Abstract
Using classical molecular dynamics simulations, we study ion-ion interactions in water. We study the potentials of mean force (PMF) for the full set of alkali halide ion pairs, and in each case, we test different parameter sets for modeling both the water and the ions. Altogether, we compared 300 different PMFs. We also calculate association equilibrium constants (KA) and compare them to two types of experiments. Of additional interest here was the proposition of Collins called the ‘law of matching water affinities’, where the relative affinity of ions in solution depends on the matching of cation and anion sizes. From observations on the relative depths of the free energies of the contact ion pair (CIP) and the solvent-shared ion pair (SIP), along with related solvent structure analyses, we find a good correlation with this proposition: small-small and large-large should associate in water and small-large should be more dissociated.
1 Introduction
Solvated ions are a common component of nearly all biological systems. The nature of how ions interact among themselves and with water has been the subject of several classical1–5 and recent studies.6–13
The solvation of individual ions in water and their effects in solution have been studied by computer simulations,8, 14–22 with comparisons among force fields and water models.23–26 Small differences in parameters are found to affect predictions of ion association27, 28 and crystallization.25, 26 There have also been previous studies of ion pairs,7, 27, 29–35 and solvent models.36, 37 Some studies have explored peculiar ionic phenomena,7, 27, 28, 34, 38 while other studies have used aggregation results in critical analyses of the validity of ion force fields used in molecular simulations.23, 25, 26
Our interest here is to perform a comprehensive analysis of ion pairing in computer simulations and explore an idea of Collins, which he calls the ‘law of matching water affinities’.39–42 Salts such as LiF, which involve both a small anion and small cation, are only sparingly soluble in water. Salts such as CsI, which involve both a large anion and cation, also have limited solubilities. However, in contrast, salts like CsF, in which one ion is large and the other is small, are highly soluble in water. Hence our interest here is not just in a particular ion pair, but in comparing the pairings of different ions of different sizes. Here we do extensive comparative tests using molecular dynamics simulations to study such questions of water structuring around two ions as they approach each other in water. We study different cation-anion pairs, different force fields and water parameters, full potentials of mean force, and detailed solvent structure analyses at different stages of the ion pairing process. From the potentials of mean force, we are able to extract ion association constants, allowing us to quantitatively assess the long-time behavior of the possible ion pairs as a function of ion parameter and water model. Through this combined study, we explore the physical forces that are common to all the models in addition to the individual features that arise from different parameters among force fields.
2 Methods
We studied the potential of mean force (PMF) for each group I and VII univalent ion pair. Ions were modeled using three different parameter sets: 1) the OPLS all atom forcefield,43 2) a set derived from the work of Dang and others,30, 37, 44–47 herein referred to as the Dang forcefield, and 3) a newer set of ion parameters developed by Jensen and Jorgensen,48 herein referred to as the JJ forcefield. Water was represented using the SPC,49 SPC/E,50 TIP3P,51 TIP4P-Ew,52 and TIP5P-E53 forcefields. The Lennard-Jones and electrostatic parameters of the relevant ions and water models are listed in Table 1.
Table 1.
Lennard-Jones and electrostatic parameters for the ion and water forcefields.
| q (or qH) |
σLJ (Å) |
εLJ (kcal/mol) |
|||||
|---|---|---|---|---|---|---|---|
| OPLS | Dang | JJ | OPLS | Dang | JJ | ||
| Li+ | 1 | 2.12645 | 2.337 | 2.87 | 0.018278 | 0.16013 | 0.0005 |
| Na+ | 1 | 3.33045 | 2.876 | 4.07 | 0.002772 | 0.12467 | 0.0005 |
| K+ | 1 | 4.93463 | 3.250 | 5.17 | 0.000328 | 0.12467 | 0.0005 |
| Rb+ | 1 | 5.62177 | 3.348 | 5.60 | 0.000171 | 0.12467 | 0.0005 |
| Cs+ | 1 | 6.71600 | 3.526 | 6.20 | 0.000081 | 0.12467 | 0.0005 |
| F− | −1 | 2.73295 | 3.143 | 3.05 | 0.720000 | 0.16714 | 0.7100 |
| Cl− | −1 | 4.41724 | 3.785 | 4.02 | 0.117790 | 0.12467 | 0.7100 |
| Br− | −1 | 4.62376 | 3.896 | 4.28 | 0.090000 | 0.12467 | 0.7100 |
| I− | −1 | 5.40000 | 4.168 | 4.81 | 0.070000 | 0.12467 | 0.7100 |
|
| |||||||
| SPC | 0.41000 | 3.16549 | 0.15532 | ||||
| SPC/E | 0.42380 | 3.16549 | 0.15532 | ||||
| TIP3P | 0.41700 | 3.15061 | 0.15210 | ||||
| TIP4P-Ew | 0.52422 | 3.16435 | 0.16275 | ||||
| TIP5P-E | 0.24100 | 3.09700 | 0.17800 | ||||
2.1 Simulation details
All our molecular dynamics calculations were performed using version 3.3.1 of the GROMACS molecular dynamics package.54 All systems consisted of 2 ions and 864 water molecules in cubic simulation boxes under periodic boundary conditions. The leap-frog algorithm with a time step of 2 fs was used to integrate the equations of motion. The isothermal-isobaric ensemble was used to maintain a temperature 300 K and a pressure of 1 bar. The weak coupling algorithms of Berendsen and co-workers55 were used for both the thermostat and barostat with coupling constants of 0.1 and 1.0 ps respectively. Electrostatics were handled using the smooth version of the particle mesh Ewald (PME) method56 under tin-foil boundary conditions with a grid spacing of 0.12, a PME order of 4, real-space cutoff of 9 Å, and an Ewald screening parameter of 0.347 Å−1. The Lennard-Jones interactions shared the same cutoff, and energy and pressure tail corrections were included. The Lennard-Jones mixing rules followed the convention dictated by each ion force field; all sets of ion parameters use a geometric mean for εLJ, while an arithmetic mean is used for the Dang σLJ parameters and a geometric mean is used for the OPLS and JJ σLJ parameters.
To calculate the PMFs, the method of constraint molecular dynamics was used to sample the ion pair systems at fixed interparticle separations.13, 24, 57 The SHAKE algorithm was used to constrain the ions at distances starting between 2.0 and 3.0 Å depending on the size of the ions and extending out to 12 Å. To enhance the detail and smoothness of the resulting PMF, the separation step was varied down to values as low as 0.05 Å around sharp features after generating a coarse PMF with a 0.4 Å step size. Following this procedure, each resulting PMF was composed of approximately 50 separate simulations. Several simulation run times ranging from 0.1 to 20 ns at each constrained separation length were tested to determine the sampling required to obtain fully converged PMFs. While all PMFs for the same salt system overlapped within error, simulation times of 2 ns were indistinguishable from the 20 ns simulation times aside from a slightly greater error. With this lower boundary in mind, we selected a data accumulation time of 3 ns for separation distances less than 10 Å and 6 ns of simulation time for distances greater than 10 Å.
