regime (Fig. 2).
Figure 2.
Internal disulphide bond prevents the expulsion of the hinge from β-sheet A. Close up of the internally oriented disulphide bond (AT coloured and oriented as in Fig. 1), between the hinge region and strand 5A. The RCL before energy minimization is coloured in yellow, and the stereo view allows the depth of the disulphide bond (green) to be appreciated. Energy minimization of the RCL results in the full expulsion of the hinge in the absence of the disulphide bond (magenta), while no expulsion is permitted for the variant after the same energy minimization regime (cyan).
Kinetics of protease inhibition
From our study on the externally oriented disulphide bond mutant15 and previous work on a S380C variant11, we expected the novel 375–380 disulphide bond to have minimal effects on the kinetics of protease inhibition in the absence of heparin. This is indeed what we observed (Table 2). The second-order rate constant of thrombin inhibition was not affected, when accounting for a small increase in stoichiometry of inhibition (SI) observed for the S380C and double variants. The V375C mutation had no effect on the rate of factor Xa inhibition in the absence or presence of the pentasaccharide. In contrast, the S380C variant inhibited factor Xa 3.7-fold slower than control in the absence of heparin, consistent with the mutation stabilizing the native state of the hinge, as observed previously11. The pentasaccharide-accelerated rate of fXa inhibition for the S380C variant was reduced 2.3-fold relative to the control. The double variant inhibited factor Xa at a rate 6.8-fold reduced relative to the S137A control in the absence of heparin, but only 1.8-fold reduced relative to the single Cys mutant S380C, indicating that the disulphide bond did not appreciably alter the basal rate of factor Xa inhibition. However, pentasaccharide acceleration of factor Xa inhibition by the V375C/S380C variant was totally abolished, even at saturating pentasaccharide concentrations. This indicates quantitative formation of the disulphide bond, as observed previously for the externally oriented disulphide bonded variant15. We conclude from the kinetic data that the designed disulphide bond is quantitatively formed, and that its presence does not affect the overall structure or activity of native AT.
Table 2.
Thrombin and factor Xa inhibition by the AT variants.
| Thrombin | fXa | ||||||
|---|---|---|---|---|---|---|---|
| AT | H5 | SI | kapp (mM−1s−1) | kapp x SI | SI | kapp (mM−1s−) | kapp x SI |
| S137A | − | 1.04± 0.01 | 8.23 ± 0.21 | 8.56 ± 0.17 | 1.04± 0.05 | 5.59± 0.10 | 5.81± 0.21 |
| + | 1.19±0.01 | 756± 9 | 900±9 | ||||
| V375C | − | 1.08± 0.01 | 7.06 ± 0.32 | 7.62 ± 0.25 | 1.13± 0.02 | 7.12± 0.15 | 8.05± 0.16 |
| + | 1.24±0.04 | 664± 15 | 823±23 | ||||
| S380C | − | 1.66± 0.06 | 5.47 ± 0.09 | 9.08 ± 0.25 | 1.63± 0.01 | 0.961± 0.025 | 1.57± 0.30 |
| + | 2.03± 0.05 | 194± 1 | 394±6 | ||||
| V375C/S380C | − | 2.11± 0.06 | 3.08 ± 0.05 | 6.50 ± 0.15 | 1.99± 0.01 | 0.430± 0.015 | 0.856± 0.021 |
| + | 2.08± 0.03 | 0.331± 0.036 | 0.688±0.053 | ||||
Pentasaccharide-induced fluorescence enhancement
A 40% enhancement of the intrinsic fluorescence of AT is commonly used as a marker of heparin binding. The bulk of the fluorescence enhancement at 340nm (74%) has been shown to be the result of change in the environment of two tryptophan residues, Trp225 and Trp307, both of which are located in the upper domain of AT20. Trp225 is of particular importance as it lies directly under the inserted hinge region between strands 3 and 5A. Trp189 contributes only about 8%, and Trp49, located on helix A in the heparin binding site, contributes 18%. Interestingly, the wavelength of maximum fluorescence (λmax) is red-shift by 3–5 nm for Trp225 and Trp307, and is unchanged for Trp189 upon heparin binding. Therefore, the 1nm blue-shift normally associated with AT binding to the pentasaccharide is due entirely to a 15nm blue-shift of Trp4920.
