Abstract
Conformationally constrained CIS-amide chimeric inhibitors of Hsp90 have been synthesized and evaluated for their Hsp90 inhibitory activity. These new compounds exhibited Hsp90 ATPase inhibition and induced Hsp90-dependent client protein degradation in a dose-dependent manner. Biological data reported herein suggests that amide bond isomerization of geldanamycin derivatives plays an important role in affinity for the heteroprotein complex present in cancer cells.
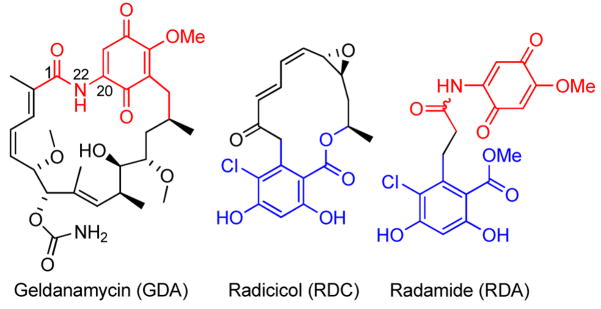
The 90 kDa heat shock proteins (Hsp90) are ATP-dependent molecular chaperones overexpressed in response to cellular stress and necessary for the folding and rematuration of nascent and denatured polypeptides, respectively.1,2,3,4 Two natural products, geldanamycin5 (GDA, Figure 1) and radicicol6 (RDC), bind the Hsp90 N-terminal binding pocket and competitively inhibit ATP binding resulting in degradation of Hsp90-dependent client proteins via the ubiquitin-proteasome pathway.7 Hsp90 clientele play key roles in all six hallmarks of cancer8, therefore inhibition of the Hsp90 protein folding machinery results in simultaneous disruption of all six mechanisms of oncogenesis.9,10 Consequently, not only has Hsp90 emerged as a promising anti-cancer target11, but GDA and RDC have proven to represent excellent models for which the development of new Hsp90 inhibitors can be pursued for drug development and mechanistic studies.12
Figure 1.
Inhibitors of Hsp90.
Although RDC and GDA bind Hsp90 with high affinity, their modes of binding and inhibitory activities are different. Radicicol exists in the same bent conformation when bound and unbound to Hsp90 and produces a favorable entropy of 8.3 cal/mol upon binding. Not surprisingly, the predisposition of RDC to the bent conformation is believed to be responsible for its similar activity in both cellular and recombinant assays.13
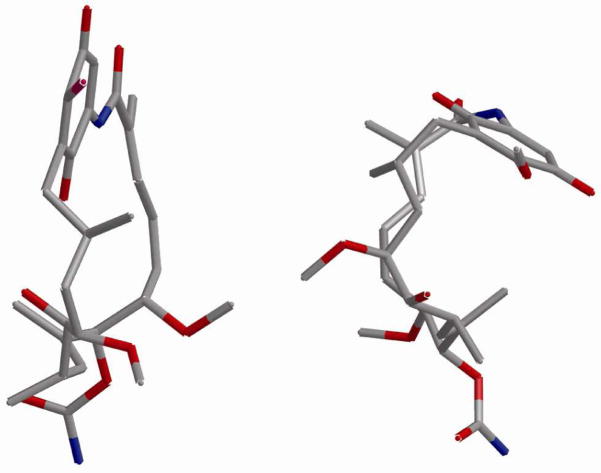
In contrast to the bent, cis-amide conformation of GDA when bound to Hsp90, both solution and crystal structures have demonstrated that this natural product exists in an extended, trans-amide conformation when unbound to Hsp90 (Figure 2).14 Multiple studies have demonstrated that prior to binding Hsp90, GDA must undergo two conformational changes; the ansa ring must rotate over the benzoquinone moiety and the amide bond must isomerize from trans to cis by rotation about C1–N22 and C20–N22 (Figure 2). The first event is reported to occur spontaneously; however isomerization of the amide bond is estimated to exceed 20 kcal/mol.15 Accordingly, isothermal titration calorimetry (ITC) experiments have shown that GDA exhibits an entropic penalty of −6.4 cal/mol upon binding Hsp90.16,17
Figure 2.
