Summary
Systems biology is the comprehensive and quantitative analysis of the interactions between all of the components of biological systems over time. Systems biology involves an iterative cycle, in which emerging biological problems drive the development of new technologies and computational tools. These technologies and tools then open new frontiers that revolutionize biology. Innate immunity is well suited for systems analysis, because the relevant cells can be isolated in various functional states and their interactions can be reconstituted in a biologically meaningful manner. Application of the tools of systems biology to the innate immune system will enable comprehensive analysis of the complex interactions that maintain the difficult balance between host defense and inflammatory disease. In this review, we discuss innate immunity in the context of the systems biology concepts, emergence, robustness, and modularity, and we describe emerging technologies we are applying in our systems-level analyses. These technologies include genomics, proteomics, computational analysis, forward genetics screens, and analyses that link human genetic polymorphisms to disease resistance.
Keywords: systems biology, innate immunity, Toll-like receptors, gene regulation, genomics, proteomics
Systems biology
We define systems biology as a comprehensive, quantitative analysis of the manner in which all of the components of a biological system interact functionally over time. To practice systems biology, one must capture and integrate global sets of biological data from as many hierarchical levels of information as possible. These could include DNA sequences, RNA and protein measurements, protein-protein and protein-DNA interactions, biomodules, signaling and gene regulatory networks, cells, organs, individuals, populations, and ecologies. The data are then transferred to comprehensive databases, where they are warehoused and annotated. Human minds are incapable of inferring the emergent properties of a system from thousands of data points, but we have evolved to intelligently interpret an enormous amount of visual information. The data are therefore transferred to visualization programs. This is the initiation point for the formulation of detailed graphical or mathematical models, which are then refined by hypothesis-driven, iterative systems perturbations and data integration. In this manner, the phenotypic features of the system are tied directly to the behavior of the protein and gene regulatory networks. Cycles of iteration will result in a more accurate model; ultimately, these models will explain the systems or emergent properties of the biological system of interest. Once the model is sufficiently accurate and detailed, it will allow biologists to accomplish two tasks never possible before: (1) predict the behavior of the system given any perturbation and (2) redesign or perturb the gene regulatory networks to create completely new emergent systems properties. This latter possibility lies at the heart of preventative medicine. Thus, systems biology is hypothesis driven, global, quantitative, iterative, integrative, and dynamic.
Systems analysis can only be executed by an interdisciplinary team of investigators that is also capable of developing required technologies and computational tools. In this model, biology dictates what new technology and computational tools should be developed. This too is an iterative cycle, however, as these tools open new frontiers in biology that go well beyond the original question. Thus, biology drives technology and computation, and, in turn, technology and computation revolutionize biology (Fig. 1). Creative interaction within an interdisciplinary team raises specific challenges. Biological systems, as opposed to engineered man-made systems, are not the result of a rational design process, but rather the result of a random evolutionary process that selects whatever works. For this reason, reverse-engineering approaches that require a rational underlying design will often be thwarted by biology. This is at variance with the approaches and culture of non-biologists.
Fig. 1. The iterative cycle of systems biology.

Biology dictates what new technology and computational tools must be developed to answer specific questions. Newly developed technologies and tools in turn open new frontiers, revolutionizing biology and generating new fields of inquiry.
Basic concepts crucial to understanding complex biological systems: emergence, robustness, and modularity
Emergence
Complex systems display properties, often called `emergent properties' that are not demonstrated by their individual parts and cannot be predicted even with a full understanding of the parts alone. The arch is an example of an emergent property that arises from simple constituents (Fig. 2). A comprehensive analysis of the physical properties of rocks (or monitors) will not predict that they give rise to an arch when assembled in a specific context. Life, too, is emergent and not inherent in its individual components. Mixing DNA, RNA, protein, carbohydrate, and lipid does not result in a functional biological system. Life is a consequence of their organization and interactions. A systems approach is necessary to understand how the emergent properties that underlie life are derived from the individual components of a biological system.
Fig. 2. Emergent properties.

Biological functions, like the arch, emerge from context-specific interactions of the constituent elements. Emergent functional properties cannot be understood by analyzing the constituents in isolation (adapted from an image posted anonymously at http://cache.gizmodo.com/gadgets/images/monitor_arch.jpg).
Robustness
Biological systems maintain phenotypic stability in the face of diverse perturbations imposed by the environment, stochastic events, and genetic variation. Robustness often arises as an emergent property through positive and negative feedback loops and other forms of control that constrain gene outputs. This feedback insulates the system from fluctuations imposed on it by the environment. Robustness also emerges biologically through redundancy of pathways that achieve the same function.
Modularity
A biologist defines a module in a network as a set of nodes that have strong interactions and a common function. Modularity can contribute to robustness by confining damage to independent parts, preventing the spread of damage to the entire network. Modularity can also contribute to evolution of the system, where adaptation can be achieved by rewiring connections between modules rather than reconstituting the modules themselves.
A systems biology approach to studying innate immunity
The complex interactions within the innate immune system that result in effective host defense under normal conditions and inflammatory disease when perturbed can only be dissected in a comprehensive way by systems biology approaches. Immunology is particularly well suited for such analysis, as the cells can be isolated in various functional states and can be reconstituted in a biologically meaningful manner. This contrasts with most other systems; for example, it is very hard to dissociate the brain into individual neurons and reconstitute the appropriate cellular interactions in vitro.
We have attempted to understand the mechanisms that enable macrophages to formulate appropriate responses to pathogens. In this review, we highlight the salient features of the system, discuss the tools and approaches we and others have developed and are applying to gain novel insights into it, and illustrate their application in uncovering gene regulatory networks that control Toll-like receptor (TLR)-induced signaling pathways.
The role of pattern recognition receptors in innate immunity
As this volume is focused on pattern recognition receptors (PRRs) in the innate immune system, a comprehensive discussion of the specifics of these molecules and their signaling pathways would be redundant. Rather, we describe some features of the innate immune system in the context of emergence, robustness, and modularity. In the later section, we discuss how we are using the tools of systems biology to obtain a preliminary understanding of the complex interactions that gives rise to host defense.
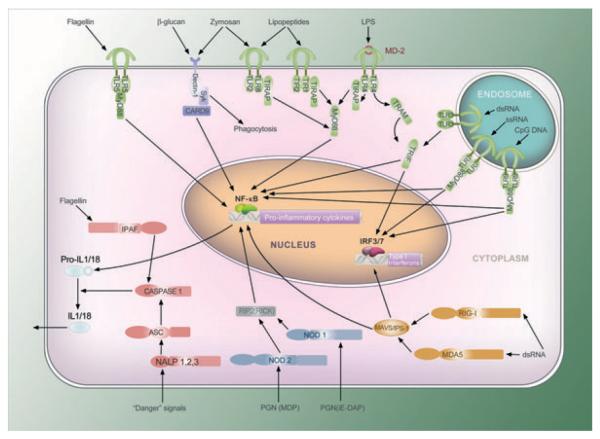
The recognition, phagocytosis, and presentation of specific pathogens by macrophages represent emergent properties that arise from the concerted action of a number of receptors and signaling pathways. Specific pathogen-derived molecules are detected by chemotactic receptors on the macrophage, leading to alterations in the cytoskeleton that culminate in directed movement. The macrophage then uses its PRRs, which include the TLRs, the nucleotide binding and oligomerization domain (NOD)-like receptors (NLRs), and the retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), to identify the nature of the pathogen by recognizing specific pathogen-associated molecular patterns (PAMPs). Fig. 3 shows all currently known TLRs, NLRs, and RLRs and their cognate adapters (adapted from 1). Phagocytic receptors, such as the Fc receptor, the complement receptor, and DECTIN, bind the particle and activate signaling pathways that lead to its internalization (2). Upon internalization, the pathogen is degraded in a process that culminates in the presentation of peptide fragments via major histocompatibility complex class II (MHC II) as well as activation of additional PRRs by pathogen breakdown products (3). It is not possible to predict the complex behavior underlying chemotaxis, phagocytosis, and antigen presentation by having a complete understanding of each individual receptor and its cognate signaling pathway in isolation. Systems biology approaches will enable an understanding of how the crosstalk between these pathways results in the emergent properties that give rise to these functional responses.
Fig. 3. Currently known TLR, NLR, and RLR receptors, their cognate adapters, and primary agonists.
Membrane-bound TLRs and their adapters are shown in green. Cytoplasmic NLRs are shown in blue (for NOD1 and NOD2) and red (for the inflammasome NLRs and adapters). Cytoplasmic RLRs are shown in yellow. The phagocytic PRR Dectin-1 is shown in purple (figure adapted from 1).
Crosstalk between phagocytic receptors and PRRs
It has long been known that phagocytosis can be uncoupled from the induction of an inflammatory response (4, 5). For example, phagocytosis of latex beads is not accompanied by the production of arachidonic acid metabolites unless the macrophages are primed with bacterial lipopolysaccharide (LPS), in which case a synergistic response is observed (6). Similar synergy also occurs for Fc receptor and zymosan-induced phagocytosis but not for complement-induced phagocytosis, which will not induce arachidonic acid metabolite release even with LPS priming (6). This paradigm was further extended by the demonstration that phagocytosis of yeast by macrophages is triggered by DECTIN, while TLR2 stimulates the accompanying inflammatory response (7, 8). These interactions are even more subtle when considering the internalization of bacteria. When macrophages internalize Gram-negative bacteria, tumor necrosis factor (TNF) is only produced in the presence of TLR4. By contrast, TLR2 is required for TNF production during phagocytosis of Gram-positive bacteria (7). Additional subtlety exists as TLR2 signals exclusively via the adaptor myeloid differentiation primary response gene-88 (MyD88), whereas TLR4 activates both MyD88 and TRIF [Toll/interleukin-1 (IL-1) receptor (TIR) domain-containing adapter protein inducing interferon-β (IFN-β)] pathways, resulting in more complex, pathogen-specific inflammatory responses (9).
