Abstract
Nuclear pore complexes are large aqueous channels that penetrate the nuclear envelope, connecting the nuclear interior with the cytoplasm. Until recently, these macromolecular complexes were viewed as static structures whose only function was to control the molecular trafficking between the two compartments. It has now become evident that this simplistic scenario is inaccurate and that nuclear pore complexes are highly dynamic multiprotein assemblies involved in diverse cellular processes ranging from the organization of the cytoskeleton to gene expression. In this review, we will discuss the most recent developments in the nuclear pore complex field, focusing in the assembly, disassembly, maintenance and function of this macromolecular structure.
Introduction
A hallmark of eukaryotic cells is the compartmentalization of the genetic material inside the nucleus. By restricting the accessibility of cytoplasmic proteins to DNA with the physical barrier of the nuclear envelope (NE), eukaryotic cells have achieved a complexity in transcriptional regulation not found in prokaryotes. Furthermore, the NE provides additional levels of regulation of gene expression such as the selective export of newly synthesized mRNA into the ribosome-containing cytoplasm and the establishment of higher-order levels of organization of the nuclear genome.
The NE comprises two concentric lipid bilayers, the outer and inner nuclear membranes (ONM and INM respectively) [1]. The ONM is continuous with the endoplasmic reticulum (ER) and studded with ribosomes, whereas the INM is characterized by a set of integral membrane proteins [1]. Large multiprotein structures known as nuclear pore complexes (NPCs) penetrate the NE at sites where the INM and the ONM are fused [1]. NPCs act as gatekeepers of the nucleus, performing the essential cellular role of mediating the exchange of molecules between the nucleoplasm and the cytoplasm [2]. Ions and small metabolites can diffuse through NPCs; however, molecules possessing a mass greater than 40–60 kDa need to be actively transported. Nucleocytoplasmic transport is a complex process carried out by a large family of transport receptors known as karyopherins or importins/exportins, the latter named depending on their direction of transport [3]. The molecular mechanism of nuclear transport has recently been the subject of several major reviews and thus will not be discussed [2, 4]. Here, we focus on the latest advances in our understanding of nuclear pore assembly, disassembly, maintenance and function.
NPC structure
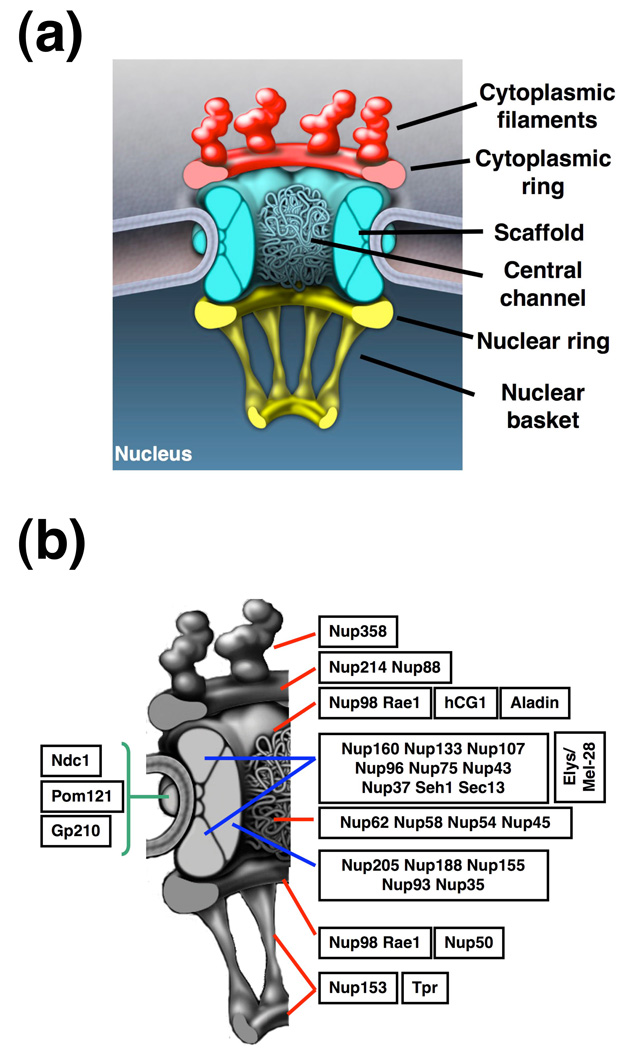
Owing to their function as exclusive nucleocytoplasmic transport channels and unique structural features, NPCs have been actively studied since their discovery in the 1950s. Although the initial descriptions of the overall NPC structure were performed more than 40 years ago [5], it was not until recently that new techniques, such as cryo-electron tomography, field-emission in-lens scanning electron microscopy (FEISEM), 4pi and atomic force microscopy, as well as improved cell fixation protocols, could provide a detailed picture of its three-dimensional (3D) organization [6–11]. Overall, the NPC is an eight-fold-symmetrical structure comprising a NE-embedded scaffold that surrounds a central transport channel and two rings – the cytoplasmic and nuclear rings – to which eight filaments are attached (Figure 1a). While the cytoplasmic filaments have loose ends, the nuclear filaments are joined in a distal ring, forming a structure known as the nuclear basket. Even though the size of the NPC varies between species, its overall structure is evolutionarily conserved from yeasts to mammals [12, 13].
Figure 1. Nuclear pore complex (NPC) structure and composition.
(a) Schematic illustration of the NPC structure (b) Predicted localization of subcomplexes and nucleoporins within the NPC. The members of the Nup214 complex (Nup214, Nup88), Nup98 complex (Nup98, Rae1), Nup107–160 complex (Nup160, Nup133, Nup107, Nup96, Nup75, Nup43, Nup37, Sec13, Seh1), Nup62 complex (Nup62, Nup58, Nup54, Nup45), and Nup93–205 complex (Nup205, Nup188, Nup155, Nup93, Nup35) are enclosed in the same box. Green lines show the location of the three transmembrane nucleoporins, red lines the location of peripheral components and blue lines indicate the location of scaffold subcomplexes.
Despite their high molecular mass of ~60–125 MDa in mammals and ~40–60 MDa in yeasts, proteomic analysis has revealed that NPCs contains only ~30 different proteins, known as nucleoporins or nups [13–16] (Table 1). Except for three transmembrane proteins that are believed to anchor the NPC to the NE [17, 18], all other nucleoporins are soluble. Owing to the eightfold symmetry of pores, each nucleoporin is present in copies of eight or multiples of eight, resulting in ~ 500–1000 nups per pore. Remarkably, nucleoporins have a very limited set of domains, restricted to β-propellers, α-solenoids, phenylalanine-glycine (FG) repeats, coiled-coiled and transmembrane domains [19, 20]. Most of these proteins associate in biochemically stable subcomplexes that are believed to act as the ‘building blocks’ of the NPC (Figure 1b).
Table 1.
Mammalian, S. cerevisiae and C. elegans nucleoporins homologuesa,b
| Mammalian | S. cerevisiae | C. elegans |
|---|---|---|
| Nup35 | Nup53p | Npp-19 |
| Nup37 | - | - |
| Nup43 | - | C09G9.2 |
| Nup50 | Nup2p | Npp-16 |
| Nup54 | Nup57p | Npp-1 |
| Nup58/45 | Nup49p | Npp-4 |
| Nup62 | Nsp1p | Npp-11 |
| Nup75 | Nup85p | Npp-2 |
| Nup88 | Nup82p | - |
| Nup93 | Nic96p | Npp-13 |
| Nup96 | Nup145Cp | Npp-10 |
| Nup98 | Nup145Np Nup100p Nup116p |
Npp-10 |
| Nup107 | Nup84p | Npp-5 |
| Nup133 | Nup133p | Npp-15 |
| Nup153 | Nup1p Nup2p Nup60p | Npp-7 |
| Nup155 | Nup157p Nup170p | Npp-8 |
| Nup160 | Nup120p | Npp-6 |
| Nup188 | Nup188p | - |
| Nup205 | Nup192p | Npp-3 |
| Nup214 | Nup159p | Npp-14 |
| Nup358/RanBP2 | - | Npp-9 |
| Sec13R | Sec13p | Npp-20 |
| Seh1 | Sehp | Npp-18 |
| Pom121 | - | - |
| Gp210 | - | Npp-12 |
| Ndc1 | Ndc1p | Npp-22 |
| Tpr | Mlp1p Mlp2p | Npp-21 |
| RAE1 | Gle2p | Npp-17 |
| ALADIN | - | - |
| NLP1/hCG1 | Nup42p | - |
Recently, a detailed three-dimensional model for the position and abundance of each nup in the S. cerevisiae NPC structure was proposed based on experimental data obtained from molecular, biochemical and structural information of the NPCs and their components [20, 21]. In the modeled structure, the scaffold of the NPC is formed by two main protein subcomplexes that, through linker proteins, anchor a set of FG-containing nucleoporins [20]. The FG-rich nucleoporins, which can contain 4 to 48 GLFG, FxFG, PxFG or SxFG repeats, represent a third of the total pore proteins and fill the central channel of the NPC, extending into the cytoplasmic and nucleoplasmic sides. The FG domains have an unfolded structure and are responsible for interaction with transport receptors [22–26]. Notably, it was demonstrated recently that, at high concentrations, the FG repeats of the S. cerevisiae nucleoporin Nsp1p formed a three-dimensional hydrogel that reproduced the permeability properties of the NPC [27, 28]. These results indicate that the FG nucleoporins form a sieve-like meshwork through weak hydrophobic interactions that acts as a barrier restricting the passage of molecules through the NPC and support the ‘selective phase’ model for the pore permeability barrier [29, 30] (Box 1). To carry cargoes through the NPC, the transport receptors overcome this barrier by interacting with the FG-repeats, locally dissolving the FG–FG meshwork.
