Summary
Aurora kinase family members coordinate a range of events associated with mitosis and cytokinesis. Anti-cancer therapies are currently being developed against them. Here, we evaluate whether Aurora kinase-1 (TbAUK1) from pathogenic Trypanosoma brucei might be targeted in anti-parasitic therapies as well. Conditional knockdown of TbAUK1 within infected mice demonstrated its essential contribution to infection. An in vitro kinase assay was developed which used recombinant trypanosome histone H3 (rTbH3) as a substrate. Tandem MS identified a novel phosphorylation site in the carboxyl-tail of rTbH3. Hesperadin, an inhibitor of human Aurora B, prevented the phosphorylation of substrate with IC50 of 40 nM. Growth of cultured bloodstream forms (BF) was also sensitive to Hesperadin (IC50 of 50 nM). Hesperadin blocked nuclear division and cytokinesis, but not other aspects of the cell cycle. Consequently, growth arrested cells accumulated multiple kinetoplasts, flagella and nucleoli; similar to the effects of RNAi-dependent knockdown of TbAUK1 in cultured BF cells. Molecular models predicted high affinity binding of Hesperadin to both conserved and novel sites in TbAUK1. Collectively, these data demonstrate that cell cycle progression is essential for infections with T. brucei, and that parasite Aurora kinases can be targeted with small-molecule inhibitors.
Keywords: Trypanosoma brucei, Aurora kinase, mitosis, histone H3, histone H2B, Hesperadin, therapy
Introduction
Human African trypanosomiasis (HAT) is a vector borne disease caused by two sub-species of Trypanosoma brucei. HAT is invariably lethal when untreated, and spreads rapidly through populations when surveillance and treatment programs are disrupted (Smith et al., 1998). Current therapies can be costly, difficult to administer and have considerable risks of toxicity. The problem is aggravated by the growing incidence of drug-resistant trypanosomes, making the need for new therapies acute. The current study tests the broad hypothesis that regulatory proteins of the cell cycle are rational and druggable targets for therapy. Here we focus on the T. brucei Aurora kinase-1 (TbAUK1) because it is essential for mitotic progression in cultured trypanosomes; and as we report in this study; is essential for infection in a mouse model. Additionally, inhibitors of Aurora kinase family members are actively being pursued as therapies against cancer (Reviewed in Matthews et al., 2006; Mountzios et al., 2008; Carpinelli and Moll, 2008).
Aurora kinases regulate key events associated with chromatin condensation, spindle function and cytokinesis (Andrews et al., 2003; Carmena and Earnshaw, 2003). Yeast contain a single Aurora kinase homologue (Ipl1), while mammals contain three (Aurora A, B, and C). Aurora A is localized to the centrosomal region from prophase to telophase and is important for centrosome maturation, segregation, and the assembly of the mitotic spindle. The activity of Aurora A is mediated indirectly by the small G protein Ran, and directly by TPX2; a substrate and binding partner (Tsai et al., 2003; Eyers et al., 2003). Aurora A activity is also attenuated by PP1 (Francisco and Chan, 1994; Katayama et al., 2001). Aurora B and the yeast Ipl1 are each considered chromosomal passenger proteins (Vagnarelli and Earnshaw, 2004; Vader et al., 2006; Jeyaprakash et al., 2007). Early in mitosis Aurora B phosphorylates Ser-10 on histone H3. This event is detectable with antibodies, and is widely used as a biomarker for mitotic progression. The function of Ser-10 phosphorylation is unclear (Prigent and Dimitrov, 2003). In Drosophila, but not in humans, it contributes towards chromosome condensation (Giet and Glover, 2001; Hauf et al., 2003). The phosphorylated H3 has been identified among the chromosome passenger proteins (Liping et al, 2007), and in conjunction with methylation of Lys-9, displaces heterochromatin protein-1 (HP-1) during mitosis (Fischle et al., 2005; Hirota et al., 2005). During metaphase, Aurora B and Ipl1 form protein complexes at the kinetochores and help establish bi-orientation and tension of the kinetochore fibers (Biggins et al., 1999; Biggins and Murray, 2001; Cheeseman et al., 2002; Murata-Hori and Wang, 2002; Tanaka et al., 2002). They also serve as a checkpoint to stall mitosis until the spindle orientation and tension are correct (Pinsky et al., 2006). This latter function appears to involve recruitment of BubR1 to the anaphase promoting complex/cyclosome (APC/C) (Morrow et al., 2005) and phosphorylation of MCAK in humans and Dam1p in yeast (Cheeseman et al., 2002; Kang et al., 2001; Andrews et al., 2004). Later in the mitotic process, Aurora B migrates to the spindle midbody and helps orient the cleavage furrow during cytokinesis. Aurora B phosphorylates the intermediate filament protein vimentin in vitro and regulates vimentin filament segregation during cytokinesis (Goto et al., 2003). The function of Aurora C is less well understood, but it appears to play a role in meiosis and can overlap with some Aurora B functions during mitosis (Sasai et al., 2004; Yan et al., 2005).
Aurora kinases contribute to neoplastic transformations and this has fueled the development of anti-cancer therapies directed against them (Reviewed in Matthews et al., 2006; Mountzios et al., 2008; Carpinelli and Moll, 2008). Aurora A can affect the activity of oncogenes such as p53, BRCA1 or Lats2 (Katayama et al., 2004; Ouchi et al., 2004; Toji et al, 2004). The overexpression of Aurora kinases has been linked to loss of mitotic checkpoint control, chromosome instability, aneuploidy and the formation of invasive tumors (Wang et al, 2006; Ota et al., 2002; Anand et al., 2003; Hauf et al., 2003; Giet et al., 2005). The amplification of wild-type or mutated Aurora kinase genes are found in ovarian, esophageal, breast, gastric and colorectal cancers (Ewart et al., 2005; Gritsko et al, 2003; Tatsuka et al., 2005; Tchatchou et al., 2006; Wang et al., 2006; Sun et al., 2004; Ju et al., 2006; Katayama et al., 1999; Bischoff et al., 1998). Presently, an estimated 20 pharmaceutical agents are in pre-clinical, Phase I, or Phase II trials. Several pharmacophores have proven to be effective at disrupting Aurora kinase activity, including: substituted pyrimidines, the quinazolines, indolinones, and aminothizaoles to name a few (Tarri et al., 2007; D'Alise et al., 2008; Harrington et al., 2004; Hauf et al., 2003; Ditchfield et al., 2003; Francelli et al., 2005; Heron et al., 2006; Carpinelli et al., 2007; Foote et al., 2008; Andersen et al., 2008). Regardless of the starting pharmacophore, the current inhibitors insert into the ATP-binding pocket and adjacent hydrophobic pocket, and interact with the hinge region between the two kinase domains. Specificity for the Aurora kinases derives in part from a bulky aliphatic “gatekeeper” residue which blocks access to the hydrophobic pocket in most kinases, but is a smaller leucine residue in Aurora A and B. Inhibitors whose substituent groups are either too short to extend into this pocket, or too large, are not able to inhibit Aurora A (Andersen et al., 2008). Collectively, these data demonstrate that Aurora kinases are druggable targets.
In the present study we begin to evaluate whether TbAUK1 might serve as a target for novel antiproliferative therapies. An evaluation of the trypanosome kinome identified three Aurora kinase paralogs (Parsons et al., 2005). RNAi revealed that only TbAUK1, but not TbAUK2 or TbAUK3 was required for mitotic progression (Jetton et al., 2005; Tu et al., 2006). We, and others, have shown that loss of TbAUK1 inhibits nuclear division, cytokinesis and growth in cultured infectious bloodstream forms (BF) and insect stage procyclic forms (PF) (Jetton et al., 2005, Tu et al., 2006; Li and Wang, 2006). Similar to the situation in other organisms, the disruption in mitosis is coupled to an inability to form the spindle apparatus. The mechanism by which TbAUK1 modulates spindle formation is not understood. Many of the conserved kinetocore proteins that would ordinarily bind or activate Aurora kinases are missing from the genome of T. brucei (Berriman et al., 2005). In C. elegans, the Tousled kinase interacts with Aurora B (Air2) and modulates spindle assembly (Riefler et al., 2008). In T. brucei, a Touseled-like kinase (TbTLK1) also binds to TbAUK1 and is required for spindle assembly (Li et al., 2007). Recently, an elegant affinity tag analysis identified two kinesin-like proteins (TbKIN-A and TbKIN-B) along with two hypothetical proteins (TbCPC-1 and TbCPC-2) that along with TbAUK1, form a chromosomal passenger complex (Li et al., 2008). Each of these proteins shifts distribution at different stages in mitosis. When cells are depleted of TbAUK1 with RNAi, the other chromosomal passenger proteins disperse. Although TbAUK1 can be localized near the site where cytokinesis initiates, its role in the process is not known. A classical actin ring does not appear to be necessary to pinch the cell in two (Garcia-Salcedo et al., 2004). Dynamin can pinch vesicles and organelles in mammalian cells. Conditional knockdown of Tb-dynamin-like protein arrests trypanosomes midway through cytokinesis (Chanez et al., 2006). We have shown that complete cleavage furrow ingression also requires the signal anchor protein TbRACK1 (Rothberg et al., 2006; Regmi et al., 2008). Interestingly, TbRACK1 seems to exert its effects through the translation process (Regmi et al., 2008).
The present study builds upon these observations and seeks to identify small molecule inhibitors of TbAUK1. We demonstrate that TbAUK1 is essential for infection within a mammalian host, indicating that TbAUK1 is a viable target for therapeutic intervention. Our molecular models predict that TbAUK1 has conserved and novel high affinity binding sites for the inhibitor Hesperadin. High affinity binding is confirmed with in vitro kinase assays, where an IC50 value of 40 nM is reported. Cell growth of cultured BF is also very sensitive to Hesperadin with an IC50 of 48 nM. Growth of insect stage PF is more refractory with IC50 of 550 nM. Within 24 hr of drug addition, cells exhibit morphological changes that phenocopy RNAi of TbAUK1. These include a cessation of nuclear division and cytokinesis, and accumulation of nucleoli, kinetoplasts and flagella. Altogether, these data demonstrate that both the activity of TbAUK1 and growth of cultured BF are equally sensitive to inhibition by Hesperadin. Since conserved paralogs of TbAUK1 are found in T. cruzi and Leishmania, it may be possible to develop broad-spectrum therapies against this protein.