Phagocytosis of fungal zymosan provides an example of how phagocytic and PRR pathways can function as interlocking pieces in their regulation of the macrophage response (reviewed in 10). Zymosan is recognized by both TLR2 and DECTIN. TLR2 signaling induces inflammatory cytokines through the MyD88 pathway and activation of NF-κB but does not induce reactive oxygen species (ROS), phagocytosis, and only weak arachidonic acid release. DECTIN, which recognizes β-glucan, activates Syk kinase, induces zymosan phagocytosis, ROS induction, and weak arachidonic acid release. When both TLR2 and DECTIN are activated, inflammatory cytokine induction, ROS production, and arachidonic acid metabolism are all synergistically enhanced.
Interactions between PRR signaling and phagocytic pathways extend beyond internalization and inflammation. TLR signaling has been implicated in the enhanced maturation of phagosomes (11), although this finding is controversial (12). More importantly, the presence of TLR ligands within a dendritic cell phagosome markedly enhances the MHC class II-mediated presentation of antigens within that phagosome (13). Thus, the entire set of functional macrophage responses to pathogens are shaped and modulated by complex interactions between PRR, phagocytic, and other pathways.
Crosstalk between PRRs
Macrophages are not confronted with purified PAMPs in nature. Rather, they interact with complete pathogens that present a cocktail of agonists to the numerous PRRs they express (2, 14). These combinations of PAMPs enable the innate immune cell to carry out `multiparameter analysis', which permits far greater accuracy in the determination of the threat. For example, if TLR4 and the NLR IL-1β-converting enzyme protease-activating factor (IPAF) are simultaneously activated, the cell can compute that it has encountered a Gram-negative flagellated bacterium that contains a type III secretion system (15).
Additional complexity is illustrated by the flagellin detection system which comprises TLR5, a sensor for extracellular flagellin (16), and the inflammasome-activating IPAF, a sensor for cytosolic flagellin (15, 17, 18, reviewed in 19). Binding of flagellin to TLR5 causes it dimerize and induces TIR domain interactions that recruit MyD88, which induces activation of IL-1 receptor-associated kinase-4 (IRAK4) and IRAK1 kinases, several mitogen-activated protein kinases (MAPKs), and inhibitor of κB(IκB) kinases that culminate in NF-κB-dependent inflammatory gene expression. Flagellin is also secreted into the cytoplasm via the type III secretion systems of certain bacteria (15). In the absence of cytoplasmic flagellin, IPAF resides in a state in which the leucine-rich repeat (LRR) domain binds to the NOD domain and inhibits oligomerization with the NOD domains of other IPAF molecules. During infection, the LRR binds flagellin, exposing the NOD domain, allowing oligomerization and formation of a scaffold for recruitment (20, 21) and activation of the caspase-1 inflammasome (22), which processes IL-1β and IL-18 for secretion (23).
Dual sensing of flagellin by TLR5 and IPAF opens up the possibility for a complex, two-step process of regulation. When a macrophage encounters a Salmonella bacterium, TLR5 is initially stimulated by flagellin (in addition to activation of TLR4 by LPS). This signal induces, among others, the mRNAs encoding IL-1β and IL-18 and their precursor proteins. Once the bacterium is in the phagosome, flagellin is injected into the cytoplasm via the type III secretion system, and IPAF is subsequently activated. This activity stimulates the assembly of an IPAF inflammasome that activates caspase-1, leading to the processing and secretion of mature IL-1β and IL-18 (23). This mechanism may be a general one, as other NLRs, for example Nalp1-3, have also been shown to induce processing of IL-1β that had previously been induced by a TLR (for example TLR4) (24-27). Conceptually, TLR signaling in the absence of NLRs may constitute a `yellow alert', indicating that microbes have penetrated the physical barrier of the epithelial layer. The inflammasome NLRs, when activated in conjunction with the TLRs, may then trigger a `red alert', alerting the immune system to the presence of pathogens which harbor more threatening virulence factors such as the type III secretion system (19). Signaling by TLRs alone or by NLRs alone does not initiate the red alert, and thus the red alert emerges from the convergent activation of the two pathways. Greater complexity is exhibited by the system, because the IL-1R signals exclusively through the adapter MyD88 (28), and thus signaling pathways activated by IL-1β parallel those activated by the TLRs. IL-1β is not known to be capable of activating the inflammasome itself, and thus paracrine IL-1β signaling can propagate the yellow alert but not the red alert, which is reserved for the infected macrophage. A similar distinction between the reserved red alert status of the infected cell and the yellow alert status for neighboring cells activated by paracrine cytokine signaling has been postulated for viral nucleic acid detection (29): cytotoxic lymphocytes and natural killer cells must be able to distinguish between virus-infected cells that should be targeted for apoptosis and cells that have been activated into an antiviral state by paracrine type I IFN signaling.
The system is even more complex than described due to crosstalk arising from simultaneous engagement of multiple TLRs and other receptor families. For example, flagellin is highly immunogenic, being recognized by both antibodies and CD4+ T cells over the course of Salmonella infection (30). Thus, in the late stages of infection, antibody-flagellin complexes can engage macrophage Fc receptors at the same time that TLR5 is engaged by flagellin and TLR4 is engaged by LPS. Furthermore, under these conditions both TNF and IL-1β will be produced and activate shared components of the TLR pathway via an autocrine mechanism. Simultaneous engagement of multiple TLRs has been shown to give rise to a number of synergistic responses (reviewed in 14). For example, TLR7 can synergize with TLR3 or TLR4 for IL-12p70 production in DCs. Viral RNA is recognized by at least five PRRs [TLR3, TLR7, TLR8, melanoma differentiation-associated gene 5 (MDA5), and RIG-I], and it is interesting to speculate on how convergent detection can lead to synergistic, virus-specific responses. Recent results from the Akira laboratory (31) suggest that the adjuvant effects of the double-stranded RNA (dsRNA) analog polyinosinic-polycytidylic acid (polyI:C) arise from cooperative activation of TLR and cytoplasmic RLR pathways. Thus, pathogen recognition by the innate immune system is perhaps best considered as a process in which activation of several PRR pathways in combination gives rise to an emergent, pathogen-specific response that seeks to neutralize the threat, alert neighboring cells to the presence of microbes, and initiate an appropriate adaptive immune response.
Robustness and modularity in innate immunity
While combinatorial PAMP detection by PRR pathways allows macrophages to accurately determine threat levels posed by invading pathogens, as described above, it also illustrates two additional key properties of the innate immune system: robustness and modularity.
To provide protection, the innate immune system must be robust: pathogens must be detected and the immune system alerted, even as evolution favors development of pathogen strategies to evade detection. The large number of PAMPs that may be detected by macrophage PRRs thus constitutes a robust, `fail-safe' detection system: if a particular PRR fails to detect a pathogen, or if a pathogen evolves a strategy to evade a particular PRR, it nevertheless will be detected by all of the relevant remaining PRRs expressed by the cell. This level of robustness is revealed by gene-targeting studies, in which specific PRR knockouts or knockdowns fail to exhibit phenotypes. For example, TLR3, which detects viral dsRNA, when ablated does not result in universally enhanced susceptibility to viral infection (32), presumably because signaling by other viral RNA detectors (RIG-I, MDA5, and TLR7) is sufficient for protection. Similarly, we have demonstrated that inflammasome activation in response to Listeria monocytogenes involves detection by three or more cytoplasmic receptors: IPAF, NALP3, and at least one other NLR utilizing the adapter ASC (apoptosis-associated speck-like protein containing a C-terminal caspase recruitment domain) (33).
Modularity in the PRR pathways is typified by the modularity in the structures of the PRRs themselves. In the TLR family, for example, a less conserved N-terminal LRR domain is coupled to a more highly conserved C-terminal TIR domain (34) by a single transmembrane domain. The LRR domains are so variable that they cannot be aligned over large evolutionary distances; alignment can only be accomplished using the TIR domains. The TIR domain couples the TLR to the restricted set of adapters [the linker adapters, MyD88 adapter-like protein (MAL) and translocating chain-associating membrane protein (TRAM), and the major signaling adapters, MyD88 and TRIF], whereas the LRR domain is responsible for PAMP recognition. Evolution of LRRs has resulted in an extraordinary diversity in ligands detected by the TLRs, giving rise to six major TLR families in vertebrates (34). Recent structural studies of TLR- ligand complexes have revealed diversity in LRR ligand binding mechanisms (reviewed in 35). While TLR2/TLR1 heterodimer binding of Pam3CSK4 is achieved by hydrophobic interactions at the boundary between central and C-terminal domains (36), TLR3 dimers bind dsRNA at two regions near the N-terminal and C-terminal ends (37).
Robustness in the innate immune system emerges not only from the modularity of the PRRs and the pathways they activate but also from the feedback architectures of the pathways themselves. Type I IFN induction by cytoplasmic viral sensors in fibroblasts is an example of a positive feedback loop which results in robust induction of an antiviral state (reviewed in 38). Cytoplasmic detection of viral RNA by the RLRs RIG-I or MDA5 results in type I IFN induction by activated IFN regulatory factor-3 (IRF3) and IRF7 transcription factors (TFs). The type I IFN then feeds back on the cells in an autocrine manner to induce IRF7 to high levels. IRF7 then induces additional type I IFN species and increases the expression of the sensors RIG-I and MDA5 themselves, which presumably renders the cell more sensitive to viral RNA. On the other hand, precise control and robustness to intracellular noise is partly achieved in PRR pathways by negative feedback loops. For example, TLRs induce the expression of many genes that negatively regulate the TLR pathways (reviewed in 39). In particular, the ubiquitin-editing protein A20 (Tnfaip3) is both induced by and is a negative regulator of TLR, RLR, and NLR pathways (40-44), acting directly on key adapter molecules such as tumor necrosis factor receptor-associated factor 6 (TRAF6), TRIF, and receptor-interacting protein 2 (RIP2). A second example of this type of regulation is illustrated by the TF-activating transcription factor 3 (ATF3), which is induced by TLRs and then inhibits gene induction stimulated by the same TLRs (45) (discussed in detail below).