GDA conformations in solution (left) and bound to Hsp90 (right).
As a consequence of these thermodynamic data, Santi and coworkers hypothesized that GDA analogues containing a predisposed cis-amide bond will result in ~1000 fold increase in Hsp90 affinity through reduction of entropic penalties. Such postulations have inspired subsequent studies aimed at determining the effect of trans/cis isomerization of the GDA-amide moiety.18,19,20 However, to the best of our knowledge, no analogues have been synthesized that exhibit a predisposed cis-amide functionality.
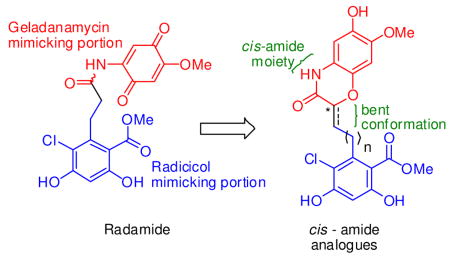
Recently, chimeric inhibitors of Hsp90 were disclosed that contained both the quinone ring from GDA and the resorcinol moiety of RDC in an attempt to mimic the hydrogen bonding interactions exemplified by the two natural products when bound to the Hsp90 N-terminal nucleotide-binding pocket. This approach has produced novel scaffolds for Hsp90 inhibition, however, none of the reported analogues exhibit conformational characteristics exhibited by either natural product when bound to Hsp90.21,22,23 Analysis of the seco derivative, radamide (RDA, Figure 1), revealed the potential to introduce conformational aspects of both natural products when bound to Hsp90. Herein we present the first series of conformationally constrained chimeric Hsp90 inhibitors exhibiting both a cis-amide moiety and a bent conformation.
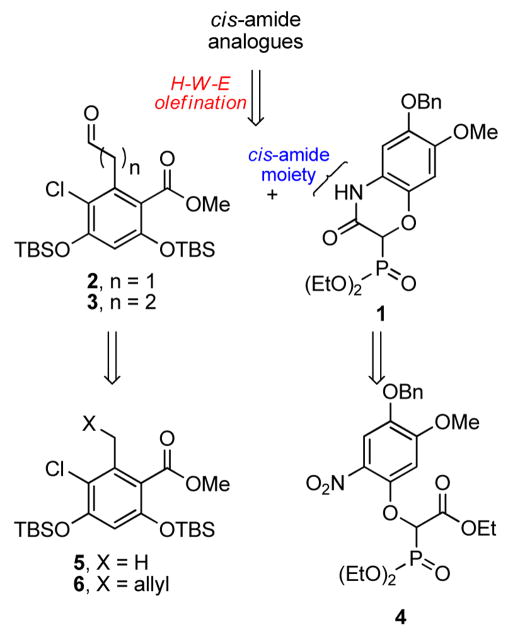
Retrosynthetically, we envisioned the desired analogues could be obtained via a Horner–Wadsworth–Emmons (HWE) olefination reaction between cyclic phosphonate 1 and homologated aldehydes, 2 and 3 (Scheme 1). Compound 1 was proposed to result from tandem reduction/intramolecular cyclization of compound 4, which could be obtained from commercially available 4-benzyloxy-3-methoxybenzaldehyde in 4 steps. Aldehydes 2 and 3 could be prepared directly from 521 and 6, respectively.
Scheme 1.
