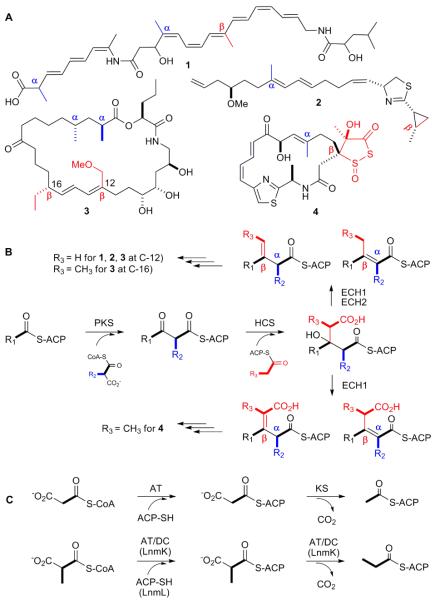
Both α- and β-alkylations contribute to the vast structural diversity displayed by polyketide natural products (Figure 1A).1 While the α-alkyl branches are typically derived from the extender units, the choice of which is dictated by the acyltransferase (AT) domain of modular polyketide synthases (PKSs),1 the βalkyl branches often result from the activities of hydroxymethylglutaryl-CoA (HMG-CoA) synthase homologs (HCSs).2 For a βmethyl branch, HCS catalyzes condensation of acetyl-S-acyl carrier protein (ACP) with the βcarbonyl group of the PKS-ACP-tethered growing polyketide intermediate to afford an HMG-S-ACP intermediate, which is subsequently dehydrated and decarboxylated by two enoyl-CoA hydratase homologs (ECH1 and ECH2) sequentially to afford a βmethylated intermediate in either olefinic form (Figure 1B). This pathway has been experimentally confirmed for the biosynthesis of bacillaene (1),3 curacin (2),4 and myxovirescin A (also known as TA) (3)5, and a dedicated set of three proteins - an ACP, an AT, and a ketosynthase homolog (KS) - has been identified that derives acetyl-SACP from malonyl-CoA for this pathway (Figure 1C).2-5
Figure 1.
(A) Selected polyketides bacillaene (1), curacin (2), myxovirescin A (3), and leinamycin (4) with a- (blue) or β-alkyl (red) branches; (B) a unified pathway for β-alkylation utilizing both acetyl-S-ACP and propionyl-S-ACP as substrates; and (C) distinct pathways for acetyl-S-ACP and propionyl-S-ACP biosynthesis.
A parallel pathway replacing acetyl-S-ACP with propionyl-S-ACP could be envisaged for βethyl branch introduction, and this proposal has been supported for 3 using chemoenzymatically prepared propionyl-S-ACP as a substrate (Figure 1B).5b However, counterparts for propionyl-S-ACP biosynthesis from methylmalonyl-CoA, such as the AT and KS enzymes required to generate acetyl-S-ACP from malonyl-CoA, are absent from gene clusters known to encode biosynthesis of polyketides with βethyl branches (Table 1); the origin of propionyl-S-ACP remains unknown.2-5
Table 1.
Enzymes that generate acetyl-S-ACP and propionyl-S-ACP and incorporate them into polyketides with β-alkyl branch (methyl for 1, 2, and 3 at C-12 or ethyl for 3 at C-16 and propionyl for 4).2-6
| Compd | AT/DC | AT | KS | ACP | HCS | ECH1 | ECH2 |
|---|---|---|---|---|---|---|---|
| 1 | - | PksC | PksF | AcpK | PksG | PksH | PksI |
| 2 | - | - | CurC | CurB | CurD | CurE | CurF |
| 3 (C-12) | - | TaV | TaK | TaB | TaC | TaX | TaY |
| 3 (C-16) | TaD | - | - | TaE | TaF | TaX | TaY |
| 4 | LnmK | - | - | LnmL | LnmM | LnmF | - |
Leinamycin (LNM, 4), a potent antitumor antibiotic, possesses a β-branched C3 unit, which is a part of its unique five-membered 1,3-dioxo-1,2-dithiolane moiety. We have previously cloned, sequenced, and characterized the lnm biosynthetic gene cluster from Streptomyces atroolivaceus S-140.6 Close examination of the lnm cluster revealed a subset of four genes - lnmL, lnmM, lnmF, and lnmK - encoding an ACP (LnmL), an HCS (LnmM), an ECH1 (LnmF), and a protein of unknown function (LnmK). Counterparts of LnmL, LnmM, and LnmF are present in biosynthetic clusters of polyketides with both β-methyl and βethyl branches,2-6 supporting the proposal that the C3 βbranch of 4 is likely installed by LnmL/LnmM/LnmF in a mechanistic analogy to the βmethyl branch for 1, 2 and 3. Homologs of LnmK however can only be found in gene clusters encoding the biosynthesis of ethyl branch-bearing polyketides, suggesting LnmK as a candidate for propionyl-S-ACP biosynthesis (Table 1) (Figure 1C). Here we report the characterization of LnmK as a bifunctional acyltransferase/decarboxylase (AT/DC) that derives propionyl-S-ACP from methylmalonyl-CoA. Hence, LnmK represents a new family of AT/DC enzymes supplying a key substrate for βalkylation in polyketide biosynthesis.