Box 1. Models of NPC selectivity
NPCs can perform 1000 translocations events per second, shuttling a mass of ~100MDa/second [29]. Yet these structures efficiently restrict the passages of large molecules that lack nuclear transport signals. Although it has long been known that the major diffusion barrier of the NPC resides in its central channel, how nuclear pores achieve their transport selectivity is still a matter of debate. Several models have been proposed to explain the NPC selectivity taking into account that active transport through the pore occurs by facilitated diffusion and requires the interaction of transport receptors with bind the FG-nucleoporins.
The virtual gate model [33] predicts that the highly dense FG nucleoporins of the central channel generate an entropic barrier that prevents the passage of inert molecules. For a molecule that is freely diffusing in the cytoplasm entering the overcrowded central channel of the pore implies an immediate restriction of its liberty of movement and a drop of its entropy. The entropic price that a molecule has to pay to move though the pore increases with its size lowering its probability of crossing the channel. The interaction of a molecule with the FG-nucleoporins increases its probability to enter and move though the pore. This model proposes the existence of an energetic barrier and not a physical barrier controlling NPC permeability.
The spaghetti oil model [25] proposes that extended FG repeats fill and obstruct the pore. The idea is that the FG-repeats are constantly moving at physiological temperature and can be pushed to one side by translocation complexes. In this model, transport receptors move though the FG-spaghetti by a binding/release mechanism. Because the FG-transport receptor interactions are weak, the carriers would bind the FG repeats only transiently and once released they would be free to diffuse thought the pore until finding another nucleoporin to bind. The continuous binding/release/diffusion cycle would allow the transporter to move through the pore by a facilitated diffusion.
In contrast to the previous models, the selective phase model [29] suggests that the FG-nucleoporins from the central channel form a sieve-like meshwork through weak hydrophobic interactions. This gel-like network prevents the passage of inert molecules larger than its pore size. In this model, the transport receptors would selectively partition into the semi-liquid pore phase and move through the central channel by competing the FG-FG interactions locally dissolving the meshwork. Thus, by interacting with the FG nucleoporins, the carriers would help molecules carrying nuclear transport signals cross the pore by increasing their solubility in the central channel. This model proposes the existence of a physical barrier that determines NPC selectivity, where the exclusion size of the pore is defined by the pore size of the sieve-like meshwork.
Finally, the Reduction of dimensionality [26] model proposes that transport receptors act as ferries that can slide on the surface of an FG-surface. In this model a continuous FG-surface extends from the cytoplasmic filaments to the nucleoplasmic basket and throughout the walls of the central channel. Transport receptors would bind to FG-surface in the cytoplasmic or nucleoplasmic side of the NPC and move through the pore by a two-dimensional walk. The model predicts the existence of a selectivity filter in the central channel also generated by the FG repeats with an unobstructed narrow tube in the center that would allow the diffusion of small molecules. The transport receptor-cargo complexes would overcome the filter because they can bind and enter the FG-surface through its extremes. On the other hand, inert molecules that are too large to diffuse through the central tube and cannot bind the FG-repeats would no be able to enter filter and will not be able to move across the pore.
Although the existence of a three-dimensional hydrogel has yet to be proven in vivo, the formation of the FG-meshwork is supported by a study showing that the FG nucleoporins located at the NPC central channel interact through their FG motifs in vitro and in vivo [31]. Nevertheless, the FG repeats of the nucleoporins that constitute the cytoplasmic and nuclear filaments were found not to interact with the FG domains of the nucleoporins from the central channel, suggesting that the peripheral FG-nucleoporins could act as “entropic bristles” that prevent the entry of molecules to the pore. Acting as “entropic bristles” means that, by movement, the filaments would reduce the available space to access the pore channel generating in this way an entropic barrier that would decrease the molecules probability of entering the pore without physically repelling them. To access this limited space and enter the pore a molecule would have to reduce its entropy, which is energetically unfavorable. Supporting this idea, it was recently reported that the FG domains of Nup153, a main component of the nuclear basket, form a brush-like structure in vitro with entropic repulsion properties [32]. According to this, the filaments would help to maintain the permeability barrier, in agreement with the ‘virtual gate’ model [33] (Box 1), which proposes that the probability of a molecule to overcome the entropic cost of entering a limited space packed with non-interacting nucleoporins decreases with increasing size (Box 1). The interaction of transport receptors with the FG repeats would allow large molecules to overcome the energy loss and move through the entropic barrier. It is worth mentioning that in the selective-phase model, the formation of the sieve-like meshwork would also generate an entropic barrier for those molecules that can enter the pores of the FG-network. This barrier would reduce the probability of these molecules to enter the pores but would not define the permeability limit of the NPC, which would be determined by the meshwork pore size.
Although these findings point towards the FG-network as the main mechanism for pore selectivity they are not yet sufficient to completely discard other proposed models for NPC permeability (see Box 1) [25, 26, 29, 33]. In the end, it is possible that a combination of them would be required to explain all the reported NPC properties.
Dynamic organization of NPCs
A remarkable, yet largely uncharacterized, feature of the NPC is their dynamic molecular organization. Using a systematic approach, where 19 GFP-tagged nucleoporins were studied by fluorescence recovery after photobleaching (FRAP), it was shown that the residence times of different nups at the NPC varied from a few seconds to more than 70 hours depending on their location/function [34]. While the proteins that form the NPC scaffold, such as members of the Nup107–160 complex, are stably embedded in the NE during interphase (having residence times longer than the average cell cycle), the more peripheral components, such as Nup153 and Nup50, are highly dynamic having residence times of seconds to minutes. A third class of nucleoporins, which are believed to work as linkers between the scaffold and the peripheral nucleoporins, have intermediate residence times.
The functional significance of ‘dynamic nucleoporins’ is still unclear. It has been suggested that mobile nucleoporins could help to deliver cargo to the NPC. Supporting this idea, the mobility of two RNA-binding nucleoporins, Nup153 and Nup98, is transcription dependent, suggesting that they could assist newly transcribed RNAs to reach the pore and be exported to the cytoplasm [35]. While a transport-dependent function of dynamic nups seems plausible, it is also possible that these proteins have pore-independent functions (see below). A dynamic organization of NPCs could also be indicative of changes in protein composition in response to altered transport requirements. The presence of tissue and developmental-specific nucleoporins has been reported [36–39]; however, there is no evidence that NPCs of different composition coexist in the same cell.
The dynamic properties of NPCs can also be observed at the structural level. Early cryo-EM studies found that the NPCs were present in either an open or closed conformation, the latter characterized by the presence of a central plug or transporter [40]. Historically, the central plug was considered to be a structural component of the NPC, but recent data indicate that it represents translocating cargo. Transmission electron microscopy (TEM) and FEISEM have identified different conformations of the NPC nuclear basket in response to the export of large ribonucleoprotein particles [41]. Cryoelectron tomography has shown two different structural states of the cytoplasmic filaments and variability in the position of the narrowest constriction of the central channel [7]. Furthermore, the NPC has been shown to alter its conformation in response to Ca2+, glucocorticoids and ATP [42–47].