Retrosynthetic analysis for the synthesis of cis-amide chimeric analogues
Commencing with commercially available 4-benzyloxy-3-methoxybenzaldehyde (7, Scheme 2), phenol 8 was formed via Dakin oxidation.24 Treatment of 8 with diazophosphonate 9, resulted in the phenolic ether 10, upon a rhodium carbenoid mediated O-H insertion.25 Regioselective nitration of 10 was accomplished under mild conditions enlisting ammonium nitrate and trifluoroacetic anhydride. Treatment of 4 with tin(II) chloride under refluxing conditions resulted in not only reduction of the nitro group to the corresponding aniline, but also cyclization to give the desired lactam, 1. The protected resorcinol precursors were prepared by treatment of 5 with lithium diisopropylamide at −78°C to generate the benzylic anion, which was quenched upon addition of dimethylformamide to afford the aldehyde product, 2, or with allyl bromide to give 6 (Scheme 3). Oxidation of 6 with osmium tetraoxide gave the corresponding diol, which was cleaved in situ with sodium periodate to yield the homologated aldehyde, 3.
Scheme 2.
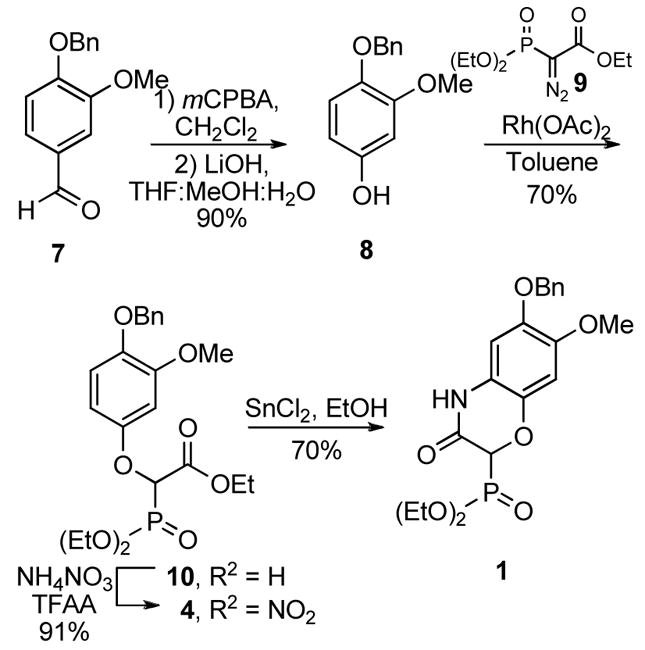
Synthesis of cis-amide cyclic phosphonate
Scheme 3.
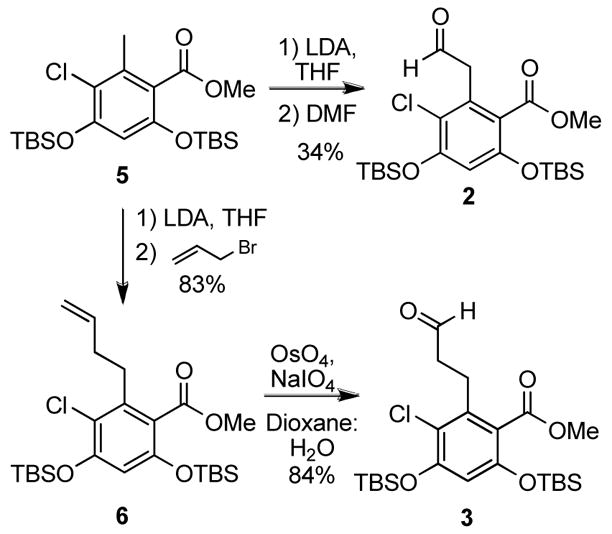
Synthesis of homologated aldehydes.