Generating the PMFs from these constrained simulations involves integrating the average mean force values over the ion separation distances. Errors in the mean force values were accumulated from the limiting value of block averages.58 Before integrating the mean force values, the entropic force due to the increase in phase space with ion separation was added as described elsewhere.24 The errors in the PMFs were determined by integrating the mean force variances, and the average error for points along each PMF was approximately 0.06 kcal/mol. It is important to note that a PMF obtained through this integration procedure will originate at 0 kcal/mol, while the true PMF should show a weak attractive interaction at the distant ion separations. We have approximated this 4 distant interaction with a simple screened Coulomb potential,59, 60
| (1) |
where λB is the Bjerrum length, and κ is the Debye-Hückel screening parameter, which for low number density (ρi) univalent salt solutions is,
| (2) |
2.2 Structural analysis
We calculated the local water occupancy and orientation around various ion pairs, the relative strength the surrounding water-water hydrogen bond network, the numbers of waters released from the first solvation shells upon ion pairing, and the free energy penalty for solvent caging about the ion pair. These quantities were determined from 20-ns simulations of systems with ion-constraint separations at the three primary minima and first maximum in the PMF, as well as when the ions are at a 12 Å separation distance. The three minima are the contact ion pair (CIP), solvent-shared ion pair (SIP), and solvent-separated ion pair (2SIP) states.61 The maximum corresponds to the approximate center of the transition state ensemble between the CIP and SIP states.62
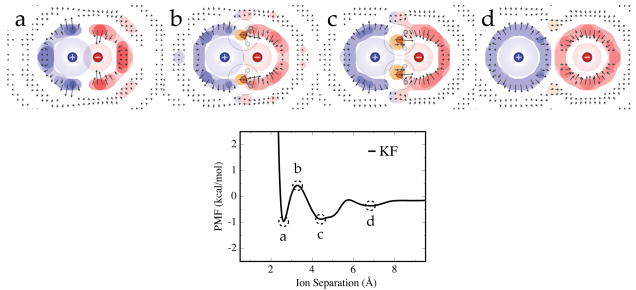
The local water occupancy and orientation maps were generated by gridding up the space around the ions into 0.5 Å edge-length cubes and binning each water molecule’s position and orientation within this network of cells. Cylindrical symmetry about the inter-ion axis was utilized in mapping the resulting occupancies onto a two-dimensional surface, and these occupancies were converted to free energies to generate free energy maps of solvent around the ion pairs. The average dipole vector magnitude and direction relative to the ion pair are shown as arrows, where longer arrows correspond to more orientationally constrained water molecules. Only cells with an occupancy greater than the ideal background water density are given arrows for clarity. See Figure 6 for an example series of these structure maps.
Figure 6.
Free energy surfaces about the KF ion pair at the a) CIP, b) 1st maximum, c) SIP, and d) 2SIP states as labeled on the PMF. The arrows indicate the average water dipole vector orientation and magnitude. The blue, red, and orange contours indicate the cation, anion, and bridging water occupancy regions respectively, and the free energy tolerances from lightest to darkest tone are -0.25, -0.5, -0.75, and -1.0 kcal/mol. Water molecule illustrations are shown at particularly stable bridging water regions. The rings around the water illustrations and the shaded circles around the ions depict the Lennard-Jones spheres of the particles. The ions were modeled using the OPLS parameters in TIP5P-E water.
To assess the perturbation of the hydrogen bonding network in the surrounding solvent, we used a similar spatial grid as above, only with smaller 0.25 Å edge-length cubes. Here, we calculated water-water interaction energies for all solvent particles in these simulations and binned the magnitude of these interactions at the midpoints of the interaction lines. For a measure of the strength of the average bulk water-water hydrogen bond, we performed a separate simulation containing only water, and calculated the energy pair distribution function.51 Interactions stronger than the minimum of this curve (−2.5 kcal/mol for TIP5P-E under the stated simulation conditions) were taken to represent hydrogen bonds, and this provided a tolerance for selecting which interactions were included in the binning procedure. The resulting plots (see Figure 8) indicate the spatial location and average strength of hydrogen bonds in the surrounding network relative to the strength of bulk water hydrogen bonds.
Figure 8.
The average hydrogen bond strength about a) LiF, b) CsI, and c) LiI in the CIP and SIP states. Blue contours indicate water-water hydrogen bonds weaker than the bulk (white) values, while red contours indicate stronger water-water hydrogen bonds. The contours are drawn in 0.2 kcal/mol levels, with the lightest red level 0.2 kcal/mol stronger and the lightest blue level 0.2 kcal/mol weaker than bulk water-water hydrogen bonds. Here, the ions were represented using the OPLS parameters and TIP5P-E was used to model water.
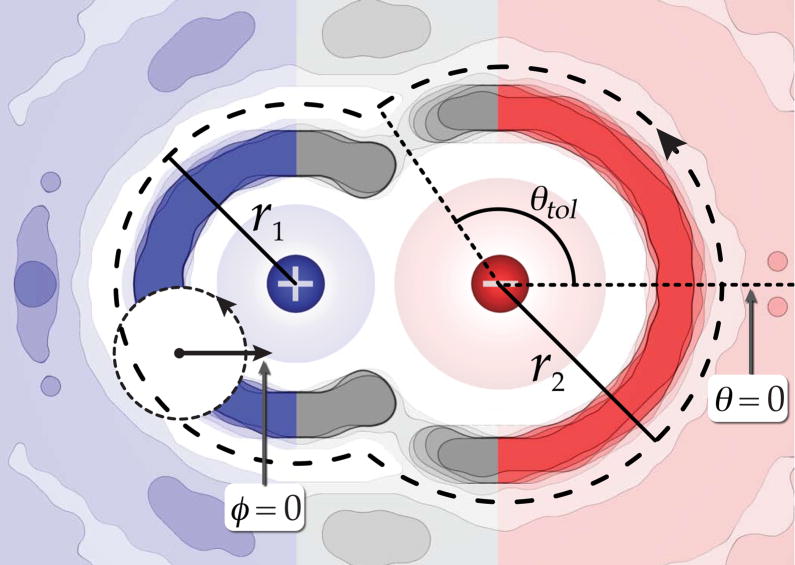
To calculate the number of water molecules in the first solvation shell of the ion pairs, spatially restricted radial distribution functions (g(r)s) were performed about each ion. Figure 1 illustrates the spatial regions included in these functions when the cation and anion are ions 1 and 2 respectively. By restricting the solvent space included in the g(r)s, we avoid artifacts arising from the excluded volume of the opposing ion. The distances to the first minimum of each g(r) are indicated as r1 and r2, and all water molecules within the combined volume traced out by these radii are taken to constitute the first solvation shell of the ion pair. The nrel value is simply the difference between first shell water counts in the paired state of interest and that of the furthest separation distance considered in the PMF (12 Å in these simulations).
Figure 1.
A two dimensional map of the water oxygen atom occupancy about an ion pair. The blue coloring indicates the region used in the spatially restricted g(r) about the cation, the red coloring indicates the region included in the spatially restricted g(r) about the anion, and the gray colored region is neglected by both functions. The radial distances to the first minimum of each g(r) are r1 and r2, θtol is an angle tolerance used in determining the Kullback-Leibler free energy, and the dashed line boundary indicates the region defining the first solvation shell about the ion pair.
To estimate the free energy penalty due to caging of solvent in the first-shell about the ion pair, we computed the Kullback-Leibler entropy
| (3) |
where p(θ, φ) is the probability distribution of water occupancy around an ion as a function of the angles θ and φ as shown in Figure 1, and p0(θ, φ) is this same quantity for a chosen reference state (an ion separation of 12 Å in our case). For each θ value, we calculate an average probability density from the ion surface out to the first minimum of the water density (r1 or r2 in Figure 1). Due to cylindrical symmetry about the inter-ion axis, θ ranges from 0 to π around fully solvated ions. To avoid double counting the occupancy in states where the first solvent shells of the ions are shared, we define an angle (θtol) to the line extending from more weakly solvated ion through the minimum occupancy region where the solvent shells intersect (see Figure 1). Here, θ ranges from 0 to θtol and the probability distribution is set to zero from θtol to π. It should be noted that the excluded volume region will not contribute substantially to ΔSKL, as p(θ) will be zero in this region. In each one of these θ wedges, we accumulate the orientational probability distributions of the encompassed water, angle φ. The φ angle is a measure of the water dipole vector direction relative to the inter-ion axis, and it ranges from 0 to 2π. Thus, summing over both φ and θ give us an estimate of both the orientational and translational entropy change of the first-shell waters upon ion pairing. The total ΔSKL is taken to be the sum of the individual values calculated about each ion.
Because of the a priori probability quantity p0(θ, φ) in the Kullback-Leibler quantity, the quantity
| (4) |
represents a free energy of a given probability distribution relative to a reference probability distribution. We use this quantity FKL as a simple measure of the free energy of structuring the first shell around an ion complex, relative to the separated ions. It should be noted that the FKL we calculate here will underestimate the full entropy change due to solvent caging about the ions, primarily because of the local (first shell) spatial restriction. Our interest in using this measure is simply to explore trends in local solvent confinement as ions pair in solution.