Results
TbAUK1 is essential for infection in mice
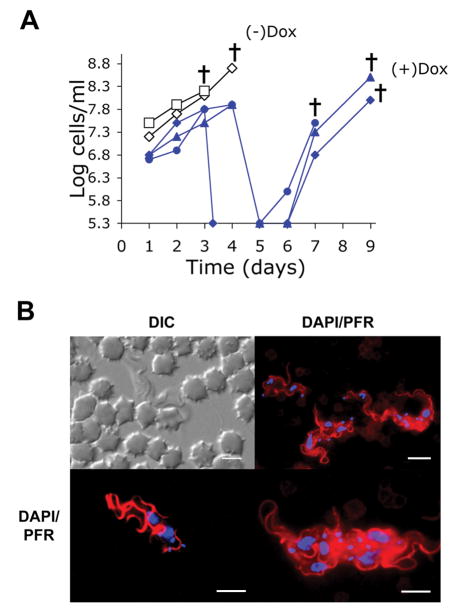
Small molecule inhibitors are currently being produced against Aurora kinases. These inhibitors are of value for two separate reasons: they have therapeutic potential as antiproliferative agents, and they can also be useful research tools. To determine whether TbAUK1 is essential for infection within a mammalian host, mice were inoculated with BF TbAUK1 RNAi cells (Fig. 1A). The mice were either untreated, or treated with 1 mg/ml of doxycycline added to the drinking water in order to induce RNAi. Parasitemia was quantified in peripheral tail blood at the times indicated. Each curve plots the progress of infection in a single mouse. The detection limit for this assay was 2×105 trypanosomes per ml of blood. The two control mice achieved infections greater than 1×108 trypanosomes per ml within 3 days post infection and each succumbed by days 4 and 5. In contrast, the doxycycline treated mice at day 4 did not attain 1×108 trypanosomes per ml, and by day 5, the parasitemias in each mouse had fallen below the detectable limit of the assay. Eventually the parasitemia returned in each of the doxycycline treated mice. A similar phenomenon has been reported with in vivo knockdown of the transcription factor TbXPD from T. brucei (Lecordier et al., 2005). We have previously shown that doxycycline by itself does not affect the outcome of trypanosome infections (Rothberg et al., 2006). To confirm that TbAUK1 affects the trypanosome cell cycle during infection as it does in culture, trypanosomes were collected from a separate mouse after 3-days of infection. The trypanosomes were fixed, permeabilized and stained for the paraflagellar rod protein (PFR) and for DNA (Fig. 1B). Trypanosomes with single nuclei and multiple flagella and kinetoplasts were identified. The TbAUK1 RNAi cells from the doxycycline treated mice had the same phenotype as the cultured BF after treatment with tetracycline (compare Fig. 1B and Fig. 6). Taken as a whole, these data indicate that TbAUK1 is essential for infection in mice. Moreover, within the mammalian host, TbAUK1 is required for cell cycle progression and in its absence, nuclear division is uncoupled from that of kinetoplasts and flagella; as was observed in culture.
Fig. 1.
TbAU1K is essential for infection in mice. Mice were inoculated ip with 3×106 TbAUK1 RNAi cells. One group containing three mice received 1 mg/ml doxycycline in the water at day 0 (+ Dox). The control group comprised of two mice received water without doxycycline (- Dox).
(A) Parasiemia was monitored in peripheral blood at the times indicated. Each line plots the infection in a single mouse. The crosses indicate that the mice died within 24 hours of the last recorded parasitemia. The detection limit of the assay is 2×105 cells/ml.
(B) After 3 days of growth in a separate doxycycline treated mouse, trypanosomes were examined in the blood by DIC, or were labeled with antibodies against PFR (red) and counterstained with DAPI (blue). The cells at upper right were viewed at 60× while cells in the lower panels were viewed at 100×. The lower right panel shows two cells, each of which has multiple flagella and multiple kinetoplasts. The bars are size markers of 10 μm. Substrate specificities of TbAUK1 and TbAUK3.
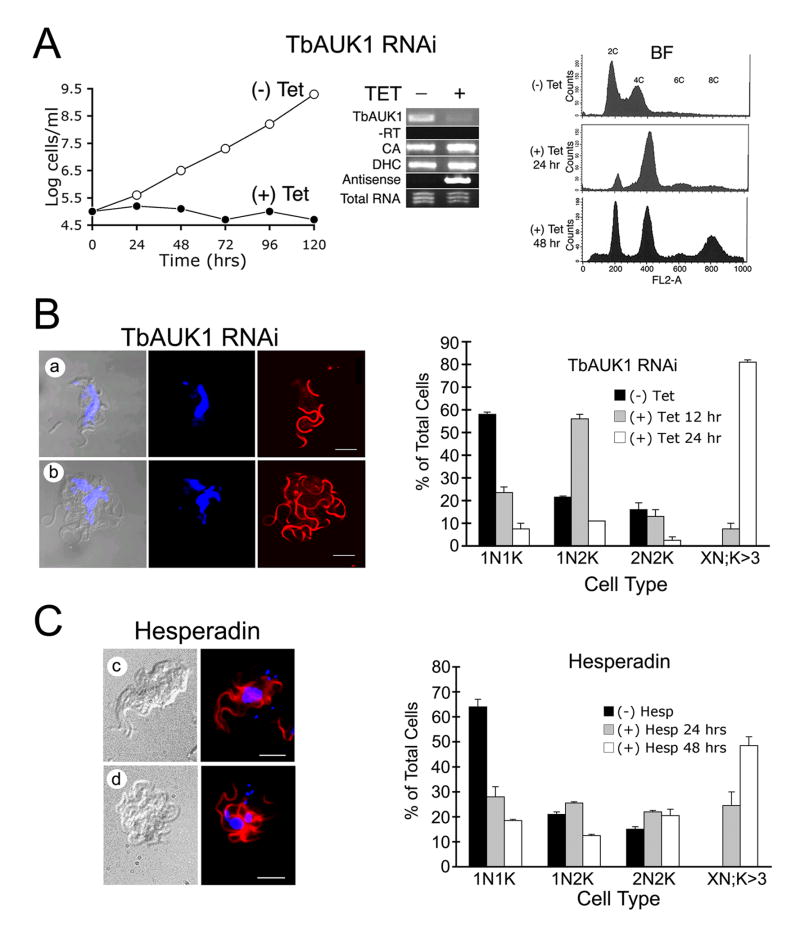
Fig. 6.
Treatment with Hesperadin phenocopies RNAi knockdown of TbAUK1.
(A) RNAi of TbAUK1 disrupts cell cycle progression in BF. The left panel shows a growth curve of TbAUK1 RNAi cells diluted to 1×105 cells/ml with (+) or without (-) tetracycline. The middle panel is RT-PCR using template RNA isolated from the TbAUK RNAi BF cells after growth for 72 hours with or without tetracycline. Primer pairs were designed to amplify the following: TbAUK1; the upstream gene carbonic anhydrase (CA); the downstream gene dynein heavy chain (DHC); and antisense TbAUK1. Also shown is the PCR control without reverse transcriptase (-RT) and the loading control of ethidium bromide stained total RNA (1 μg). The right panel shows flow cytometry of BF cells at increasing times after RNAi induction. A shift in population to 4C and 8C DNA content is observed.
(B) Changes in cell cycle progression induced by RNAi of TbAUK1. The left panels show morphological changes in BF after 24 hr induction with tetracycline. Cells were viewed by DIC, or stained for nuclei (TOTO) and flagella (PFR) (panels a-b). The right panel shows cell cycle progression in the RNAi cells. BF cultures were untreated, or induced with tetracycline for the amount of time indicated. After DAPI staining, cells were evaluated microscopically for their number of nuclei (N) and kinetoplasts (K). Cells designated 1N1K have one nucleus and one kinetoplast. Each time point is the evaluation of at least 200 cells (n=2, ±SE).
(C) Changes in cell cycle progression induced by Hesperadin. The left panels show morphological changes in cells treated with 100 nM Hesperadin for 24 hr (panels c-d); The right panel shows cell cycle progression of Hesperadin treated cells. The methods are the same as described in panel B.
Activity of TbAUK1
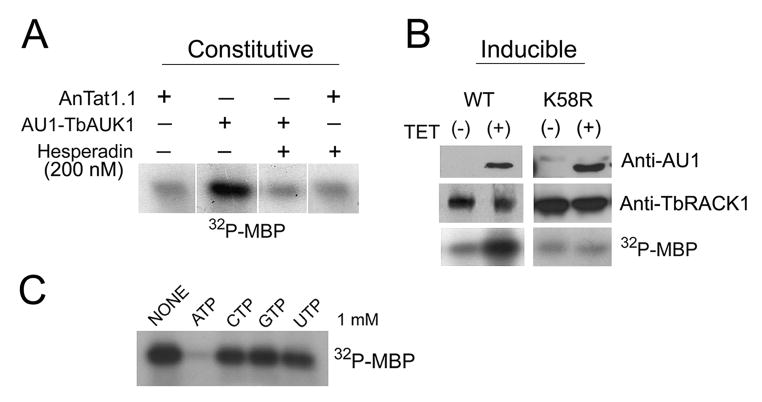
An in vitro kinase assay was developed. Cultured PF were transformed with AU1-tagged TbAUK1 and kinase was immunopreciptiated with anti-AU1-Sepharose beads (Fig. 2). In one set of experiments, wild-type AU1-TbAUK1 in pHD496 was constitutively expressed in AnTat1.1 PF (Fig. 2A). Pull-down assays with homogenates from these transformants yielded a kinase that phosphorylated myelin basic protein (MBP), whereas equivalent assays with the parental AnTat1.1 cells only produced a background kinase activity. Hesperadin is an inhibitor that inserts into the ATP-binding pocket of Aurora A and B. It inhibits Aurora B with IC50 of 250 nM, but has IC50 values in the range of 1.2 μM to >10 μM for Cdk1/cyclin B or Cdk2/cyclin E, respectively (Hauf et al., 2003). At a concentration of 200 nM, Hesperadin lowered the activity of the immunoprecipitated kinase to the background level. When the pull-down fraction from parental AnTat1.1 was treated with Hesperadin, the kinase activity was not significantly inhibited (inhibition of 4±4%; n=3). These data show that enzyme activity is dependent upon expression of a tagged Aurora kinase and is sensitive to the inhibitor Hesperadin.
Fig. 2.
In vitro kinase assay of TbAUK1.
(A) Constitutive expression of AU1-tagged TbAUK1. Parental AnTat1.1 PF were transformed with the constitutive expression vector pTSA-AU1.TbAUK. When pulled down with anti-AU1 Sepharose beads, transformants phosphorylated myelin basic protein (MBP), in a manner that was inhibited by Hesperadin. Parental cells phosphorylated MBP at a background level.
(B) Inducible expression of AU1-tagged wild-type TbAUK1 and the K58R mutated TbAUK1. The tagged proteins were detected in cell homogenates by western blot with antibodies against AU1 (upper panels). TbRACK1 was used as a loading control (middle panels). The immunoprecipitated proteins phosphorylated MBP (lower panel).
(C) Nucleotide specificity of TbAUK1. Unlabeled nucleotides (1 mM each) were added to the standard reaction mix. Only unlabeled ATP prevented phosphorylation of MBP.
A separate tetracycline inducible construct in pLEW100 had an AU1 epitope tag added to the carboxyl-terminus (Fig. 2B). The inducible construct allowed us to compare the activity of immunoprecipitated tagged kinase from the same cloned cell line; with the only difference being the presence or absence of tetracycline. Western blot revealed that TbAUK1.AU1 was only present in the cells induced with tetracycline (Fig. 2B, upper panel). TbRACK1 was used as a loading control. The pull-down fraction plus tetracycline was able to phosphorylate MBP significantly above the background level (lower panel).
Finally, the kinase dead TbAUK1 was constructed to verify that activity in the pull-down assay resulted from TbAUK1 rather than from other co-precipitating kinases. The K58R mutation generates a non-functioning TbAUK1 (Li and Wang, 2006), and was made here with an AU1-epitope tag at the amino terminus. It was cloned into the tetracycline inducible expression vector pLEW100, and transfected into PF 29-13 cells. Expression of the kinase was induced with tetracycline (Fig. 2B; right panel), however no kinase activity above the background was pulled down with the anti-AU1-Sepharose beads. Collectively these data demonstrate that the kinase activity precipitated in these studies derived from TbAUK1. The kinase specificity for nucleotide was evaluated by adding 1 mM of unlabeled nucleotides to the reaction mix (Fig. 2C). Only unlabeled ATP was able to compete with [γ-32P]ATP and prevent phosphorylation of MBP, while CTP, GTP and UTP were without effect.