The molecular basis of the structural changes in pore architecture and its physiological role is still a mystery. One possibility is that the location of nucleoporins is actively reorganized for functional purposes within the NPC structure. Supporting this idea, two nucleoporins, Nup153 and Nup214, have been found in different regions of the NPC in a transport-dependent manner [48, 49]. Moreover, the yeast NPC has been shown to undergo cell-cycle-specific rearrangements [50]. It was observed that, by changing interacting partners, some nucleoporins modify their location at the yeast NPC, affecting specific nuclear transport pathways [50]. Another possibility is that structural changes of the NPC are a consequence of alterations in nucleoporin conformation and not in their location. Recently, the crystal structure of α-helical regions of Nup58/45, a nucleoporin from the pore central channel, was solved. It was found that the α-helical domains formed tetramers through the interaction of two antiparallel dimers [51]. The authors described the presence of different tetramer conformations that were laterally displaced along the axis of the dimer–dimer interface. Thus, they proposed that eight Nup58/45 tetramers would be built around the central channel and regulate the aperture/diameter of the NPC by sliding along the dimer interface. Also supporting a nucleoporin conformational change, Nup153 has been reported to collapse reversibly into a compact conformation when its FG-repeats bind to the transport receptor Karyopherin/Importin β [52].
NPC life cycle
Assembly
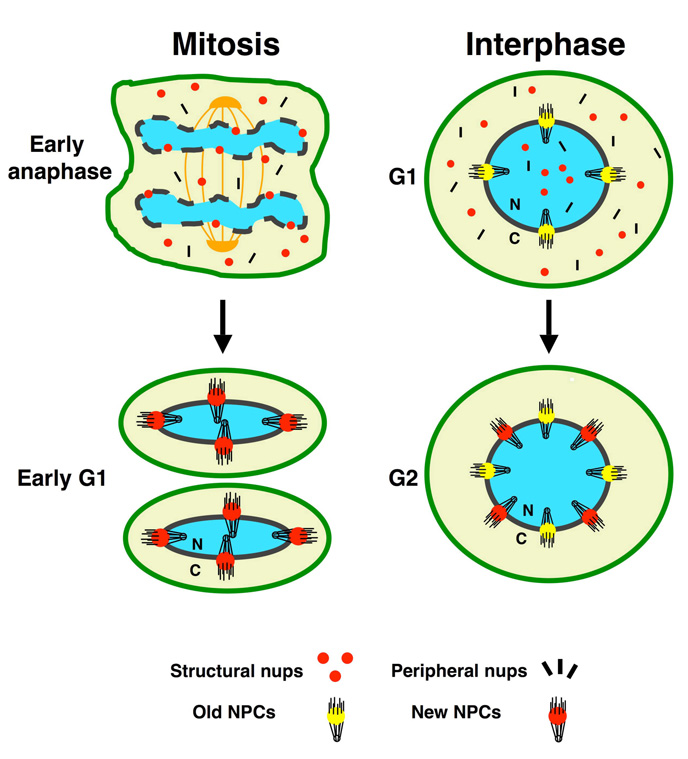
The biogenesis of nuclear pores is essential for cell survival and proliferation. There are two phases during the metazoan cell cycle when NPCs need to be assembled: first, at the end of mitosis, when the NE reforms around the segregated chromosomes, and second during interphase when the cells double their number of pores preparing for the next round of division. Even though both processes lead to the same final structure, they occur under very different conditions. Mitotic NPC assembly takes place concomitantly with reformation of the nuclear membrane around segregated chromosomes. At this time, NPCs are rebuilt from disassembled subcomplexes that were dispersed into the cytoplasm during breakdown of the nuclear envelope (Figure 2). By contrast, NPC assembly during interphase occurs in an intact NE using newly synthesized nups and in a cellular environment where the nucleus and the cytoplasm are physically separated (Figure 2). Interphase assembly is the only existing mechanism in organisms, such as yeasts, that undergo closed mitosis (i.e. their NE does not break down during cell division). Although accumulating evidence suggests that both types of pore assembly might occur through similar processes, owing to their very different environments it is likely that differences will be uncovered.
Figure 2. NPC assembly during mitosis and interphase.
Mitotic NPC assembly occurs concomitantly with the formation of new nuclear envelopes (NEs) around chromatin. During this time, NPCs assemble by recycling subcomplexes that were dispersed into the cytoplasm during NPC and NE breakdown. Note that, during mitosis, the cytoplasmic and nuclear contents are mixed together. Mitotic assembly is a step-wise process that begins with the recruitment of structural nups to chromatin during early anaphase. During interphase, by contrast, NPCs assemble into an intact NE when the nucleus and cytoplasm are physically separated. During this process, NPCs utilize newly synthesized nucleoporins present on both sides of the nuclear envelope.
Mitotic assembly
As mentioned above, NPC assembly during mitosis occurs when the NEs are forming around the chromatin of the two nascent daughters [53, 54]. Data obtained from mammalian cells and an in vitro nuclear assembly system based on Xenopus egg extracts have shown that mitotic NPC assembly is a highly organized step-wise process. Several reports have shown that nucleoporins are recruited to chromatin in a sequential manner starting in early anaphase [55–59]. Recently, in an elegant systematic analysis of nucleoporin recruitment in vivo, Dultz and colleagues analyzed the kinetics of assembly of eight different NPC subcomplexes in living cells [60]. Taken together, these results indicate that mitotic NPC assembly begins on chromatin with the recruitment of the scaffold Nup107–160 complex, known to be essential for NPC assembly [57], followed by the recruitment of a small fraction of the total Nup153 and Nup50 proteins. It is generally assumed, although not truly demonstrated, that the early recruited nucleoporins (Elys/Mel-28, Nup107–160 complex, Nup153, Nup50 and possibly other not yet analyzed nucleoporins) form a chromatin-bound intermediate, sometimes referred to as ‘prepore’, that acts as a binding platform for the recruitment of transmembrane nucleoporins and, later, the more peripheral proteins. Notably, the recruitment of several nucleoporins occurs after NPCs have started to transport, supporting the idea that not all nups are essential for pore function [60, 61] or that different NPC functions or transport pathways get activated at different times of assembly.
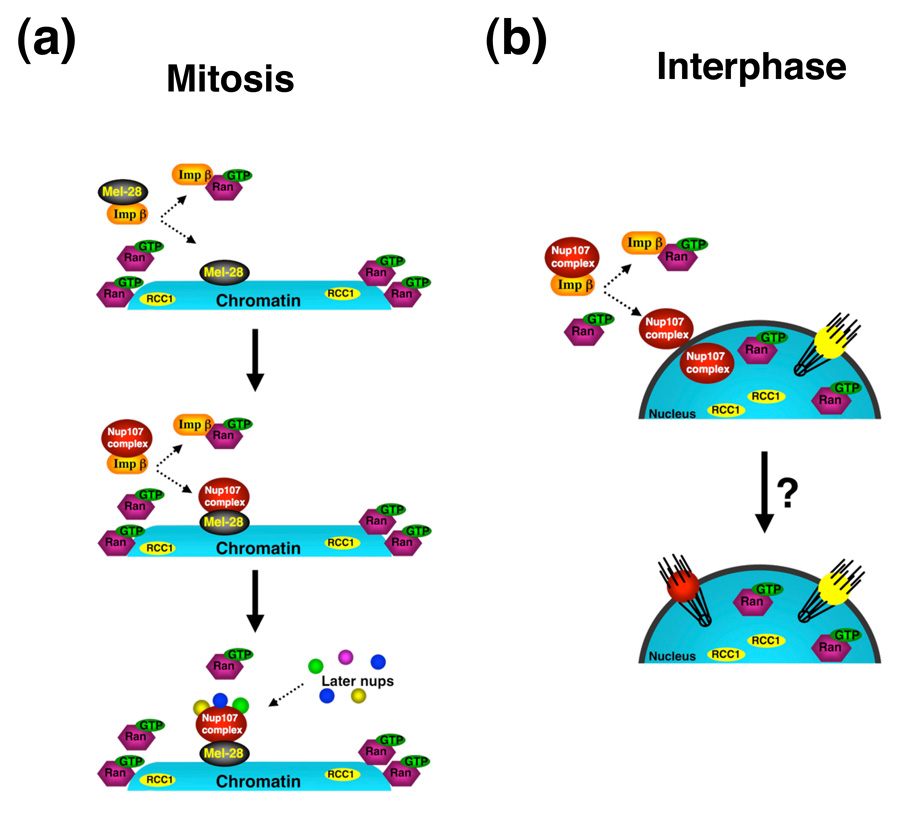
The step-wise mechanism of NPC assembly and the existence of intermediate structures on chromatin is supported by EM studies [62–65]. Evidence from several recent reports indicates that the early recruitment of the Nup107–160 complex to chromatin is mediated by Elys/Mel-28, which was initially discovered as an AT-hook-containing transcription factor [66–69]. The chromatin associations of nups as well as the interactions between many of them are regulated by the ratio between Importin β and the small GTPase Ran [58, 70]. During mitosis, Importin β associates with and sequesters a subset of nups, including Elys/Mel-28 and the Nup107–160 complex, preventing their association with chromatin and interaction with other nups [58, 68] (Figure 3a). The RanGTP-dependent release of nucleoporins from the import receptor is an early step in NPC assembly, and, because RanGTP is concentrated around chromosomes, occurs in the proximity of the chromosomal DNA. In this way, Importin β and RanGTPase coordinate the spatial positioning of the assembly of NPCs during mitosis.