2.3 Association constants
The equilibrium constant, KA, for ion-ion association in water can be expressed as
| (5) |
| (6) |
where C+ are the free cations, A− are the free anions, and CA are the paired ions. In order to compute these quantities from molecular simulations, we take the Bjerrum approach for defining the KA, which is a 2-state method where the ions are considered either associated or dissociated.61, 63 With strongly interacting multivalent ions, the SIP and 2SIP have been measured to contribute significantly to the overall association constant, and the multistep Eigen-Tamm mechanism is often used to interpret experimental analysis of ion pairing in such systems.9, 61, 64, 65 For univalent ions, it is unclear how appreciably these states contribute, so it is often convenient to use simplified association expressions like Eq. 6 in analyzing ion pairing.26, 34, 65
If the associated condition is taken to be composed entirely of the CIP state, from contact out to the first peak of the PMF, we obtain the following
| (7) |
We consider all other states to be dissociated, so the concentration of free cations and anions is simply the total salt concentration less the CIP concentration. The relative CIP concentration was determined by inverting the PMF into a g(r), integrating the curve out to the first minimum, and calculating the ratio of this integral to the full system integral, which has a known concentration. The error in KA was determined by standard error propagation following integration of the variance of the PMF to obtain the error in the CIP concentration. Additionally, a standard cubic spline interpolation was used to smooth the original PMF; however, integration of the original and interpolated curves produced the same KA results within error. We chose to display the spline interpolated curves in all the PMF graphics and KA values in the relevant tables.
To check that our simulations were not affected by finite size effects (i.e., artifactual interactions from image charges), we calculated PMFs for two test systems, each with twice the number of water molecules, having half the effective salt concentrations of our typical systems. The resulting PMFs had the same shape as the smaller systems, and the KA values were identical within error, indicating that our results do not suffer from substantial finite-size artifacts.
3 Results and Discussion
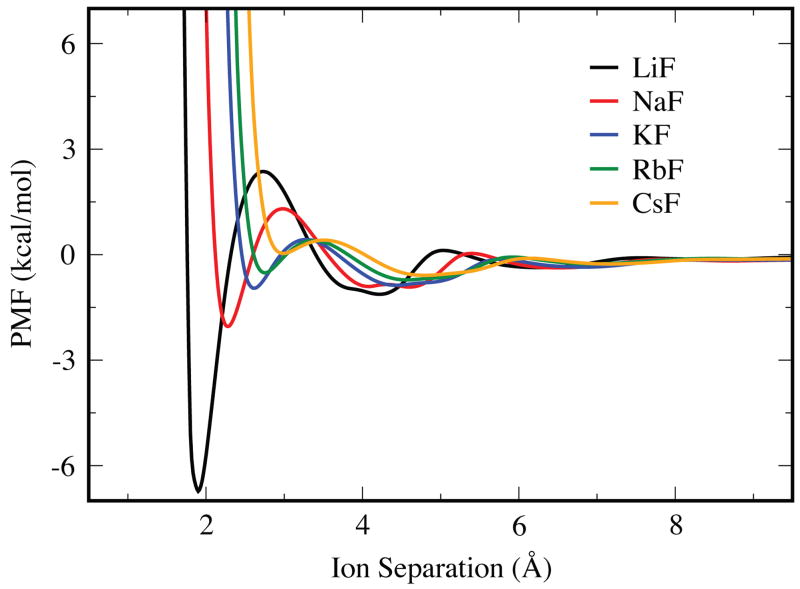
Figure 2 shows a series of PMFs for the five different cations all paired with a given anion, fluoride in this case. Certain general trends are clear from this figure. First, of course, the contact minimum shifts to larger distances with increasing ion radius. Second, the contact minimum is the most stable for the smallest ions (i.e., having the highest charge density). Ion-ion contacts become less stable as the size of the cation increases. The LiF ion pair has a very short contact distance (1.9 Å) and the energy well is deep (−6.8 kcal/mol). Though not shown here, similar trends are seen for the series of anions, with a fixed cation (see the supplementary material for plots of other PMFs).
Figure 2.
PMF curves for the fluoride ion paired with different cations, calculated using the OPLS force field and the TIP3P water model.
3.1 PMFs depend on the force-field parameters
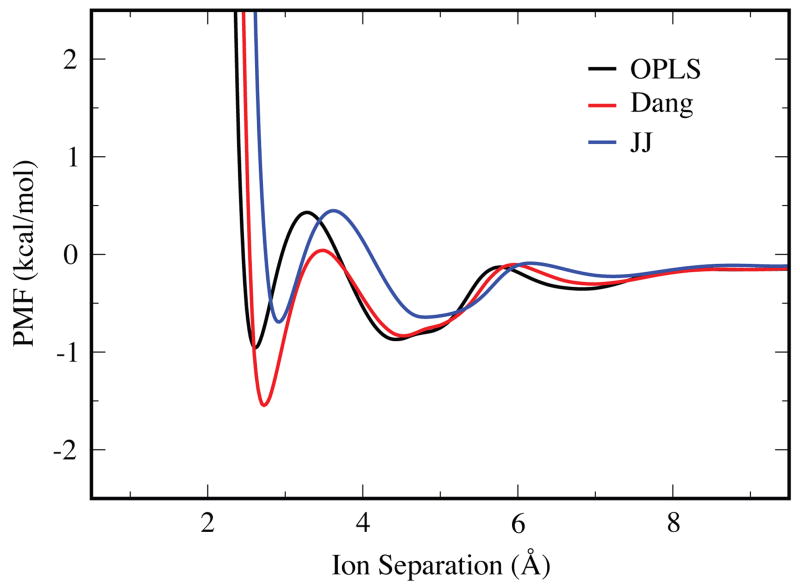
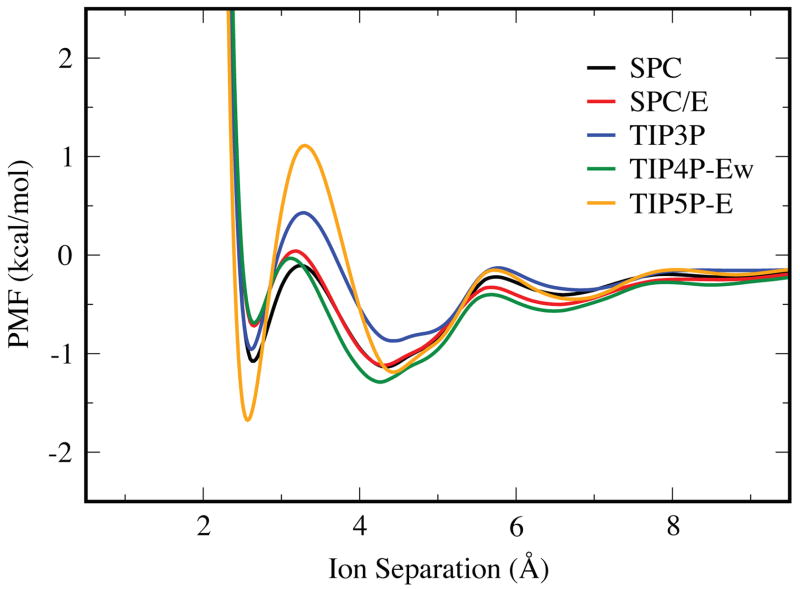
Figure 3 shows how the PMFs depend on the force field, for a given salt (KF) and a given water model (TIP3P). Correspondingly, Figure 4 shows the dependence on the water model for a given salt (KF) and a given force field (OPLS). When moving from the OPLS to the Dang ion force field parameters, the locations of the CIP, SIP, and 2SIP states all shift to slightly larger separations, and the CIP well is more negative by 0.6 kcal/mol. The dependence on water models has been noted before,36 and in that particular case was explained by the larger dipole moment of SPC/E over its predecessor. The higher dielectric constants of TIP5P-E and TIP3P, in the 90’s for both models,53, 66 cause their PMFs to decay more rapidly at larger separations than the other water models, which have dielectric constants in the 60’s.52, 66 Also, the barrier between the CIP and SIP states is much larger for TIP5P-E than for the other water models, indicating a more significant structural change upon transition between these two states.