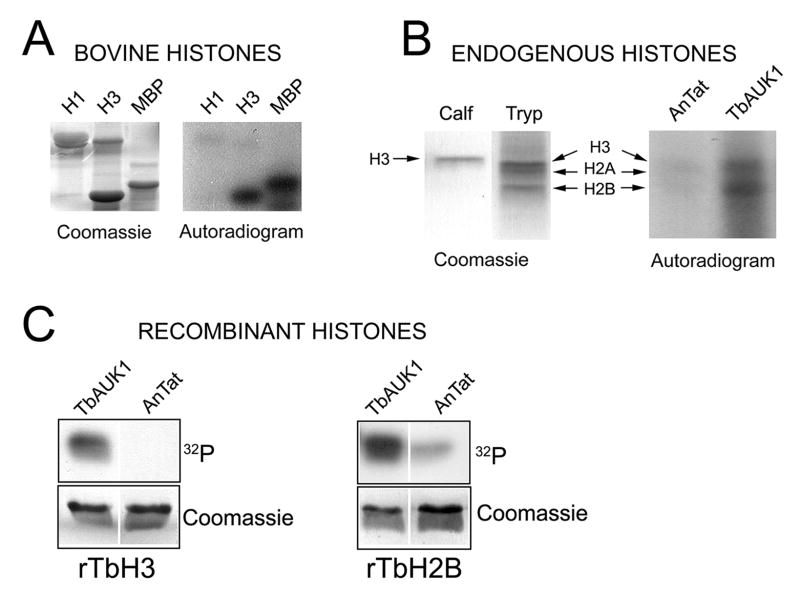
Mammalian Aurora B phosphorylates histone H3 on Ser-10 and Ser-28; where Ser-10 phosphorylation in particular is detected with antibodies and is a convenient biomarker for Aurora kinase activity in vivo (Hauf et al., 2003). Here we evaluate whether histone phosphorylation might be a useful biomarker for TbAUK1 activity. TbAUK1 phosphorylated the heterologous substrates MBP and bovine histone H3; but not bovine histone H1 (Fig. 3A). Particulate fractions from T. brucei were acid extracted and precipitated with acetone (Fig. 3B). When incubated with TbAUK1 two proteins in the extract were phosphorylated; a broad band at 15 kDa and another protein of 12 kDa. By contrast, the background kinase from the control parental homogenate (AnTat1.1) was not able to phosphorylate any proteins in the acid extract (Fig. 3B). LC/MS/MS analysis of the two bands revealed a complex mixture of proteins, including TbH3 (Mr of 14.7 kDa) and TbH2B (Mr of 12.5 kDa). To determine whether TbAUK1 could phosphorylate TbH3 or the novel substrate TbH2B, recombinant proteins were bacterially expressed and used as substrate (Fig. 3C). Both the recombinant TbH3 and TbH2B were phosphorylated by TbAUK1, but not by cell homogenates that lacked AU1-tagged TbAUK1 (AnTat). The Coomassie stained gels show that equivalent amounts of substrate were present in each reaction.
Fig. 3.
TbAUK1 phosphorylates trypanosome histones.
(A) TbAUK1 can phosphorylate mammalian histone H3 and MBP, but not H1. Coomassie stain reveals equivalent substrate in each reaction.
(B) Phosphorylation of endogenous histones by TbAUK1. Trypanosome histones were acid extracted from a particulate fraction and TbH3, TbH2A and TbH2B were tentatively identified based upon their molecular weights of 14.7 kDa, 14.2 kDa and 12.5 kDa, respectively. The histones were incubated with immunoprecipitated proteins from control cultures (AnTat); or from cultures expressing AU1.TbAUK1. TbAUK1 phosphorylated proteins with sizes equivalent to H3 and H2B.
(C) TbAUK1 can phosphorylate recombinant TbH3 and TbH2B. TbH3 (Tb927.1.2530) and TbH2B (Tb10.406.0350) were amplified from genomic DNA, cloned in pQE80 and used as substrate in assays where the kinase source was either precipitated from control cultures (AnTat), or precipitated from cultures expressing AU1.TbAU1K.
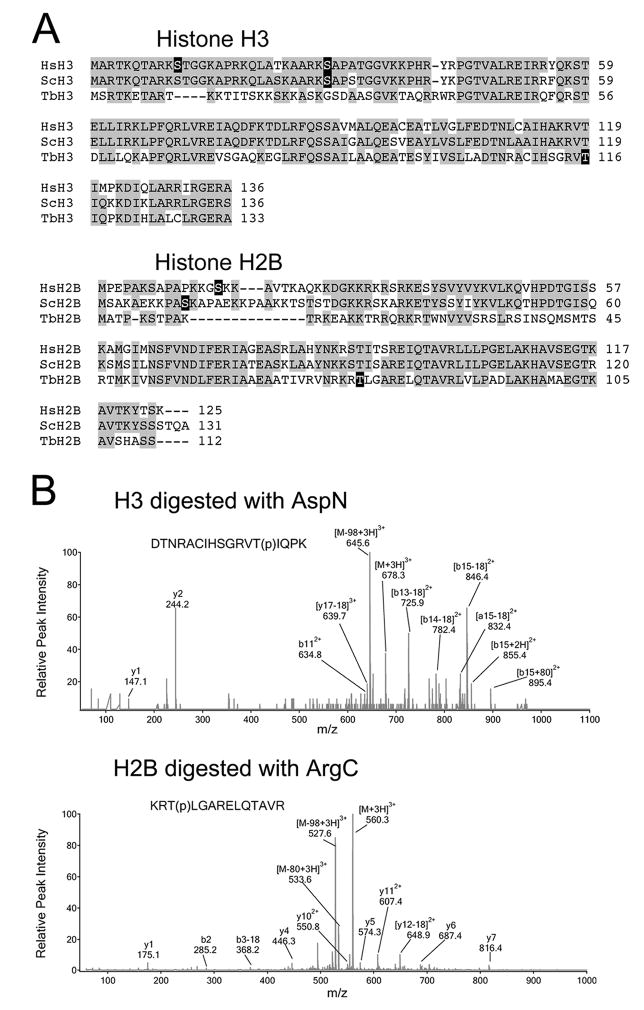
The amino terminal tail of TbH3 is divergent and lacks phosphorylatable residues corresponding to Ser-10 or Ser-28, although alternative phosphorylatable sites are in the vicinity (Fig. 4A). Mammalian H2B is also phosphorylated in the amino terminal tail (Fig. 4B), but this event has not been attributed to Aurora kinase activity. Consequently, the phosphorylation sites in trypanosome TbH3 and TbH2B were evaluated by LC/MS/MS (Fig. 4C). In each case, the phosphorylation site was identified within the carboxyl region of the protein, but not in the flexible amino terminal tail region. TbH3 was phosphorylated on T116 within the peptide DTNRACIHSGRVT(p)IQPK. This residue corresponds to T118 in human and S. cerevisae histone H3. TbH2B was phosphorylated on T77 within the peptide KRT(p)LGARELQTAVR. This residue corresponds to T88 in human and T90 in S. cerevisae. Attempts were made to verify that these sites were utilized in vivo. We could not detect this phosphorylation by LC/MS/MS of histones that had been acid extracted from chromatin or following acid extraction of a whole cell homogenate. Our methods cannot rule out the possibility that phosphorylation occurs within a small region of the chromatin, and only transiently at one stage of the cell cycle. Nonetheless, phosphorylation of TbH3 is used as a substrate for our in vitro kinase assay to measure sensitivity of TbAUK1 to the small molecule inhibitor Hesperadin.
Fig. 4.
Identification of phosphorylation sites within TbH3 and TbH2B.
(A) Sequence alignments of histone H3 and H2B from human (Hs), yeast (Sc) and T. brucei (Tb). Phosphorylation sites are blocked in black.
(B) LC/MS/MS spectra of the triply-charged phosphorylated peptides detected in TbH3 and TbH2B. TbH3 digested with AspN endopeptidase. Peptide DTNRACIHSGRVT(p)IQPK (amino acids 104–120) had a mass-to-charge ratio (m/z) of 678.3. The MS/MS spectrum revealed multiple fragment ions including neutral loss fragments [M-H3PO4+3H]3+ at m/z 645.6. Wild-type b112+ was detected, together with [b13-18]2+, [b14-18]2+, [b15-18]2+ and [b13+18]2+. The b-ion series indicated that the phosphorylation occurred at Thr116. TbH2B was digested with ArgC endopeptidase. Triply charged peptide KRT(p)LGARELQTAVR (amino acids 164–177) appeared at 560.3 in the MS scan. The MS/MS spectrum revealed several fragment ions including [M-H3PO4+3H]3+ at m/z 527.6, and y4, y5, y6, y7, y112+, [y12-18]2+. The MS/MS data demonstrated that Thr166 was phosphorylated.
Hesperadin inhibits TbAUK1 activity and growth of BF and PF cultures
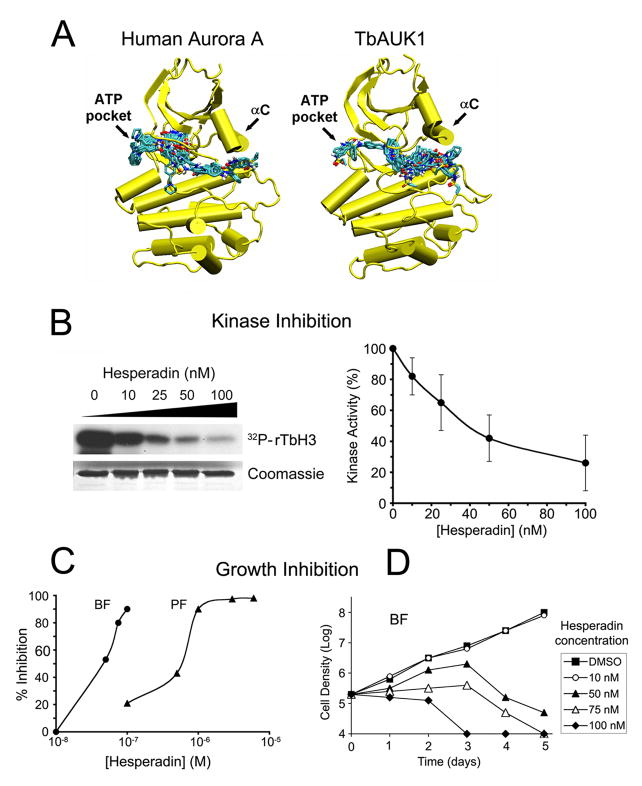
Hesperadin is an indolinone inhibitor of Aurora B. Its sulfonamide group extends beyond the ATP pocket and into the adjacent hydrophobic pocket (Hauf et al., 2003). To evaluate binding of Hesperadin to TbAUK1, molecular models were generated. The crystal structure of Xenopus Aurora B with Hesperadin bound in the ATP pocket was used as a template (PDB 2BFY; Sessa et al., 2005). As a control for our methods, we also modeled human Aurora A using the same Xenopus Aurora B crystal structure as template (Fig. 5A). Hesperadin was included in the template during modeling, but it was removed before the models were allowed to relax by use of a conjugant gradient energy minimization routine in the NAMD molecular dynamics suite (Phillips et al., 2005). The minimized structures were then used in Hesperadin docking experiments. Of the 25 highest affinity Hesperadin dockings to the human Aurora A model, we observed that 22 bound to the ATP-pocket (Fig. 5A). These results are consistent with the crystal structures obtained with Aurora B. By contrast, only 3 of the 25 highest affinity Hesperadin dockings localized to the ATP-pocket in the TbAUK1 model (Figure 5B). The majority of dockings were near the αC helix. The calculated affinities for these interactions varied in the range of 0.2-1.1 μM for the human Aurora A model and 1.4-3.6 μM for the TbAUK1 model. These values are not significantly different due to the known limitations associated with estimating binding affinities from in silico docking calculations (± 2 kcal/mol; Huey et al. 2007). These data suggest that small molecule inhibitors can bind to conserved and novel sites in TbAUK1 when compared with the human host proteins.