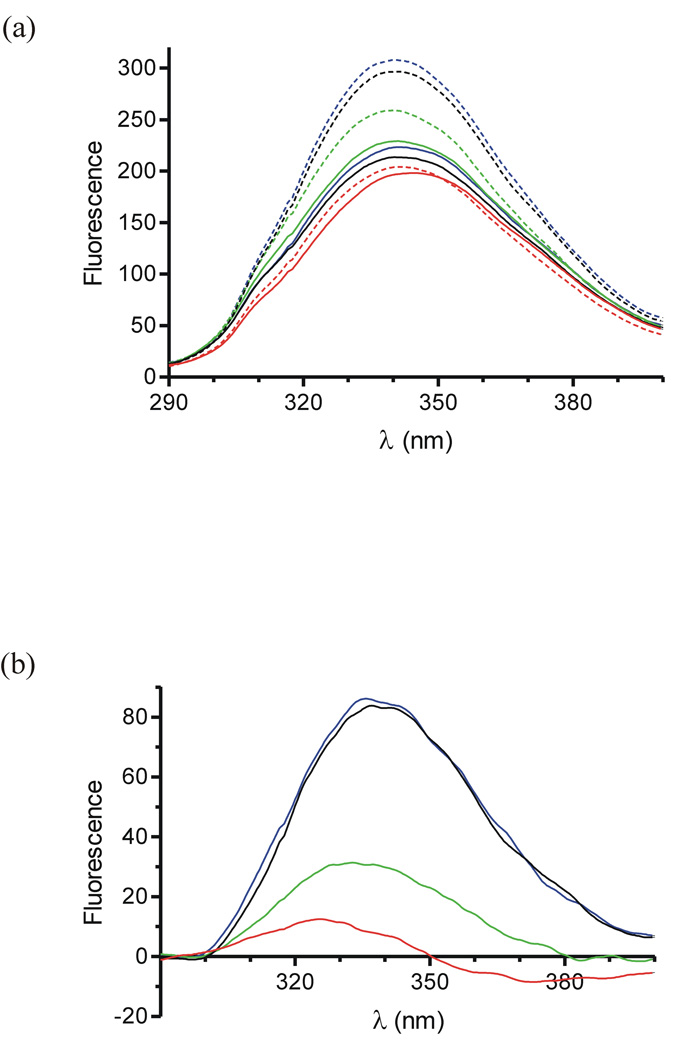
Fluorescence spectra were collected for the control, two single Cys, and disulphide bonded variants of AT to assess their ability to undergo the native to activated conformational change upon pentasaccharide binding, and the results are given in Fig. 3a and Table 3. The spectra of the native control and single Cys variants were essentially superimposable, indicating that the individual mutations did not appreciably affect the fluorescence properties of native AT. Upon pentasaccharide binding the fluorescence of the control and V375C variants were identically enhanced by ∼38%. The S380C variant had a reduced fluorescence enhancement of 13%, consistent with previous studies and suggesting a larger population of a native-like hinge when saturated with the pentasaccharide11. This interpretation is supported by a pentasaccharide-accelerated rate of factor Xa inhibition which is lower for S380C than for the control. The intensity of the fluorescence spectrum of the native disulphide bonded variant is slightly reduced relative to the controls. The fluorescence enhancement upon saturation with the pentasaccharide was a mere 3.6% at 340 nm, and the largest effect was a 3 nm blue-shift in λmax.
Figure 3.
Fluorescence spectra of AT variants. (a) The intrinsic fluorescence spectra of control (black), V375C (blue), S380C (green) and V375C/S380C (red) ATs in the absence (solid lines) and presence (dashed lines) of the pentasaccharide. (b) Difference spectra of curves shown in (a) reveal the fluorescence change due to the binding of the pentasaccharide to the variants of AT (coloured as in (a)).
Table 3.
Fluorescence properties of AT variants with and without the pentasaccharide.
| % Enhancement | ||||||
|---|---|---|---|---|---|---|
| AT | H5 | λmax (nm) | F at λmax | Δλmax (nm) | at λmax | at 340nm |
| S137A | − | 340.25 | 213.6 | +0.5 | 38.9 | 38.9 |
| + | 340.75 | 296.7 | ||||
| V375C | − | 341.25 | 223.4 | −1.0 | 37.8 | 38.0 |
| + | 340.25 | 307.9 | ||||
| S380C | − | 340.75 | 229.3 | −0.75 | 13.0 | 13.0 |
| + | 340 | 259.0 | ||||
| V375C/S380C | − | 344.5 | 198.3 | −2.75 | 2.9 | 3.6 |
| + | 341.75 | 204.1 | ||||
F is the fluorescence intensity.
To determine which of the Trp residues responds to pentasaccharide binding in the disulphide bonded variant, we subtracted the native fluorescence spectrum from the pentasaccharide-bound spectrum for each variant (Fig. 3b). The difference spectra are identical for the control and the V375C variant, and that of the S380C variant is reduced in magnitude by about 3-fold and is blue shifted by 3 nm. The disulphide bonded mutant exhibits 10-fold lower fluorescence change than the control, and a 7.5 nm blue shift. The small fluorescence enhancement and large blue shift upon pentasaccharide saturation of the disulphide bonded variant correspond closely to the fluorescence changes attributed to Trp49 alone20.
Contribution of hinge region expulsion to pentasaccharide affinity
We determined equilibrium dissociation constants (Kd) for all AT variants by titrating the pentasaccharide into a solution of AT and following change in intrinsic fluorescence (Table 4). At physiological ionic strength (I=0.15) pentasaccharide affinity for V375C was identical to that of the control, and for S380C was reduced by 35-fold, consistent with the earlier report where full-length heparin was used11. The reduced pentasaccharide affinity for the S380C variant, coupled with the fluorescence and kinetic results, suggest that the Cys side chain stabilises the hinge region inserted state in the absence and presence of the pentasaccharide, providing evidence that hinge region expulsion contributes substantially to the energetics of pentasaccharide binding.
Table 4.
Pentasaccharide binding properties of AT variants.
| Antithrombin | Ionic Strength | Kd (nM) | Z* | KNI (µM)* |
|---|---|---|---|---|
| S137A | 0.150 | 3.1 ± 0.8 | 4.28 | 2.41 |
| 0.225 | 13.2 ± 3.5 | |||
| 0.300 | 26.8 ± 1.9 | |||
| 0.375 | 92.5 ± 6.8 | |||
| V375C | 0.150 | 3.1 ± 0.1 | 3.96 | 1.42 |
| 0.225 | 9.4 ± 1.4 | |||
| 0.300 | 29.8 ± 3.8 | |||
| 0.375 | 60.7 ± 6.7 | |||
| S380C | 0.075 | 10.9 ± 0.7 | 4.03 | 64.3 |
| 0.1125 | 35.6 ± 8.2 | |||
| 0.150 | 108.2 ± 12.7 | |||
| 0.200 | 346.1 ± 61.4 | |||
| V375C/S380C | 0.050 | 5.4 ± 5.2 | 4.08 | 203.7 |
| 0.075 | 24.9 ± 0.9 | |||
| 0.1125 | 133.2 ± 25.5 | |||
| 0.150 | 321.3 ± 14.0 | |||
Z and KNI are the number of ionic interactions and the non-ionic dissociation constant, respectively, as defined in the Methods section.