Figure 3.
The PMF curves for KF using the OPLS, Dang, and JJ force field parameter sets with the TIP3P water model.
Figure 4.
The PMF curves for KF using the SPC, SPC/E, TIP3P, TIP4P-Ew, and TIP5P-E water models in conjunction with the OPLS force field parameters.
3.2 PMFs show the asymmetric solvation response of water
There have been several studies exploring the asymmetric response of explicit water, in regard to solvation of both charged18, 67–71 and neutral molecules.72 These reflect on an early observation by Latimer et al. that hydrated cation radii must be significantly larger than the hydrated anion radii to fit ion hydration data to the Born expression.73 This size asymmetry comes about because water’s charge distribution is asymmetric. The localized positive charges on the hydrogens interact more strongly with the anion than the more diffuse negative charge on the oxygen interacts with the cation. Can this favorability of anion solvation over cation solvation be seen in ion pair PMFs?
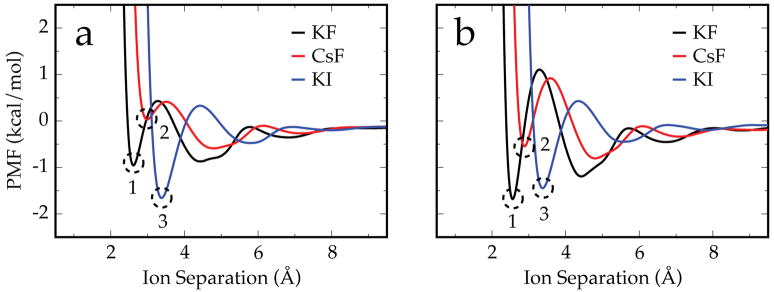
Figure 5a is a collection of PMFs using the same water model (TIP3P) and ion-parameter set (OPLS), while Figure 5b is the same study but using the TIP5P-E water model. Here, the ion-ion electrostatic interaction between the potassium and fluoride ions (which have similar crystal radii74) is weakened by independently replacing them with a larger ion. With TIP3P, substituting potassium with cesium weakens the CIP by ~ 1 kcal/mol. With a weaker ion-ion interaction, cesium is less capable of competing with water for close interactions with the fluoride than the more strongly interacting potassium. When the fluoride is replaced with an iodide, the CIP interaction is ~ 0.7 kcal/mol stronger, despite the overall weaker ion-ion interaction strength! This comes about because TIP3P has more favorable interactions with the fluoride than the potassium, despite their similar crystal radii. With a larger cation, there is less competition for these favorable water-anion interactions, and water can more readily separate the ion pair. With a larger anion, the water-anion interactions are weakened more than the ion-ion interaction. This has a net effect of the surrounding water “squeezing” the KI ion pair together more than the KF ion pair, hence the lower PMF energy. As expected and observed by others,72, 75 this asymmetric solvation response is weaker with TIP5P-E than TIP3P because of its more centralized charge distribution with respect to the Lennard-Jones sphere. It is weakened such that the CIP state of KI (point 3 in Figure 5b) is no longer lower than that of KF (point 1). However, it should be noted that considering the relative depths of the CIP and SIP wells, the equilibrium distribution of KI is still favors more associated states than KF. The specifics of ion association are discussed in greater detail in Section 3.5.
Figure 5.
Free energy surfaces and PMFs for KF, CsF, and KI using OPLS parameters and the a) TIP3P and b) TIP5P-E water models assembled together to illustrate the asymmetric solvation response of water. Replacing the potassium with cesium depletes the CIP state (point 2 is higher than point 1), while replacing fluoride with iodide lowers the energy of and populates the CIP state (point 3 is lower than the other labeled points). As expected, TIP3P shows a stronger asymmetric response than TIP5P-E.
3.3 PMF features reflect the structure of the surrounding water
How can we interpret the ion-ion PMFs in terms of the solvation-shell water structure? Figure 6 shows the results of simulations in which we fixed the ions to be at (a) the CIP, (b) the 1st maximum, (c) the SIP, and (d) the 2SIP, and explored the distribution of water densities and orientations around them. The blue color in the figure indicates water density around the cation. The red color indicates water density around the anion. Orange indicates the density of the bridging waters. The arrows indicate the directions and magnitudes of the average water dipole vectors. The long arrows of the bridging waters indicate that these waters are highly orientationally constrained.
Figure 6, from right to left, (d) to (a), shows the sequence of water structurings as ions approach each other. At step (c), waters enter between the two ions, and are electrostatically ordered. Step (b) is the point of maximal free energy, where the waters are squeezed out from between the ions. Figure (a) shows the CIP. Interestingly, it shows how the water is highly structured on the backside of the anion due to the cation, and on the backside of the cation due to the presence of the anion. We note the average numbers of first shell waters in these states: 9.9 (CIP), 10.6 (1st PMF maximum), and 11.7 (SIP). There is an increase in first shell waters in moving from the CIP to the 1st PMF maximum that is noticeably less than the increase going from this maximum to the SIP state. The form of the PMF is due to a combination of the marginal number of additional ion-water interactions vs. separation, the formation of highly confined and oriented bridging waters, and effects on the cage structure of water. All these energetic costs are relaxed in the SIP state, resulting in the stable trough in the PMF, a picture that agrees with the assertion of Geissler et al. that ion pair dissociation is driven by the insertion of one “bulk” water into the 1st solvent shell about the ion pair.62 By the time the ions reach the 2SIP state separation distance, they have fully occupied 1st shells and the PMF features are due to secondary effects like enhanced hydrogen bonding between the water solvation shells about the separated ions.
Ion pairing leads to orientational restrictions imposed upon neighboring waters. This is seen in Figures 6b and 6c, where the favorably populated bridging water regions exhibit orientation vectors with large magnitudes, indicating that they are significantly more constrained than bulk waters. The relative structuring of the first shell water occupancy in these states is also clear through the FKL values (see Table 2), and it follows an increasing trend going from the more separated 2SIP state (0.38 kcal/mol) to the closely associated CIP state (2.27 kcal/mol). This caging is an important element of the overall interaction between ions in particulate water and helps give rise to the features seen in the PMFs.
Table 2.
The Kullback-Leibler free energy FKL in kcal/mol of and number of waters released nrel from the first solvent shell upon ion pairing for the listed salts and pairing states using the TIP5P-E water model with the OPLS ion parameters.