Fig. 5.
Hesperadin inhibits TbAUK1 activity and growth of BF and PF trypanosomes.
(A) Molecular models of human Aurora A and TbAUK1 were generated based upon the crystal structure of Xenopus Aurora B (2BFY). The top 25 docks to Hesperadin are shown for the models. Each of the bound Hesperadin molecules is represented as a stick figure.
(B) Dose response for Hesperadin inhibition of TbAUK1. The left panel shows an autoradiogram with 32P incorporation into TbH3. The stained Coomassie gel shows that an equivalent amount of TbH3 substrate was loaded onto each gel lane. The right panel shows densitometry of the autoradiograms (n=4; ±SE).
(C) Hesperadin inhibits growth of BF and PF cultures. BF or PF were grown in the presence of increasing concentrations of Hesperadin for 24 hr or 48 hr, respectively. The percent growth inhibition was recorded.
(D) Time course of cell growth in the presence of increasing concentrations of Hesperadin. BF cultures were treated at time 0 with the indicated concentrations of Hesperadin and cell density was followed for 5 days. The limit of detection was 1×104 cells/ml.
Hesperadin was tested with the in vitro assay. It inhibited the TbAUK1-mediated phosphoryation of TbH3 in a dose dependent manner (Fig. 5B). Each reaction contained increasing concentrations of Hesperadin up to 100 nM. The reaction products were separated by SDS-PAGE and 32PO4 incorporation into TbH3 was assessed by densitometry of the autoradiograms (Fig. 5B, right panel). An average IC50 value of 40 nM was obtained.
The ability of Hesperadin to affect cell growth was tested (Fig. 5C-D). For the dose-response analysis, BF cultures were grown for 24 hr in the presence of increasing concentrations of drug, and compared with a control culture. Percent inhibition was recorded (Fig. 5C). Sensitivity to Hesperadin varied with the lifecycle stage. Hesperadin was effective at inhibiting growth of BF cultures with IC50 of 50 nM, while the inhibition of PF growth required approximately 11-fold more Hesperadin, with IC50 of 550 nM. To further assess the effects of Hesperadin on BF cultures, a time course of growth inhibition was evaluated over a 5-day period (Fig. 5D). The detection limit of this assay was 1×104 cells/ml. Hesperadin at 50-100 nM slowed culture growth for a period of 48-72 hr and this was followed by a decline in cell density. Hesperadin at 10 nM was without effect on culture growth. These data suggest that low doses of Aurora kinase inhibitor over a relatively short period of time are sufficient to kill cultured BF cells.
Hesperadin alters cell morphology and inhibits cell cycle progression similar to the RNAi knockdown of TbAUK1
The effects of Hesperadin on cell growth and morphology were compared with changes induced when cellular levels of TbAUK1 were depleted with RNAi. BF cells were transformed with plasmid pZJM containing a 532 base-pair fragment of TbAUK1. When induced with tetracycline, the dually opposed T7 promoters produced RNAi (Djikeng et al., 2004). RT-PCR was used to assess knockdown of TbAUK1 (Fig. 6A). A near complete loss of TbAUK1 transcript was observed (middle panel). The linearized vector was designed to integrate into the rDNA intergenic region, however it could also aberrantly integrate as a closed circle into the TbAUK1 gene locus (Motyka et al., 2004). Should this occur, the dual promoters would not produce antisense RNA for the targeted gene. Instead upstream genes would be knocked down by the read-through production of antisense RNA while the downstream genes would be upregulated. RT-PCR was used to demonstrate that transcript levels for flanking genes (carbonic anhydrase and dynein heavy chain) did not change and that tetracycline inducible antisense for TbAUK1 was observed (middle panel). Additionally, independent transformations and multiple clones for each transformation gave the same results. Taken as a whole, these data demonstrate that the effects of RNAi reported in this paper result from knockdown of TbAUK1. The depletion of TbAUK1 in BF had a rapid effect on cell growth. BF cells ceased to divide within 24 hours and remained alive but without population increase for at least 120 hours (left panel). Despite the absence of cell growth, the FACS analysis revealed that the cells continued to reinitiate S phase (right panel). Consequently, after 48 hours of RNAi induction, polyploid cells with 8C DNA content increased indicating that DNA replication continued despite the inhibition of mitosis. These results are consistent with published observations (Li and Wang, 2006).
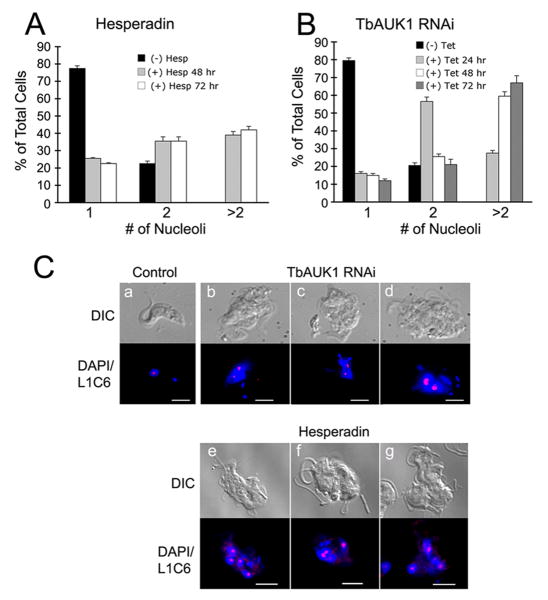
To assess whether the growth inhibitory effects of Hesperadin might have resulted from the in vivo inhibition of TbAUK1, cell morphology of treated BF was compared with changes induced by RNAi knockdown of TbAUK1 (Fig. 6B). The RNAi of TbAUK1 in BF produces a distinctive phenotype in which nuclear division is halted, but duplication of kinetoplast DNA (kDNA) and flagella continues (Fig. 6B, panels a-b). Despite the overall appearance, the cells are motile and metabolically active. Here we use this phenotype as a biomarker for in vivo activity of TbAUK1. After 24 hr exposure of BF cultures to 100 nM Hesperadin, cells contained a multi-lobed nucleus, numerous kDNA and numerous flagella (Fig. 6C, panels c-d); a pattern that phenocopied the loss of TbAUK1 with RNAi. The changes in cell population were quantified (Fig. 6B, right panel). In a wild-type BF population, approximately 60% of cells are in the 1N1K configuration, defined by a single nucleus (N) and a single kinetoplast (K). Within 24 hr of TbAUK1 depletion with RNAi, 1N1K cells declined to 8% of the population, while cells with the unusual configuration of more than 3K and an indeterminate number of nuclei (XN; K>3) increased to 81% of the population. After 24 hr exposure to 200 nM Hesperadin, cells with a 1N1K configuration dropped to 28% of the population, while cells with XN; K>3 increased to 25% of the population. Within 48 hr, cells with a XN; K>3 configuration increased to 48% of the population (Fig. 6C, right panel).
Although the nuclei in BF TbAUK1 RNAi cells failed to divide, the cells continued to reinitiate S phase (Fig. 6A). The increase in DNA can be detected using nucleoli as a cytological marker (Fig. 7A). Here, replication and segregation of nucleoli were monitored with the monoclonal antibody L1C6 (a gift from K. Gull). The majority of BF control cells (78%) contained a single nucleolus (Fig 7A, C). However, within 48 hr post-induction of RNAi, this value dropped to 15% of the population. The number of cells with 2 or more nucleoli increased to 85% of the population. When BF cells were treated with 200 nM Hesperadin for 48 hr, only 26% of the population had a single nucleolus, while cells with two or more nucleoli increased to 74% of the population (Fig. 7B, C). Therefore, BF cells depleted of TbAUK1 by RNAi or treated with Hesperadin each exhibited the same phenotypic changes.
Fig. 7.
Proliferation of nucleoli following treatment with Hesperadin or depletion of TbAUK1 with RNAi.
(A) Nucleoli in Hesperadin treated cells. Cells were treated with 200 nM Hesperadin for the times indicated. The nucleus was stained with DAPI, and nucleoli were labeled with antibody L1C6 and secondary antibodies coupled to Cy3. At least 200 cells were analyzed at each time (n=2; ±SE).
(B) Nucleoli in cells depleted of TbAUK1 by RNAi. The number of nucleoli in TbAUK1 RNAi cells was evaluated at different times after induction with tetracycline. At least 200 cells were analyzed at each time (n=2; ±SE).
(C) Nucleolar labeling with antibody L1C6. Trypanosomes were left untreated (panel a); were examined after 24 hours of induction with tetracycline (+ Tet) (panels b-d); or examined after 24 hr treatment with 200 nM Hesperadin (panels e-g). With RNAi and Hesperadin, note the disruption in nuclear division leading to swollen, multilobed nuclei and multiple nucleoli. The bars are size markers of 10 μm.
Overall, we have demonstrated that TbAUK1 is essential for infection in a rodent host. An in vitro kinase assay revealed that TbAUK1 phosphorylates TbH3 and TbH2B on residues that had not previously been reported to serve as Aurora kinase phosphorylation sites. Phosphorylation of TbH3 was sensitive to the small molecule inhibitor Hesperadin. Hesperadin at 100-200 nM had a strong effect on cell growth and mitotic progression. The phenotypic changes generated by Hesperadin inhibition matched those of TbAUK1 RNAi. Molecular models predict that the ATP-binding pocket of TbAUK1 is accessible to the small molecule inhibitor Hesperadin; but that docking to other sites is possible. Anti-cancer drugs currently being developed against Aurora kinases might affect a wide range of protozoan pathogens.
Discussion
It has recently been proposed that protein kinases might serve as drug targets in the treatment of infections caused by trypanosomes and Leishmania (Naula et al., 2005). Emphasis was placed on conserved CDKs and MAP kinases. The current study investigates the therapeutic potential of Aurora kinase. TbAUK1 was chosen for analysis, in part because it is essential for cell cycle progression and in part because its mammalian homologues have already been shown to be sensitive to small molecule inhibitors. A variety of pharmaceutical programs seek to develop anti-cancer therapies directed against the Aurora kinases (Reviewed in Matthews et al., 2006; Mountzios et al., 2008; Carpinelli and Moll, 2008). The economic contingency of trypanosome-afflicted populations is such that it is especially attractive to be able to piggy-back onto therapies being developed for other purposes. The structure of TbAUK1 is conserved among other trypanosomatids, including 80% identity with T. cruzi TcAUK1 (Tc00.1047053508817.80) and 76% identity with Leishmania LmAUK1 (LmjF28.0520). Therefore, therapies developed against this target might be broad spectrum and affect a range of kinetoplastid infections. The current study tests the hypothesis that TbAUK1 is essential for infection in the mammalian host, and can be targeted with small molecule inhibitors.