Since the V375C variant does not affect pentasaccharide binding affinity, the predicted additive affect of the V375C/S380C mutations would be identical to the S380C variant on its own (i.e. 35-fold reduction in binding affinity). However, the affinity of the double variant for the pentasaccharide was reduced by over 100-times relative to the control, reflecting the independent contribution of the new disulphide bond. Based on the determined Kds we calculated the free energies of binding to be −11.6 kcal/mol for the control and −8.8 kcal/mol for the V375C/S380C variant, yielding a 2.8 kcal/mol difference. In the simple two-step model of pentasaccharide binding to AT (Scheme 1), the Kd of the initial interaction (K1) for our control (recombinant β-glycoform) has been determined to be on the order of 5 µM, and the conformational change step reduces the overall Kd to 3 nM21. The free energy difference of −4.4 kcal/mol is thus the total contribution of the induced-fit conformational change, and our results show that −2.8 kcal/mol of which (64%) is due to the expulsion of the hinge and the subsequent closure of β-sheet A.
Ionic and non-ionic contributions to pentasaccharide binding
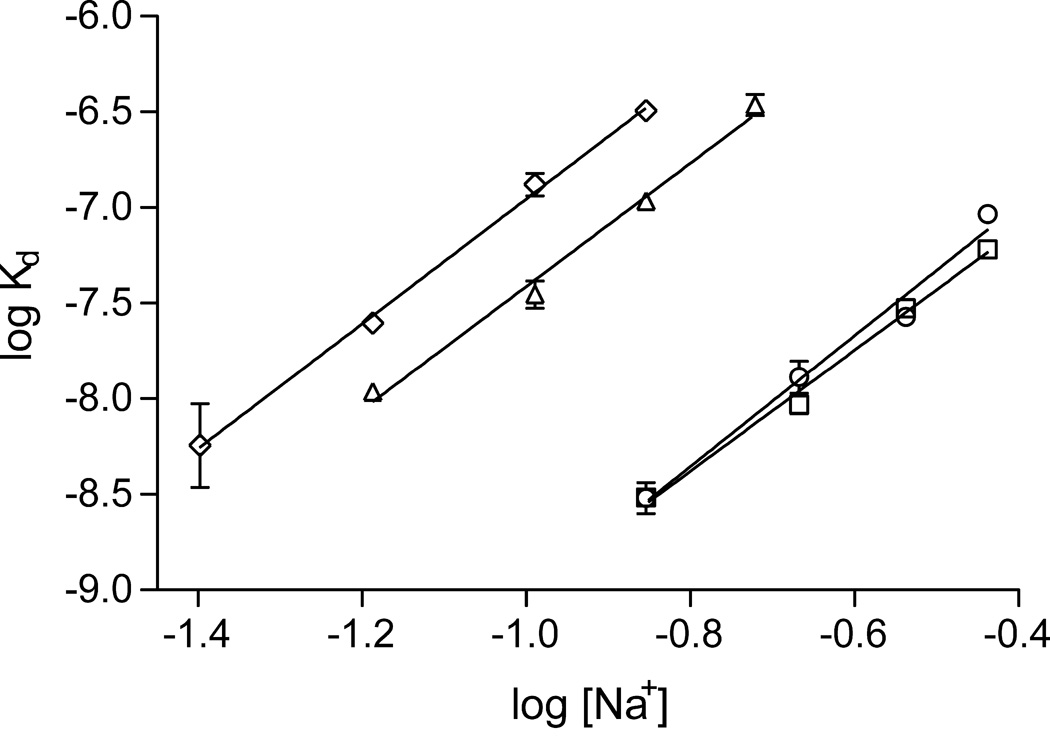
In order to determine how the blocking of hinge region expulsion affects the affinity of AT for the pentasaccharide, we determined dissociation constants at a range of ionic strengths. The slope of the double log plot of Kd vs. sodium ion concentration (Fig. 4) yields the number of ionic interactions (Z), and the y-intercept corresponds to the dissociation constant at 1M NaCl (KNI) (Table 4). The results of the analysis indicate that the number of ionic interactions is not affected by preventing hinge region expulsion and the associated conformational changes. This is supported by the crystal structures of AT-pentasaccharide complexes in the latent, intermediate and fully activated conformations that show equivalent binding interactions. The difference in pentasaccharide affinity for the variants is accounted for solely by a weakening of the non-ionic contribution (27-fold for S380C, and 85-fold or 143-fold for V375C/S380C when compared to the control or V375C, respectively). KNI has been interpreted as the contribution of direct non-ionic interactions between AT and the pentasaccharide, however, our results suggest that the major contributor to the term is the conformational change throughout AT.
Figure 4.
Dependence of pentasaccharide affinity on Na+ concentration reveals the ionic and non-ionic contributions to binding. Linear regression of the double log plot gave identical slopes for all AT variants (control, circle; V375C, square; S380C, triangle; V375C/S380C, diamond), whereas the y-intercepts were significantly different for the S380C and double mutants, indicating a weaker non-ionic contribution to binding.
Discussion
Although it has been known for several years that hinge region expulsion is a consequence of pentasaccharide binding, it was not clear how or to what extent this event contributed to the free energy of binding. The initial two step model assumed a concerted conformational change affecting the whole of AT and leading to a ∼1000-fold improvement in heparin affinity. This model was called into question by two crystal structures revealing a pentasaccharide-bound state with a native-like hinge. The crystal structure of plasma AT bound to the natural pentasaccharide, reported here, strengthens the case that hinge region expulsion is decoupled from the other conformational changes associated with heparin binding. The observed intermediate crystal structures are consistent with the two step model only if hinge region expulsion and the consequent closure of β-sheet A and extension of helix D are energetically silent, and the intermediate and activated states are in rapid equilibrium. Alternatively, the intermediate may be energetically distinct from the activated state, and therefore represent another step in the induced-fit mechanism.