We first overproduced both LnmL and LnmK in Escherichia coli BL21(DE3) and purified them to near homogeneity (Figure S1). The purified LnmL was eluted as a single peak upon HPLC analysis (Figure 2A, panel I) and confirmed to be in its apo-form by ESI-MS analysis (Table S1). In vitro phosphopantetheinylation was carried out by incubating apo-LnmL with CoA in the presence of the known promiscuous phosphopantetheinyltransferase Svp,7 and the resultant holo-LnmL was confirmed by HPLC (Figure 2A, panel II) and ESI-MS (Table S1) analyses.
Figure 2.

(A) HPLC analysis of LnmK-catalyzed formation of propionyl-S-LnmL: (I) apo-LnmL (•), (II) holo-LnmL (▼), (III) holo-LnmL and propionyl-S-LnmL (♦), (IV) methylmalonyl-SLnmL (◊), (V) propionyl-S-LnmL. (B) LnmK-catalyzed loading of acyl-CoAs to holo-LnmL and (C) LnmK-catalyzed self-acylation as judged by (I) 4-15% SDS-PAGE and (II) autoradiogram: lane 1, molecular weight standards; lane 2, [1,3-14C2]methylmalonyl-CoA; lane 3, [1,3-14C2]malonyl-CoA; lane 4, [1-14C]propionyl-CoA; lane 5, [1-14C]acetyl-CoA.
We then established that LnmK is a bifunctional AT/DC catalyzing the formation of propionyl-S-LnmL. Holo-LnmL was incubated with [1-14C]acetyl-, [1-14C]propionyl-, [1,3-14C2]malonyl- or [1,3-14C2]methylmalonyl-CoA in the presence of LnmK, and the reaction mixtures were subjected to SDS-PAGE and phosphorimaging. LnmK specifically and efficiently catalyzed the loading of methylmalonyl-CoA to holo-LnmL, and no loading was observed with the other acyl-CoAs tested (Figure 2B). To verify the molecular identity of the acyl-S-LnmL species, the reaction was repeated with cold methylmalonyl-CoA, and the resultant product was subjected to HPLC and ESI-MS analyses. A distinct new product was formed (Figure 2A, panel III), ESIMS analysis of which remarkably revealed it as propionyl-S-LnmL (Table S1); LnmK apparently acts as bifunctional AT/DC, catalyzing both methylmalonyl transfer to form the methylmalonyl-S-LnmL intermediate and its subsequent decarboxylation to yield propionyl-S-LnmL (Figure 1C).
We finally determined the precise timing of acyl transfer and decarboxylation events catalyzed by LnmK. The fact that LnmK cannot decarboxylate methylmalonyl-CoA and only loads methylmalonyl-CoA, but not propionyl-CoA, to holo-LnmL, indicates that decarboxylation most likely occurs on methylmalonyl-S-LnmL. To directly verify this mechanism, we prepared methylmalonyl-S-LnmL via in vitro phosphopantetheinylation by incubating apo-LnmL with methylmalonyl-CoA in the presence of Svp.7 Methylmalonyl-S-LnmL formation was monitored by HPLC (Figure 2A, panel IV) and confirmed by ESI-MS (Table S1) analyses. Incubation of methylmalonyl-S-LnmL with LnmK allowed us to investigate LnmK's DC activity directly. LnmK catalyzes specific and efficient decarboxylation of methylmalonyl-S-LnmL to yield propionyl-S-LnmL whose identity was confirmed by HPLC (Figure 2A, panel V) and ESI-MS (Table S1) analyses. Taken together, these results unambiguously established that LnmK first transfers methylmalonyl from methylmalonyl-CoA to holo-LnmL to form methylmalonyl-S-LnmL and then decarboxylates the latter to form propionyl-S-LnmL (Figure 1C).