Systems approaches and innate immunity
In the above, we described how innate immunity exhibits the key attributes which motivate a systems approach, namely emergence, robustness, and modularity. Yet the biological insights discussed above, minus the case of ATF3, were not obtained through systems biology approaches. What role, then, does systems biology play in the future of innate immunity research? We believe it is fourfold. (i) Filling in the gaps to create an integrated picture of PRR pathways and their interactions. While a great deal is known about pathogen recognition, it is largely piecemeal and not comprehensive. Systems-level approaches, as described below, will flesh out the networks and interactions to create a more complete understanding of innate immunity. (ii) Enhancing the rate at which discoveries are made. While conventional single-gene methods will always be necessary to validate and extend predictions, systems-level approaches can accelerate the speed of hypothesis development, and the identification of novel components of innate immune pathways. (iii) Clarifying how functional emergent properties arise from biomolecular networks. As innate immunity pathways are delineated in greater and greater detail, it becomes increasingly difficult to interpret them intuitively and to understand how network interactions control functional responses. Systems biology provides a quantitative framework in which the complex interactions may be represented formally and system behavior may be investigated computationally. (iv) Predicting the effects of genetic or environmental perturbations on innate immune responses. As descriptions of innate immune pathways become more and more comprehensive and quantitative, systems biology tools will make it possible to both predict and understand how subtle variations in gene sequences, for example, give rise to variations in human infections and inflammatory disease susceptibility. In the paragraphs below, we describe emerging technologies in the context of their utility for facilitating a systems-level understanding of innate immunity.
Technologies for the systems analysis of innate immunity
As discussed above, in the systems biology paradigm, biology drives technology and then technology revolutionizes biology (Fig. 1). In the present section, we describe novel technologies that will further enable a systems-level understanding of innate immunity.
Genomics: DNA sequencing
The emergence of DNA sequencing and the impact it has had on biology exemplifies the synergistic cycle of biology and technology described above. A question regarding antibody diversity was the original motivation for developing the first automatic DNA sequencer (46). However, once it was built, it ultimately enabled the Human Genome Project. Similarly, improvements to automated DNA sequencing, with the objective of making complete genome sequencing faster and cheaper, has led to the development of `next generation' sequencing technologies (NextGen sequencing). NextGen sequencing will have a fundamental impact on the analysis of genomes as well as transcriptomes and gene regulatory networks (described below).
NextGen sequencing represents a fundamental change in sequencing approach from Sanger sequencing, which has been in use for the past 30 years, and was highly optimized and automated at an industrial scale for completion of the Human Genome Project (47). Commercially available NextGen sequencers are dominated by three systems: the 454 GS FLX Genome Analyzer (Roche Applied Sciences, Indianapolis, IN, USA), the Solexa 1G sequencer (Illumina, San Diego, CA, USA), and SOLiD (Applied Biosystems, Foster City, CA, USA). The specific technologies employed differ between the platforms, but generally they all involve a massively parallel analysis of individually amplified DNA molecules (48). The sequencing output, in terms of read number and length, also differs between the platforms. While modern Sanger sequencers can read about 800 bp of 100 DNA samples in parallel, the 454 GS FLX produces about half a million reads that are 250 bp in length, and the SOLiD and Solexa systems generate tens of millions of reads that are about 35 bp in length. These differences in sequencing output dictate the suitability of the various technologies for different applications, as described below.
While the shorter read lengths from NextGen sequencing make assembly of genomic sequences de novo more difficult than with conventional sequencing, results of ambitious sequencing projects using these technologies are appearing in the literature. 454 GS FLX has been used for several ancient genome sequencing projects, including the wooly mammoth (49) and the Neanderthal (50). The 454 GS FLX system is well suited for the analysis of ancient genomes given that its read length is comparable with the DNA sequence lengths recovered from fossils and that it can generate hundreds of thousands of reads per run, which becomes necessary given the high level of DNA contamination found in fossils (50). NextGen genome sequencing will undoubtedly enable a much deeper understanding of immunology. For example, comparison of the Neanderthal genome sequence with that of human will provide insights into the recent evolution of the human immune system that would otherwise be impossible to obtain. In other sequencing applications, the 454 GS FLX system has been used to identify thousands of macaque single nucleotide polymorphisms (SNPs) (51), which will enable functional association analyses in this important immunological model organism. Novel immunologically relevant human DNA variations undoubtedly will be revealed by the data produced in the 1000 Genomes Project (http://www.1000genomes.org/), which aims to sequence the genomes of 1000 people from around the world. All three major next generation sequencing companies are participating in this project, each having agreed to produce 75 billion base pairs of sequence during the pilot phase.
Genomics: transcriptome analysis
Traditionally, transcriptome studies in innate immunity have used microarrays with probes targeting the 3′-regions of the interrogated mRNAs. The motivation for this approach was largely a practical one related to feature size constraints of the chips. Recent developments in oligonucleotide array fabrication and the emergence of NextGen sequencing, however, have resulted in the 3′-arrays being superseded by exon-level microarrays as well as transcriptome analysis by sequencing.
Given the importance of alternative splicing events in the immune system (52), replacement of conventional arrays with exon-level microarrays in typical transcriptomic analysis is a key step for the analysis of gene regulation in immune cells. On the Affymetrix GeneChip Mouse Exon 1.0 ST Array (Affymetrix, Inc., Santa Clara, CA, USA), for example, feature sizes have been shrunk from 11 μm (for the GeneChip Mouse Genome 430 2.0 Array) to 5 μm, allowing over one million exons to be tiled on a single array. Exon arrays can be analyzed in two frameworks: `gene level', where all the exon-specific information is integrated into a single expression measurement for each gene, and at the `exon level'. In both cases, the exon arrays have significantly added value compared with conventional arrays. The gene-level framework gives expression values that are more representative of the overall gene behavior. This is because it is an integrated measure of the expression of all of the exons, not just the 3′-end. By contrast, gene expression measurements at the exon level allow detection of novel transcriptional events and transcript isoforms. Exon arrays are fundamentally different from custom splicing arrays, which are designed to detect specific exon-exon junctions (53-56), and are therefore constrained to known cases of alternative splicing. Exon arrays have been applied in diverse studies ranging from colon cancer (57), neuronal systems (58), to embryonic stem cells (59). In two recent immunological studies, exon arrays were used to identify candidate target genes of a novel CD45 splicing regulator identified using an siRNA screen (60) or N-ethyl-N-nitrosourea (ENU) mutagenesis (Wu et al., unpublished data).
Despite the value-added and novel opportunities provided by exon arrays, it is likely that they will be superseded by transcriptomic application of NextGen sequencing (RNA-Seq) (61). RNA profiling by hybridization has two major limitations: (i) prior knowledge of the transcript sequences to be measured is required and (ii) hybridization-based signals have inherent limits in dynamic range (62). These limitations make it especially challenging to detect novel or rare transcripts by microarray analysis. Transcriptome analysis by NextGen sequencing does not have these limitations: sequences are detected de novo and transcript quantitation is digital, with a five orders of magnitude dynamic range already being reported (61). The strength of next generation sequencing for transcriptome analysis derives from the very attribute that render its application in genome assembly a challenge: millions of short reads. Most immunological transcriptome studies will involve analysis of a model organism for which a high-quality genome sequence - a reference genome - is available, and thus interpretation of next generation transcriptome data simply involves mapping the millions of sequences to unique genomic locations. Two recent RNA-Seq studies of poly-adenylated transcriptomes, one in differentiating human embryonic stem cells using the ABI SOLiD platform (63) and one in several mouse tissues using the Illumina Solexa platform (61), demonstrate the power of this approach. Coupling NextGen sequencing with cap analysis gene expression (CAGE) allows simultaneous detection of transcript levels and transcription start site positions (reviewed in 64). Furthermore, NextGen sequence analysis can be applied to detect novel micro-RNAs (miRNAs) (65) and other functional non-coding RNA species, which is of special relevance given the increasingly important role miRNAs have been shown to play in immune regulation (reviewed in 66).
Genome-wide location analysis
The regulatory networks controlling innate immune cell behavior during pathogen recognition will not be revealed by an analysis of transcriptomes alone. To define this complex circuitry, the dynamic localization of transcriptional regulators and epigenetic chromatin modifications during the response to infection must be measured. Genome-wide location analysis refers to a maturing set of technologies that specifically provides this information. ChIP-on-chip is one type of genome-wide location analysis that combines standard chromatin immunoprecipitation (ChIP) with microarray technology (67, 68). This method permits the definition of the entire complement of genes that are activated by a specific TF under a specific set of conditions. ChIP-on-chip and similar genome-wide location analyses have been applied in countless studies, revolutionizing our understanding of how TFs and epigenetic modifications control gene expression (reviewed in 68-71). Application of genome-wide location analysis to gain insight into immune cell gene regulation includes studies of forkhead box protein 3 (FOXP3) binding in regulatory T cells (72, 73), signal transducer and activator of transcription-1 (STAT-1) and STAT-2 binding in a cell line stimulated with IFN-α or IFN-γ (74), and LPS-induced promoter binding by NF-κB in cell lines (75, 76).
As in the case of transcriptome analysis, microarray-based ChIP-on-chip will probably be superseded by NextGen sequencing using a process called ChIP-Seq (77). This approach does not require a priori designation of candidate DNA binding locations and thus can provide a truly global, unbiased view of genome-wide TF localization. Additionally, ChIP-Seq may actually require less input DNA than ChIP-on-chip (78), giving cost advantages, permitting analysis of rare cell types, and reducing reliance on sequence amplification methods that may skew or corrupt underlying sequence distributions. A wide variety of ChIP-Seq applications have already been reported, including analysis of IFN-γ-induced STAT-1 binding in a cell line (78), transcriptional regulatory circuits in mouse embryonic stem cells (79, 80), and histone methylation in human CD4+ T lymphocytes (81). Integration of next generation sequencing-based ChIP-Seq and RNA-Seq will yield fundamental insights into the genomic regulatory programs governing the innate immune response to infection.