|
FKL |
nrel |
|||||||
|---|---|---|---|---|---|---|---|---|
| CIP | Peak | SIP | 2SIP | CIP | Peak | SIP | 2SIP | |
| LiF | 1.36 | 2.44 | 2.80 | 0.86 | 4.5 | 3.0 | 1.7 | −0.1 |
| LiI | 1.72 | 2.98 | 1.27 | 0.55 | 4.6 | 3.0 | 1.3 | −0.1 |
| KF | 2.27 | 1.54 | 1.53 | 0.38 | 3.5 | 2.8 | 1.6 | 0.0 |
| KI | 1.12 | 0.99 | 0.75 | 0.35 | 3.4 | 2.3 | 1.2 | 0.0 |
| CsF | 1.10 | 1.27 | 1.13 | 0.29 | 3.5 | 2.7 | 1.5 | 0.0 |
| CsI | 0.78 | 0.89 | 0.68 | 0.42 | 3.4 | 2.3 | 1.2 | 0.0 |
3.4 An ion size comparison
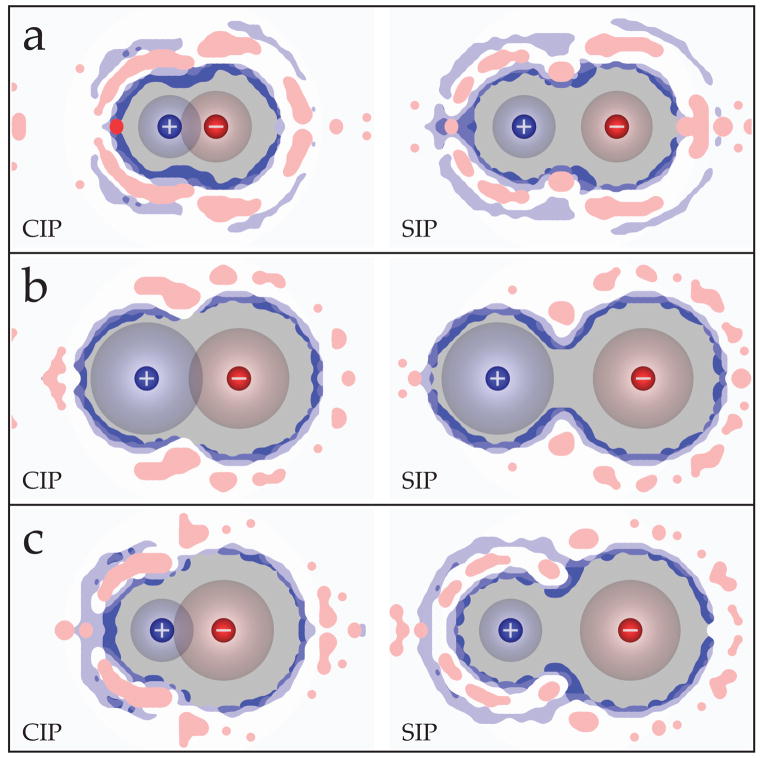
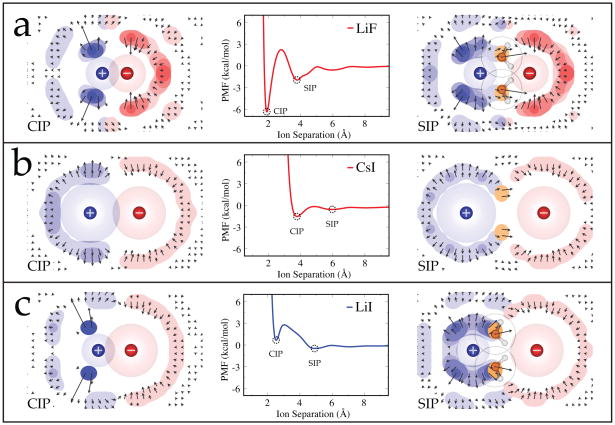
Here we compare ions of different sizes. We analyzed the solvent structuring about representative small-small (LiF), large-large (CsI), and small-large (LiI) ion pairs in greater detail. Figure 7 shows the solvent free energy maps for the CIP and SIP states alongside the PMFs for these salts when using the OPLS ion parameters and TIP5P-E water model. Also of interest is the local perturbation of the water-water hydrogen bonding network due to the presence of the ions in both of these states. To visualize this, we have binned the midpoint and strength of the water-water hydrogen bonds about these ion pairs in the CIP and SIP states, and the results are shown in Figure 8.
Figure 7.
Free energy surfaces for the (left) CIP and (right) SIP states of a) LiF, b) CsI, and c) LiI alongside their potential of mean force curves. The PMF curves for LiF and CsI indicate an enhanced associative behavior, while the PMF curve for LiI shows a preference for the dissociated states. These ions were represented using the OPLS parameters and TIP5P-E was used to model water.
The PMFs in Figure 7 show that the CIP state has a lower minimum than the SIP state in LiF and CsI, whereas the SIP is deeper than the CIP for LiI. The respective depths of these states reflects on the relative associativity of the ions in solution, with a deeper CIP indicative of more associative and a deeper SIP indicative of less associative ion pairs. Collins et al. noted that LiF (small-small) and CsI (large-large) tend to have a greater affinity for one another experimentally in solution than mixed size ion pairs such as LiI (small-large), and they developed a simple physical picture based on these observations to rationalize why LiF and CsI have lower water solubilities (0.1 M and 2.9 M respectively) than salts like LiI (small-large) which has a rather high solubility (12.3 M).39, 42 The associative trends seen in our PMFs is consistent with the physical model they proposed.
Here is the explanation from the simulations. As ions are moved apart from each other in water, there are various energetic components contributing to the overall shape of the PMF. First, the direct electrostatic interaction between the ions will weaken. Second, there will be some favorable water-ion interactions due to the added water molecules that enter the space between the ions. But third, there can also be unfavorable interactions, due to orientational restrictions of first-shell waters beyond the restrictions they would have had in the bulk. Here, we make qualitative estimates of those factors. Why is the CIP state favored relative to SIP when both ions are small (Figure 7a)? Of course, the electrostatic interaction is most favorable in the CIP state because the ion-ion separation distance is smaller than for the SIP state. In addition, water structuring effects, in this case, also stabilize the CIP state. This is reflected in the FKL values for LiF as a free energy increase due to loss of water occupancy entropy moving from the CIP (1.36 kcal/mol) to the SIP (2.80 kcal/mol), a process coupled with three additional waters entering the first shell (see the FKL and nrel values in Table 2). This increase in FKL is primarily due to confinement of these additional waters as bridging waters (shown as gold regions in the SIP plots of Figure 7), which are highly restricted in the SIP state. The water organization around the backsides of the ions are otherwise similar in the CIP and SIP states. Both these small ions act to structure the surrounding hydrogen bonding network (Figure 8a). This structuring corresponds with the occupancy regions in Figure 7a, extending beyond the first hydration shell, and it becomes more extensive as the ions are separated.
In the case of pairs of large ions, such as CsI, the difference in direct electrostatic interactions between CIP and SIP is small, and the water cage-structuring effects are also small. Only two waters are inserted in the first shell here, from CIP to SIP, and this does not contribute much to additional stabilization of the SIP state. Most of the changes due to ion pairing of CsI appear to occur in the hydrogen bonding network of the surrounding water (Figure 8b). The blue contour regions show weakened hydrogen bonding relative to the bulk value (white regions), and these regions locate at the surface of the ions and indicate that the network is looser here because first shell waters have difficulty interacting with each other when they are coordinated to a nearby ion. Possibly of more interest are the red contour regions, where hydrogen bonding is stronger than in the bulk. As the ions are pulled apart (from CIP to SIP), there is a decrease in the population of this network enhancement about the cation, particularly noticeable around where the ions meet. These plots indicate that the pairing of large ions enthalpically stabilizes the surrounding hydrogen bond network.
Finally, we consider the energetics of pairs of ions of mixed sizes, one small and one large. In this case, Table 2 shows that the SIP is stabilized relative to CIP by the gain of 3 waters (for LiI) and also by water cage structures that have lower free energy (FKL). The lesser degree of water structuring in the SIP states can be seen in Figure 7c; compare the higher degree of water localization (dark blue and dark red density contours) in the CIP states than in the SIP states. As expected, this localization is seen in the hydrogen bond network as tight structuring on the backside of lithium in the CIP state and a more continuous/diffuse structuring in the SIP state.
3.5 KA values encapsulate the trends and sensitivities of ion pairing
One way to study ion-ion pairing is through their association constants, KA, in solution.9, 10, 61, 76–80 There are several experimental techniques for determining KA; however, when KA is smaller than 2 M−1, there are errors in measurement, and detection of ion pairing is questionable.61 Such problems often arise for strong electrolytes like the alkali halide salts studied here. This has led to the view that such salts are always entirely dissociated in solution, consistent with early electrolyte theories of Debye, Hückel, and Onsager.2–4 Here, we test this computationally.
Rather than to compute KA from molecular simulations, which would involve huge simulations, here we use the PMF as a simpler surrogate measure of association. It has been shown that KA does not depend on the salt concentration of the simulation.34 To investigate the validity of this method for determining KA values, we compared a small subset of these to values calculated using cluster analysis results. Chen and Pappu have recently performed such an analysis on chloride and bromide salts of Na, K, Rb, and Cs with the TIP3P water model.34 Their KA values are shown in Table 3 alongside the results from this study. While the current numbers tend to be a little lower, all of the trends are reproduced and the results often overlap within the reported errors. This verification indicates that the KA values calculated here are useful measures of ion association.