Figure 3. Importin β and RanGTPase regulation of NPC assembly.
(a) During mitosis, Importin β binds and sequesters the Elys/Mel-28 nucleoporin (Mel-28), preventing its interaction with chromatin. When the Importin-β—Mel-28 complexes are in the proximity of DNA, where there is a high concentration of RanGTP due to the chromatin association of the Ran GDP—GTP exchange factor (regulator of chromosome condensation, RCC1), RanGTP binds to the transport receptor, releasing Mel-28 and allowing it to bind to chromatin. Following the same mechanism, the Importin-β-bound Nup107–160 complex is released by RanGTP in the proximity of DNA and recruited to chromatin through Mel-28. The chromatin-bound Nup107–160 complex can then recruit other nucleoporins in a step-wise manner. (b) NPC assembly during interphase requires the RanGTP-dependent release of the Nup107–160 complex from Importin β on the cytoplasmic and nuclear side of the NE. How the released complexes coordinate the formation of a functional NPC from both sides of the nuclear envelope is still unclear.
Interphase assembly
During interphase, the number of pores doubles to prepare the cells for re-entering mitosis. Previously, it was believed that NPC doubling was restricted to S-phase [71]; however, recent studies suggest that NPCs are assembled continuously from G1 to G2 phases [72, 73]. Little is known about the mechanism of interphase NPC assembly. Genetic studies in S. cerevisiae have identified nucleoporins essential for the formation of NPCs [74–76] as well as a requirement for Karyopherin/Importin β and RanGTP [77, 78]. However, the cellular mechanism used to assemble NPCs into an intact NE remains unclear. In metazoans, the assembly of NPCs into the NE also requires Importin β and RanGTP [79]. Moreover, RanGTP as well as the essential Nup107–160 complex is required from both sides of the NE, indicating that interphase NPC assembly requires the coordination of nuclear and cytoplasmic events (Figure 3b).
EM studies in Drosophila embryos showed that new NPCs can form in an intact nuclear envelope that lacks pre-existing pores [80]. Consistent with this finding, assembly of NPCs in regions devoid of pores has been observed in HeLa nuclei by live imaging [79]. Furthermore, experiments using in vitro assembled nuclei indicated that NPCs can form de novo, without using subunits from pre-existing pores [79].
Taken together, these results have led to the current view of interphase NPC assembly that suggests that nuclear pore complexes form independently of pre-existing pores and from both sides of the nuclear envelope, following a mechanism that is at least partially conserved with mitotic NPC assembly. This raises the question as to whether the chromatin-bound intermediates observed during mitosis indeed represent that part of the pore that in interphase is inserted into the NE from the nucleoplasmic side. If so, the mitotic and interphase assembly mechanisms would involve a coordinated interaction between chromatin-bound subcomplexes, cytoplasmic subcomplexes and transmembrane NPC components to assemble the multiprotein NPC into the double membrane of the NE.
Disassembly
NPC disassembly has only been described during mitosis, and there is no evidence of pores being dismantled during interphase. Similar to the assembly process, NPC disassembly takes place through an ordered process. Partially disassembled pores have been observed in Drosophila embryos [80] and by in vitro NE disassembly studies [81]. In Drosophila embryos, only one kind of disassembly intermediate was found during prophase, indicative of a very synchronized process, the existence of other extremely unstable intermediates or the existence of only one intermediate in this organism [80]. By contrast, a mixed population of different partially disassembled NPCs was observed in the same disassembling nuclei using the in vitro xenopus system, suggesting the existence of an asynchronous mechanism [81].
The step-wise disassembly of NPCs has been followed during NE breakdown of starfish oocytes. In these cells, the process starts with the release of the basket nucleoporins Nup98 and Nup153, and is followed by the release of Nup214 from the cytoplasmic ring. The dissociation of peripheral nucleoporins into the cytoplasm leads to a gradual loss of selective nuclear permeability.
Using a kinetic assay in mammalian cells [60], it has been shown that NPC disassembly occurs synchronously, being faster than assembly and, intriguingly, not following its exact reverse order. For example, nucleoporins that are recruited to chromatin early during assembly, such as members of the Nup107–160 complex, are released before nucleoporins that are recruited later, such as Nup58.
Supporting these results, it was observed that in Drosophila the scaffold Nup107 protein is also released from NPCs after the basket components Nup153 and Mtor (the homologue of the mammalian Tpr protein) but before the central channel nucleoporin Nup58 [82].
In the yeasts S. cerevisiae and S. pombe, which divide without NE breakdown, no disassembly of pores has been reported; however, in other fungi undergoing closed mitosis, such as Aspergillus nidulans, a partial disassembly of pores also occurs [83]. In these cells, the cytoplasmic dispersal of several FG nucleoporins starting in prophase leaves partially disassembled pores at the NE that have an increase in permeability and allow mitotic regulators to access the nucleus.
Although the molecular mechanisms of pore disassembly have not been investigated in detail, all the current evidence points towards mitotic phosphorylation as being the trigger of the disassembly process. Several groups have reported mitotic-specific hyperphosphorylation of nucleoporins both in vivo and in vitro [61, 84–87]. The Ser/Thr kinase CDC2 (Cdk1) seems to be a universal mitotic kinase involved in nucleoporin phosphorylation [83, 84, 88, 89]; however, the mammalian protein kinase A (PKA) and protein kinase C (PKC), the S. cerevisiae casein kinase and the A. nidulands Ser/Thr kinase NIMA have also been shown to phosphorylate NPC components [39, 83, 88, 90]. How mitotic phosphorylation initiates NPC disassembly is still unclear, but it is possible that it induces the dissociation of subcomplexes and prevents their association, as nucleoporins have to be dephosphorylated to reassemble the pore [58].
Maintenance
The mitotic disassembly–reassembly cycle of NPCs in metazoans ensures that every nuclear pore gets renewed during each cell-division cycle. Yet, how NPCs are maintained during interphase is unknown. As mentioned above, several nucleoporins are dynamic and can come on and off the pore, indicating that they are exchanged/recycled from NPCs during interphase. By contrast, the NPC scaffolds are very stable and only exchange once per cell cycle, when the NE breaks down. This raises the question of how NPC scaffold proteins are maintained when the cells stop dividing (e.g. after differentiation). Are old pores disassembled and replaced by new NPCs? This would imply the existence of post-mitotic disassembly mechanism. As mention above no evidence of such mechanism has been described so far. Or do the NPC scaffolds remain embedded in the NE for the entire life span of the cell? If this is the case, then in some long-lived cells, such as neurons, the core of the NPCs would remain in the NE for extensive periods of time. Because of this, the pores could be subject to age-dependent damage that could affect their function and possibly compromise nuclear integrity. In favor of this idea, alterations in the nuclear structure and mislocalization of nuclear proteins have been linked recently to the aging phenomena [91, 92].
Transport-independent functions of nups
As described above, the main function of NPCs is to control the passage of molecules between the nucleus and the cytoplasm. A tight regulation of nucleocytoplasmic transport is essential for cell homeostasis. Consequently, alterations in NPC members or the nuclear transport process have a strong impact on cell growth and survival – and, not surprisingly, have been associated with several diseases such as cancer [93, 94] and the rare autosomal recessive disorder triple A syndrome [95].
Besides acting as gatekeepers of the nucleus, it has become evident that NPCs and nucleoporins are implicated in many other biological functions [1]. NPCs have been shown to anchor and modulate the activity of sumoylating and desumoylating enzymes [96]. Interestingly, Nup358, the main component of the cytoplasmic filaments, is an active E3 ligase in the sumoylation reaction [97, 98]. Thus, through the SUMO pathway, NPCs are indirectly involved in the regulation of numerous cellular processes such as gene trasncription, DNA replication, DNA damage/repair, chromosome segregation, genome stability, cell death and senescence.
Three nucleoporins – Nup153, Nup358 and gp210 – have been associated with nuclear envelope breakdown [1, 99, 100]. While Nup153 and Nup358 contribute to this process by recruiting the coatomer COPI complex to the NE, the role of gp210 in NE breakdown is still unclear. Conversely, several other nucleoporins have been reported to play a role in NE assembly. These include the transmembrane nucleoporins Pom121 and NDC1, and the soluble nucleoporins Elys/Mel-28, Nup35 and Nup155, the latter two being part of the same subcomplex [17, 99, 101–103].