Proteomics
Obtaining a comprehensive understanding of the macrophage response will require more than delineation of the gene regulatory circuits that are activated upon pathogen recognition. To determine the functional roles of innate immune-regulated genes, we must determine how they are post-translationally modified, where they localize subcellularly, and what other proteins they associate with in space and over time. Given that it is generally not possible to predict these properties directly from protein sequence, a systems-level approach which measures them directly is required. Proteomics refers to a broad set of heterogeneous and rapidly evolving technologies that provide this information.
Quantitative mass spectrometry
The proteomics technologies we employ in our analysis of macrophages rely heavily on quantitative mass spectrometry (MS). Several different techniques have been developed and we will only briefly review them here. The isotope-coded affinity tag (ICAT) method (82) enriches for isotopically encoded, cysteine-containing peptides and has the advantage of decreasing sample complexity prior to MS analysis. Isobaric tags for relative and absolute quantitation (iTRAQ) is a multiplexed strategy that allows four samples to be analyzed simultaneously in the same experiment (83). iTRAQ differs from other isotopic labeling strategies in that the peptides from different conditions labeled with the four iTRAQ reagents remain of equal mass through the liquid chromatography step because the reporter ions are balanced. Peptide ion peaks therefore are detected simultaneously by the MS, and the relative peptide abundance in the different conditions is indicated by the relative intensities at the four mass-to-charge ratios of the reporter ions. Stable isotope labeling with amino acids in cell culture (SILAC) provides the maximum protein isotopic labeling efficiency combined with the minimum number of sample handling steps and is one of the most sensitive quantitative MS approaches (84). In SILAC, two populations of cells are cultured under identical conditions except that the culture medium for one population contains all 20 essential amino acids in their naturally occurring isotopic forms (light), while the other population is grown in medium where amino acids are replaced by stable, heavy isotope-labeled analogs (heavy). SILAC allows for all peptides within the sample to be analyzed, increasing the accuracy of identification and quantitation. One disadvantage with SILAC is that, because cells must be cultured in the presence of the isotope-labeled analogs for approximately 2 weeks, it is not suitable for experiments where prolonged culture is not possible.
Surface and secreted proteins
Macrophages and dendritic cells execute their innate immune functions inlarge part by dynamically modifying the complement of proteins expressed ontheir surfaces and secreted into the extracellular milieu. For this reason,we are directing substantial effort towards the definition of the macrophage secretome and surface proteome. Given the historic importance of the cluster of differentiation (CD) markers for the field of immunology, we anticipate that there will be immense value in the new surface markers for classifying various stages of macrophage and dendritic cell responses to pathogensthat we will identify. Similarly, we anticipate great value in the identification of novel proteins secreted by or surface proteins shed by innate immune cells during the response to pathogens, as these may play key roles in combating infection, triggering/resolving inflammation, and priming the subsequent adaptive immune response.
Analyses of the macrophage surface proteome and secretome both present significant technical challenges. For characterization of the surface proteome, the greatest challenge is contamination of samples purified by subcellular fractionation/centrifugation methods with proteins derived from other cellular membranes. For the secretome, the greatest challenges are the low abundance of secreted proteins, contamination by cellular proteins released during accidental cell lysis, and contamination with serum proteins (85). To address the issues of contamination and specificity, a selective surface ⁄ secreted protein labeling and affinity purification strategy, glycoproteomics, was developed. This approach exploits the fact that both cell membrane-associated and secreted proteins are typically glycosylated on asparagine residues (86, 87). Use of a membrane-impermeable biotinylation reagent that covalently modifies extracellular glycosylation sites allows selective purification of surface and/or secreted proteins. These selected protein subsets can then be analyzed with quantitative MS technology to define the secretome and surface proteome.
Identifying signaling complexes
Most signaling networks are activated by the assembly of protein complexes. The analysis of the complex constituents and formation dynamics pose many technological challenges. While various affinity purification methodologies have been applied successfully in conjunction with MS to study stable covalently linked complexes, many immunologically relevant signaling interactions are transient or weak and are not suitable for analysis by this method. A newly developed tagging system, which utilizes concatenated affinity tags (HB) for the purification of protein complexes after formaldehyde cross-linking (88), may overcome this limitation. This approach is fully compatible with the quantitative proteomics approaches described above, such as SILAC or iTRAQ (88).
Transcription is regulated by complex enhanceosomes that assemble on the promoters of target genes. Recent advances in proteomic analysis have permitted the identification of novel components of these transcriptional complexes by taking advantage of the promoter-specific binding of the enhanceosome (89). In this approach, the promoter sequence of interest is coupled to a bead which serves as an affinity matrix for TFs and associated transcriptional regulatory factors. The technique, initially perfected in yeast, has now been extended to detect dynamic changes in transcriptional complexes in mammalian cells (90, 91).
Multiple reaction monitoring
Blood biomarkers have recently gained significant attention because of their potential for early diagnosis and stratification of disease. These measurements are also critical in the immune context given the search for correlates of immunity during vaccine development. A recently developed proteomic method, multiple reaction monitoring (MRM), allows for high throughput, multiplexed measurement of complex protein mixtures, such as those found in blood (92). The first phase of the study uses shotgun proteomics and is directed at identification of all protein constituents of the system, in this case blood. These proteins and their associated MS parameters are then placed within a database called the PeptideAtlas (http://www.peptideatlas.org/, 93). These predefined ion lists then enable specific peptides of interest to be targeted for quantitation by MRM (94). As MRM is scalable, thousands of proteins in multiple samples can be rapidly analyzed. State-of-the-art instruments operating in the MRM mode are expected to quantitate at least 500 peptides in a single run, and recent work has demonstrated that MRM-based approaches can detect plasma proteins in the sub-ng/ml range (95).
Computation
The most significant challenge in systems biology is computation; the data have to be stored in databases, annotated, analyzed, and interpreted. Computation in systems biology is highly heterogeneous and is carried out by scientists with diverse backgrounds ranging from information technology and computer science to statistical analysis and engineering. One unifying theme for all successful computational approaches, however, is that they are ultimately motivated by the biology, be it development of intuitive interfaces that facilitate interpretation of complex data sets in the appropriate biological context or application of modeling approaches to answer specific biological questions. An astonishing number of databases, analysis tools, and algorithms for computational systems biology analysis are published each year and it is beyond the scope of the present work to review them here. Instead, we describe a subset of computational tools that have been developed at the Institute for Systems Biology (ISB) and elsewhere that we actively use to meet the computational needs of our analysis of the innate immune system. Our policy at the ISB is that all software is freely downloadable and open source, which will facilitate the rapid development and distribution of computational tools. ISB-developed software can be found at http://www.systemsbiology.org/Resources_and_Development/Downloadable_Software.
Computational analysis of transcriptome data
The majority of systems biology tools have been developed to facilitate analysis of transcriptome data sets. One inescapable computational challenge in transcriptome analysis is making complex data sets easily accessible and interpretable for biologists, regardless of whether those biologists have programming experience. As the scale of the data sets increases, so does the difficulty in distributing the data in a useful way. While most biologists with fluency in spreadsheet programs can easily navigate and interpret conventional microarray data sets, spreadsheets alone are not suitable for interpretation of exon-level microarray data sets or microarray data sets spanning hundreds of conditions. How complex data sets can be made accessible depends on the nature of the data sets themselves. Our Systems Biology Experiment Analysis Management System (SBEAMS)-Microarray pipeline (http://www.sbeams.org/Microarray) (96), a module in SBEAMS (http://www.sbeams.org), is used for normalization and standard analysis of conventional 3′-microarrays and gene-level analysis of exon arrays. Data analysis usually begins with the discovery of contextual patterns of gene regulation; our tool, cMonkey (http://baliga.systemsbiology.net/cmonkey/download/) (97), facilitates complex biclustering analysis of gene-level expression measurements, grouping genes, and experimental conditions. Our approach to exon-level expression data is evolving, and analysis is largely performed in statistical/engineering programing environments such as R (http://www.r-project.org/) and MATLAB (The Mathworks, Inc., Natick, MA, USA). This analysis is best carried out in an interdisciplinary environment that permits creative interaction between biologists and computer scientists/statisticians/engineers.
Additional systems biology tools facilitate deeper analysis of transcriptomes that may identify novel regulators and putative networks. It is easiest to discuss these tools in the context of concrete biological problems. Below, we describe two studies aimed at unraveling the gene regulatory programs that underlie TLR-induced macrophage activation. In the first example, we show how systems analysis led us to identify ATF3 as a novel negative regulator of the innate immune response (45). In the second example, we describe a novel computational approach we used to generate hypotheses about the dynamic transcriptional regulatory networks controlling macrophage activation (98).
ATF3 is a negative regulator of TLR signaling
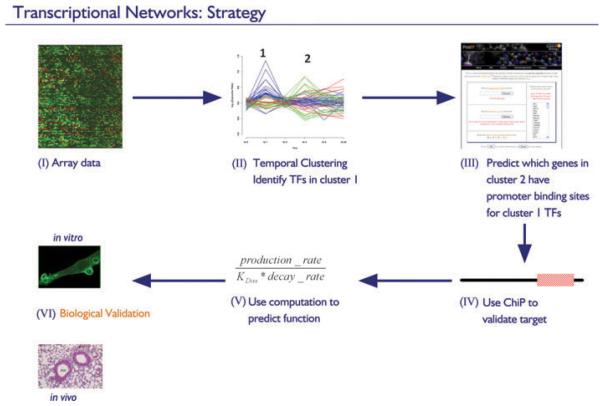
In this case study, we used the tools of systems biology to unravel the transcriptional circuitry leading to the TLR4-activated state in macrophages; our strategy is shown in Fig. 4. Briefly, temporal activation of macrophages by LPS was analyzed using microarrays (I). These data were then clustered to reveal regulated `waves' of transcription, in which different sets of genes exhibit sequential peaks in expression (II). Given that transcriptional programs are propagated by sequential cascades of TFs, it is reasonable to expect that the TFs induced in early clusters will regulate genes expressed in subsequent clusters. Furthermore, co-expressed genes are often co-regulated and are therefore likely to have cis-regulatory elements in common. Based on these arguments, we inferred putative regulatory links by applying computational motif scanning to identify which genes in cluster 2 contain promoter binding sites for TFs in cluster 1 (III). These regulatory predictions were validated and the kinetics of promoter occupancy was defined using ChIP (IV). These kinetic data allowed mathematical modeling of the transcriptional circuitry, which, in turn, enabled the prediction of novel functions that are not easily identified using conventional approaches (V). The functional predictions were then tested in cell culture systems and in mice (VI).