To distinguish between these two possibilities, we designed and produced an AT variant with an internally oriented disulphide bond between the P14 residue (S380C) and an adjacent residue on strand 5A (V375C). We showed that the disulphide bond was quantitatively formed and that it prevented the conformational change that is reported by Trp307 and Trp225 in the upper domain of AT. The heparin binding properties of the variant indicate that hinge region expulsion is not energetically silent, but that it contributes more than half of the energy of pentasaccharide binding provided by the conformational change of AT. Thus, the crystal structures of pentasaccharide-bound AT with native-like hinge regions represent a conformation that is energetically distinct from the fully activated form. We hypothesize that this conformation is an actual intermediate on the pathway to AT activation for three reasons. Firstly, there is no direct link between the heparin binding site and the hinge region, and so it is likely that local conformational changes near heparin will precede changes in more remote regions. Secondly, the crystal structure and biochemical results reported here show that the intermediate conformation is a distinct higher energy state than the fully activated conformation. Thirdly, we know from previous analysis of the native, intermediate, activated and latent states that helix D extension cannot occur until β-sheet A has adopted the closed five-stranded state. It is also evident that β-sheet A cannot close until the hinge has been expelled.
From a functional viewpoint it also makes sense to have an intermediate between the native and activated states. AT requires RCL expulsion and extension to achieve full activity towards factors IXa and Xa, yet it also requires rapid RCL incorporation into β-sheet A to inhibit proteases. If pentasaccharide binding really locked AT in the hinge region expelled state, then it would also prevent or slow hinge region reinsertion, effectively turning AT from a protease inhibitor into a substrate. A hinge region inserted intermediate on the pathway to activation suggests a hinge region inserted intermediate on the pathway to full incorporation of the RCL. Our study shows that the energy difference between these two states is significant, but far less than between the native and activated states, and would predictably aid in allowing the reinsertion of the hinge. Evidence that AT is finely balanced between the substrate and inhibitory pathways is provided by studies showing higher SIs in the presence of heparin at physiological ionic strength22.
The heparin binding mechanism of AT can now be understood to involve three steps (Scheme 2). In the first step AT interacts weakly with the pentasaccharide (H) to form an initial encounter complex (AT:H) with a Kd in the µM range (K1). Conformational changes in AT result in a medium affinity intermediate complex (AT+:H) with a Kd of about 300 nM. This intermediate state sets the stage for the final and activating conformational change of hinge region expulsion, closure of β-sheet A and extension of helix D, resulting in a further 100-fold increase in binding affinity. The incremental nature of this three step induced-fit mechanism allows us to address how the binding of the pentasaccharide along helix D effects the expulsion of the hinge region from β-sheet A.
Scheme 2.
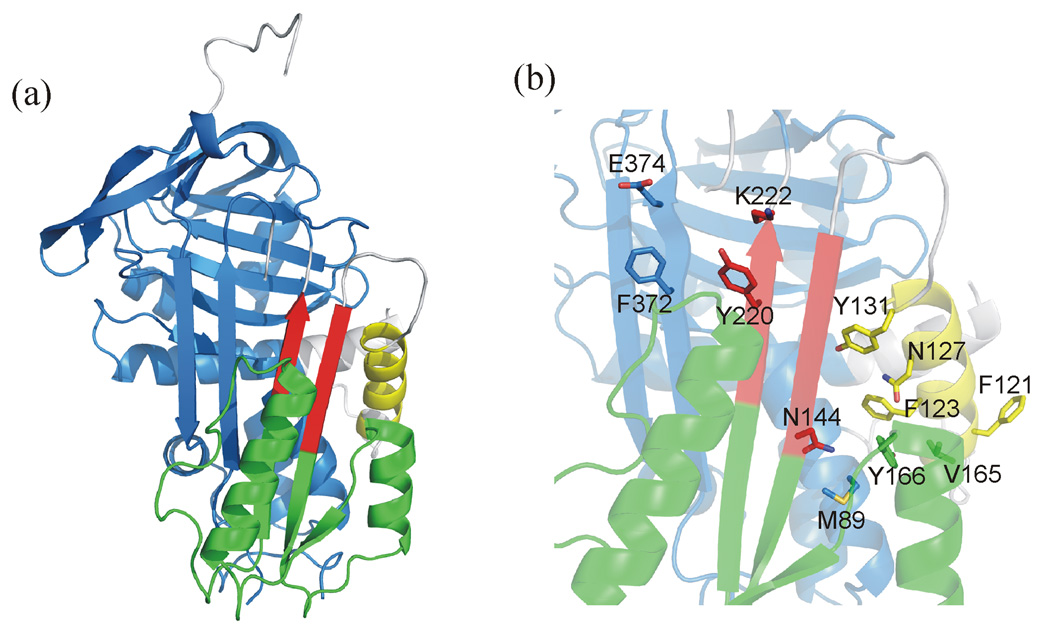
In a previous analysis of the native and activated states, AT was divided into several rigid substructures that moved in a concerted fashion upon pentasaccharide binding23. The major ‘domain’ was composed of 167 residues, corresponding to the top and left-hand side of AT (in the classic orientation, as in Fig. 1a), and all other substructure movements were calculated relative to this constant domain. The current analysis includes the intermediate structure, and addresses how the initial conformational change facilitates the expulsion of the hinge. We superimposed the constant domains and reassessed the size and composition of the rigid fragments based on structural differences between native (1E05) and intermediate (1NQ9), and between intermediate and activated (1E03). We find that AT can be divided into four groups, three of which move essentially as rigid bodies relative to the large constant domain (Fig. 5). Group 1 is the constant domain, and consists of 238 residues (55–107, 224–378, 401–430), including the C-terminal half of helix A, helices B, C, G and H, the entirety of β-sheets B and C, and strands 5 and 6 of β-sheet A. The second largest fragment, group 2 (145–217), is composed of helices E and F, strand 1A and the bottom halves of strands 2 and 3A. The third group (119–131) is the core of helix D, and the fourth group (139–144, 218–222) is composed of the top halves of strands 2 and 3A.