To validate TbAUK1 as a drug target, we used conditional gene silencing during the infection cycle within a rodent host. This experimental approach was first developed by Lecordier et al., 2005 to demonstrate that TbXPD was essential for trypanosome survival in the mammalian host. We later used the same methods to demonstrate the essential nature of TbRACK1 (Rothberg et al., 2006). In the current studies, mice were infected with BF trypanosomes containing a tetracycline inducible RNAi construct for TbAUK1. Within three days of induction, trypanosomes appeared in the blood that phenocopied RNAi of TbAUK1 in cultured cells. In the absence of other biomarkers, such as the phosphorylation of histone H3 on Ser-10, the appearance of cells with multiple kinetoplasts, multiple flagella and large multi-lobed nucleus provides a good indication that the gene for TbAUK1 was knocked down in situ. By day five post-infection, the population of TbAUK1-deficient cells declined below detectable levels. The transitory nature of RNAi production in trypanosomes likely accounted for recovery of the parasites (Lecordier et al., 2005). Overall, these data demonstrated a role for TbAUK1 in cell cycle control within the mammalian host and validated TbAUK1 as a rational drug target.
An in vitro assay was devised to measure sensitivity of TbAUK1 to the small molecule inhibitor Hesperadin. The ability of Hesperadin to inhibit TbAUK1 is important at two levels: as a research tool and as a step towards therapy design. In some kinetoplastid parasites, where RNAi methods do not work, a selective inhibitor of Aurora kinase would aid in the study of cell division. Additionally, TbAUK1 exists in a protein complex, and its depletion by RNAi affects the distribution of TbKin-A, TbKin-B, TbCPC1 and TbCPC2 (Li et al., 2008). Therefore inhibition within the context of a functional complex is important. Inhibitors of TbAUK1 are also rational therapeutic agents. To determine whether Hesperadin inhibits TbAUK1, an in vitro assay was devised with TbH3 as the phosphoryl acceptor. In mammalian cells, the phosphorylation of H3 on Ser-10 (H3S10p) is of unknown function (Prigent and Dimitrov, 2003), although it may play a role in displacement of HP-1 from chromatin during mitosis (Fischle et al., 2005; Hirota et al., 2005). The ease of detecting H3S10p with antibodies makes this event a good biomarker for Aurora B activity. In trypanosomes, sequence divergence at the amino-terminus of TbH3 makes it unclear whether its phosphorylation might serve as a biomarker for TbAUK1 activity. In the present report, TbAUK1 phosphorylated recombinant TbH3 and TbH2B. MS/MS revealed that phosphorylation occurred within the carboxy-terminal tail (T116 in TbH3 and T77 in TbH2B). Recently, mammalian Aurora kinase B was shown to phosphorylate histone H2A on its carboxy-tail (Brittle et al., 2007). The study relied upon immunolocalization with specific antibodies. Only mitotic cells exhibited this post-translational modification, and only in the centromeric region. Our bulk extraction methods would not have detected an event of this limited temporal and spatial distribution. The role of this unusual phosphorylation is unknown. Selective antibodies against the trypanosome histones will be required to identify whether trypanosomes utilize the unusual phosphorylation sites for TbH3 and TbH2B in vivo, and establish whether it is a true biomarker of TbAUK1 activity. The only other known target of TbAUK1 is the TbTousled-like kinase, but this target has not been validated in vivo (Li et al., 2007).
We used TbH3 phosphorylation to monitor TbAUK1 activity in the presence of Hesperadin. Hesperadin was initially identified as an indolinone that produced polyploidy in cultured human cells (Hauf et al., 2003). Extension of its sulfonamide into the adjacent hydrophobic pocket may account for its specificity towards the Aurora kinase family (Sessa et al., 2005). Hesperadin inhibits recombinant human Aurora B kinase with IC50 of 250 nM when tested with an in vitro kinase assay. It is significantly less effective against Cdk1/cyclin B or Cdk2/cyclin E where the IC50 ranges from 1.2 μM to >10 μM, respectively. When added to mammalian cells, Hesperadin prevented chromosome alignment and segregation, and phosphorylation of Ser-10 on histone H3 (Hauf et al., 2003). Interestingly, Hesperadin became 5-fold more effective when added to cell cultures compared with purified enzyme. When we tested Hesperadin in an in vitro kinase assay, TbAUK1 was more sensitive than the reported values for mammalian Aurora kinase B (IC50 of 40 nM versus 250 nM, respectively). When applied in culture, both trypanosomes and HeLa cells were equally sensitive to Hesperadin (IC50 around 50 nM). In the current report, cultured BF trypanosomes rapidly developed morphological changes that phenocopied those observed for RNAi of TbAUK1. Notably, the cells ceased to divide, and arrested with swollen multi-lobed nuclei, multiple nucleoli, multiple kinetoplasts and multiple flagella. The disruption of CYC6/CRK3 with RNAi can also generate a similar phenotype (Hammarton et al., 2003). However, neither of the related Cdk1 and Cdk2 of humans is inhibited by Hesperadin in the nanomolar range (Hauf et al., 2003).
As a step towards the identification of other selective inhibitors against TbAUK1, we made computer models of TbAUK1 and the human Aurora A protein sequences using the Xenopus Aurora B backbone for three-dimensional alignment. The ATP-pocket and adjacent hydrophobic pocket of Aurora A and Aurora B are currently being targeted in anti-cancer therapies. Amino acids that line the ATP-pocket are identical in TbAUK1 and human Aurora A (E211, V147. A160 and L194; using the numbering for Aurora A). Only the gatekeeper to the adjacent hydrophobic pocket differs. It is Leu-210 in Aurora A and Met-106 in TbAUK1. We chose the Aurora B structure for the alignment of our backbone because of the high amino acid sequence homology to TbAUK1 and since both TbAUK1 and Aurora B have been shown to be chromosomal passenger proteins (Vagnarelli and Earnshaw, 2004; Vader et al., 2006; Jeyaprakash et al., 2007; Li et al., 2008). For comparison, the human Aurora A amino acid sequence was also modeled in exactly the same way. Interestingly, the top 25 Hesperadin dockings observed for the two models had somewhat different preferences. Along with docking within the ATP pocket, TbAUK1 exhibited an additional docking site near the αC helix. Conservation of structure can confer sensitivity of TbAUK1 to inhibitors directed against mammalian Aurora kinases, however, selective inhibition may also be possible.
In summary, the present study demonstrates that TbAUK1 is essential for infection in the mammalian host, and can be targeted with small molecule inhibitors. Anti-cancer drugs directed against mammalian Aurora kinases appear to also inhibit TbAUK1. Structural similarities between TbAUK1 and its homologues from T. cruzi and Leishmania raise the specter of broad-spectrum therapies aimed at Aurora kinase.
Experimental Procedures
Cell cultures
PF T. brucei strains AnTat 1.1E and 29-13 (Wirtz et al., 1999) were grown in SDM-79 with 15% tetracycline-deficient fetal bovine serum (BD Biosciences) at 27°C and 6.5% CO2. 29-13 cells were grown in media supplemented with 15 μg/ml G418 and 50 μg/ml hygromycin B to maintain selective pressure on the tetracycline repressor and T7 polymerase genes. Bloodstream-forms of T. brucei strain 90-13 (Wirtz et al., 1999) were grown at 37°C in HM19 medium with 10% FBS (BD Biosciences) and 10% serum plus (JRH Biosciences). The medium was supplemented with G418 (2.5 μg/ml) and hygromycin B (5 μg/ml).
Infections in mice
An exponentially growing culture of BF TbAUK1 RNAi cells was washed 1× in PBSG (Invitrogen) and suspended in the same buffer. Mice were injected ip with 3×106 cells on day 0. One group of three mice received 1 mg/ml doxycycline in the water to induce the TbAUK1 RNAi, A control group of two mice received water without doxycycline. Each day, the parasitemia was monitored in peripheral blood as described by others (Herbert and Lumsden, 1976). To determine whether cells in the blood phenocopied the cultured RNAi cells, a separate mouse was harvested after three days of infection. The trypanosomes were concentrated from the blood by centrifugation and collected in the buffy layer prior to fixation. The fixed and permeabilized cells were labeled with antibodies against PFR and counterstained with DAPI as described below. The use of animals in this study complied with all relevant federal guidelines and institutional policies.
Cloned genes in trypanosomes
Genomic DNA was used as a template for PCR amplification. A list of specific primer pairs is presented in Table 1. Full-length TbAUK1 (Tb11.01.0330) was PCR amplified with an AU1 epitope tag encoded by the forward primer. The product was cloned into the HindIII/BamHI site of the constitutive trypanosome expression vector pHD496. TbAUK1 with a carboxyl-terminal AU1 tag was cloned into the HindIII/BamHI site of the tetracycline inducible expression vector pLEW100. The kinase dead K58R mutation was produced with a mutagenic forward primer that extended from the BssHII site (residue 159). The primer introduced the arginine codon, which also generated a new FspI site. The reverse primer was pHD496.AU1-TbAUK1 (Table 1). A fragment of TbAUK1 from the BssHII site to the BamHI site was excised and replaced with the mutagenic fragment. The full-length gene with AU1 tag at the 5′ end was amplified with the forward and reverse pHD496.AU1-TbAUK1 primers and cloned into pHD496. RNAi was generated with pZJM (Wang et al., 2000). A 532 base-pair fragment of TbAUK1 was cloned into the XhoI/HindIII site. The pZJM vector has dually opposed tetracycline sensitive promoters flanking the insertion site, and generates dsRNA when de-repressed with tetracycline. To transform trypanosomes, the NotI linearized vectors (pZJM, pHD496 or pLEW100) were electroporated into T. brucei PF cells (pHD496.AU1-TbAUK1, pLEW100.TbAUK1-AU1) or BF (pZJM.TbAUK1) as described previously (Rothberg et al., 2006). The vectors were designed to integrate into the rDNA spacer region of T. brucei. Where appropriate, PF cultures were selected with hygromycin (50 μg/ml), G418 (15 μg/ml) and phleomycin (2.5 μg/ml). BF transformants were selected with G418 (2.5 μg/ml), hygromycin B (5 μg/ml) and phleomycin (2.5 μg/ml). Limiting dilution was used to generate cloned cell lines. Throughout this study, the induction of RNAi was initiated with 1 μg/ml tetracycline.
TABLE 1. Primer pairs used for RNAi and RT-PCR analyses.