Fig. 4. Strategy for unraveling transcriptional networks.
(I) Microarrays are used to profile temporal gene expression responses in stimulated macrophages. (II) Clustering reveals groups of genes with different expression kinetics. (III) Candidate regulatory links between genes are made by searching for enrichment of regulatory elements of transcription factors expressed in early groups (no. 1) in the promoters of genes expressed in later groups (no. 2). (IV) Regulatory predictions are validated by chromatin immunoprecipitation. (V) Mathematical modeling is used to predict the functional nature of validated interactions (activation, repression, etc.). (VI) Functional predictions are tested in vitro and in vivo.
We stimulated bone marrow-derived macrophages (BMMs) with LPS and collected expression profiles measuring the response from 20 min to 24 h. The microarray data were uploaded into SBEAMS-Microarray (96) and processed using the BioConductor (99) and VERA/SAM (100) software packages. A set of 1232 differentially expressed genes were clustered into 11 distinct temporal profiles. One cluster with expression profiles peaking 1 h poststimulation contained a large number of TFs, including several with established roles in inflammatory gene regulation such as NF-κB (REL), activator protein 1 (AP1) (JUN, FOS), and early growth response 1 (EGR1), and candidate novel inflammatory regulators (ATF3, EGR2). Scanning the promoters of genes in this cluster for TF binding sites from TRANSFAC (101) using MotifLocator (102) revealed over-representation of ATF3/cyclic adenosine monophosphate-responsive element binding protein (CREB) family binding sites. This enrichment provided additional support for a role of ATF3 in regulating the response, and we decided to investigate the role of this TF in greater detail.
To identify proteins that ATF3 may interact with over the course of the inflammatory response, we visualized ATF3 protein-protein interactions from HPRD (http://www.hprd.org) (103) using Cytoscape (http://www.cytoscape.org) (104). Remarkably, ATF3 interacts at the protein level with components of the pro-inflammatory TFs it clustered with, namely NF-κB (NFKB1) and AP-1 (JUN, JUND). Given that these TFs were expressed together early in the LPS response and that they interact at the protein level, we hypothesized that they function together in regulating target gene expression at later time points. We therefore searched the promoter regions of 165 genes in the subsequent temporal cluster for ATF3/CREB, NF-κB, and AP1 binding sites. We observed enrichment of binding sites for these TF families and, moreover, identified 30 genes, including the cytokines IL-6 and IL-12p40, for which predicted ATF3 binding sites occur within 100 bp of an NF-κB binding site and within 500 bp of the transcription start site. This result suggested that ATF3 cooperates with NF-κB to regulate the expression of pro-inflammatory genes, including IL-6 and IL-12p40. We used ChIP to directly investigate the temporal binding of ATF3 and NF-κB (REL) to IL-6 and IL-12p40 promoters during the LPS response. While we observed rapid, transient LPS-induced promoter binding by NF-κB, ATF3 promoter binding in response to LPS was delayed and sustained.
To estimate the relative contributions of ATF3 and NF-κB promoter binding to IL-6 and IL-12p40 gene expression, we analyzed the data using the Inferelator (105), which generates predictive kinetic gene regulatory models with parameters quantitating the strength and sign (activator/repressor) of the interactions. The Inferelator analysis predicted that NF-κB is a positive regulator of IL-6 and IL-12p40 expression (as expected), but that ATF3 should function as a negative regulator. This prediction of an anti-inflammatory role for ATF3 was confirmed by analysis of BMMs derived from ATF3-/- mice, in which LPS-induced production of IL-6, IL-12p40, and a number of other genes was substantially greater than that observed in wildtype (WT) BMMs. Furthermore, an anti-inflammatory role for ATF3 in vivo was demonstrated by an analysis of ATF3-/- mice, which succumb to LPS-induced shock much more rapidly than WT mice (45). This case study illustrates the power of an integrated experimental and computational approach to generate novel insights into gene regulation.
Transcription networks controlling macrophage activation
We have shown that the macrophage response involves dynamic expression changes for nearly 100 TFs (106). To extend the network we modified our approach described above (98).
We identified the complement of genes differentially expressed in WT BMM over time in response to a panel of TLR stimuli. Expression profiling the responses to multiple stimuli provided additional strength for inferring regulatory networks because pathway-specific patterns of TF activation were revealed. In these experiments, we identified approximately 2000 genes differentially expressed in response to at least one TLR ligand, of which 80 were TFs with DNA binding motifs in TRANSFAC (101).
We complemented our set of expression profiles in WT BMMs with profiles from BMMs derived from several mutant strains and performed cluster analysis to identify gene groups that respond similarly over time to the different stimuli in the various genetic backgrounds. The clusters we obtained exhibited diversity in both timing and ligand specificity of response: while some showed peak induction within 1 h of stimulation, most exhibited responses within 2-4 h. Some clusters contained genes that were not induced by polyI:C or were induced by polyI:C with a lag compared with other stimuli. Groups of genes emerged that were specifically induced/repressed by specific pathways. We found that many cytokines clustered together into co-regulated groups: TNF, CCL3, CCL4, CXCL1, and CXCL2 clustered together, while CXCL10 and IL-10 clustered together, and IL-1β, IL-6, and IL-12p40 clustered together. Clusters consisting of downregulated genes were enriched for cell cycle processes. Finally, we found that gene clusters induced early were enriched for TFs.
As a first step to make regulatory links in the network, we performed time-lagged correlation (TLC) analysis of TF and candidate target gene expression profiles. The set of regulating TFs was defined as those 80 differentially expressed TFs that also have binding site information in TRANSFAC (101). Candidate target genes for a given TF were defined as the set of differentially expressed genes with binding sites for that TF in their promoters. TLC directly accounts for the temporal delay that results between changes in the expression profile of a TF and the expression profiles of its targets, and therefore takes advantage of temporal ordering of expression to identify potentially causal relationships.
As a complementary step towards inferring the underlying transcriptional network, we tested for over-representation of TF binding sites in the promoters of the genes in each cluster of co-regulated genes. We found enrichments for DNA regulatory elements recognized by the innate immune TFs NF-κB and IRF in upregulated clusters, with IRF elements being particularly enriched in TRIF-dependent clusters, while E2F and MYCMAX binding sites were over-represented in downregulated gene clusters.
We integrated the TLC and binding site enrichment results to obtain a putative transcriptional regulatory network controlling the macrophage response to PRR activation. While the TLC analysis and binding site enrichment analyses each identify potential transcriptional regulatory interactions, both approaches have limitations, and the number of false positives is greatly reduced by focusing on the intersection of the results of the two methods. Using these criteria, we identified a total of 90 statistically significant interactions involving 36 TF genes and 27 clusters.
To partially validate our predicted transcriptional regulatory network, we performed targeted ChIP-on-chip experiments. NF-κB/p50 and IRF1were predicted to regulate three and two clusters of co-regulated genes, respectively. We found that two of the three predicted NF-κB/p50 target clusters were significantly enriched for NF-κB/p50 ChIP-on-chip binding, while both of the predicted IRF1 target clusters were enriched for IRF1 ChIP-on-chip binding. These results suggest that our computational analysis method predicts true regulatory interactions in the macrophage response.
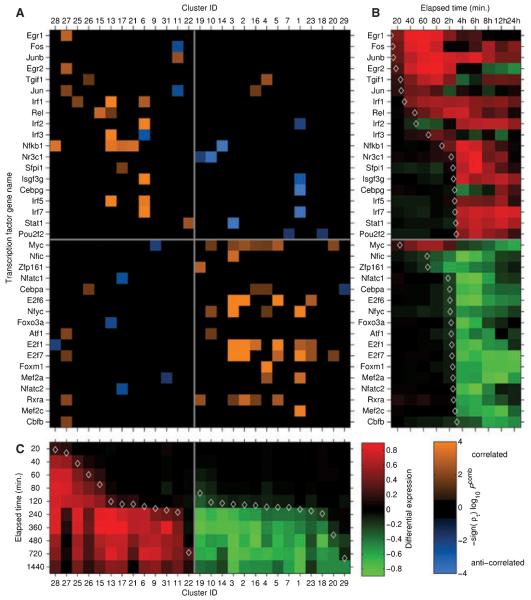
The resulting transcriptional regulatory network implicates large numbers of established and novel TFs in the innate immune response. The most highly connected TFs in the network (MYC and E2F) are involved cell cycle regulation. Other highly connected TFs, such as NFYC [involved in transforming growth factor (TGF)-β and other pathways] and RXRA (a component of nuclear receptor heterodimers involved in inflammation and cholesterol metabolism), may play novel roles in the system. The NF-κB family members REL and NF-κB/p50 were predicted to regulate a large number of genes, as were IRF family members and STAT-1. The connectivity in the network is approximately scale free in a global sense, which is consistent with known mammalian regulatory networks (107, 108). One novel TF identified in our analysis is TG-interacting factor-1 (TGIF1), a transcriptional repressor in the TGF-β signaling pathway (109). TGIF1 is associated with a cluster containing ubiquitin cycle and immune response genes, and is in particular predicted to negatively regulate CSF2 and CXCL3. We validated the TGIF1 expression profile from the arrays and confirmed its induction after 1 h of stimulation by LPS or Pam3CSK4. This predicted transcriptional regulatory network, identified through application of the tools of systems biology, is a hypothesis-generating framework which will direct our future experiments into innate immune gene regulation (98) (Fig. 5).
Fig. 5. Hypothesis-generating transcriptional regulatory network controlling the macrophage response to TLR activation.