Figure 5.
Illustration of rigid fragments and important side chains. (a) A ribbon diagram of the native structure of AT (oriented as in Fig. 1) reveals the four rigid groups by colour: blue for group 1, green for group 2, yellow for group 3 and red for group 4. Plastic regions that do not move as part of any rigid body are coloured gray. For simplicity, the flexible N-terminus and N-terminal region of the RCL are not shown. (b) The side chains that are predicted to play important roles in both conformational changes are shown as sticks on a semitransparent ribbon background (coloured as before).
It is readily apparent from Supplemental Video 1 of the morph between the three states that the majority of the conformational change occurs during the first step, with the second step affecting only group 4 and plastic regions such as the C-terminus of helix D and the hinge. The final conformational changes must be sequential for steric reasons, with expulsion of the hinge allowing the sliding movement of the tops of strands 2 and 3, which in turn permits the elongation of helix D. This deconstruction of the AT conformational changes allows us to address the issue of how the first conformational change promotes the expulsion of the hinge region from β-sheet A.
It is natural to consider propagation of conformational change in terms of rigid fragments bumping into one another in a sequential way, akin to dominos tumbling. For the heparin-induced conformational change of AT, it is safe to assume that the conformational change must start at the heparin binding site and eventually affect the gap between strands 3 and 5 of sheet A and thus the environment of the hinge. Two of the three rigid fragments, groups 2 and 3, move prior to hinge expulsion (native to intermediate structures). Group 3 is the principal heparin binding site consisting of the core of helix D, and it is here that initial conformational changes are likely to take place. As shown in Supplemental Video 2, helix D rotates in response to pentasaccharide binding to align the basic residues and form what has been called a binding groove. Residues 120–124 on helix D make multiple van der Waals contacts with residues 161–166 of helix E. The 17° rotation of helix D in response to the initial heparin interaction pivots on the side chain of Phe123 and clashes into helix E. There are several important consequences for the packing of side chains, including the large movement of Tyr166. This and other changes directly affect the bottom of β-sheet A, resulting in a significant closure of the gap between strands 3 and 5A (Supplemental Video 3).
It is thought that the hinge is in rapid equilibrium between inserted and expelled states, and that the effect of heparin binding is to trap the hinge in the expelled state by the closure of β-sheet A. Thus, the question is not how do the initial conformational changes set the stage for hinge region expulsion, rather, how they prepare AT for rapid and stable closure of the remaining gap between strands 3 and 5A. Supplemental Video 4 addresses this question by showing a close-up of the hinge during the initial and final conformational changes. Movement of Tyr131 closes the middle of sheet A, bringing Tyr220 on s3A into van der Waals contact with Phe372 on s5A. These residues maintain contact in the final step, to achieve a perpendicular π stack. Electrostatics also play a role in effecting the rapid closure of the gap at the top of sheet A. Lys222 on strand 3A and Glu374 on s5A move into ionic bonding distance in the final step, with Glu374 also hydrogen bonding with the side chain hydroxyl group of Tyr220. Along with the additional β-sheet hydrogen bonds between strands 3 and 5A, the side chain interactions will stabilize the closed conformation and keep the hinge in an expelled state. Mutation of any of these residues would be expected to affect either the rate of final conformational change, k3, or the rate of its reversal, k−3. In addition, the subsequent extension of the C-terminus of helix D is likely to help keep sheet A closed, since blocking helix D extension has been shown to selectively increase koff (k−2 in the original two step model)24.
In summary, the data presented here provide important new insight into the role of conformational change in the induced-fit heparin binding mechanism of AT, and reveal a plausible mechanism for the propagation of conformational change from the heparin binding site to the distant hinge region of the RCL, required for the allosteric activation of AT.
Materials and Methods
Materials
Human factor Xa was obtained from Enzyme Research Laboratories (Swansea, UK), and human alpha thrombin from Haematologic Technologies (Essex Junction, VT). Linear polyethylenimine (25000 Da) was obtained from Polysciences Inc (Eppelheim, Germany). Human Embryonic Kidney cells expressing the Epstein Barr Nuclear Antigen 1 were obtained from ATCC (293 c18, CRL-10852). The heparin pentasaccharide (fondaparinux) was kindly provided by Maurice Petitou. Chromogenic substrates S2238 and S2222 were purchased from Chromogenix. All crystallization reagents were purchased from Hampton Research. Plasma AT was purified from outdated fresh-frozen plasma as before25. The latent form of AT was produced by the glycerol method as previously described26.