Table 3.
Ion pairing association constants in units of m−1 calculated from PMFs when using the OPLS and TIP3P parameters compared with the same values obtained from the clustering analysis in Ref. 34.
| salt | KA:this work | KA:Ref. 34 |
|---|---|---|
| NaCl | 0.60(4) | 0.76(5) |
| NaBr | 0.61(4) | 0.72(7) |
| KCl | 0.52(2) | 0.57(4) |
| KBr | 0.53(2) | 0.65(7) |
| RbCl | 0.46(2) | 0.52(5) |
| RbBr | 0.58(3) | 0.60(6) |
| CsCl | 0.42(2) | 0.48(5) |
| CsBr | 0.54(2) | 0.55(4) |
Tables 1 and 2 of the supporting information show the calculated KA values with error for all water models and ion parameter sets considered. For clarity, we have plotted smaller subsets of this data in Figures 9 and 10. Interested readers are encouraged to explore the results in the supporting information for details on more specific cases.
Figure 9.
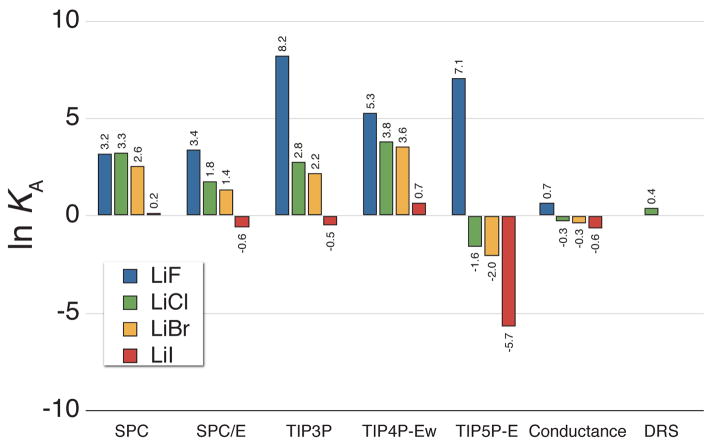
The natural logarithm of the KA values for lithium salts calculated from simulations using all five water models with the OPLS ion parameters alongside the values determined from analysis of conductance [Ref. 78] and dielectric relaxation spectroscopy (DRS) data [Ref. 81].
Figure 10.
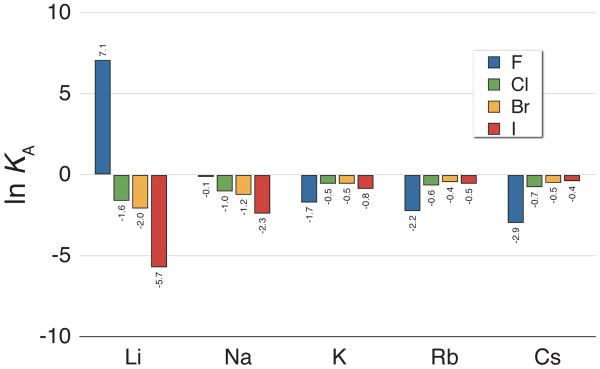
The natural logarithm of the KA values for the alkali halide salts calculated from simulations using the OPLS ion parameters with the TIP5P-E water model.
In Tables 1 and 2 of the supporting information, the salts with the largest KA values are lithium salts, and a portion of these results are shown in Figure 9. This figure highlights some of the differences in ion association as a function of water model, and similar differences are seen between different ion parameter sets (see the supporting information). While all the ion parameters utilized in this study were originally devised in conjunction with specific water models,48, 82 there have been several investigations into the transferability of the various water models in molecular simulations, which have resulted in both opposition to and support for such replacement.18, 36, 72, 83, 84 The results displayed here indicate that while in some cases the water model can be substituted with little change in the physics (e.g. replacing SPC with SPC/E when solvating LiF), many salt combinations will show significantly different ion pairing behavior. For example, using TIP4P-Ew tends to result in more associative ion behavior than other water models. In general, the Dang and JJ ion parameters are more resistant to changes arising from water model replacement than the OPLS parameters. Care should be taken when altering the water model to avoid changing the ion pairing physics in unexpected ways.
One extreme example of altered ion pairing physics can be seen by swapping any of the 3- or 4-point models for TIP5P-E in lithium salt simulations. With the simpler models, LiCl or LiBr show a high propensity to form ion aggregates as the free energy of ion association is always well below kBT, while with TIP5P-E they are highly dissociated. This comes about because in TIP5P-E, electron lone pair sites are located nearer to the surface of the Lennard-Jones sphere than the 3- and 4-point models, which have a single negative charge buried near the center.85 The reduced distance between water’s negative charges and lithium’s central point charge leads to TIP5P-E water having a greater affinity for the lithium ion than the simpler water models. This increase in affinity between TIP5P-E and lithium outweighs the direct electrostatic interactions in LiCl and LiBr, causing these salts to readily dissociate in solution. Here, the tetrahedral geometry of TIP5P-E may result in a more realistic behavior, as one would expect the electron cloud around the nearby water oxygen atoms to become more localized near small cations, similar to the separate discrete electron lone pairs of TIP5P-E. The static nature of the electrostatics in the simulations performed here limits the certainty of such explanations, and this is likely a case where ab initio simulations could provide additional insight.
Considering these results, it is not surprising that ion cluster structures have been observed in simulations of lithium salts using TIP3P and the OPLS parameters.28 The large KA values seen for this water model in Figure 9 are a signature for ion aggregation behavior, and these values are much larger than those seen with TIP5P-E or determined using experimental conductance or dielectric relaxation spectroscopy (DRS) data. One question posed by this difference in lithium association is “what ion pairing behavior is more realistic: highly associated or dissociated?” Large association constants like those seen when using the OPLS and JJ parameters for lithium along with a 3- or 4-point water model should be readily observable with experimental techniques like DRS, and the current experimental studies of LiCl have favored a primarily dissociated picture.81 This indicates that in the case of LiCl, the aggregate structures observed in molecular simulations27, 28 are likely an artifact of chosen modeling parameters.
The connection between these ion pairing results and the specific choice of ion parameters and water model presents a unique challenge for those developing improved forcefields for molecular simulations. As ion parameters are typically developed with a particular water model in mind, an alternative direction would be to tether these options within the chosen forcefield. This is the tactic has recently been taken by Joung and Cheatham in their recent work on correcting ion aggregation issues in the AMBER forcefield,86 and looks to provide a more consistent depiction of ion behavior within this particular forcefield.
Despite the sensitivity of these results to choice of modeling parameters, general trends due to ion size are consistent for any given water model and ion parameter set. Figure 10 is an example case visualizing these trends which follow the ‘law of matching water affinities’ discussed in the previous section. When the cation and anion are both small (LiF), their affinity for one another is much greater than for the surrounding water, causing them to associate (large lnKA). Individually increasing the size of either the cation or anion weakens the ion-ion affinity relative to the ion-water affinities, causing them to dissociate more readily in solution. Finally, the associative behavior can be enhanced by increasing the size of both ions, weakening the overall ion-water affinities relative to the water-water affinities, causing an increased association of large ion pairs. This result is not surprising given that KA is derived from the depth of the CIP state relative to the whole of the PMF, and we saw how the PMF shape and water structure give rise to these trends in the previous section. It is also interesting to note that the NaF and KF KA results seen here and in the supporting information correspond well with previous simulation and experimental results on the preference of sodium ions over potassium ions for interacting with high charge density regions on protein surfaces.87–89
KA values derived from experimental conductance and DRS data at 298 K are shown in the last columns of Table 2 of the supporting information. In these columns, the ion size trend described above is less clear than the stated salt solubilities. In the conductance results, the salts do become more dissociated as ion sizes increase; however, the opposing desolvation effect seen in pairs of large ions is less pronounced. This could be partly do to the somewhat arbitrary convention used in choosing what separation distance tolerance (rmax) defines an ion pair in the conductance data analysis.78–80, 90 It has been shown that the resulting KA values are not independent of the choice of rmax in experiments at standard temperature and pressure.61, 91 This uncertainty in the ion pairing distance definition in conductance measurements makes justification of simulation results based on exact agreement to these numbers somewhat premature. The effects of these definition differences are visible when comparing the conductance numbers to the simulation results, particularly in the case of the lithium and sodium salts. The conductance KA values are generally larger than those seen in the simulations, with the exception of the highly associative lithium OPLS cases shown in Figure 9.