The computational analyses used to generate this predicted regulatory network are described in the main text. (A) Matrix defining potential interactions between transcription factors (TFs) and clusters of co-expressed genes. Each column represents a cluster of co-expressed genes, while each row represents a TF potentially controlling gene expression in the network. Clusters and TFs are ordered according to the kinetics of their responses to LPS stimulation. An orange solid rectangle indicates that the TF is potentially an activator for the genes in the cluster, while a blue solid rectangle indicates that the TFis potentially a repressor for the genes in the cluster. (B) Heat-map depicting gene expression profiles for TFs in the network in response to LPS stimulation. Red indicates upregulation, while green indicates downregulation. (C) Heat-map depicting median expression profile of genes in each co-regulated cluster in response to LPS stimulation. Red indicates upregulation, while green indicates downregulation (figure reproduced from 98).
Additional computational tools
There are many computational tools in use at the ISB in addition to the ones mentioned in the case studies above. BioTapestry (http://www.biotapestry.org/) (110) is a convenient tool for representing networks where regulatory interactions have directionality and complex feedback loops are present, such as gene regulatory networks. BioTapestry lays out `wiring diagram' representations of the networks, capable of visualizing hundreds of genes and thousands of interactions. Pointil-list (http://magnet.systemsbiology.net/software/Pointillist/) (111) is an algorithm and software framework developed for the purpose of integrating heterogeneous large-scale data sets in a statistically principled manner. cMonkey (97), mentioned above, facilitates data integration by allowing genes to be grouped based on similarity in expression profiles and attributes such as functional associations or the presence of shared TF binding sites. The systems biology workbench environment Gaggle (http://gaggle.systemsbiology.net/docs/) (112) was developed to facilitate integration between visualization and data analysis programs. A wide variety of tools have been integrated into the Gaggle, including Cytoscape, BioTapestry, TIGR Multiexperiment Viewer (http://www.tm4.org/mev.html), and the EMBL String database (http://string.embl.de/) (113). Although not developed at the ISB, the tool model-based analysis of tiling arrays (MAT) (http://liulab.dfci.harvard.edu/MAT/) (114) coupled to the Integrated Genome Browser (http://www.affymetrix.com/support/developer/tools/download_igb.affx, Affymetrix Inc.) is widely used for analysis and interpretation of ChIP-on-chip data. Higher level Dizzy (http://magnet.systemsbiology.net/software/Dizzy/) (115) was developed for analysis of biochemical network dynamics in either deterministic (differential equation) or stochastic frameworks. Computational analysis of quantitative proteomics data is a field unto itself, requiring substantial infrastructure. The necessary tools for quantitative proteomics analysis, such as SEQUEST, PeptideProphet, ASAPRatio, and XPRESS, have been integrated into a software package called the Trans-Proteomic Pipeline (TPP) (http://tools.proteomecenter.org/wiki/) (116). The TPP provides tools to interpret experimental results in a straightforward manner. The Aebersold groups at ISB and ETH-Zurich continuously maintain and upgradethe TPP.
Screens
Forward genetics provides an approach to unraveling innate immune pathways that is complementary to the experimental and computational methodologies described above. In forward genetics, the genome is perturbed in an unbiased manner to identify the genes required to maintain a particular well-defined phenotype (TLR activation-induced TNF production, resistance to identify the immune-relevant function of a novel differentially expressed gene or gene isoform. Forward genetics screens can be applied to both in vitro and in vivo phenotypes, utilizing small interfering RNA (siRNA) libraries and ENU mutagenesis, respectively. Interaction partners and greater context for the genes identified using these approaches can be identified using the genomics, proteomics, and computational tools described above.
In vitro screens: siRNA
A forward genetics approach can be taken to identify genes controlling cellular processes in vitro through the use of large-scale siRNA libraries. Two recent siRNA screen studies identified human host genes involved in human immunodeficiency virus (HIV) and West Nile virus infection (117, 118). In both studies, a siRNA library targeting approximately 20 000 human genes was transfected into human cell lines that were then infected with virus. Over 250 genes were identified that facilitate HIV infection, including many novel genes such as those involved in Golgi and nuclear transport (118). In the West Nile Virus screen, 283 genes were identified that support viral infection, while 22 genes were found to inhibit viral infection (117).
In vivo screens: ENU random mutagenesis
Forward genetics is especially powerful when the phenotypic screens are combined with germ-line mutagenesis: the genome is randomly perturbed and the resulting mutants are tested for immune phenotypes in vivo and/or in vitro. Use of the germ-line mutagen, ENU, has led to the identification of many novel genes playing critical roles in immunity, and several excellent reviews have been published on the topic (119, 120). The contribution of ENU mutagenesis techniques to our understanding of innate immunity has been significant: for example, the Lps2 mutation identified TRIF as the master adapter in MyD88-independent TLR4 signaling (121). The Triple d mutation, which affects UNC93B1, a gene of previously unknown function, impairs signaling by nucleic acid-sensing TLRs, confers susceptibility to murine cytomegalovirus infection, and causes defects in exogenous antigen presentation (122). UNC93B1 has recently been shown to play a specific role in TLR trafficking (123). ENU mutagenesis screens for autoimmune phenotypes have also revealed critical roles for novel genes. For example, the Sanroque mutation in the gene ROQUIN causes a lupus-like syndrome (124). Mechanistic analysis revealed that ROQUIN inhibits autoimmunity by destabilizing inducible costimulator (ICOS) mRNA (125). ENU mutagenesis screens can also identify novel functions or properties of established immunologically relevant genes. While knockout alleles in some genes may be lethal, it is possible that ENU mutant alleles in the same genes will be viable (126). ENU mutagenesis has revealed unexpected specificity in the coupling between the master adapter MyD88 and TLRs. The Pococurante missense mutation, for example, abolishes all signaling except that activated by TLR2/6, while the Lackadaisical mutation selectively impairs signals activated by TLR7 and TLR9 (127).
Limitations
In spite of their power for linking genes to phenotype, forward genetics approaches do have limitations; for example, only genes with non-redundant functions are identified. Given evolutionary pressures to buttress critical functions with redundancy, the complete set of non-redundant genes for any immune phenotype can be expected to be a small subset of the entire complement of genes involved in any innate immune process. It has been estimated that only 10-20 nonredundant genes linking TLR signaling to TNF activity await discovery (119). For more complex phenotypes, such as resistance to murine cytomegalovirus infection, the 'resistome' may be larger, but still relatively small (about 300 genes) (128). To gain a holistic understanding of innate immunity, we must extend beyond the critical core of non-redundant genes. Another drawback, restricted to in vitro screens, is that genes with general cellular functions may be identified as playing critical roles in the investigated phenotype. For example, genes identified in an in vitro screen for West Nile virus infection were enriched for intracellular protein traffic and cell adhesion functions (117). As many essential cellular processes are likely to require intracellular protein trafficking, the genes identified in such screens are not necessarily specific for virus infection and may not constitute good targets for antiviral drugs. Core cellular genes are less frequently identified in ENU mutagenesis screens, because mutations in these genes are more likely to be lethal. Contrasted with their power for discovery, however, these limitations to forward genetics are relatively minor. Forward genetics will continue to play a critical role in the holistic analysis of innate immunity, especially when coupled to the genomics, proteomics, and computational approaches described above.
Human genetics
There is mounting evidence that genetic polymorphisms regulate innate immune function in humans. Most of these associations have been identified through a candidate gene approach, although genome-wide association studies are also being pursued (reviewed in 129). In the following paragraphs, we described several examples from our laboratory and those of our collaborators that link human disease susceptibility to molecular function of TLRs.
We identified a stop codon polymorphism in the ligandbinding domain of TLR5, TLR5392STOP, that renders it unable to mediate flagellin signaling (130). This polymorphism acts in a dominant fashion and is associated with increased susceptibility to pneumonia caused by the flagellated bacterium Legionella pneumophila.
In an investigation of links between TLR2 signaling and leprosy, we demonstrated that macrophages produce TNF in response to Mycobacterium leprae in a TLR2-dependent manner (131). Furthermore, we have shown that a human polymorphism of TLR2 associated with lepromatous leprosy, TLR2Arg677Trp, impairs its ability to activate NF-κB in response to M. leprae and M. tuberculosis.
To investigate the role of TLR4 polymorphisms in Legionnaires' disease (LD) susceptibility, we used a case- control study to compare the allele frequencies of two TLR4 SNPs, A896G and C1196T (132). Surprisingly, we found that both of the SNPs were protective for LD, despite the fact that some previous studies suggested that these SNPs are associated with increased susceptibility to other infections. These results demonstrate that genetic TLR variations can be either beneficial or deleterious to resistance, depending on the pathogen.
Polymorphisms in TLR adapter proteins can also influence human disease susceptibility. We identified an SNP in the adapter TIR domain-containing adapter protein (TIRAP) [also known as MyD88 adapter-like (MAL)], which links TLR2 and TLR4 to MyD88 (9), and mediates signals from TLRs activated by mycobacteria. This SNP, C558T, was found to influence susceptibility to both meningeal and pulmonary tuberculosis, but by different immune mechanisms (133).
The human studies described above show that polymorphisms in TLRs are associated with changes in susceptibility or resistance to various infectious diseases. These influences involve many genes and are relatively subtle and are therefore distinct from those studied using mouse knockout models. While mouse knockouts have clearly been invaluable in dissecting out the functions of various components of the innate immune system, they are, however, binary in nature: the gene is either present or absent. The analogous situation in human populations is very rare (134). In contrast to mouse knockout models, the mathematical modeling approaches discussed above can capture the small quantitative differences in signaling that arise from mutant molecules. Such systems biology approaches, when coupled to functional studies of human polymorphisms, have the potential to enable a deeper understanding of how human immune responses are influenced by genetic variation.
Conclusions
The innate immune system lies at a critical crossroads in the immune system, maintaining a balance between resistance to infection and inflammatory disease. Enormous progress in defining the molecular pathways of innate immunity has been made in recent years. Application of the tools of systems biology, particularly the emerging technologies described above, will enable a comprehensive understanding of these increasingly complex networks - understanding that will ultimately bridge the gap between molecular variability and human disease susceptibility.