In the past few years, many components of the NPC have been described as having important roles during mitosis. Nup170p, the yeast homolog of mammalian Nup155, was the first nucleoporin linked to kinetochore function and chromosome segregation [104]. Subsequently, Nup358 was found anchored to kinetochores and spindles through the export receptor CRM1. Depletion of Nup358 in cells has revealed its essential role in kinetochore assembly and interaction with microtubules [100, 105]. More recently, the Nup107–160 complex and Elys/Mel-28 were found at kinetochores during mitosis [106–108]. The recruitment of the Nup107–160 complex to these structures was shown to be dependent on the Ndc80 complex and CENP-F and upstream of Nup358. Cells depleted of several members of the Nup107–160 complex together, or the Seh1 component alone, showed a mitotic delay and failed to attach microtubules to kinetochores properly, resulting in abnormal chromosome congression. Although these cells showed activation of the spindle checkpoint and longer spindles in metaphase, no significant defects in spindle assembly were observed [107]. In a different study, the Nup107–160 complex was shown to be required for assembly of the bipolar spindle in vitro, although no activation of the spindle checkpoint was observed in extracts depleted of the complex [109]. The differences observed could be attributable to the fact that the Nup107–160 complex strongly localizes to spindles assembled in vitro but only transiently associates with cellular spindles during prometaphase [109]. A role for the Nup107–160 complexes in spindle assembly and function is supported by the findings that fission yeasts lacking Nup120p, a homologue of the human Nup160 protein, show abnormal spindles [110]. Rae1 is a shuttling nucleoporin that has also been associated with spindle formation [14, 15, 111, 112]. It was found that this protein is part of a large ribonucleoprotein complex that controls microtubule dynamics and has an essential role in spindle assembly [112]. Finally, RNAi experiments in C. elegans have revealed a role for several nucleoporins in the regulation of spindle orientation [113].
Very recently, a small fraction of the Nup358 nucleoporin, the main constituent of the NPC cytoplasmic filaments, was found in the cytoplasm of cells colocalizing with microtubules during interphase. In this study, Nup358 was found to interact with and regulate the dynamics of interphase microtubules directly, thus playing a role in organization of the cytoskeleton [114].
Last but not least, several reports have associated NPCs with chromatin organization and the regulation of gene expression (reviewed in [115, 116]), especially in lower eukaryotes. The nuclear periphery has historically been associated with gene silencing and repression [reviewed in [117]]. The presence of heterochromatin patches, gene-poor chromosomal regions and silent genomic domains adjacent to the NE support this hypothesis. In agreement with this idea, NPCs were initially associated with silent chromatin regions [118, 119]. In budding yeast Nup145p is responsible for tethering silent telomeric chromatin to the NPCs through the MLP1 and MLP2 proteins [118], although the role of the MLP proteins in telomere silencing and anchoring has been recently challenged [120]. Opposing the classical view of the nuclear periphery there is now increasing evidences indicating that this region also acts as a gene-activating domain. In a screening for proteins with chromatin boundary activity the Nup2p nucleoporin was found to block the propagation of heterochromatin in specific domains, thus preventing their silencing, and is thought to do so by recruiting chromatin to pores [121]. These studies suggest that chromatin activity can be modulated by relocation to the NPC. Chromatin immunoprecipitations (CHIP) studies with antibodies against NPC components have shown that many nucleoporins preferentially associate with transcriptionally active genes [122]. The results of this technique do not directly connect active genes to the NPC structure and it remains a possibility that the DNA binding of the nucleoporins occurs entirely in the nuclear interior. However, the fact that the NPCassociated protein Sus1 has been described to be a member of the SAGA histone acetylase complex involved in transcriptional initiation [123] and to be required for the confinement of GAL genes to NPCs during transcriptional activation in S. cerevisiae [124] directly links nuclear pores to active transcription. The relocation of genes to pores during activation has been reported for several other genes, including the HXK1 (encoding hexokinase-1), INO1 and several mating-response genes, although there are some controversial findings in the nucleoporin requirements for gene tethering (reviewed in [115, 125]). Also supporting a role of NPCs in transcriptional activation, experiments in which the Nup2p was fused to the microccocal nuclease revealed that the nuclease cleavage occurred mainly in promoter regions indicating the association of Nup2p with actively transcribed genes [126].
How genes are activated at the NPC is not well understood, but the Rap1 transcription factor seems to have an important role. CHIP analyses have indicated that most nups preferentially associate with genes containing Rap1-binding sites [122]. Furthermore, the Rap1-dependent activation in yeast was shown to occur through the Nup84p complex (the yeast Nup107–160 complex homologue) at the NPC [127]. This result led to the idea of a ‘reverse recruitment mechanism’ that suggests that, during activation, genes move to a platform tethered to the NPC containing the preassembled transcriptional machinery [127].
In higher eukaryotes, very little is known about the connection between the NPC and regulation of gene expression, but the facts that the SAGA-dependent anchorage of genes to the NPC has been reported in Drosophila [128] and that two nucleoporins are linked to the transcriptional regulation of dosage compensation in this organism [129] suggest that the mechanism might be conserved. Interestingly, a recent report shows the association of Nup93, a stable component of the NPC scaffold, with several regions of the human genome [130]. But in this case, the chromosome regions associated with Nup93 were enriched in markers of heterochromatin circling back to first findings in budding yeast suggesting the association of NPCs with silent chromatin. Notably, the chromosome-association of Nup93 varied depending on the global histone acetylation state of chromatin.
The association of chromatin to NPCs together with the fact that in higher eukaryotes NPCs are immobile [55] suggest that pores could act as positional markers at the NE for the organization and maintenance of the genome architecture. The relative location of several genes/chromosome regions within the cell nucleus has been shown to vary in response to metabolic changes and during cell differentiation. The findings that the DNA/NPC interactions are dynamic and change depending on the chromatin state, suggests that NPCs could be involved, at least in part, in regulating these genomic rearrangements. Supporting a role of NPCs in cell differentiation, Nup133, a member of the scaffold Nup107–160 complex, has recently been described to be important for neural differentiation during embryogenesis potentially opening a new aspect of NPC/nucleoporin function [131].
Concluding remarks
Owing to its interesting properties and increasing repertoire of functions, the biology of nuclear pore complexes has become an area of focus for many fields. Despite the substantial progress that has been made in the understanding of NPC biogenesis and function in the past decade, there are still large gaps to fill. The actual location of every nucleoporin inside the NPC structure, the detailed molecular mechanisms of assembly, disassembly and translocation through the pore, and how NPCs perform their transport-independent functions are just some of the mysteries that might be clarified in the coming years.
Acknowledgements
We apologize to all colleagues whose work could not be cited directly owing to space limitation. We thank the Hetzer lab and Marcela Raices for helpful suggestions and critically reading the manuscript.