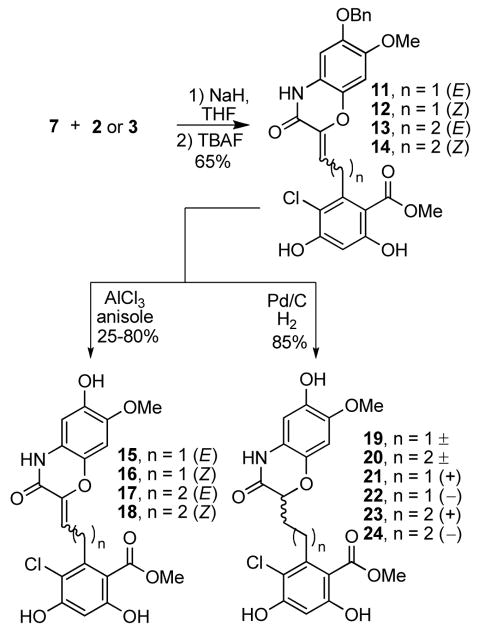
With the synthons in hand, the fragments were joined via a Horner–Wadsworth–Emmons olefination reaction (3:1, E:Z) and subsequent removal of the protecting groups with tetrabutylammonium fluoride to give compounds 11–14 (Scheme 4).
Scheme 4.
Fragment coupling and deprotection.
Originally, we were interested in the saturated analogues, but realizing the olefinated intermediates exhibited higher conformational rigidity than the saturated analogues, we attempted to selectively remove the benzyl ether in the presence of the unsaturated amide with FeCl3, Pd(OAc)2/Et3SiH/Et3N, and NaI/TMSCl. However, none of these conditions afforded the desired products. Eventually, conditions were employed utilizing aluminum(III) chloride in anisole that effectively cleaved the benzyl ether without alteration of the α,β-unsaturated amide to afford compounds 15–18 (Scheme 4).
Standard hydrogenation conditions with palladium on carbon under a hydrogen atmosphere yielded racemic products 19–20, which were subjected to chiral HPLC to afford the enantiopure analogues, 21–24 (Scheme 4).
After completing the synthesis of 10 analogues, biological investigation was undertaken. Anti-proliferation studies with compounds 15–24 were conducted with MCF7 and SKBR3 breast cancer cell lines. As shown in table 1, the E-olefin is more active than the Z-olefin for both linker lengths and the (+)-enantiomer is more active than the (−)-enantiomer.
Table 1.
Anti-proliferation and ATPase activity of cis-amide analogues. IC50 values are expressed in μM concentrations unless noted otherwise.
| compound | MCF7 | SKBR3 | ATPase |
|---|---|---|---|
| 15 | 3.2 ± 0.1 | 1.8 ± 0.1 | 2.4 ± 0.1 |
| 16 | 78.9 ± 11.3 | 57.1 ± 8.4 | – |
| 17 | 1.5 ± 0.1 | 1.2 ± 0.1 | 1.6 ± 0.0 |
| 18 | 9.8 ± 1.9 | 15.9 ± 4.1 | – |
| 19 | 4.8 ± 1.1 | 7.3 ± 0.2 | – |
| 20 | 6.3 ± 0.9 | 7.8 ± 0.2 | – |
| 21 | 3.6 ± 0.1 | 2.1 ± 0.2 | 1.5 ± 0.1 |
| 22 | 14.9 ± 0.2 | 15.4 ± 0.8 | – |
| 23 | 2.7 ± 0.5 | 2.1 ± 0.7 | 1.2 ± 0.0 |
| 24 | 78.0 ± 4.4 | 15.4 ± 0.8 | – |
| Radamide | 18.6 ± 0.9a | 23.7 ± 1.7a | 5.9b |
| GDA | 9.8 ± 0.1 nM | 8.5 ± 1.1 nM | 2.5 ± 0.4 |
| RDC | 47.7 ± 2.6 nM | 37.5 ± 4.0 nM | 0.4 ± 0.0 |
IC50 = concentration needed to produce 50% inhibition,
= results obtained from recently submitted manuscript,
= obtained from reference #21.