PCR primers are listed 5′ to 3′ with restriction sites in bold, the AU1 tag underlined and mutations italicized in lower case. When gene fragments were cloned, the sequence positions of the primers are listed.
| Primer Pairs | Sequence |
|---|---|
| pZJM.TbAUK1 | |
| Fwd: XhoI 105-127 | ATCGCTCGAGTGGCGGCGGCAATTACGGGGATG |
| Rev: HindIII 615-637 | ATCGAAGCTTTGCCCAGGCACCAAAGGTCAGCAC |
| pHD496.AUK1-TbAUK1 | ATCGAAGCTTATGGACACGTACCGCTACATTAGGTCAAC |
| Fwd: HindIII | TGAGGTCGGGCGTGTT |
| Rev: BamHI | ATCGGGATCCTCAATTCTCTTTCCCTGCAGTTGGCTCTGC |
| K58R TbAUK1 | |
| Fwd: BssHII 159-211 | TTTGTTTGCGCGCTGcgcAGGTTGTCCATTTAAAAAACTTG CGGATTTTGACAT |
| pLEW100.TbAUK1-AU1 | |
| Fwd: HindIII | ATCGAAGCTTATGAGGTCAACTGAGGTCGGGCGTG |
| Rev: BamHI | ATCGGGATCCTCAAATGTAGCGGTACGTGTCATTCTCTTT CCCTGCAGTTGGCT |
| TbAUK1-RT | |
| Fwd: 654-677 | ATCGTCTCGTTGGTAAAACACCGTTTGT |
| Rev: 834-857 | ATCGGGGAGATAATAATACTTGAGAAGG |
| Carbonic Anhydrase-RT | |
| Fwd: 84-107 | ATCGCACGGCAGGAATGGGGCGGGCGGA |
| Rev: 284-307 | ATCGCGCCGTTGTCGATTTTTAGGGAAG |
| Dynein Heavy Chain-RT | |
| Fwd: 313-336 | ATCGCGCCGCCGCCGACTATACGAATCG |
| Rev: 494-517 | ATCGCGGCGGGAAGGTAGCGACCAGCGG |
| Alpha Tubulin-RT | |
| Fwd: 11-34 | ATCGCTATCTGCATCCACATTGGTCAGG |
| Rev: 516-539 | ATCGAGCCGTCGACACCTGCGGTGATGG |
Kinase Assays
Epitope tagged TbAUK1 was pulled down from cell homogenates with anti-AU1 Sepharose beads (Covance). Logarithmically growing PF cultures (1×108 cells) were washed two times in PBS and suspended in 400 μl of lysis buffer (50 mM Hepes, pH 7.4, 100 mM KCl, 25 mM NaF, 0.5% NP-40, 1 mM Na3VO4, 1 mM DTT) containing protease inhibitor cocktail (Sigma, P-8340). After 15 min incubation on ice, the lysate was centrifuged at 10,000 xg for 15 min at 4°C. The supernatant was pre-cleared for 2 hrs at 4°C with 80 μl of a 50% slurry of Sephadex G-25 beads. The beads had previously been washed in lysis buffer minus NP-40. Following centrifugation, the pre-cleared supernatant was collected and 50 μl of a 50% slurry of AU1-conjugated beads were added. The beads were incubated overnight at 4°C, washed and collected by centrifugation. Kinase reactions were in 50 μl containing 10 μl of beads, 20 mM Hepes, pH 7.4, 150 mM KCl, 5 mM MgCl2, 5 mM NaF, and 1 mM DTT, and 250 μg of substrate protein [histone H1 from calf thymus (Calbiochem), histone H3 from calf thymus (Roche), myelin basic protein (Sigma), recombinant (His)6-TbH3 or (His)6-TbH2B]. Upon addition of 8 μM ATP (4 μCi of [γ-32P] ATP) the reaction was incubated for 30 min at 30°C, mixed with an equal volume of 2× Laemmli buffer and products were separated by SDS-PAGE. The gels were exposed to X-ray film for 24 hr. The intensity of silver grains was calculated using the SpotDenso function of an Alpha Innotech Imaging system.
Purification of trypanosome histones
BF clone M110 (4×1010 cells) was suspended in 80 ml of lysis buffer (10 mM Na-glutamate, 250 mM sucrose, 2.5 mM CaCl2, 0.1% TritonX-100, and protease inhibitor cocktail (Sigma P2714)). The homogenate was centrifuged at 16,000 xg for 15 min. The pellet was washed once in lysis buffer without TritonX-100 and two additional times in lysis buffer without TritonX-100 and without sucrose. The pellet was acid extracted with 0.3 N HCl for 2 hr at 4°C and centrifuged at 12,000 xg for 15 min at 4°C. The supernatant was precipitated with 8 volumes of acetone. The pellet was washed three times with acetone containing 0.1M HCl (10:1 v:v), and twice with pure acetone. The final precipitate was vacuum dried.
Recombinant trypanosome histone H3 and H2B
Trypanosome histone H3 (Tb927.1.2530) and H2B (Tb10.406.0350) were cloned into the BamHI/HindIII sites of pQE80. TbH3 and TbH2B were extracted from inclusion bodies with Buffer B (100 mM NaH2PO4, 10 mM Tris, 6 M guanadinium HCl, pH 8.0) as described (Luger, K. et al., 1999). All incubations were at room temperature. After a 1 hr extraction, the lysate was centrifuged at 10,000 xg for 30 min. The supernatant was mixed with Ni-NTA resin for 1 hr and then washed with Buffer B in which 8 M urea replaced the 6 M guanadinium HCl. The column was washed with Buffer C (100 mM NaH2PO4, 10 mM Tris, 8 M urea, pH 6.3) and the protein eluted with Buffer E (100 mM NaH2PO4, 10 mM Tris, 8 M urea, pH 4.5). The eluted samples were dialyzed twice against H20 with 2 mM β-mercaptoethanol at 4°C.
Detection of phosphorylation sites in TbH2B and TbH3 by LC/MS/MS
ArgC and AspN (Roche, Penzberg, Germany) were used to digest 1D-SDS PAGE slices of H2B and H3, respectively. The digests were analyzed by nano-LC/MS/MS with a Dionex LC-Packings HPLC (Sunnyvale, CA) coupled to a QStar XL mass spectrometer (Applied Biosystems, Foster City, CA). Peptides were first desalted on a 300 μm × 5 mm PepMap C18 trap column with 0.1 % formic acid in HPLC grade water at a flow rate of 30 μl/min. After desalting for 5 min, peptides were flushed onto a LC-Packings 75 μm × 15 cm C18 nano column (3 micron, 100 A) at a flow rate of 250 nl/min. Peptides were eluted with a 30 min gradient of 3-35% acetonitrile in 0.1% formic acid. Mass ranges for the MS survey scan and MS/MS were m/z 300-2000 and m/z 50-2000, respectively. The scan time for MS and MS/MS were 1.0 sec and 2.0 sec, respectively. The top three multiply-charged ions with MS peak intensity greater than 30 counts/scan were chosen for MS/MS fragmentation with a precursor ion dynamic exclusion of 60 sec.
Reverse transcriptase-PCR
To verify that RNAi resulted in the selective disruption of the mRNA for TbAUK1, Reverse Transcription (RT)-PCR was performed. Total RNA was extracted from T. brucei using TRIzol reagent (Invitrogen). Following DNAse treatment for 3 hours at 37°C, the total RNA was used as a template for RT-PCR. The RT reactions were completed with the Access RT-PCR kit (Promega) according to the manufacturer's instructions. Identical reactions were set up without RT to serve as a control for undigested DNA contamination. The specific primers used here were different from those used to amplify the fragment of TbAUK1 for the RNAi construct. The RT-PCR primers are outlined in Table 1.
Cell growth studies
Growth studies were initiated by diluting logarithmically growing cells to a starting density of 1×105 cells/ml (BF) or 1×106 cells/ml (PF). Cell density was measured with a Neubauer hemocytometer.
Cell cycle analysis
Cells were analyzed by flow cytometry for DNA content following induction of RNAi. Cells were collected by centrifugation at 2,500 xg for 10 minutes and washed in cold PBS containing Dulbecco's salts (Invitrogen). The cell pellets were suspended in 100 μl PBS and mixed with 200 μl of 10% ethanol/5% glycerol in PBS. Another 200 μl of 50% ethanol/5% glycerol was added prior to incubation on ice for 5 min. One ml of 70% ethanol/5% glycerol was added and the fixed cells were left overnight at 4°C. Cells were washed in PBS and and incubated for 30 min at room temperature in 1 ml of PBS containing 10 μg/ml RNase A and 20 μg/ml propidium iodide. Fluorescence analysis was performed with the FACSCalibur flow cytometer (Becton Dickinson). Cell populations were quantified with the CellQuest software.
Microscopy
Immunolocalizations were as described previously (Rothberg et al., 2006). Briefly, cells in culture were fixed in 4% paraformaldehyde for 60 min at room temperature, and were washed in 50 mM Tris-HCl, 150 mM NaCl, pH 7.5. The concentrated cells were allowed to settle onto Fisher (+) Gold positively charged microscope slides. Following a 3 minute permeabilization step with 0.1% Igapal (Sigma), the slides were washed in PBS (Invitrogen). Cells were incubated with rat antibodies against paraflagellar rod protein (αPFR 1:200; kindly provided by T. Seebeck, University of Berne) or mouse antibodies against nucleolar protein (Ab L1C6: 1:200; kindly provided by K. Gull, Oxford University). Secondary antibodies were Cy3 (Jackson Labs). The cells were counterstained with 4,6-Diamidino-2-phenylindole (DAPI) contained in the antifade (Vectorshield) or with TOTO (1:1000; Molecular Probes). To quantify the number of nuclei, kinetoplasts or nucleoli in each cell, 200 BF were evaluated in each of 2 separate experiments. Results are the average ± SE. The cells were visualized with the Nikon C1 Digital Eclipse Confocal E600 microscope. Images were collected with Metamorph or EZ-C1 software (Nikon).
Homology Modeling of the TbAUK1 and human Aurora A proteins
The TbAUK1 and human Aurora A protein sequences were individually aligned to the sequence of Xenopus Aurora B using the ClustalW alignment program (Thompson et al., 1994). Homology models were then built using modeller9v2 (Fiser and Sali, 2003) with the X-ray crystallographic structure of Xenopus Aurora B in complex with Hesperadin and activated by INCENP (PDB 2BFY; Sessa et al., 2005). Hesperadin was included in the template of these modeling experiments, while INCENP was not. After removal of the bound Hesperadin from the models, the low energy conformation of either the resultant TbAUK1 or human Aurora A structures was then relaxed using a conjugant gradient energy minimization routine implemented in the NAMD molecular dynamics program suite (Kale et al., 1999; Phillips et al., 2005). Virtual docking of Hesperadin to the minimized TbAUK1 homology model was then performed with a fixed protein using autodock4 (Huey et al., 2007; Goodsell et al., 1996; Morris et al., 1998). Models were visualized and figures were produced using the VMD program from Humphrey et al. (1996).
Acknowledgments
The authors wish to thank N. Kraut from the Department of Lead Discovery, Boehringer-Ingleheim for the generous gift of Hesperadin. We are also grateful to E. Pays for the PF AnTat cultures; GAM Cross for the BF 90-13 cultures and pLEW100 vector; P. Englund for the pZJM vector; T. Seebeck for antibodies against PFR; and K. Gull for antibody L1C6 against the nucleolus. The authors also thank Joe Gargiula and the SMU ITS department for providing computers and especially Justin Ross for Linux cluster support. This work was supported by NIH grant AI051531.