References
- 1.D'Angelo MA, Hetzer MW. The role of the nuclear envelope in cellular organization. Cell Mol Life Sci. 2006;63:316–332. doi: 10.1007/s00018-005-5361-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 3.Mosammaparast N, Pemberton LF. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 2004;14:547–556. doi: 10.1016/j.tcb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 5.Gall JG. Octagonal nuclear pores. J Cell Biol. 1967;32:391–399. doi: 10.1083/jcb.32.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hueve J, Wesselmann R, Kahms M, Peters R. 4Pi microscopy of the nuclear pore complex. Biophys J. 2008 doi: 10.1529/biophysj.107.127449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck M, Forster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeister W, Medalia O. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306:1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- 8.Beck M, Lucic V, Forster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–615. doi: 10.1038/nature06170. [DOI] [PubMed] [Google Scholar]
- 9.Lim RY, Aebi U, Fahrenkrog B. Towards reconciling structure and function in the nuclear pore complex. Histochem Cell Biol. 2008;129:105–116. doi: 10.1007/s00418-007-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoffler D, Feja B, Fahrenkrog B, Walz J, Typke D, Aebi U. Cryo-electron tomography provides novel insights into nuclear pore architecture: implications for nucleocytoplasmic transport. J Mol Biol. 2003;328:119–130. doi: 10.1016/s0022-2836(03)00266-3. [DOI] [PubMed] [Google Scholar]
- 11.Maco B, Fahrenkrog B, Huang NP, Aebi U. Nuclear pore complex structure and plasticity revealed by electron and atomic force microscopy. Methods Mol Biol. 2006;322:273–288. doi: 10.1007/978-1-59745-000-3_19. [DOI] [PubMed] [Google Scholar]
- 12.Suntharalingam M, Wente SR. Peering through the pore: nuclear pore complex structure, assembly, and function. Dev Cell. 2003;4:775–789. doi: 10.1016/s1534-5807(03)00162-x. [DOI] [PubMed] [Google Scholar]
- 13.Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- 14.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reichelt R, Holzenburg A, Buhle EL, Jr, Jarnik M, Engel A, Aebi U. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J Cell Biol. 1990;110:883–894. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mansfeld J, Guttinger S, Hawryluk-Gara LA, Pante N, Mall M, Galy V, Haselmann U, Muhlhausser P, Wozniak RW, Mattaj IW, et al. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol Cell. 2006;22:93–103. doi: 10.1016/j.molcel.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 18.Stavru F, Hulsmann BB, Spang A, Hartmann E, Cordes VC, Gorlich D. NDC1: a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J Cell Biol. 2006;173:509–519. doi: 10.1083/jcb.200601001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwartz TU. Modularity within the architecture of the nuclear pore complex. Curr Opin Struct Biol. 2005;15:221–226. doi: 10.1016/j.sbi.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 21.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- 22.Isgro TA, Schulten K. Cse1p-binding dynamics reveal a binding pattern for FG-repeat nucleoporins on transport receptors. Structure. 2007;15:977–991. doi: 10.1016/j.str.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 23.Isgro TA, Schulten K. Association of nuclear pore FG-repeat domains to NTF2 import and export complexes. J Mol Biol. 2007;366:330–345. doi: 10.1016/j.jmb.2006.11.048. [DOI] [PubMed] [Google Scholar]
- 24.Denning DP, Patel SS, Uversky V, Fink AL, Rexach M. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc Natl Acad Sci U S A. 2003;100:2450–2455. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters R. Translocation through the nuclear pore complex: selectivity and speed by reduction-of-dimensionality. Traffic. 2005;6:421–427. doi: 10.1111/j.1600-0854.2005.00287.x. [DOI] [PubMed] [Google Scholar]
- 27.Frey S, Gorlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130:512–523. doi: 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 28.Frey S, Richter RP, Gorlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–817. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- 29.Ribbeck K, Gorlich D. Kinetic analysis of translocation through nuclear pore complexes. Embo J. 2001;20:1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fried H, Kutay U. Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci. 2003;60:1659–1688. doi: 10.1007/s00018-003-3070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel SS, Belmont BJ, Sante JM, Rexach MF. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- 32.Lim RY, Huang NP, Koser J, Deng J, Lau KH, Schwarz-Herion K, Fahrenkrog B, Aebi U. Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. Proc Natl Acad Sci U S A. 2006;103:9512–9517. doi: 10.1073/pnas.0603521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rout MP, Aitchison JD, Magnasco MO, Chait BT. Virtual gating and nuclear transport: the hole picture. Trends Cell Biol. 2003;13:622–628. doi: 10.1016/j.tcb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 34.Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- 35.Griffis ER, Craige B, Dimaano C, Ullman KS, Powers MA. Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription-dependent mobility. Mol Biol Cell. 2004;15:1991–2002. doi: 10.1091/mbc.E03-10-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storr HL, Clark AJ, Priestley JV, Michael GJ. Identification of the sites of expression of triple A syndrome mRNA in the rat using in situ hybridisation. Neuroscience. 2005;131:113–123. doi: 10.1016/j.neuroscience.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 37.Olsson M, Scheele S, Ekblom P. Limited expression of nuclear pore membrane glycoprotein 210 in cell lines and tissues suggests cell-type specific nuclear pores in metazoans. Exp Cell Res. 2004;292:359–370. doi: 10.1016/j.yexcr.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 38.Fan F, Liu CP, Korobova O, Heyting C, Offenberg HH, Trump G, Arnheim N. cDNA cloning and characterization of Npap60: a novel rat nuclear pore-associated protein with an unusual subcellular localization during male germ cell differentiation. Genomics. 1997;40:444–453. doi: 10.1006/geno.1996.4557. [DOI] [PubMed] [Google Scholar]
- 39.Cai Y, Gao Y, Sheng Q, Miao S, Cui X, Wang L, Zong S, Koide SS. Characterization and potential function of a novel testis-specific nucleoporin BS-63. Mol Reprod Dev. 2002;61:126–134. doi: 10.1002/mrd.1139. [DOI] [PubMed] [Google Scholar]
- 40.Akey CW. Structural plasticity of the nuclear pore complex. J Mol Biol. 1995;248:273–293. doi: 10.1016/s0022-2836(95)80050-6. [DOI] [PubMed] [Google Scholar]
- 41.Kiseleva E, Goldberg MW, Daneholt B, Allen TD. RNP export is mediated by structural reorganization of the nuclear pore basket. J Mol Biol. 1996;260:304–311. doi: 10.1006/jmbi.1996.0401. [DOI] [PubMed] [Google Scholar]
- 42.Erickson ES, Mooren OL, Moore D, Krogmeier JR, Dunn RC. The role of nuclear envelope calcium in modifying nuclear pore complex structure. Can J Physiol Pharmacol. 2006;84:309–318. doi: 10.1139/y05-109. [DOI] [PubMed] [Google Scholar]
- 43.Stoffler D, Goldie KN, Feja B, Aebi U. Calcium-mediated structural changes of native nuclear pore complexes monitored by time-lapse atomic force microscopy. J Mol Biol. 1999;287:741–752. doi: 10.1006/jmbi.1999.2637. [DOI] [PubMed] [Google Scholar]
- 44.Rakowska A, Danker T, Schneider SW, Oberleithner H. ATP-Induced shape change of nuclear pores visualized with the atomic force microscope. J Membr Biol. 1998;163:129–136. doi: 10.1007/s002329900377. [DOI] [PubMed] [Google Scholar]
- 45.Perez-Terzic C, Pyle J, Jaconi M, Stehno-Bittel L, Clapham DE. Conformational states of the nuclear pore complex induced by depletion of nuclear Ca2+ stores. Science. 1996;273:1875–1877. doi: 10.1126/science.273.5283.1875. [DOI] [PubMed] [Google Scholar]
- 46.Shahin V, Ludwig Y, Schafer C, Nikova D, Oberleithner H. Glucocorticoids remodel nuclear envelope structure and permeability. J Cell Sci. 2005;118:2881–2889. doi: 10.1242/jcs.02429. [DOI] [PubMed] [Google Scholar]
- 47.Paulillo SM, Powers MA, Ullman KS, Fahrenkrog B. Changes in nucleoporin domain topology in response to chemical effectors. J Mol Biol. 2006;363:39–50. doi: 10.1016/j.jmb.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 48.Fahrenkrog B, Maco B, Fager AM, Koser J, Sauder U, Ullman KS, Aebi U. Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J Struct Biol. 2002;140:254–267. doi: 10.1016/s1047-8477(02)00524-5. [DOI] [PubMed] [Google Scholar]