To evaluate this series for inhibition of ATPase activity,26 recombinant yeast Hsp90 was overexpressed and purified.27 The purified protein was incubated with ATP in the presence of 15, 17, 21, and 23. As shown in Table 1, the IC50 values were similar to those found in the anti-proliferation studies, suggesting equipotency of these analogues for the heteroprotein complex present in cancer cells and the purified homodimeric species.
To confirm the cytotoxic nature of these compounds resulted from Hsp90 inhibition, compounds 17 and 23 were incubated with MCF7 cells for 24 h and western blot analyses were performed with the cell lysates (see supporting information). As expected, the immunoblots confirmed concentration-dependent degradation of Hsp90-dependent client proteins, Her-2 and Raf.
In conclusion, we have designed, synthesized, and provided biological data for a series of analogues that not only contain hydrogen bonding moieties exhibited by GDA and RDC, but also exhibit the conformational biases adopted by the natural products when bound to Hsp90 via inclusion of a bent conformation and a cis-amide. In accord with Santi and co-workers, introduction of a cis-amide moiety increased Hsp90 inhibitory activity (~10 fold). As indicated by the biological results, introduction of both the cis-amide and the bent conformation to the chimeric inhibitors does not influence activity against purified recombinant Hsp90, however the activity against cancer cells increased ~10-fold with respect to the seco derivative, radamide.
These results coincide with the ideas posed by multiple groups, whom suggest a mechanistic difference between the heteroprotein complex present in cancer cells versus purified recombinant Hsp90, which is reminiscent of the form found in normal cells.15,28,29 In addition, the data suggests the heteroprotein complex is more sensitive to amide isomerization and conformational predisposition. Further SAR studies and co-crystallization experiments are ongoing in hopes to delineate the interactions of this class of compounds with Hsp90 and will be reported in due course.
Supplementary Material
Acknowledgments
The authors greatfully acknowledge the Prisinzano laboratory for instrumentation use. The authors also thank the Madison and Lila Self Graduate Fellowship and the American Foundation of Pharmaceutical Education (A.F.P.E.) for financial support. This project was funded by NIH/NCI CA109265.
Footnotes
Supporting Information Available Experimental procedures and characterization for all compounds. This material is avalible free of charge via the internet at http://pubs.acs.org.
References
- 1.Freeman BC, Morimoto RI. EMBO J. 1996;15:2969. [PMC free article] [PubMed] [Google Scholar]
- 2.Walter S, Buchner J. Angew Chem Int Ed. 2002;41:1098. doi: 10.1002/1521-3773(20020402)41:7<1098::aid-anie1098>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 3.Schneider C, Sepp LL, Nimmesgern E, Ouerfelli O, Danishefsky S, Rosen N, Hartl FU. Proc Natl Acad Sci USA. 1996;93:14536. doi: 10.1073/pnas.93.25.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schumacher RJ, Hansen WJ, Freeman BC, Alnemri E, Litwack G, Toft DO. Biochemistry. 1996;35:14889. doi: 10.1021/bi961825h. [DOI] [PubMed] [Google Scholar]