As a final point in comparisons to experimentally measured ion association constants, the sparseness of the DRS results are a result of the instrumental precision not being sufficient to reliably measure KA in these types of electrolyte solutions.10, 81, 92 The only verification that can come from these values is that the ions do not aggregate extensively in solution, and the available salts have KA values less than 3 M−1 (with error bars of similar magnitude to the reported KA values). DRS is a promising technique that has had considerable success in measuring ion pairing,9, 10, 61 and hopefully future improvements will help clarify the degree of ion association in these systems.
4 Conclusions
We have studied the PMFs of different ion pairs in water. We compare different force fields and different water models. We have looked at the different equilibrium states: the contact ion pair (CIP), the free energy peak, the solvent-shared ion pair (SIP) and the solvent-separated ion pair (2SIP). We have also looked at the ion association constants of the full series of alkali halide salts. Our simulations give a microscopic interpretation for the empirical observation of salt pairing effects that are captured by Collins’s ‘law of matching water affinities’. Our simulations are consistent with the view that: (1) small-small ion pairs associate strongly in water because of the strong electrostatics due to the small separations and because water caging stabilizes the CIP, (2) large-large ion pairs tend to associate because the electrostatics is weak and they are held together by hydrophobic-like water caging, and (3) small-large ions readily dissociate in water because the smaller ion interacts with water more strongly than the ion-ion or water-water interactions, leading to a SIP that is more stable than the CIP.
Supplementary Material
Acknowledgments
The authors would like to thank David L. Mobley (University of New Orleans) for insightful discussions and comments on an early version of this manuscript. We also appreciate the financial support provided by NIH grant GM63592 and the Slovenian Research Agency (Research Program 0103-0201).
Footnotes
Supporting Information Available Plots of all the calculated ion pair PMFs and the full tables of ion association constants can be found in the related supporting information. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Arrhenius S. Z Phys Chem. 1887;1:631–648. [Google Scholar]
- 2.Debye P, Hückel E. Phys Z. 1923;24:185–206. [Google Scholar]
- 3.Onsager L. Phys Z. 1926;27:388–392. [Google Scholar]
- 4.Onsager L. Phys Z. 1927;28:277–298. [Google Scholar]
- 5.Bernal JD, Fowler RH. J Chem Phys. 1933;1:515–548. [Google Scholar]
- 6.Rashin AA. J Phys Chem. 1989;93:4664–4669. [Google Scholar]
- 7.Lyubartsev A, Laaksonen A. J Phys Chem. 1996;100:16410–16418. [Google Scholar]
- 8.Hribar B, Southall NT, Vlachy V, Dill KA. J Am Chem Soc. 2002;124:12302–12311. doi: 10.1021/ja026014h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchner R, Chen T, Hefter G. J Phys Chem B. 2004;108:2365–2375. [Google Scholar]
- 10.Wachter W, Kunz W, Buchner R, Hefter G. J Phys Chem A. 2005;109:8675–8683. doi: 10.1021/jp053299m. [DOI] [PubMed] [Google Scholar]
- 11.Dill KA, Truskett TM, Vlachy V, Hribar-Lee B. Annu Rev Biophys Biomol Struct. 2005;34:173–199. doi: 10.1146/annurev.biophys.34.040204.144517. [DOI] [PubMed] [Google Scholar]
- 12.Jungwirth P, Tobias D. Chem Rev. 2006;106:1259–1281. doi: 10.1021/cr0403741. [DOI] [PubMed] [Google Scholar]
- 13.Hess B, Holm C, van der Vegt N. Phys Rev Lett. 2006;96:147801. doi: 10.1103/PhysRevLett.96.147801. [DOI] [PubMed] [Google Scholar]
- 14.Chandrasekhar J, Spellmeyer DC, Jorgensen WL. J Am Chem Soc. 1984;106:903–910. [Google Scholar]
- 15.Lybrand TP, Ghosh I, McCammon JA. J Am Chem Soc. 1985;107:7793–7794. [Google Scholar]
- 16.Jungwirth P, Tobias D. J Phys Chem B. 2002;106:6361–6373. [Google Scholar]
- 17.Grossfield A, Ren P, Ponder J. J Am Chem Soc. 2003;125:15671–15682. doi: 10.1021/ja037005r. [DOI] [PubMed] [Google Scholar]
- 18.Rajamani S, Ghosh T, Garde S. J Chem Phys. 2004;120:4457–4466. doi: 10.1063/1.1644536. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh T, Kalra A, Garde S. J Phys Chem B. 2005;109:642–651. doi: 10.1021/jp0475638. [DOI] [PubMed] [Google Scholar]
- 20.Zangi R, Hagen M, Berne B. J Am Chem Soc. 2007;129:4678–4686. doi: 10.1021/ja068305m. [DOI] [PubMed] [Google Scholar]
- 21.Hess B, van der Vegt NFA. J Chem Phys. 2007;127:234508. doi: 10.1063/1.2812547. [DOI] [PubMed] [Google Scholar]
- 22.Jagoda-Cwiklik B, Vacha R, Lund M, Srebro M, Jungwirth P. J Phys Chem B. 2007;111:14077–14079. doi: 10.1021/jp709634t. [DOI] [PubMed] [Google Scholar]
- 23.Patra M, Karttunen M. J Comp Chem. 2004;25:678–689. doi: 10.1002/jcc.10417. [DOI] [PubMed] [Google Scholar]
- 24.Hess B, Holm C, van der Vegt N. J Chem Phys. 2006;124:164509. doi: 10.1063/1.2185105. [DOI] [PubMed] [Google Scholar]
- 25.Auffinger P, Cheatham TE, Vaiana AC. J Chem Theory Comput. 2007;3:1851–1859. doi: 10.1021/ct700143s. [DOI] [PubMed] [Google Scholar]
- 26.Chen AA, Pappu RV. J Phys Chem B. 2007;111:11884–11887. doi: 10.1021/jp0765392. [DOI] [PubMed] [Google Scholar]
- 27.Degrève L, Mazzé FM. Mol Phys. 2003;101:1443–1453. [Google Scholar]
- 28.Thomas A, Elcock A. J Am Chem Soc. 2007;129:14887–14898. doi: 10.1021/ja073097z. [DOI] [PubMed] [Google Scholar]
- 29.Dang LX, Pettitt BM. J Phys Chem. 1990;94:4303–4308. [Google Scholar]
- 30.Dang LX. J Chem Phys. 1992;96:6970–6977. [Google Scholar]
- 31.Pratt LR, Hummer G, Garcia AE. Biophys Chem. 1994;51:147–165. doi: 10.1016/s0301-4622(99)00018-6. [DOI] [PubMed] [Google Scholar]
- 32.Degreve L, da Silva FLB. J Mol Liq. 2000;87:217–232. [Google Scholar]
- 33.Zhang Z, Duan Z. Chem Phys. 2004;297:221–233. [Google Scholar]
- 34.Chen AA, Pappu RV. J Phys Chem B. 2007;111:6469–6478. doi: 10.1021/jp0708547. [DOI] [PubMed] [Google Scholar]
- 35.Lenart PJ, Jusufi A, Panagiotopoulos AZ. J Chem Phys. 2007;126:044509. doi: 10.1063/1.2431169. [DOI] [PubMed] [Google Scholar]