LnmK homologs are known but to date were all annotated as hypothetical proteins (Figure S2).2-5 We now propose LnmK to represent a new family of AT/DC enzymes supplying substrates for β-alkylation in polyketide biosynthesis. To further probe the catalytic mechanism of this newly discovered family of AT/DC enzymes, LnmK was incubated with [1,3-14C2] methylmalonyl-CoA in the absence of holo-LnmL, and the reaction mixtures were subjected to SDS-PAGE and phosphorimaging. Specific and efficient loading of [1,3-14C2]methylmalonyl-CoA onto LnmK was observed (Figure 2C), indicative of a transient acyl-LnmK intermediate in LnmK catalysis. This is reminiscent of ATs with Ser at their active sites,8 although no conserved AT or DC active site motif is apparent in LnmK (Figure S2).
In summary, LnmK has been characterized as a bifunctional AT/DC that catalyzes the formation of propionyl-S-ACP from methylmalonyl-CoA, accounting for the missing link for the β-ethyl or propionyl branch in polyketide biosynthesis. LnmK therefore could be exploited by combinatorial biosynthesis methods to engineer novel polyketides, especially those with β-alkyl branches. LnmK also represents an emerging family of novel AT/DC enzymes.
Supplementary Material
Acknowledgment
We thank Kyowa Hakko Kogyo Co. Ltd., Tokyo, Japan, for the wild-type S. atroolivaceus S-140 strain and the Analytical Instrumentation Center of the School of Pharmacy, UW-Madison for support in obtaining MS data. This work is supported in part by NIH grants CA106150 and CA113297.
Footnotes
Supporting Information Available: Full experimental details, Figures S1, S2, Table S1 are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Staunton J, Weissman KJ. Nat. Prod. Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]; (b) Fischbach MA, Walsh CT. Chem. Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 2.Calderone CT. Nat. Prod. Rep. 2008;25:845–853. doi: 10.1039/b807243d. [DOI] [PubMed] [Google Scholar]
- 3.Calderone CT, Kowtoniuk WE, Kelleher NL, Walsh CT, Dorrestein PC. Proc. Natl. Acad. Sci. U.S.A. 2006;103:8977–8982. doi: 10.1073/pnas.0603148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu L, Jia J, Liu H, Hakansson K, Gerwick WH, Sherman DH. J. Am. Chem. Soc. 2006;128:9014–9015. doi: 10.1021/ja0626382. [DOI] [PubMed] [Google Scholar]
- 5.(a) Simunovic V, Müller R. ChemBioChem. 2007;8:497–500. doi: 10.1002/cbic.200700017. [DOI] [PubMed] [Google Scholar]; (b) Simunovic V, Müller R. ChemBioChem. 2007;8:1273–1280. doi: 10.1002/cbic.200700153. [DOI] [PubMed] [Google Scholar]; (c) Calderone CT, Iwig DF, Dorrestein PC, Kelleher NL, Walsh CT. Chem. Biol. 2007;14:835–846. doi: 10.1016/j.chembiol.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Cheng Y, Tang G, Shen B. J. Bacteriol. 2002;184:7013–7024. doi: 10.1128/JB.184.24.7013-7024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cheng Y, Tang G, Shen B. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3149–3154. doi: 10.1073/pnas.0537286100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Tang G, Cheng Y, Shen B. Chem. Biol. 2004;11:33–45. doi: 10.1016/j.chembiol.2003.12.014. [DOI] [PubMed] [Google Scholar]; (d) Cheng Y, Tang G, Shen B. J. Nat. Prod. 2006;69:387–393. doi: 10.1021/np050467t. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Tang G, Cheng Y, Shen B. J. Bio. Chem. 2007;282:20273–20282. doi: 10.1074/jbc.M702814200. [DOI] [PubMed] [Google Scholar]
- 7.Du L, Shen B. Chem. Biol. 2001;6:507–517. doi: 10.1016/S1074-5521(99)80083-0. [DOI] [PubMed] [Google Scholar]
- 8.(a) Reeves CD, Murli S, Ashley GW, Piagentini M, Hutchinson CR, Mcdaniel R. Biochemistry. 2001;40:15464–15470. doi: 10.1021/bi015864r. [DOI] [PubMed] [Google Scholar]; (b) Ridley CP, Lee HY, Khosla C. Proc. Natl. Acad. Sci. U.S.A. 2008;105:4595–4600. doi: 10.1073/pnas.0710107105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.