- 5.Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Proc Natl Acad Sci USA. 1994;91:8324. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schulte TW, Akinaga S, Soga S, Sullivan W, Stensgard B, Toft D, Neckers LM. Cell Stress & Chap. 1998;3:100. doi: 10.1379/1466-1268(1998)003<0100:arbttn>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binder RJ, Blachere NE, Srivastava PK. J Biol Chem. 2001;276:17163. doi: 10.1074/jbc.M011547200. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. Cell. 2000;100:57. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 9.Maloney A, Workman P. Expert Opin Biol Ther. 2002;2:3. doi: 10.1517/14712598.2.1.3. [DOI] [PubMed] [Google Scholar]
- 10.Neckers L. Curr Top Med Chem. 2006;6:1163. doi: 10.2174/156802606777811979. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhury S, Welch TR, Blagg BSJ. ChemMedChem. 2006;1:1331. doi: 10.1002/cmdc.200600112. [DOI] [PubMed] [Google Scholar]
- 12.Hadden MK, Lubbers DJ, Blagg BSJ. Curr Top Med Chem. 2006;6:1173. doi: 10.2174/156802606777812031. [DOI] [PubMed] [Google Scholar]
- 13.Roe SM, Prodromou C, O’Brien R, Ladbury JE, Piper PW, Pearl LH. J Med Chem. 1999;42:260. doi: 10.1021/jm980403y. [DOI] [PubMed] [Google Scholar]
- 14.Thepchatri P, Eliseo T, Cicero DO, Myles D, Snyder JP. J Am Chem Soc. 2007;129:3127. doi: 10.1021/ja064863p. [DOI] [PubMed] [Google Scholar]
- 15.Lee Y, Marcu MG, Neckers L. Chem & Biol. 2004;11:991. doi: 10.1016/j.chembiol.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 16.Chiosis G, Huezo H, Rosen N, Mimnaugh E, Whitesell L, Neckers L. Mol Cancer Ther. 2003;2:123. [PubMed] [Google Scholar]
- 17.Schulte TW, Neckers LM. Cancer Chemother Pharmacol. 1998;42:273. doi: 10.1007/s002800050817. [DOI] [PubMed] [Google Scholar]
- 18.Onuoha SC, Mukund SR, Coulstock ET, Sengerova B, Shaw J, McLaughlin SH, Jackson SE. J Mol Biol. 2007;372:287. doi: 10.1016/j.jmb.2007.06.065. [DOI] [PubMed] [Google Scholar]
- 19.Onodera H, Kaneko M, Takahashi Y, Uochi Y, Funahashi J, Nakashima T, Soga S, Suzuki M, Ikeda S, Yamashita Y, Rahayu ES, Kanda Y, Ichimura M. Bioorg & Med Chem Lett. 2008;18:1577. doi: 10.1016/j.bmcl.2008.01.072. [DOI] [PubMed] [Google Scholar]
- 20.Zhang MQ, Gaisser S, Nur-E-Alam M, Sheehan LS, Vousden WA, Gaitatzis N, Peck G, Coates NJ, Moss SJ, Radzom M, Foster TA, Sheridan RM, Gregory MA, Roe SM, Prodromou C, Pearl L, Boyd SM, Wilkinson B, Martin CJ. J Med Chem. 2008;51:5494. doi: 10.1021/jm8006068. [DOI] [PubMed] [Google Scholar]
- 21.Clevenger RC, Blagg BSJ. Org Lett. 2004;6:4459–4462. doi: 10.1021/ol048266o. [DOI] [PubMed] [Google Scholar]
- 22.Shen G, Blagg BSJ. Org Lett. 2005;7:2157. doi: 10.1021/ol050580a. [DOI] [PubMed] [Google Scholar]
- 23.Shen G, Wang M, Welch TR, Blagg BSJ. J Org Chem. 2006;71:7618. doi: 10.1021/jo061054f. [DOI] [PubMed] [Google Scholar]
- 24.Li S, Chen L, Tsai J, Tzeng J, Tsai I, Chi E. Tet Lett. 2007;48:2139. [Google Scholar]
- 25.Haigh D. Tetrahedron. 1994;50:3177. [Google Scholar]
- 26.Avila C, Kornilayev BA, Blagg BSJ. Bioorg Med Chem. 2006;14:1134. doi: 10.1016/j.bmc.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 27.Richeter K, Muschler P, Hainzl O, Buchner J. J Biol Chem. 2001;276:33689. doi: 10.1074/jbc.M103832200. [DOI] [PubMed] [Google Scholar]
- 28.Chiosis G, Neckers L. ACS Chem Biol. 2006;1:279. doi: 10.1021/cb600224w. [DOI] [PubMed] [Google Scholar]
- 29.Kamal A, Thao L, Sensintaffar J, Zhang L, Boehm MF, Fritz LC, Burrows FJ. Nature. 2003;425:407. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.