References
- 1.Ishii KJ, et al. Host innate immune receptors and beyond: making sense of microbial infections. Cell Host Microbe. 2008;3:352–363. doi: 10.1016/j.chom.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Underhill DM, Ozinsky A. Phagocytosis of microbes: complexity in action. Annu Rev Immunol. 2002;20:825–852. doi: 10.1146/annurev.immunol.20.103001.114744. [DOI] [PubMed] [Google Scholar]
- 3.Herskovits AA, Auerbuch V, Portnoy DA. Bacterial ligands generated in a phagosome are targets of the cytosolic innate immune system. PLoS Pathog. 2007;3:e51. doi: 10.1371/journal.ppat.0030051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aderem AA, Scott WA, Cohn ZA. A selective defect in arachidonic acid release from macrophage membranes in high potassium media. J Cell Biol. 1984;99:1235–1241. doi: 10.1083/jcb.99.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aderem AA, et al. Ligated complement receptors do not activate the arachidonic acid cascade in resident peritoneal macrophages. J Exp Med. 1985;161:617–622. doi: 10.1084/jem.161.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aderem AA, et al. Bacterial lipopolysaccha-rides prime macrophages for enhanced release of arachidonic acid metabolites. J Exp Med. 1986;164:165–179. doi: 10.1084/jem.164.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Underhill DM, et al. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature. 1999;401:811–815. doi: 10.1038/44605. [DOI] [PubMed] [Google Scholar]
- 8.Gantner BN, et al. Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. J Exp Med. 2003;197:1107–1117. doi: 10.1084/jem.20021787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 10.Goodridge HS, Underhill DM. Fungal recognition by TLR2 and Dectin-1. Handb Exp Pharmacol. 2008;183:87–109. doi: 10.1007/978-3-540-72167-3_5. [DOI] [PubMed] [Google Scholar]
- 11.Blander JM, Medzhitov R. Regulation of phagosome maturation by signals from toll-like receptors. Science. 2004;304:1014–1018. doi: 10.1126/science.1096158. [DOI] [PubMed] [Google Scholar]
- 12.Yates RM, Russell DG. Phagosome maturation proceeds independently of stimulation of toll-like receptors 2 and 4. Immunity. 2005;23:409–417. doi: 10.1016/j.immuni.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Blander JM, Medzhitov R. Toll-dependent selection of microbial antigens for presentation by dendritic cells. Nature. 2006;440:808–812. doi: 10.1038/nature04596. [DOI] [PubMed] [Google Scholar]
- 14.Trinchieri G, Sher A. Cooperation of Toll-like receptor signals in innate immune defence. Nat Rev Immunol. 2007;7:179–190. doi: 10.1038/nri2038. [DOI] [PubMed] [Google Scholar]
- 15.Miao EA, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi F, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 17.Amer A, et al. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J Biol Chem. 2006;281:35217–35223. doi: 10.1074/jbc.M604933200. [DOI] [PubMed] [Google Scholar]
- 18.Franchi L, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 19.Miao EA, et al. TLR5 and Ipaf: dual sensors of bacterial flagellin in the innate immune system. Semin Immunopathol. 2007;29:275–288. doi: 10.1007/s00281-007-0078-z. [DOI] [PubMed] [Google Scholar]
- 20.Geddes BJ, et al. Human CARD12 is a novel CED4 / Apaf-1 family member that induces apoptosis. Biochem Biophys Res Commun. 2001;284:77–82. doi: 10.1006/bbrc.2001.4928. [DOI] [PubMed] [Google Scholar]
- 21.Poyet JL, et al. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J Biol Chem. 2001;276:28309–28313. doi: 10.1074/jbc.C100250200. [DOI] [PubMed] [Google Scholar]
- 22.Masumoto J, et al. ASC is an activating adaptor for NF-kappa B and caspase-8-dependent apoptosis. Biochem Biophys Res Commun. 2003;303:69–73. doi: 10.1016/s0006-291x(03)00309-7. [DOI] [PubMed] [Google Scholar]
- 23.Dinarello CA. Interleukin-1 beta, interleukin-18, and the interleukin-1 beta converting enzyme. Ann NY Acad Sci. 1998;856:1–11. doi: 10.1111/j.1749-6632.1998.tb08307.x. [DOI] [PubMed] [Google Scholar]
- 24.Bruey JM, et al. PAN1 / NALP2 / PYPAF2, an inducible inflammatory mediator that regulates NF-kappaB and caspase-1 activation in macrophages. J Biol Chem. 2004;279:51897–51907. doi: 10.1074/jbc.M406741200. [DOI] [PubMed] [Google Scholar]
- 25.Agostini L, et al. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20:319–325. doi: 10.1016/s1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- 26.Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38:240–244. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- 27.Kanneganti TD, et al. Critical role for Cryopyrin / Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem. 2006;281:36560–36568. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- 28.Adachi O, et al. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 29.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 30.Salazar-Gonzalez RM, McSorley SJ. Salmonella flagellin, a microbial target of the innate and adaptive immune system. Immunol Lett. 2005;101:117–122. doi: 10.1016/j.imlet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Kumar H, et al. Cutting edge: cooperation of IPS-1- and TRIF-dependent pathways in poly IC-enhanced antibody production and cytotoxic T cell responses. J Immunol. 2008;180:683–687. doi: 10.4049/jimmunol.180.2.683. [DOI] [PubMed] [Google Scholar]
- 32.Edelmann KH, et al. Does Toll-like receptor 3 play a biological role in virus infections? Virology. 2004;322:231–238. doi: 10.1016/j.virol.2004.01.033. [DOI] [PubMed] [Google Scholar]
- 33.Warren SE, et al. Multiple Nod-like receptors activate caspase 1 during Listeria monocytogenes infection. J Immunol. 2008;180:7558–7564. doi: 10.4049/jimmunol.180.11.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roach JC, et al. The evolution of vertebrate Toll-like receptors. Proc Natl Acad Sci USA. 2005;102:9577–9582. doi: 10.1073/pnas.0502272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin MS, Lee JO. Structures of TLR-ligand complexes. Curr Opin Immunol. 2008;20:414–419. doi: 10.1016/j.coi.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Jin MS, et al. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 37.Liu L, et al. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Honda K, Takaoka A, Taniguchi T. Type I interferon (corrected) gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 39.Liew FY, et al. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–4458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- 40.Boone DL, et al. The ubiquitin-modifying enzyme A20 is required for termination Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 41.Wang YY, et al. A20 is a potent inhibitor of TLR3- and Sendai virus-induced activation of NF-kappaB and ISRE and IFN-beta promoter. FEBS Lett. 2004;576:86–90. doi: 10.1016/j.febslet.2004.08.071. [DOI] [PubMed] [Google Scholar]
- 42.Saitoh T, et al. A20 is a negative regulator of IFN regulatory factor 3 signaling. J Immunol. 2005;174:1507–1512. doi: 10.4049/jimmunol.174.3.1507. [DOI] [PubMed] [Google Scholar]
- 43.Lin R, et al. Negative regulation of the retinoic acid-inducible gene I-induced antiviral state by the ubiquitin-editing protein A20. J Biol Chem. 2006;281:2095–2103. doi: 10.1074/jbc.M510326200. [DOI] [PubMed] [Google Scholar]
- 44.Hitotsumatsu O, et al. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity. 2008;28:381–390. doi: 10.1016/j.immuni.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gilchrist M, et al. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441:173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- 46.Smith LM, et al. Fluorescence detection in automated DNA sequence analysis. Nature. 1986;321:674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 47.Schuster SC. Next-generation sequencing transforms today's biology. Nat Methods. 2008;5:16–18. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- 48.Bubnoff A. Next-generation sequencing: the race is on. Cell. 2008;132:721–723. doi: 10.1016/j.cell.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 49.Poinar HN, et al. Metagenomics to paleogenomics: large-scale sequencing of mammoth DNA. Science. 2006;311:392–394. doi: 10.1126/science.1123360. [DOI] [PubMed] [Google Scholar]
- 50.Green RE, et al. Analysis of one million base pairs of Neanderthal DNA. Nature. 2006;444:330–336. doi: 10.1038/nature05336. [DOI] [PubMed] [Google Scholar]
- 51.Malhi RS, et al. MamuSNP: a resource for Rhesus Macaque (Macaca mulatta) genomics. PLoS ONE. 2007;2:e438. doi: 10.1371/journal.pone.0000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lynch KW. Consequences of regulated pre-mRNA splicing in the immune system. Nat Rev Immunol. 2004;4:931–940. doi: 10.1038/nri1497. [DOI] [PubMed] [Google Scholar]
- 53.Srinivasan K, et al. Detection and measurement of alternative splicing using splicing-sensitive microarrays. Methods. 2005;37:345–359. doi: 10.1016/j.ymeth.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 54.Boutz PL, et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ule J, et al. Nova regulates brain-specific splicing to shape the synapse. Nat Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- 56.Wells CA, et al. Alternate transcription of the Toll-like receptor signaling cascade. Genome Biol. 2006;7:R10. doi: 10.1186/gb-2006-7-2-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gardina PJ, et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McKee AE, et al. Exon expression profiling reveals stimulus-mediated exon use in neural cells. Genome Biol. 2007;8:R159. doi: 10.1186/gb-2007-8-8-r159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yeo GW, et al. Alternative splicing events identified in human embryonic stem cells and neural progenitors. PLoS Comput Biol. 2007;3:1951–1967. doi: 10.1371/journal.pcbi.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oberdoerffer S, et al. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science. 2008;321:686–691. doi: 10.1126/science.1157610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mortazavi A, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 62.Shendure J. The beginning of the end for microarrays? Nat Methods. 2008;5:585–587. doi: 10.1038/nmeth0708-585. [DOI] [PubMed] [Google Scholar]
- 63.Cloonan N, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 64.de Hoon M, Hayashizaki Y. Deep cap analysis gene expression (CAGE): genome-wide identification of promoters, quantification of their expression, and network inference. Biotechniques. 2008;44:627–628. 630, 632. doi: 10.2144/000112802. [DOI] [PubMed] [Google Scholar]
- 65.Morin RD, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baltimore D, et al. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 67.Ren B, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 68.Wu J, et al. ChIP-chip comes of age for genome-wide functional analysis. Cancer Res. 2006;66:6899–6902. doi: 10.1158/0008-5472.CAN-06-0276. [DOI] [PubMed] [Google Scholar]