Recombinant AT expression and purification
The V375C, S380C and V375C/S380C mutations were constructed on the recombinant β-glycoform S137A AT background in order to reduce glycosylation heterogeneity and facilitate purification27. S137A is sometimes referred to in the text as ‘control’ and the S137A/V375C, S137A/S380C and S137A/V375C/S380C variants are referred to as V375C, S380C and V375C/S380C (or double variant or disulphide-bonded variant) for clarity. Site directed mutagenesis of the pCEP4-S137A/AT plasmid was performed using the Stratagene QuikChange Site-Directed Mutagenesis kit and the appropriate primers for the mutations. Human Embryonic Kidney cells expressing the Epstein Barr Nuclear Antigen 1 (HEK-EBNA) were grown to 60% confluence at 37°C, 5% CO2, in DMEM with GlutaMAX-I medium (Invitrogen) supplemented with 5% fetal bovine serum (Sigma). Transfection was performed by addition of 1/10 volume of pCEP4/AT plasmid (3µg/ml) preincubated for 15 minutes in DMEM with polyethylenimine (15µg/ml). After 24 hours the cells were washed with PBS and exchanged into CD-CHO medium (Invitrogen) supplemented with 4mM L-glutamine and 0.25mg/ml Geneticin (Invitrogen). Cells were grown to confluence and media was harvested after 72 hours. The double variant expressed at levels similar to wild-type and the two single Cys controls. Purification was achieved by a combination of affinity chromatography on heparin-Sepharose and anion exchange chromatography using Q-Sepharose. Purified material was stored at −80°C prior to use. Concentration of AT was determined by absorbance at 280 nm and was judged to contain no significant fraction of unreactive material (e.g. latent or polymers) by its ability to react with thrombin by SDS-PAGE (data not shown). Quantitative formation of designed disulphide bond was confirmed by the absolute inability of the pentasaccharide to improve the rate of fXa inhibition (see Results).
Energy minimization
To verify that the designed disulphide bond would prevent hinge region expulsion, 5000 cycles of energy minimization was conducted on a model of the variant (and on wild-type) using CNS (version 1.2)28 with all residues, except 376–400 corresponding to the RCL, fixed.
Crystallization, data collection and refinement
Crystals were grown by mixing 10 mg/ml plasma native and latent ATs (in 20mM Tris, pH 8.0) at a 1:1 ratio, followed by the addition of a small excess (1.2 molar) of the pentasaccharide. The previous study used the synthetic high-affinity pentasaccharide (Idraparinux); we used the synthetic version of the natural heparin pentasaccharide (Fondaparinux). Drops containing 2µl each of protein solution and precipitant solution (200 mM NH4F, 20% PEG 3350) were mixed by pipette on a cover-slip, and placed on a Linbro hanging-drop plate. Crystals grew within one week at room temperature. Data were collected from a single flash-cooled crystal (100K) after cryoprotection in 20% glycerol at Daresbury Synchrotron Radiation Source (Warrington, Cheshire, UK) station 14.1. Data were processed using Mosflm, Scala and Truncate29, and the structure was solved by molecular replacement using the program Phaser30 with 1NQ9 as the search model. Refinement was conducted with the program CNS28 (version 1.2) employing non-crystallographic symmetry restraints throughout, and XtalView31 was used for model building. Data processing and refinement statistics are given in Table 1. Coordinates and structure factors have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Databank with accession code 3EVJ. Molecular morphing was conducted using the program LSQMAN32 followed by geometry refinement using XtalView31. Molecular images were created using Pymol33.
Stoichiometries of inhibition
Stoichiometries of protease inhibition (SI) by the AT variants were determined for thrombin in the absence of heparin and for factor Xa both in the presence and absence of equimolar concentrations of pentasaccharide. Proteases were incubated at room temperature with increasing concentrations of AT in 20 mM sodium phosphate, 100 mM NaCl, 0.1 mM EDTA, 0.1% PEG 8000, 0.1% bovine serum albumin, pH 7.4 (PBSA). Reactions (30 µl final volume) were allowed to go to completion and subsequently 200 µl of chromogenic substrate (300 µM S-2222 or S-2238 for factor Xa and thrombin, respectively) was added. Residual protease activity was determined by monitoring absorbance at 405 nm on a Thermomax plate reader (Molecular Devices, Winnersh, UK). The fractional residual protease activity was plotted against the AT/protease ratio, and the SI was taken as the x-intercept.
Rates of inhibition
The rates of inhibition of fXa and thrombin were determined under pseudo first-order conditions in AT, in PBSA essentially as before34. Briefly, 10 µl of protease (final concentration 30 nM) was incubated with an equal volume of AT so that the final inhibitory concentration of AT (accounting for the SI) was 10-times that of the protease. Reactions were incubated for between 15 seconds and 90 minutes at room temperature in the wells of a microtitre plate. At the end of the incubation period reactions were quenched by the addition of 200 µl of 300 µM chromogenic substrate, and residual protease activity was determined as above. Observed pseudo first-order rate constants (kobs) were taken as the slope of the linear plot of the natural log of residual protease activity versus time. The second-order rate constants (kapp) were calculated by dividing kobs by the total AT concentration. Pentasaccharide catalyzed second-order rate constants were derived by determining kobs at a range of pentasaccharide concentrations (5 nM to 1 µM) and calculating the slope of the linear plot of kobsversus heparin concentration. Pentasaccharide binding affinities were accounted for as previously34. No acceleration of fXa inhibition was observed for the disulphide bonded variant, even at saturating pentasaccharide concentrations.
Fluorescence studies
Fluorescence spectra were recorded for the AT variants (0.3µM) in the absence and presence of saturating pentasaccharide (10µM) on a PerkinElmer Life Sciences 50B spectrofluorimeter, exciting at 280 nm with slits set to 2.5 nm. Five scans were averaged and smoothed. Equilibrium dissociation constants (Kd) for the AT-pentasaccharide interaction were determined essentially as described previously21. Briefly, change in intrinsic fluorescence of AT (15–150 nM) upon titration of the pentasaccharide was monitored at 340 nm for controls and at 332 nm for the V375C/S380C variant. Band widths of 3.5 nm for both excitation and emission were used. All titrations were carried out at room temperature in 20 mM sodium phosphate, 0–325 mM NaCl, 0.1 mM EDTA, 0.1% PEG 8000, pH 7.4. Fluorescence emission intensity was taken as the average of 100 measurements recorded at 1 s intervals for each addition of pentasaccharide (longer for the disulphide variant). Data were fitted as previously described35. The double log plot of Kd against [Na+] provided the dissociation constant at 1M salt (KNI) and the number of ionic interactions (Z), according to the equation logKd= logKNI+Zψlog[Na+], where ψ for heparin is 0.836.