References
- Anand S, Penrhyn-Lowe S, Venkitaraman AR. Aurora-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell. 2003;3:51–62. doi: 10.1016/s1535-6108(02)00235-0. [DOI] [PubMed] [Google Scholar]
- Andersen CB, Wan Y, Chang JW, Riggs B, Lee C, Liu Y, Sessa F, Villa F, Kwiatkowski N, Suzuki M, Nallan L, Heald R, Musachhio A, Gray NS. Discovery of selective aminothiazole aurora kinase inhibitors. ACS Chem Biol. 2008;3:180–192. doi: 10.1021/cb700200w. [DOI] [PubMed] [Google Scholar]
- Andrews PD, Knatko E, Moore WJ, Swedlow JR. Mitotic mechanics: the auroras come into view. Curr Opin Cell Biol. 2003;15:672–683. doi: 10.1016/j.ceb.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L, Swedlow JR. Aurora B regulates MCAK at the mitotic centromere. Dev Cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, Böhme U, Hannick L, Aslett MA, Shallom J, Marcello L, Hou L, Wickstead B, Alsmark UC, Arrowsmith C, Atkin RJ, Barron AJ, Bringaud F, Brooks K, Carrington M, Cherevach I, Chillingworth TJ, Churcher C, Clark LN, Corton CH, Cronin A, Davies RM, Doggett J, Djikeng A, Feldblyum T, Field MC, Fraser A, Goodhead I, Hance Z, Harper D, Harris BR, Hauser H, Hostetler J, Ivens A, Jagels K, Johnson D, Johnson J, Jones K, Kerhornou AX, Koo H, Larke N, Landfear S, Larkin C, Leech V, Line A, Lord A, Macleod A, Mooney PJ, Moule S, Martin DM, Morgan GW, Mungall K, Norbertczak H, Ormond D, Pai G, Peacock CS, Peterson J, Quail MA, Rabbinowitsch E, Rajandream MA, Reitter C, Salzberg SL, Sanders M, Schobel S, Sharp S, Simmonds M, Simpson AJ, Tallon L, Turner CM, Tait A, Tivey AR, Van Aken S, Walker D, Wanless D, Wang S, White B, White O, Whitehead S, Woodward J, Wortman J, Adams MD, Embley TM, Gull K, Ullu E, Barry JD, Fairlamb AH, Opperdoes F, Barrell BG, Donelson JE, Hall N, Fraser CM, Melville SE, El-Sayed NM. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Severin FF, Bhalla N, Sassoon I, Hyman AA, Murray AW. The conserved protein kinase Ipl1 regulates the microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, Chan CS, Novotny M, Slamon DJ, Plowman GD. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittle AL, Nanba Y, Ito T, Ohkura H. Concerted action of Aurora B, Polo and NHK-1 kinases in centromere-specific histone 2A phosphorylation. Exp Cell Res. 2007;313:2780–2785. doi: 10.1016/j.yexcr.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat Rev Mol Cell Biol. 2003;4:842–854. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- Carpinelli P, Ceruti R, Giorgini ML, Cappella P, Gianellini L, Croci V, Degrassi A, Texido G, Rocchetti M, Vianello P, Rusconi L, Storici P, Zugnoni P, Arrigoni C, Soncini C, Alli C, Patton V, Marsiglio A, Ballinari D, Pesenti E, Francelli D, Moll J. PHA-739358, a potent inhibitor of Aurora kinases with a selective target inhibition profile relevant to cancer. Mol Cancer Ther. 2007;6:3158–3168. doi: 10.1158/1535-7163.MCT-07-0444. [DOI] [PubMed] [Google Scholar]
- Carpinelli P, Moll J. Aurora kinases and their inhibitors: more than one target and one drug. Adv Exp Med Biol. 2008;610:54–73. doi: 10.1007/978-0-387-73898-7_5. [DOI] [PubMed] [Google Scholar]
- Chanez AL, Hehl AB, Engstler M, Schneider A. Ablation of the single dynamin of T. brucei blocks mitochondrial fission and endocytosis and leads to a precise cytokinesis arrest. J Cell Sci. 2006;119:2968–2974. doi: 10.1242/jcs.03023. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR, Chan CS, Drubin DG, Barnes G. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- D'Alice AM, Amabile G, Iovino M, Di Giorgio FP, Bartiromo M, Sessa F, Villa F, Musacchio A, Cortese R. Reversine, a novel Aurora kinases inhibitor, inhibits colony formation of human acute myeloid leukemia cells. Mol Cancer Ther. 2008;7:1140–1149. doi: 10.1158/1535-7163.MCT-07-2051. [DOI] [PubMed] [Google Scholar]
- Ditchfieldd C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock Al, Keen N, Taylor SS. Aurora B couples chromosome alignment with anaphapse by targeting BubR1, Mad2 and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng A, Shen S, Tschudi C, Ullu E. Analysis of gene function in Trypansoma brucei using RNA interference. Methods Mol Biol. 2004;270:287–298. doi: 10.1385/1-59259-793-9:287. [DOI] [PubMed] [Google Scholar]
- Ewart-Tolarnd A, Dai Q, Gao YT, Nagase H, Dunlop MG, Farrington SM, Barnetson RA, Anton-Culver H, Peel D, Ziogas A, Lin D, Miao X, Sun T, Ostrander EA, Stanford JL, Langlois M, Chan JM, Yuan J, Harris Cc, Bowman ED, Clayman GL, Lippman SM, Lee JJ, Zheng W, Balmain A. Aurora-A/STK15 T+91A is a general low penetrance cancer susceptibility gene: a meta-analysis of multiple cancer types. Carcinogenesis. 2005;26:1368–1373. doi: 10.1093/carcin/bgi085. [DOI] [PubMed] [Google Scholar]
- Eyers PA, Erikson E, Chen LG, Maller JL. A novel mechanism for activation of the protein kinase Aurora A. Curr Biol. 2003;13:691–697. doi: 10.1016/s0960-9822(03)00166-0. [DOI] [PubMed] [Google Scholar]
- Fancelli D, Berta D, Bindi S, Cameron A, Cappella P, Carpinelli P, Catana C, Forte B, Giordano P, Giorgini ML, Mantegani S, Marsiglio A, Meroni M, Moll J, Pittalà V, Roletto F, Severino D, Soncini C, Storici P, Tonani R, Varasi M, Vulpetti A, Vianello P. Potent and selective Aurora inhibitors identified by the expansion of a novel scaffold for protein kinase inhibition. J Med Chem. 2005;248:3080–3084. doi: 10.1021/jm049076m. [DOI] [PubMed] [Google Scholar]
- Fancelli D, Moll J, Varasi M, Bravo R, Artico R, Berta D, Bindi S, Cameron A, Candiani I, Cappella P, Carpinelli P, Croci W, Forte B, Giorgini ML, Klapwijk J, Marsiglio A, Pesenti E, Rocchetti M, Roletto F, Severino D, Soncini C, Storici P, Tonani R, Zugnoni P, Vianello P. 1,4,5,6-tetrahydropyrrolo[3,4-c]pyrazoles: identification of a potent Aurora kinase inhibitor with a favorable antitumor kinase inhibition profile. J Med Chem. 2006;49:7247–7251. doi: 10.1021/jm060897w. [DOI] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- Fiser A, Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- Foote KM, Mortlock AA, Heron NM, Jung FH, Hill GB, Pasquet G, Brady MC, Green S, Heaton SP, Kearney S, Keen NJ, Odedra R, Wedge SR, Wilkinson RW. Synthesis and SAR of 1-acetanilide-4-aminopyrazole-substituted quinazolines: selective inhibitors of Aurora B kinase with potent anti-tumor activity. Bioorg Med Chem Lett. 2008;18:1904–1909. doi: 10.1016/j.bmcl.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Francisco L, Chan CS. Regulation of yeast chromosome segregation by Ipl1 protein kinase and type 1 protein phosphatase. Cell Mol Biol Res. 1994;40:207–213. [PubMed] [Google Scholar]
- García-Salcedo JA, Perez-Morga D, Gijon P, Dilbeck V, Pays E, Nolan DP. A differential role for actin during the life cycle of Trypanosoma brucei. EMBO J. 2004;23:780–789. doi: 10.1038/sj.emboj.7600094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R, Glover DM. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J Cell Biol. 2001;152:669–682. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R, Petretti C, Prigent C. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol. 2005;15:241–250. doi: 10.1016/j.tcb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Goodsell DS, Morris GM, Olson AJ. Automated docking of flexible ligands: applications of AutoDock. J Mol Recognit. 1996;9:1–5. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Goto H, Yasui Y, Kawgiri A, Nigg EA, Terada Y, Tatsuka M, Nagata K, Inagaki M. Aurora B regulates the cleavage furrow-specific vimentin phosphorylation in the cytokinetic process. J Biol Chem. 2003;278:8526–85230. doi: 10.1074/jbc.M210892200. [DOI] [PubMed] [Google Scholar]
- Gritsko TM, Coppola D, Paciga JE, Yang L, Sun M, Shelley SA, Fiorica JV, Nicosia SV, Cheng JQ. Activation and overexpression of centrosome kinase BTAK/Aurora-A in human ovarian cancer. Clin Cancer Res. 2003;9:1420–1426. [PubMed] [Google Scholar]
- Hammarton TC, Clark J, Douglas F, Boshart M, Mottram JC. Stage specific differences in cell cycle control in Trypanosoma brucei revealed by RNA interference of a mitotic cyclin. J Biol Chem. 2003;278:22877–22886. doi: 10.1074/jbc.M300813200. [DOI] [PubMed] [Google Scholar]
- Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, Graham JA, Demur C, Hercend T, Diu-Hercend A, Su M, Golec JMC, Miller KM. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nature Medicine. 2004;10:262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- Hauf S, Andrews PD, Wickramasinghe S, Sleeman J, Prescott A, Lem YW, Lyon C, Swedlow JR, Lemond AI. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachments and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert WJ, Lumsden WHR. Trypanosoma brucei: A rapid matching method for estimating the host's parasitemia. Exp Parasit. 1976;40:427–431. doi: 10.1016/0014-4894(76)90110-7. [DOI] [PubMed] [Google Scholar]
- Heron NM, Anderson M, Blowers DP, Breed J, Eden JM, Green S, Hill GB, Johnson T, Jung FH, McMiken HHJ, Mortlock AA, Pannifer AD, Pauptit RA, Pink J, Roberts NJ, Rowsell S. SAR and inhibitor complex structure determination of a novel class of potent and specific Aurora kinase inhibitors. Bioorg Med Chem Lett. 2006;16:1320–1323. doi: 10.1016/j.bmcl.2005.11.053. [DOI] [PubMed] [Google Scholar]
- Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438:1176–1180. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge-based desolvation. J Comput Chem. 2007;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- Jetton N, Rothberg KG, Ruben L. Aurora Kinase TbAUK1 is essential for nuclear division in Trypanosoma brucei. Kinetoplastid Molecular Cell Biology; Conference Proceedings; Abstract 212A 2005 [Google Scholar]
- Jeyaprakash AA, Klein UR, Lindner D, Ebert J, Nigg EA, Conti E. Structure of a Survivin–Borealin–INCENP core complex reveals how chromosomal passengers travel together. Cell. 2007;131:271–285. doi: 10.1016/j.cell.2007.07.045. [DOI] [PubMed] [Google Scholar]