- 69.Hawkins RD, Ren B. Genome-wide location analysis: insights on transcriptional regulation. Hum Mol Genet. 2006;15:R1–R7. doi: 10.1093/hmg/ddl043. [DOI] [PubMed] [Google Scholar]
- 70.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat Rev Genet. 2008;9:179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Massie CE, Mills IG. ChIPping away at gene regulation. EMBO Rep. 2008;9:337–343. doi: 10.1038/embor.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng Y, et al. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445:936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- 73.Marson A, et al. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445:931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hartman SE, et al. Global changes in STAT target selection and transcription regulation upon interferon treatments. Genes Dev. 2005;19:2953–2968. doi: 10.1101/gad.1371305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schreiber J, et al. Coordinated binding of NF-kappaB family members in the response of human cells to lipopolysaccharide. Proc Natl Acad Sci USA. 2006;103:5899–5904. doi: 10.1073/pnas.0510996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lim CA, et al. Genome-wide mapping of RELA(p65) binding identifies E2F1 as a transcriptional activator recruited by NF-kappaB upon TLR4 activation. Mol Cell. 2007;27:622–635. doi: 10.1016/j.molcel.2007.06.038. [DOI] [PubMed] [Google Scholar]
- 77.Mardis ER. ChIP-seq: welcome to the new frontier. Nat Methods. 2007;4:613–614. doi: 10.1038/nmeth0807-613. [DOI] [PubMed] [Google Scholar]
- 78.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 79.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 80.Marson A, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 82.Gygi SP, et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 83.Ross PL, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 84.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 85.Chevallet M, et al. Toward a better analysis of secreted proteins: the example of the myeloid cells secretome. Proteomics. 2007;7:1757–1770. doi: 10.1002/pmic.200601024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang H, et al. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 87.Zhang H, Aebersold R. Isolation of glycoproteins and identification of their N-linked glycosylation sites. Methods Mol Biol. 2006;328:177–185. doi: 10.1385/1-59745-026-X:177. [DOI] [PubMed] [Google Scholar]
- 88.Guerrero C, et al. An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinitypurified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol Cell Proteomics. 2006;5:366–378. doi: 10.1074/mcp.M500303-MCP200. [DOI] [PubMed] [Google Scholar]
- 89.Ranish JA, et al. The study of macromolecular complexes by quantitative proteomics. Nat Genet. 2003;33:349–355. doi: 10.1038/ng1101. [DOI] [PubMed] [Google Scholar]
- 90.Himeda CL, et al. Quantitative proteomic identification of six4 as the trex-binding factor in the muscle creatine kinase enhancer. Mol Cell Biol. 2004;24:2132–2143. doi: 10.1128/MCB.24.5.2132-2143.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Himeda CL, Ranish JA, Hauschka SD. Quantitative proteomic identification of MAZ as a transcriptional regulator of muscle-specific genes in skeletal and cardiac myocytes. Mol Cell Biol. 2008;28:6521–6535. doi: 10.1128/MCB.00306-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 93.Deutsch EW, Lam H, Aebersold R. Peptide-Atlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 2008;9:429–434. doi: 10.1038/embor.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lange V, et al. Targeted quantitative analysis of Streptococcus pyogenes virulence factors by multiple reaction monitoring. Mol Cell Proteomics. 2008;7:1489–1500. doi: 10.1074/mcp.M800032-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stahl-Zeng J, et al. High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol Cell Proteomics. 2007;6:1809–1817. doi: 10.1074/mcp.M700132-MCP200. [DOI] [PubMed] [Google Scholar]
- 96.Marzolf B, et al. SBEAMS-Microarray: database software supporting genomic expression analyses for systems biology. BMC Bioinformatics. 2006;7:286. doi: 10.1186/1471-2105-7-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Reiss DJ, Baliga NS, Bonneau R. Integrated biclustering of heterogeneous genome-wide datasets for the inference of global regulatory networks. BMC Bioinformatics. 2006;7:280. doi: 10.1186/1471-2105-7-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ramsey SA, et al. Uncovering a macrophage transcriptional program by integrating evidence from motif scanning and expression dynamics. PLoS Comput Biol. 2008;4:e1000021. doi: 10.1371/journal.pcbi.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ideker T, et al. Testing for differentially-expressed genes by maximum-likelihood analysis of microarray data. J Comput Biol. 2000;7:805–817. doi: 10.1089/10665270050514945. [DOI] [PubMed] [Google Scholar]
- 101.Wingender E. The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief Bioinform. 2008;9:326–332. doi: 10.1093/bib/bbn016. [DOI] [PubMed] [Google Scholar]
- 102.Thijs G. Probabilistic Methods to Search for Regulatory Elements in Sets of Coregulated Genes. Faculty of Applied Sciences, Katholieke Universiteit Leuven; Leuven: 2003. [Google Scholar]
- 103.Mishra GR, et al. Human protein reference database - 2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cline MS, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bonneau R, et al. The Inferelator: an algorithm for learning parsimonious regulatory networks from systems-biology data sets de novo. Genome Biol. 2006;7:R36. doi: 10.1186/gb-2006-7-5-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Roach JC, et al. Transcription factor expression in lipopolysaccharide-activated peripheral-blood-derived mononuclear cells. Proc Natl Acad Sci USA. 2007;104:16245–16250. doi: 10.1073/pnas.0707757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Basso K, et al. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 108.Potapov AP, et al. Topology of mammalian transcription networks. Genome Inform. 2005;16:270–278. [PubMed] [Google Scholar]
- 109.Bartholin L, et al. TGIF inhibits retinoid signaling. Mol Cell Biol. 2006;26:990–1001. doi: 10.1128/MCB.26.3.990-1001.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Longabaugh WJ, Davidson EH, Bolouri H. Visualization, documentation, analysis, and communication of large-scale gene regulatory networks. Biochim Biophys Acta. 2008 doi: 10.1016/j.bbagrm.2008.07.014. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hwang D, et al. A data integration methodology for systems biology. Proc Natl Acad Sci USA. 2005;102:17296–17301. doi: 10.1073/pnas.0508647102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shannon PT, et al. The Gaggle: an opensource software system for integrating bioinformatics software and data sources. BMC Bioinformatics. 2006;7:176. doi: 10.1186/1471-2105-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.von Mering C, et al. STRING 7 -recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Johnson WE, et al. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci USA. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ramsey S, Orrell D, Bolouri H. Dizzy: stochastic simulation of large-scale genetic regulatory networks. J Bioinform Comput Biol. 2005;3:415–436. doi: 10.1142/s0219720005001132. [DOI] [PubMed] [Google Scholar]
- 116.Keller A, et al. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol. 2005;1:2005, 0017. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Krishnan MN, et al. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brass AL, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 119.Beutler B, et al. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu Rev Immunol. 2006;24:353–389. doi: 10.1146/annurev.immunol.24.021605.090552. [DOI] [PubMed] [Google Scholar]
- 120.Cook MC, Vinuesa CG, Goodnow CC. ENU-mutagenesis: insight into immune function and pathology. Curr Opin Immunol. 2006;18:627–633. doi: 10.1016/j.coi.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 121.Hoebe K, et al. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 122.Tabeta K, et al. The Unc93b1 mutation 3 d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol. 2006;7:156–164. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- 123.Kim YM, et al. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 124.Vinuesa CG, et al. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 2005;435:452–458. doi: 10.1038/nature03555. [DOI] [PubMed] [Google Scholar]
- 125.Yu D, et al. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature. 2007;450:299–303. doi: 10.1038/nature06253. [DOI] [PubMed] [Google Scholar]
- 126.Du X, et al. An essential role for Rxr alpha in the development of Th2 responses. Eur J Immunol. 2005;35:3414–3423. doi: 10.1002/eji.200535366. [DOI] [PubMed] [Google Scholar]
- 127.Jiang Z, et al. Details of Toll-like receptor:adapter interaction revealed by germ-line mutagenesis. Proc Natl Acad Sci USA. 2006;103:10961–10966. doi: 10.1073/pnas.0603804103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Beutler B, et al. Genetic dissection of innate immunity to infection: the mouse cytomegalovirus model. Curr Opin Immunol. 2005;17:36–43. doi: 10.1016/j.coi.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 129.Xavier RJ, Rioux JD. Genome-wide association studies: a new window into immune-mediated diseases. Nat Rev Immunol. 2008;8:631–643. doi: 10.1038/nri2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hawn TR, et al. A common dominant TLR5 stop codon polymorphism abolishes flagellin signaling and is associated with susceptibility to legionnaires' disease. J Exp Med. 2003;198:1563–1572. doi: 10.1084/jem.20031220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bochud PY, Hawn TR, Aderem A. Cutting edge: a Toll-like receptor 2 polymorphism that is associated with lepromatous leprosy is unable to mediate mycobacterial signaling. J Immunol. 2003;170:3451–3454. doi: 10.4049/jimmunol.170.7.3451. [DOI] [PubMed] [Google Scholar]
- 132.Hawn TR, et al. Toll-like receptor 4 polymorphisms are associated with resistance to Legionnaires' disease. Proc Natl Acad Sci USA. 2005;102:2487–2489. doi: 10.1073/pnas.0409831102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hawn TR, et al. A polymorphism in susceptibility to meningeal tuberculosis. J Infect Dis. 2006;194:1127–1134. doi: 10.1086/507907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Picard C, et al. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science. 2003;299:2076–2079. doi: 10.1126/science.1081902. [DOI] [PubMed] [Google Scholar]