Supplementary Material
Four videos are given as supplemental information to aid in visualizing the conformational events on the pathway to activation of AT. All are coloured according to the rigid groups detailed in the main text, with group 1 blue, group 2 green, group 3 yellow and group 4 red. Supp_vid1.mpg shows the front view of the conformational transitions between native and intermediate, and following a pause, the transition between the intermediate and activated conformers. Supp_vid2.mpg shows the same conformational changes, but from a side view, focussing on the heparin binding site. Supp_vid3.mpg is a close-up of the heparin binding site, and only shows the first conformational transition to illustrate the rotation of helix D and the movement of important side chains. Supp_vid4.mpg shows both conformational transitions and important side chains from the perspective of the hinge region and the top of sheet A.
Acknowledgements
This work was supported by an MRC Senior Non-clinical Fellowship, the National Institutes of Health (HL68629) and the Tesny Perry Trust. We thank Dr. Catherine Huntington for help establishing the HEK-EBNA expression system.
Footnotes
Accession Numbers
Coordinates and structure factors have been deposited in the Protein Data Bank with accession number 3EVJ.
References
- 1.Rau JC, Beaulieu LM, Huntington JA, Church FC. Serpins in thrombosis, hemostasis and fibrinolysis. J. Thromb. Haemost. 2007;5 Suppl. 1:102–115. doi: 10.1111/j.1538-7836.2007.02516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Boven HH, Lane DA. Antithrombin and its inherited deficiency states. Semin. Hematol. 1997;34:188–204. [PubMed] [Google Scholar]
- 3.Ishiguro K, Kojima T, Kadomatsu K, Nakayama Y, Takagi A, Suzuki M, Takeda N, Ito M, Yamamoto K, Matsushita T, Kusugami K, Muramatsu T, Saito H. Complete antithrombin deficiency in mice results in embryonic lethality. J. Clin. Invest. 2000;106:873–878. doi: 10.1172/JCI10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olson ST, Srinivasan KR, Bjork I, Shore JD. Binding of high affinity heparin to antithrombin III. Stopped flow kinetic studies of the binding interaction. J. Biol. Chem. 1981;256:11073–11079. [PubMed] [Google Scholar]
- 5.Hunt LT, Dayhoff MO. A surprising new protein superfamily containing ovalbumin, antithrombin-III, and alpha 1-proteinase inhibitor. Biochem. Biophys. Res. Commun. 1980;95:864–871. doi: 10.1016/0006-291x(80)90867-0. [DOI] [PubMed] [Google Scholar]
- 6.Huntington JA, Read RJ, Carrell RW. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000;407:923–926. doi: 10.1038/35038119. [DOI] [PubMed] [Google Scholar]
- 7.Schechter I, Berger A. On the size of the active site in proteases. I Papain. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 8.Carrell RW, Stein PE, Fermi G, Wardell MR. Biological implications of a 3 A structure of dimeric antithrombin. Structure. 1994;2:257–270. doi: 10.1016/s0969-2126(00)00028-9. [DOI] [PubMed] [Google Scholar]
- 9.Schreuder HA, de Boer B, Dijkema R, Mulders J, Theunissen HJ, Grootenhuis PD, Hol WG. The intact and cleaved human antithrombin III complex as a model for serpin-proteinase interactions. Nat. Struct. Biol. 1994;1:48–54. doi: 10.1038/nsb0194-48. [DOI] [PubMed] [Google Scholar]
- 10.van Boeckel CA, Grootenhuis PD, Visser A. A mechanism for heparin-induced potentiation of antithrombin III. Nat. Struct. Biol. 1994;1:423–425. doi: 10.1038/nsb0794-423. [DOI] [PubMed] [Google Scholar]
- 11.Huntington JA, Gettins PG. Conformational conversion of antithrombin to a fully activated substrate of factor Xa without need for heparin. Biochemistry. 1998;37:3272–3277. doi: 10.1021/bi972182o. [DOI] [PubMed] [Google Scholar]
- 12.Huntington JA, Olson ST, Fan B, Gettins PG. Mechanism of heparin activation of antithrombin. Evidence for reactive center loop preinsertion with expulsion upon heparin binding. Biochemistry. 1996;35:8495–8503. doi: 10.1021/bi9604643. [DOI] [PubMed] [Google Scholar]
- 13.Jin L, Abrahams JP, Skinner R, Petitou M, Pike RN, Carrell RW. The anticoagulant activation of antithrombin by heparin. Proc. Natl. Acad. Sci. USA. 1997;94:14683–14688. doi: 10.1073/pnas.94.26.14683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olson ST, Bjork I. Role of protein conformational changes, surface approximation and protein cofactors in heparin-accelerated antithrombin-proteinase reactions. Adv. Exp. Med. Biol. 1992;313:155–165. doi: 10.1007/978-1-4899-2444-5_16. [DOI] [PubMed] [Google Scholar]
- 15.Langdown J, Johnson DJ, Baglin TP, Huntington JA. Allosteric activation of antithrombin critically depends upon hinge region extension. J. Biol. Chem. 2004;279:47288–47297. doi: 10.1074/jbc.M408961200. [DOI] [PubMed] [Google Scholar]