- 49.Paulillo SM, Phillips EM, Koser J, Sauder U, Ullman KS, Powers MA, Fahrenkrog B. Nucleoporin domain topology is linked to the transport status of the nuclear pore complex. J Mol Biol. 2005;351:784–798. doi: 10.1016/j.jmb.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 50.Makhnevych T, Lusk CP, Anderson AM, Aitchison JD, Wozniak RW. Cell cycle regulated transport controlled by alterations in the nuclear pore complex. Cell. 2003;115:813–823. doi: 10.1016/s0092-8674(03)00986-3. [DOI] [PubMed] [Google Scholar]
- 51.Melcak I, Hoelz A, Blobel G. Structure of Nup58/45 suggests flexible nuclear pore diameter by intermolecular sliding. Science. 2007;315:1729–1732. doi: 10.1126/science.1135730. [DOI] [PubMed] [Google Scholar]
- 52.Lim RY, Fahrenkrog B, Koser J, Schwarz-Herion K, Deng J, Aebi U. Nanomechanical basis of selective gating by the nuclear pore complex. Science. 2007;318:640–643. doi: 10.1126/science.1145980. [DOI] [PubMed] [Google Scholar]
- 53.Maul GG. Nuclear pore complexes. Elimination and reconstruction during mitosis. J Cell Biol. 1977;74:492–500. doi: 10.1083/jcb.74.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burke B, Ellenberg J. Remodelling the walls of the nucleus. Nat Rev Mol Cell Biol. 2002;3:487–497. doi: 10.1038/nrm860. [DOI] [PubMed] [Google Scholar]
- 55.Daigle N, Beaudouin J, Hartnell L, Imreh G, Hallberg E, Lippincott-Schwartz J, Ellenberg J. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J Cell Biol. 2001;154:71–84. doi: 10.1083/jcb.200101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belgareh N, Rabut G, Bai SW, van Overbeek M, Beaudouin J, Daigle N, Zatsepina OV, Pasteau F, Labas V, Fromont-Racine M, et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walther TC, Alves A, Pickersgill H, Loiodice I, Hetzer M, Galy V, Hulsmann BB, Kocher T, Wilm M, Allen T, et al. The conserved Nup107–160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- 58.Walther TC, Askjaer P, Gentzel M, Habermann A, Griffiths G, Wilm M, Mattaj IW, Hetzer M. RanGTP mediates nuclear pore complex assembly. Nature. 2003;424:689–694. doi: 10.1038/nature01898. [DOI] [PubMed] [Google Scholar]
- 59.Theisen U, Straube A, Steinberg G. Dynamic Rearrangement of Nucleoporins during Fungal "Open" Mitosis. Mol Biol Cell. 2008;19:1230–1240. doi: 10.1091/mbc.E07-02-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dultz E, Zanin E, Wurzenberger C, Braun M, Rabut G, Sironi L, Ellenberg J. Systematic kinetic analysis of mitotic dis-and reassembly of the nuclear pore in living cells. J Cell Biol. 2008;180:857–865. doi: 10.1083/jcb.200707026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bodoor K, Shaikh S, Salina D, Raharjo WH, Bastos R, Lohka M, Burke B. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J Cell Sci. 1999;112(Pt 13):2253–2264. doi: 10.1242/jcs.112.13.2253. [DOI] [PubMed] [Google Scholar]
- 62.Goldberg MW, Wiese C, Allen TD, Wilson KL. Dimples, pores, star-rings, and thin rings on growing nuclear envelopes: evidence for structural intermediates in nuclear pore complex assembly. J Cell Sci. 1997;110(Pt 4):409–420. doi: 10.1242/jcs.110.4.409. [DOI] [PubMed] [Google Scholar]
- 63.Macaulay C, Forbes DJ. Assembly of the nuclear pore: biochemically distinct steps revealed with NEM, GTP gamma S, and BAPTA. J Cell Biol. 1996;132:5–20. doi: 10.1083/jcb.132.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sheehan MA, Mills AD, Sleeman AM, Laskey RA, Blow JJ. Steps in the assembly of replication-competent nuclei in a cell-free system from Xenopus eggs. J Cell Biol. 1988;106:1–12. doi: 10.1083/jcb.106.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Drummond SP, Rutherford SA, Sanderson HS, Allen TD. High resolution analysis of mammalian nuclear structure throughout the cell cycle: implications for nuclear pore complex assembly during interphase and mitosis. Can J Physiol Pharmacol. 2006;84:423–430. doi: 10.1139/y05-148. [DOI] [PubMed] [Google Scholar]
- 66.Kimura N, Takizawa M, Okita K, Natori O, Igarashi K, Ueno M, Nakashima K, Nobuhisa I, Taga T. Identification of a novel transcription factor, ELYS, expressed predominantly in mouse foetal haematopoietic tissues. Genes Cells. 2002;7:435–446. doi: 10.1046/j.1365-2443.2002.00529.x. [DOI] [PubMed] [Google Scholar]
- 67.Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc Natl Acad Sci U S A. 2006;103:17801–17806. doi: 10.1073/pnas.0608484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Franz C, Walczak R, Yavuz S, Santarella R, Gentzel M, Askjaer P, Galy V, Hetzer M, Mattaj IW, Antonin W. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 2007;8:165–172. doi: 10.1038/sj.embor.7400889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gillespie PJ, Khoudoli GA, Stewart G, Swedlow JR, Blow JJ. ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr Biol. 2007;17:1657–1662. doi: 10.1016/j.cub.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harel A, Chan RC, Lachish-Zalait A, Zimmerman E, Elbaum M, Forbes DJ. Importin beta negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol Biol Cell. 2003;14:4387–4396. doi: 10.1091/mbc.E03-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maul GG, Maul HM, Scogna JE, Lieberman MW, Stein GS, Hsu BY, Borun TW. Time sequence of nuclear pore formation in phytohemagglutinin-stimulated lymphocytes and in HeLa cells during the cell cycle. J Cell Biol. 1972;55:433–447. doi: 10.1083/jcb.55.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maeshima K, Yahata K, Sasaki Y, Nakatomi R, Tachibana T, Hashikawa T, Imamoto F, Imamoto N. Cell-cycle-dependent dynamics of nuclear pores: pore-free islands and lamins. J Cell Sci. 2006;119:4442–4451. doi: 10.1242/jcs.03207. [DOI] [PubMed] [Google Scholar]
- 73.Winey M, Yarar D, Giddings TH, Jr, Mastronarde DN. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol Biol Cell. 1997;8:2119–2132. doi: 10.1091/mbc.8.11.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vasu SK, Forbes DJ. Nuclear pores and nuclear assembly. Curr Opin Cell Biol. 2001;13:363–375. doi: 10.1016/s0955-0674(00)00221-0. [DOI] [PubMed] [Google Scholar]
- 75.Lusk CP, Makhnevych T, Marelli M, Aitchison JD, Wozniak RW. Karyopherins in nuclear pore biogenesis: a role for Kap121p in the assembly of Nup53p into nuclear pore complexes. J Cell Biol. 2002;159:267–278. doi: 10.1083/jcb.200203079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Madrid AS, Mancuso J, Cande WZ, Weis K. The role of the integral membrane nucleoporins Ndc1p and Pom152p in nuclear pore complex assembly and function. J Cell Biol. 2006;173:361–371. doi: 10.1083/jcb.200506199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ryan KJ, McCaffery JM, Wente SR. The Ran GTPase cycle is required for yeast nuclear pore complex assembly. J Cell Biol. 2003;160:1041–1053. doi: 10.1083/jcb.200209116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ryan KJ, Zhou Y, Wente SR. The karyopherin Kap95 regulates nuclear pore complex assembly into intact nuclear envelopes in vivo. Mol Biol Cell. 2007;18:886–898. doi: 10.1091/mbc.E06-06-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.D'Angelo MA, Anderson DJ, Richard E, Hetzer MW. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 2006;312:440–443. doi: 10.1126/science.1124196. [DOI] [PubMed] [Google Scholar]
- 80.Kiseleva E, Rutherford S, Cotter LM, Allen TD, Goldberg MW. Steps of nuclear pore complex disassembly and reassembly during mitosis in early Drosophila embryos. J Cell Sci. 2001;114:3607–3618. doi: 10.1242/jcs.114.20.3607. [DOI] [PubMed] [Google Scholar]
- 81.Cotter L, Allen TD, Kiseleva E, Goldberg MW. Nuclear membrane disassembly and rupture. J Mol Biol. 2007;369:683–695. doi: 10.1016/j.jmb.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 82.Katsani KR, Karess RE, Dostatni N, Doye V. In Vivo Dynamics of Drosophila Nuclear Envelope Components. Mol Biol Cell. 2008 doi: 10.1091/mbc.E07-11-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.De Souza CP, Osmani AH, Hashmi SB, Osmani SA. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr Biol. 2004;14:1973–1984. doi: 10.1016/j.cub.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 84.Macaulay C, Meier E, Forbes DJ. Differential mitotic phosphorylation of proteins of the nuclear pore complex. J Biol Chem. 1995;270:254–262. doi: 10.1074/jbc.270.1.254. [DOI] [PubMed] [Google Scholar]
- 85.Favreau C, Worman HJ, Wozniak RW, Frappier T, Courvalin JC. Cell cycle-dependent phosphorylation of nucleoporins and nuclear pore membrane protein Gp210. Biochemistry. 1996;35:8035–8044. doi: 10.1021/bi9600660. [DOI] [PubMed] [Google Scholar]
- 86.Glavy JS, Krutchinsky AN, Cristea IM, Berke IC, Boehmer T, Blobel G, Chait BT. Cell-cycle-dependent phosphorylation of the nuclear pore Nup107–160 subcomplex. Proc Natl Acad Sci U S A. 2007;104:3811–3816. doi: 10.1073/pnas.0700058104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci U S A. 2006;103:5391–5396. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lusk CP, Waller DD, Makhnevych T, Dienemann A, Whiteway M, Thomas DY, Wozniak RW. Nup53p is a target of two mitotic kinases, Cdk1p and Hrr25p. Traffic. 2007;8:647–660. doi: 10.1111/j.1600-0854.2007.00559.x. [DOI] [PubMed] [Google Scholar]