- Ju H, Cho H, Kim YS, Kim WH, Ihm C, Noh SM, Kim JB, Hahn DS, Choi BY, Kang C. Functional polymorphism 57Val>Ile of aurora kinase A associated with increased risk of gastric cancer progression. Cancer Lett. 2006;242:273–279. doi: 10.1016/j.canlet.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Kang J, Cheeseman IM, Kallstrom G, Velmurugan S, Barnes G, Chan CS. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J Cell Biol. 2001;155:763–774. doi: 10.1083/jcb.200105029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama H, Ota T, Jisaki F, Ueda Y, Tanaka T, Odashima S, Suzuki F, Terada Y, Tatsuka M. Mitotic kinase expression and colorectal cancer progression. J Natl Cancer Inst. 1999;91:1160–1162. doi: 10.1093/jnci/91.13.1160. [DOI] [PubMed] [Google Scholar]
- Katayama H, Sasai K, Kawai H, Yuan ZM, Bondaruk J, Suzuki F, Fujii S, Arlinghaus RB, Czerniak BA, Sen S. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat Genet. 2004;36:55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- Katayama H, Zhou H, Li Q, Tatsuka M, Sen S. Interaction and feedback regulation between STK15/BTAK/Aurora A kinase and protein phosphatase 1 through mitotic cell division cycle. J Biol Chem. 2001;276:46219–46224. doi: 10.1074/jbc.M107540200. [DOI] [PubMed] [Google Scholar]
- Lecordier L, Walgraffe D, Devaux S, Poelvoorde P, Pays E, Vonhamme L. Trypanosoma brucei RNA interference in the mammalian host. Mol Bio Parasit. 2005;140:127–131. doi: 10.1016/j.molbiopara.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Li Z, Gourguechon S, Wang CC. Tousled-like kinase in a microbial eukaryote regulates spindle assembly and S-phase progression by interacting with Aurora kinase and chromatin assembly factors. J Cell Sci. 2007;120:3883–3894. doi: 10.1242/jcs.007955. [DOI] [PubMed] [Google Scholar]
- Li Z, Lee JH, Chu F, Burlingame AL, Günzl A, Wang CC. Identification of a novel chromosomal passenger complex and its unique localization during cytokinesis in Trypanosoma brucei. PLoS One. 2008;3:e2354. doi: 10.1371/journal.pone.0002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang CC. Changing roles of aurora-B kinase in two life cycle stages of Trypanosoma brucei. Eukaryot Cell. 2006;5:1026–1035. doi: 10.1128/EC.00129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liping Song L, Li D, Liu R, Zhou H, Chen J, Huang X. Ser-10 phosphorylated histone H3 is involved in cytokinesis as a chromosomal passenger. Cell Biol Int. 2007;31:1184–1190. doi: 10.1016/j.cellbi.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ. Preparation of Nucleosome Core Particles from Recombinant Histones. Methods in Enzymology. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- Matthews N, Visintin C, Hartzoulakis B, Jarvis A, Selwood DL. Aurora A and B kinases as targets for cancer: will they be selective for tumors? Expert Rev Anticancer Ther. 2006;6:109–120. doi: 10.1586/14737140.6.1.109. [DOI] [PubMed] [Google Scholar]
- Morrow CJ, Tighe A, Johnson VL, Scott MIF, Ditchfield C, Taylor SS. Bub1 and aurora B cooperate to maintain BubR1-mediated inhibition of APC/CCdc20. J Cell Sci. 2005;118:3639–3652. doi: 10.1242/jcs.02487. [DOI] [PubMed] [Google Scholar]
- Motoyka SA, Zhao Z, Gull K, Englund PT. Integration of pZJM library plasmids into unexpected locations in the Trypanosoma brucei genome. Mol Biochem Parasitol. 2004;134:163–167. doi: 10.1016/j.molbiopara.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Mountzios G, Terpos E, Dimopoulos MA. Aurora kinases as targets for cancer therapy. Cancer Treat Rev. 2008;34:175–182. doi: 10.1016/j.ctrv.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Murata-Hori M, Wang YL. The kinase activity of aurora B is required for kinetochore-microtubule interactions during mitosis. Curr Biol. 2002;12:894–899. doi: 10.1016/s0960-9822(02)00848-5. [DOI] [PubMed] [Google Scholar]
- Naula C, Parsons M, Mottram JC. Protein kinases as drug targets in trypanosomes and Leishmania. Biochim Biophys Acta. 2005;30:151–159. doi: 10.1016/j.bbapap.2005.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T, Suto S, Katayama H, Han ZB, Suzuki F, Maeda M, Tanino M, Terada Y, Tatsuka M. Increased mitotic phosphorylation of histone H3 attributable to AIM-1/Aurora-B overexpression contributes to chromosome number instability. Cancer Res. 2002;62:5168–5177. [PubMed] [Google Scholar]
- Ouchi M, Fujiuchi N, Sasai K, Kayayama H, Minamishima YA, Ongusaha PP, Dengll C, Sen S, Lee SW, Ouchi T. BRCA1 phosphorylation by Aurora-A in the regulation of G2 to M transition. J Biol Chem. 2004;279:19643–19648. doi: 10.1074/jbc.M311780200. [DOI] [PubMed] [Google Scholar]
- Parsons M, Worthey EA, Ward PN, Mottram JC. Comparative analysis of the kinomes of three pathogenic trypanosomatids: Leishmania major, Trypanosoma brucei and Trypanosoma cruzi. BMC Genomics. 2005;6:127. doi: 10.1186/1471-2164-6-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nature Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent C, Dimitrov S. Phosphorylation of serine 10 in histone H3, what for? J Cell Sci. 2003;116:3677–3685. doi: 10.1242/jcs.00735. [DOI] [PubMed] [Google Scholar]
- Regmi S, Rothberg KG, Hubbard JG, Ruben L. The RACK1 signal anchor protein from Trypanosoma brucei associates with eukaryotic elongation factor1A: a role for translational control in cytokinesis. Mol Microbiol. 2008;70:724–745. doi: 10.1111/j.1365-2958.2008.06443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riefler GM, Dent SYR, Schumacher JM. Tousled-mediated activation of Aurora B kinase does not require Tousled Kinase activity in vivo. J Biol Chem. 2008;283:12763–12768. doi: 10.1074/jbc.M709034200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg KG, Burdette DL, Pfannstiel J, Jetton N, Singh R, Ruben L. The RACK1 homologue from Trypanosoma brucei is required for the onset and progression of cytokinesis. J Biol Chem. 2006;281:9781–9790. doi: 10.1074/jbc.M600133200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai K, Katayama H, Stenoien DL, Fujii S, Honda R, Kimura M, Okano Y, Tatsuka M, Suzuki F, Nigg EA, Earnshaw WW, Brinkley WR, Sen S. Aurora-C lkinase is a novel chromosomal passenger protein that can complement Aurora-B kinase function in mitotic cells. Cell Motil Cytoskeleton. 2004;59:249–263. doi: 10.1002/cm.20039. [DOI] [PubMed] [Google Scholar]
- Sessa F, Mapelli M, Ciferri C, Tarricone C, Areces LB, Schneider TR, Stukenberg PT, Musacchio A. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol Cell. 2005;18:379–391. doi: 10.1016/j.molcel.2005.03.031. [DOI] [PubMed] [Google Scholar]
- Smith DH, Pepin J, Stich AHR. Human African trypanosomiasis: an emerging public health crisis. British Medical Bulletin. 1998;54:341–355. doi: 10.1093/oxfordjournals.bmb.a011692. [DOI] [PubMed] [Google Scholar]
- Sommer JM, Cheng CL, Keller GA, Wang CC. In vivo import of firefly luciferase into the glycosomes of Trypanosoma brucei and mutational analysis of the C-terminal targeting signal. Mol Biol Cell. 1992;3:749–759. doi: 10.1091/mbc.3.7.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Miao X, Wang J, Tan W, Zhou Y, Yu C, Lin D. Functional Phe31Ile polymorphism in Aurora A and risk of breast carcinoma. Carcinogenesis. 2004;25:2225–2230. doi: 10.1093/carcin/bgh244. [DOI] [PubMed] [Google Scholar]
- Tanaka TV, Rachidi N, Jenke C, Pereira G, Galova M, Schiebel E, Stark MJ, Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase INCENP) complex promotes chromosome biorientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Tari LW, Hoffman ID, Bensen DC, Hunter MJ, Nix J, Nelson KJ, McRee DE, Swanson RV. Structural basis for the inhibition of Aurora A kinase by a novel class of high affinity di-substituted pyrimidine inhibitors. Bioorg Med Chem Lett. 2007;17:688–691. doi: 10.1016/j.bmcl.2006.10.086. [DOI] [PubMed] [Google Scholar]
- Tatsuka M, Sato S, Kitajima S, Suto S, Kawai H, Miyauchi M, Ogawa I, Maeda M, Ota T, Takata T. Overexpression of Aurora-A potentiates HRAS-mediated oncogenic transformation and is implicated in oral carcinogenesis. Oncogene. 2005;24:1122–1127. doi: 10.1038/sj.onc.1208293. [DOI] [PubMed] [Google Scholar]
- Tchatchou S, Wirtenberger M, Hemminki K, Sutter C, Meindl A, Wappenschmidt B, Kiechle M, Bugert P, Schmutzler RK, Bartram CR, Burwinkel B. Aurora kinases A and B and familial breast cancer risk. Cancer Lett. 2007;247:266–272. doi: 10.1016/j.canlet.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toji S, Yabuta N, Hosomi T, Nishihara S, Kobayashi T, Suzuki S, Tamai K, Nojima H. The centrosomal protein Lats2 is a phosphorylation target of Aurora-A kinase. Genes Cells. 2004;9:383–397. doi: 10.1111/j.1356-9597.2004.00732.x. [DOI] [PubMed] [Google Scholar]
- Tsai MY, Wiese C, Cao K, Martin O, Donovan P, Ruderman J, Prigent C, Zheng Y. A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nat Cell Biol. 2003;5:242–248. doi: 10.1038/ncb936. [DOI] [PubMed] [Google Scholar]
- Tu X, Kumar P, Li Z, Wang CC. An aurora kinase homologue is involved in regulating both mitosis and cytokinesis in Trypanosoma brucei. J Biol Chem. 2006;281:9677–9687. doi: 10.1074/jbc.M511504200. [DOI] [PubMed] [Google Scholar]
- Vader G, Medema RH, Lens SM. The chromosomal passenger complex: guiding Aurora-B through mitosis. J Cell Biol. 2006;173:833–837. doi: 10.1083/jcb.200604032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli P, Earnshaw WC. Chromosomal passengers: the four-dimensional regulation of mitotic events. Chromosoma. 2004;113:211–222. doi: 10.1007/s00412-004-0307-3. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhou YX, Qiao W, Tominaga Y, Ouchi M, Ouchi T, Deng CX. Overexpression of aurora kinase A in mouse mammary epithelium induces genetic instability preceding mammary tumor formation. Oncogene. 2006 doi: 10.1038/sj.onc.1209707. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Wang Z, Morris JC, Drew ME, Englund PT. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J Biol Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant negative generation in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- Yan X, Cao L, Li Q, Wu Y, Zhang H, Saiyin H, Liu X, Zhang X, Shi Q, Yu L. Aurora C is directly associated with Survivin and required for cytokinesis. Genes Cells. 2005;10:617–626. doi: 10.1111/j.1365-2443.2005.00863.x. [DOI] [PubMed] [Google Scholar]