- 16.Johnson DJ, Li W, Adams TE, Huntington JA. Antithrombin-S195A factor Xa-heparin structure reveals the allosteric mechanism of antithrombin activation. EMBO J. 2006;25:2029–2037. doi: 10.1038/sj.emboj.7601089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, Johnson DJ, Esmon CT, Huntington JA. Structure of the antithrombin-thrombin-heparin ternary complex reveals the antithrombotic mechanism of heparin. Nat. Struct. Mol. Biol. 2004;11:857–862. doi: 10.1038/nsmb811. [DOI] [PubMed] [Google Scholar]
- 18.Johnson DJ, Huntington JA. Crystal structure of antithrombin in a heparin-bound intermediate state. Biochemistry. 2003;42:8712–8719. doi: 10.1021/bi034524y. [DOI] [PubMed] [Google Scholar]
- 19.Johnson DJ, Langdown J, Li W, Luis SA, Baglin TP, Huntington JA. Crystal structure of monomeric native antithrombin reveals a novel reactive center loop conformation. J. Biol. Chem. 2006;281:35478–35486. doi: 10.1074/jbc.M607204200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meagher JL, Beechem JM, Olson ST, Gettins PG. Deconvolution of the fluorescence emission spectrum of human antithrombin and identification of the tryptophan residues that are responsive to heparin binding. J. Biol. Chem. 1998;273:23283–23289. doi: 10.1074/jbc.273.36.23283. [DOI] [PubMed] [Google Scholar]
- 21.Johnson DJ, Huntington JA. The influence of hinge region residue Glu381 on antithrombin allostery and metastability. J. Biol. Chem. 2004;279:4913–4921. doi: 10.1074/jbc.M311644200. [DOI] [PubMed] [Google Scholar]
- 22.Olson ST. Heparin and ionic strength-dependent conversion of antithrombin III from an inhibitor to a substrate of alpha-thrombin. J. Biol. Chem. 1985;260:10153–10160. [PubMed] [Google Scholar]
- 23.Whisstock JC, Pike RN, Jin L, Skinner R, Pei XY, Carrell RW, Lesk AM. Conformational changes in serpins: II. The mechanism of activation of antithrombin by heparindagger. J. Mol. Biol. 2000;301:1287–1305. doi: 10.1006/jmbi.2000.3982. [DOI] [PubMed] [Google Scholar]
- 24.Belzar KJ, Zhou A, Carrell RW, Gettins PG, Huntington JA. Helix D elongation and allosteric activation of antithrombin. J. Biol. Chem. 2002;277:8551–8558. doi: 10.1074/jbc.M110807200. [DOI] [PubMed] [Google Scholar]
- 25.Zhou A, Huntington JA, Carrell RW. Formation of the antithrombin heterodimer In vivo and the onset of thrombosis. Blood. 1999;94:3388–3396. [PubMed] [Google Scholar]
- 26.Carrell RW, Huntington JA, Mushunje A, Zhou A. The conformational basis of thrombosis. Thromb. Haemost. 2001;86:14–22. [PubMed] [Google Scholar]
- 27.Mushunje A, Zhou A, Carrell RW, Huntington JA. Heparin-induced substrate behavior of antithrombin Cambridge II. Blood. 2003;102:4028–4034. doi: 10.1182/blood-2003-05-1560. [DOI] [PubMed] [Google Scholar]
- 28.Brunger AT, Adams PD, Clore GM, Delano WL, Gros P, GrosseKunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. ACTA CRYST. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Leslie AWG. AnonymousJoint CCP4 and ESF-EACMB Newsletter on Protein. Crystallography. 1992:26. [Google Scholar]
- 30.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 31.McRee DE. A Visual Protein Crystallographic Software System for X11/XView. J. Mol. Graphics. 1992;10:44–46. [Google Scholar]
- 32.Kleywegt GJ. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr. D Biol. Crystallogr. 1996;52:842–857. doi: 10.1107/S0907444995016477. [DOI] [PubMed] [Google Scholar]
- 33.San Carlos, CA, USA: DeLano, W. DeLano Scientific LLC; 2002. The PyMOL Molecular Graphics System. [Google Scholar]
- 34.Olson ST, Bjork I, Shore JD. Kinetic characterization of heparin-catalyzed and uncatalyzed inhibition of blood coagulation proteinases by antithrombin. Methods Enzymol. 1993;222:525–559. doi: 10.1016/0076-6879(93)22033-c. [DOI] [PubMed] [Google Scholar]
- 35.Olson ST, Shore JD. Binding of high affinity heparin to antithrombin III. Characterization of the protein fluorescence enhancement. J Biol Chem. 1981;256:11065–11072. [PubMed] [Google Scholar]
- 36.Olson ST, Bjork I. Predominant contribution of surface approximation to the mechanism of heparin acceleration of the antithrombin-thrombin reaction. Elucidation from salt concentration effects. J. Biol. Chem. 1991;266:6353–6364. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Four videos are given as supplemental information to aid in visualizing the conformational events on the pathway to activation of AT. All are coloured according to the rigid groups detailed in the main text, with group 1 blue, group 2 green, group 3 yellow and group 4 red. Supp_vid1.mpg shows the front view of the conformational transitions between native and intermediate, and following a pause, the transition between the intermediate and activated conformers. Supp_vid2.mpg shows the same conformational changes, but from a side view, focussing on the heparin binding site. Supp_vid3.mpg is a close-up of the heparin binding site, and only shows the first conformational transition to illustrate the rotation of helix D and the movement of important side chains. Supp_vid4.mpg shows both conformational transitions and important side chains from the perspective of the hinge region and the top of sheet A.