- 89.Onischenko EA, Gubanova NV, Kiseleva EV, Hallberg E. Cdk1 and okadaic acid-sensitive phosphatases control assembly of nuclear pore complexes in Drosophila embryos. Mol Biol Cell. 2005;16:5152–5162. doi: 10.1091/mbc.E05-07-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Miller MW, Caracciolo MR, Berlin WK, Hanover JA. Phosphorylation and glycosylation of nucleoporins. Arch Biochem Biophys. 1999;367:51–60. doi: 10.1006/abbi.1999.1237. [DOI] [PubMed] [Google Scholar]
- 91.Herndon LA, Schmeissner PJ, Dudaronek JM, Brown PA, Listner KM, Sakano Y, Paupard MC, Hall DH, Driscoll M. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- 92.Haithcock E, Dayani Y, Neufeld E, Zahand AJ, Feinstein N, Mattout A, Gruenbaum Y, Liu J. Age-related changes of nuclear architecture in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2005;102:16690–16695. doi: 10.1073/pnas.0506955102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kau TR, Way JC, Silver PA. Nuclear transport and cancer: from mechanism to intervention. Nat Rev Cancer. 2004;4:106–117. doi: 10.1038/nrc1274. [DOI] [PubMed] [Google Scholar]
- 94.Poon IK, Jans DA. Regulation of nuclear transport: central role in development and transformation? Traffic. 2005;6:173–186. doi: 10.1111/j.1600-0854.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- 95.Cronshaw JM, Matunis MJ. The nuclear pore complex: disease associations and functional correlations. Trends Endocrinol Metab. 2004;15:34–39. doi: 10.1016/j.tem.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 96.Palancade B, Doye V. Sumoylating and desumoylating enzymes at nuclear pores: underpinning their unexpected duties? Trends Cell Biol. 2008;18:174–183. doi: 10.1016/j.tcb.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 97.Pichler A, Knipscheer P, Saitoh H, Sixma TK, Melchior F. The RanBP2 SUMO E3 ligase is neither HECT-nor RING-type. Nat Struct Mol Biol. 2004;11:984–991. doi: 10.1038/nsmb834. [DOI] [PubMed] [Google Scholar]
- 98.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 99.Galy V, Antonin W, Jaedicke A, Sachse M, Santarella R, Haselmann U, Mattaj I. A role for gp210 in mitotic nuclear-envelope breakdown. J Cell Sci. 2008;121:317–328. doi: 10.1242/jcs.022525. [DOI] [PubMed] [Google Scholar]
- 100.Salina D, Enarson P, Rattner JB, Burke B. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J Cell Biol. 2003;162:991–1001. doi: 10.1083/jcb.200304080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Antonin W, Franz C, Haselmann U, Antony C, Mattaj IW. The Integral Membrane Nucleoporin pom121 Functionally Links Nuclear Pore Complex Assembly and Nuclear Envelope Formation. Mol Cell. 2005;17:83–92. doi: 10.1016/j.molcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 102.Hawryluk-Gara LA, Platani M, Santarella R, Wozniak RW, Mattaj IW. Nup53 is Required for Nuclear Envelope and Nuclear Pore Complex Assembly. Mol Biol Cell. 2008 doi: 10.1091/mbc.E07-08-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Franz C, Askjaer P, Antonin W, Iglesias CL, Haselmann U, Schelder M, de Marco A, Wilm M, Antony C, Mattaj IW. Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates. Embo J. 2005 doi: 10.1038/sj.emboj.7600825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kerscher O, Hieter P, Winey M, Basrai MA. Novel role for a Saccharomyces cerevisiae nucleoporin, Nup170p, in chromosome segregation. Genetics. 2001;157:1543–1553. doi: 10.1093/genetics/157.4.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Joseph J, Liu ST, Jablonski SA, Yen TJ, Dasso M. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr Biol. 2004;14:611–617. doi: 10.1016/j.cub.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 106.Loiodice I, Alves A, Rabut G, Van Overbeek M, Ellenberg J, Sibarita JB, Doye V. The entire Nup107–160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zuccolo M, Alves A, Galy V, Bolhy S, Formstecher E, Racine V, Sibarita JB, Fukagawa T, Shiekhattar R, Yen T, et al. The human Nup107–160 nuclear pore subcomplex contributes to proper kinetochore functions. Embo J. 2007;26:1853–1864. doi: 10.1038/sj.emboj.7601642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Galy V, Askjaer P, Franz C, Lopez-Iglesias C, Mattaj IW. MEL-28, a novel nuclear-envelope and kinetochore protein essential for zygotic nuclear-envelope assembly in C. elegans. Curr Biol. 2006;16:1748–1756. doi: 10.1016/j.cub.2006.06.067. [DOI] [PubMed] [Google Scholar]
- 109.Orjalo AV, Arnaoutov A, Shen Z, Boyarchuk Y, Zeitlin SG, Fontoura B, Briggs S, Dasso M, Forbes DJ. The Nup107–160 nucleoporin complex is required for correct bipolar spindle assembly. Mol Biol Cell. 2006;17:3806–3818. doi: 10.1091/mbc.E05-11-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bai SW, Rouquette J, Umeda M, Faigle W, Loew D, Sazer S, Doye V. The fission yeast Nup107–120 complex functionally interacts with the small GTPase Ran/Spi1 and is required for mRNA export, nuclear pore distribution, and proper cell division. Mol Cell Biol. 2004;24:6379–6392. doi: 10.1128/MCB.24.14.6379-6392.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pritchard CE, Fornerod M, Kasper LH, van Deursen JM. RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. J Cell Biol. 1999;145:237–254. doi: 10.1083/jcb.145.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Blower MD, Nachury M, Heald R, Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–234. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 113.Schetter A, Askjaer P, Piano F, Mattaj I, Kemphues K. Nucleoporins NPP-1, NPP-3, NPP-4, NPP-11 and NPP-13 are required for proper spindle orientation in C. elegans. Dev Biol. 2006;289:360–371. doi: 10.1016/j.ydbio.2005.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Joseph J, Dasso M. The nucleoporin Nup358 associates with and regulates interphase microtubules. FEBS Lett. 2008;582:190–196. doi: 10.1016/j.febslet.2007.11.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brown CR, Silver PA. Transcriptional regulation at the nuclear pore complex. Curr Opin Genet Dev. 2007;17:100–106. doi: 10.1016/j.gde.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 116.Kalverda B, Roling MD, Fornerod M. Chromatin organization in relation to the nuclear periphery. FEBS Lett. 2008 doi: 10.1016/j.febslet.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 117.Shaklai S, Amariglio N, Rechavi G, Simon AJ. Gene silencing at the nuclear periphery. Febs J. 2007;274:1383–1392. doi: 10.1111/j.1742-4658.2007.05697.x. [DOI] [PubMed] [Google Scholar]
- 118.Galy V, Olivo-Marin JC, Scherthan H, Doye V, Rascalou N, Nehrbass U. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature. 2000;403:108–112. doi: 10.1038/47528. [DOI] [PubMed] [Google Scholar]
- 119.Feuerbach F, Galy V, Trelles-Sticken E, Fromont-Racine M, Jacquier A, Gilson E, Olivo-Marin JC, Scherthan H, Nehrbass U. Nuclear architecture and spatial positioning help establish transcriptional states of telomeres in yeast. Nat Cell Biol. 2002;4:214–221. doi: 10.1038/ncb756. [DOI] [PubMed] [Google Scholar]
- 120.Hediger F, Dubrana K, Gasser SM. Myosin-like proteins 1 and 2 are not required for silencing or telomere anchoring, but act in the Tel1 pathway of telomere length control. J Struct Biol. 2002;140:79–91. doi: 10.1016/s1047-8477(02)00533-6. [DOI] [PubMed] [Google Scholar]
- 121.Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 2002;109:551–562. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- 122.Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 123.Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear poreassociated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- 124.Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- 125.Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- 126.Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 127.Menon BB, Sarma NJ, Pasula S, Deminoff SJ, Willis KA, Barbara KE, Andrews B, Santangelo GM. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc Natl Acad Sci U S A. 2005;102:5749–5754. doi: 10.1073/pnas.0501768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kurshakova MM, Krasnov AN, Kopytova DV, Shidlovskii YV, Nikolenko JV, Nabirochkina EN, Spehner D, Schultz P, Tora L, Georgieva SG. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J. 2007;26:4956–4965. doi: 10.1038/sj.emboj.7601901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 130.Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 2008;22:627–639. doi: 10.1101/gad.1632708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lupu F, Alves A, Anderson K, Doye V, Lacy E. Nuclear pore composition regulates neural stem/progenitor cell differentiation in the mouse embryo. Dev Cell. 2008;14:831–842. doi: 10.1016/j.devcel.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]