- 36.Dang LX, Rice JE, Kollman PA. J Chem Phys. 1990;93:7528–7529. [Google Scholar]
- 37.Smith DE, Dang LX. J Chem Phys. 1994;100:3757–3766. [Google Scholar]
- 38.Degreve L, Carlos Borin A, Mazze FM, Rodrigues ALG. Chem Phys. 2001;265:193–205. [Google Scholar]
- 39.Collins KD. Biophys J. 1997;72:65–76. doi: 10.1016/S0006-3495(97)78647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collins KD. Methods. 2004;34:300–311. doi: 10.1016/j.ymeth.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 41.Collins KD. Biophys Chem. 2006;119:271–281. doi: 10.1016/j.bpc.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 42.Collins KD, Neilson GW, Enderby JE. Biophys Chem. 2007;128:95–104. doi: 10.1016/j.bpc.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 43.Jorgensen WL. OPLS Force Fields. In: Schleyer vR, et al., editors. The Encyclopedia of Computational Chemistry. Vol. 3. John Wiley & Sons; New York: 1998. [Google Scholar]
- 44.Dang LX, Garrett BC. J Chem Phys. 1993;99:2972–2977. [Google Scholar]
- 45.Dang LX, Smith DE. J Chem Phys. 1993;99:6950–6956. [Google Scholar]
- 46.Dang LX. J Am Chem Soc. 1995;117:6954–6960. [Google Scholar]
- 47.Koneshan S, Rasaiah J, Lynden-Bell R, Lee S. J Phys Chem B. 1998;102:4193–4204. [Google Scholar]
- 48.Jensen K, Jorgensen W. J Chem Theory Comput. 2006;2:1499–1509. doi: 10.1021/ct600252r. [DOI] [PubMed] [Google Scholar]
- 49.Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J. Simple Point Charge Water. In: Pullman B, editor. Intermolecular Forces. Reidel: Dordrecht; 1981. [Google Scholar]
- 50.Berendsen HJC, Grigera JR, Straatsma TP. J Phys Chem. 1987;91:6269–6271. [Google Scholar]
- 51.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 52.Horn HW, Swope WC, Pitera JW, Madura JD, Dick TJ, Hura GL, Head-Gordon T. J Chem Phys. 2004;120:9665–9678. doi: 10.1063/1.1683075. [DOI] [PubMed] [Google Scholar]
- 53.Rick SW. J Chem Phys. 2004;120:6085–6093. doi: 10.1063/1.1652434. [DOI] [PubMed] [Google Scholar]
- 54.Lindahl E, Hess B, van der Spoel D. J Mol Mod. 2001;7:306–317. [Google Scholar]
- 55.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 56.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 57.Guàrdia E, Rey R, Padró JA. Chem Phys. 1991;155:187–195. [Google Scholar]
- 58.Hess B. J Chem Phys. 2002;116:209–217. [Google Scholar]
- 59.Lyubartsev AP, Laaksonen A. Phys Rev E. 1995;52:3730–3737. doi: 10.1103/physreve.52.3730. [DOI] [PubMed] [Google Scholar]
- 60.Ullner M, Woodward CE, Jonsson B. J Chem Phys. 1996;105:2056–2065. [Google Scholar]
- 61.Marcus Y, Hefter G. Chem Rev. 2006;106:4585–4621. doi: 10.1021/cr040087x. [DOI] [PubMed] [Google Scholar]
- 62.Geissler PL, Dellago C, Chandler D. J Phys Chem B. 1999;103:3706–3710. [Google Scholar]
- 63.Justice MC, Justice JC. J Solution Chem. 1976;5:543–561. [Google Scholar]
- 64.Eigen M, Tamm K. Z Elektrochem. 1962;66:93–106. [Google Scholar]
- 65.Pottel R, Haller J, Kaatze U. Multistep association of cations and anions. The Eigen-Tamm mechanism some decades later. In: Kurz T, Parlitz U, Kaatze U, editors. Oscillations, Waves, and Interactions. Universitätsverlag Göttingen; Göttingen: 2007. [Google Scholar]
- 66.van der Spoel D, van Maaren P. J Chem Theory Comput. 2006;2:1–11. doi: 10.1021/ct0502256. [DOI] [PubMed] [Google Scholar]
- 67.Rashin AA, Honig B. J Phys Chem. 1985;89:5588–5593. [Google Scholar]
- 68.Roux B, Yu HA, Karplus M. J Phys Chem. 1990;94:4683–4688. [Google Scholar]
- 69.Hummer G, Pratt L, Garcia A. J Phys Chem. 1996;100:1206–1215. [Google Scholar]
- 70.Ashbaugh HS. J Phys Chem B. 2000;104:7235–7238. [Google Scholar]
- 71.Chorny I, Dill KA, Jacobson MP. J Phys Chem B. 2005;109:24056–24060. doi: 10.1021/jp055043m. [DOI] [PubMed] [Google Scholar]
- 72.Mobley DL, Barber AE, II, Fennell CJ, Dill KA. J Phys Chem B. 2008;112:2405–2414. doi: 10.1021/jp709958f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Latimer WM, Pitzer KS, Slansky CM. J Chem Phys. 1939;7:108–111. [Google Scholar]
- 74.Pauling L. J Am Chem Soc. 1927;49:765–790. [Google Scholar]
- 75.Schurhammer R, Engler E, Wipff G. J Phys Chem B. 2001;105:10700–10708. [Google Scholar]
- 76.Grunwald E. Anal Chem. 1954;26:1696–1701. [Google Scholar]
- 77.Denison JT, Ramsey JB. J Am Chem Soc. 1955;77:2615–2621. [Google Scholar]
- 78.Fuoss RM. Proc Natl Acad Sci USA. 1980;77:34–38. doi: 10.1073/pnas.77.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hefter GT, Salomon M. J Solution Chem. 1996;25:541–553. [Google Scholar]
- 80.Bešter-Rogač M, Neueder R, Barthel J. J Solution Chem. 1999;28:1071–1086. [Google Scholar]
- 81.Wachter W, Fernandez S, Buchner R, Hefter G. J Phys Chem B. 2007;111:9010–9017. doi: 10.1021/jp072425e. [DOI] [PubMed] [Google Scholar]
- 82.Åqvist J. J Phys Chem. 1990;94:8021–8024. [Google Scholar]
- 83.Nutt DR, Smith JC. J Chem Theory Comput. 2007;3:1550–1560. doi: 10.1021/ct700053u. [DOI] [PubMed] [Google Scholar]
- 84.Mobley DL, Dumont E, Chodera JD, Dill KA. J Phys Chem B. 2007;111:2242–2254. doi: 10.1021/jp0667442. [DOI] [PubMed] [Google Scholar]
- 85.Mahoney MW, Jorgensen WL. J Chem Phys. 2000;112:8910–8922. [Google Scholar]
- 86.Joung IS, Cheatham TE. J Phys Chem B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vrbka L, Vondrášek J, Jagoda-Cwiklik B, Vácha R, Jungwirth P. Proc Natl Acad Sci USA. 2006;103:15440–15444. doi: 10.1073/pnas.0606959103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Uejio JS, Schwartz CP, Duffin AM, Drisdell WS, Cohen RC, Saykally RJ. Proc Natl Acad Sci USA. 2008;105:6809–6812. doi: 10.1073/pnas.0800181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Aziz EF, Ottosson N, Eisebitt S, Eberhardt W, Jagoda-Cwiklik B, Vácha R, Jungwirth P, Winter B. J Phys Chem B. 2008;112:12567–12570. doi: 10.1021/jp805177v. [DOI] [PubMed] [Google Scholar]
- 90.Paterson R, Jalota SK, Dunsmore HS. J Chem Soc A. 1971:2116–2121. [Google Scholar]
- 91.Duer WC, Robinson RA, Bates RG. J Solution Chem. 1976;5:765–771. [Google Scholar]
- 92.Chen T, Hefter G, Buchner R. J Phys Chem A. 2003;107:4025–4031. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.