Abstract
While the recognition of microbial infection often occurs at the cell surface via Toll-like receptors, the cytosol of the cell is also under surveillance for microbial products that breach the cell membrane. An important outcome of cytosolic recognition is the induction of IFNα and IFNβ, which are critical mediators of immunity against both bacteria and viruses. Like many intracellular pathogens, a significant fraction of the transcriptional response to Mycobacterium tuberculosis infection depends on these type I interferons, but the recognition pathways responsible remain elusive. In this work, we demonstrate that intraphagosomal M. tuberculosis stimulates the cytosolic Nod2 pathway that responds to bacterial peptidoglycan, and this event requires membrane damage that is actively inflicted by the bacterium. Unexpectedly, this recognition triggers the expression of type I interferons in a Tbk1- and Irf5-dependent manner. This response is only partially impaired by the loss of Irf3 and therefore, differs fundamentally from those stimulated by bacterial DNA, which depend entirely on this transcription factor. This difference appears to result from the unusual peptidoglycan produced by mycobacteria, which we show is a uniquely potent agonist of the Nod2/Rip2/Irf5 pathway. Thus, the Nod2 system is specialized to recognize bacteria that actively perturb host membranes and is remarkably sensitive to mycobacteria, perhaps reflecting the strong evolutionary pressure exerted by these pathogens on the mammalian immune system.
Author Summary
Bacterial and viral infection stimulates production of several cytokines and chemokines that are thought to protect the host against infection. The bacterial strain known to cause tuberculosis elicits production of type I interferons, yet it was unclear how the bacteria isolated within the cell was capable of stimulating this host response. This study reveals that the bacteria use a specialized system to cause damage to these cellular compartments and release bacterial products that activate intracellular innate immune pathways. In this work, we demonstrate that Nod2, Rip2, Tbk-1, Irf3 and Irf5 proteins cooperate to produce type I interferons. Understanding how these pathways are mediated is likely to aid in the design of more effective tuberculosis vaccines.
Introduction
Mycobacterium tuberculosis (Mtb), the causative agent of human tuberculosis, is an exquisitely adapted obligate human pathogen that is thought to persist within as many as one billion individuals worldwide [1]. This bacterium's ability to survive and replicate inside a modified phagosomal compartment of host macrophages is central to the pathogenesis of this disease [2]. While residing at this site, Mtb is able to persist for decades. However, a robust cell-mediated immune response effectively inhibits bacterial replication in approximately 90% of otherwise healthy individuals, and the infection can be controlled indefinitely. Deficits in this immune response result in progressive bacterial replication, necrosis of infected lung tissue, and spread to other individuals. Thus, like many other pathogens that cause chronic infections, the long-term survival of Mtb, depends on a delicate balance between bacterial virulence and host immunity.
Immunity to tuberculosis depends on both the innate and adaptive responses of the host. Initial recognition of the bacterium is mediated by pattern recognition receptors (PRR) such as Toll-like receptors (TLRs) [3],[4] or nucleotide binding oligomerization domain (NOD)-like receptors (NLRs) [5],[6], both of which recognize conserved microbial structures known as pathogen associated molecular patterns (PAMPs). TLRs monitor the extracellular environment and endosomal compartments, and recognize a variety of microbial components including bacterial lipoprotein, peptidoglycan, CpG DNA, and double- and single-stranded RNA [4]. NLRs constitute a more diverse family of approximately 25 proteins, including the caspase-recruiting domain (CARD)-containing Nod1, Nod2 and NLRCs, the pyrin (PYR) domain-containing NLRPs and the baculovirus-inhibitor-of-apoptosis-repeats (BIRs)-containing NLRBs. Nod1 and Nod2 reside in the cytosol and recognize microbial products in this compartment [7]. While the functions of most NLR's remain undefined, the Nod1 and Nod2 proteins have been shown to respond to bacterial cell wall fragments. The Nod1 protein recognizes a fragment of peptidoglycan (PGN) containing the dipeptide γ-d-glutamyl-meso-diaminopimelic acid (iE-DAP) produced by Gram-negative and some Gram-positive bacteria. Nod2 recognizes muramyl dipeptide (MDP) present on most types of PGN [8],[9],[10],[11]. While the recognition of these common forms of peptidoglycan have been extensively studied, bacteria modify their cell walls in a myriad of ways and the effects of these modifications on Nod1/2 recognition are only beginning to be appreciated (reviewed in [12],[13],[14]). For example, Listeria monocytogenes removes a common N-acetyl moiety from the glucosamine of its peptidoglycan, which renders the cell wall resistant to host lysozyme and thereby inhibits bacterial recognition by Nod1 [15]. In contrast, mycobacteria, replace the N-acetyl group of the muramic acid of MDP with a N-glycolyl moiety[16],[17], and this modification significantly increases the potency of this compound as a Nod2 agonist (Coulombe, F. and Behr, M.A. unpublished data).
Nod1 and Nod2 functions depend on a downstream signaling component, Rip2, which belongs to a protein family currently consisting of 7 members [18]. Like the prototype Rip1, Rip2 contains an N-terminal serine threonine kinase domain followed by an intermediate region and a C-terminal caspase recruitment domain (CARD). Rip2 has been shown to be essential for cytosolic Nod1/2 signaling, and its overexpression stimulates NF-κB activity and induces apoptosis [19],[20]. We have shown that Rip2 is stably modified with ubiquitin in cells treated with the Nod2 agonist MDP [21]. This modification is required for Nod1-mediated NF-κB activation [22], indicating that stable polyubiquitination is a critical component of this signaling cascade.
Intact Mtb bacilli are recognized by both TLRs and NLRs, which cooperatively respond to infection and synergistically induce NF-κB activation [23]. However, a large fraction of the transcriptional response to Mtb, including many immunologically important proteins, such as the chemokines RANTES and IP-10, and the inducible nitric oxide synthase enzyme NOS2 that is critical for mycobacterial immunity, are induced independently of TLR2/4 and the adapter proteins MyD88, MAL and TRIF. Instead these responses rely on autocrine or paracrine signaling via type I interferons (IFNα/β), which are induced through largely undefined pathways [24].
Despite the ability of cell surface localized TLR4 to trigger IFNα and IFNβ transcription, existing evidence indicates that during genuine bacterial infections, this response instead requires the recognition of bacterial products in the cytosol. This has been most clearly demonstrated for pathogens that replicate in the host cell cytosol, such as Listeria monocytogenes and Francisella tularensis. In both cases, the bacterium must disrupt the phagosomal membrane and escape into the cytosol in order to trigger the type I IFN response in resting macrophages [25],[26],[27]. Despite its residence in the phagosome, Mtb still induces rapid and robust IFNα/β transcription, and this response depends on a specialized secretion system of the bacterium, ESX1 [28]. This system has been suggested to contribute to the perturbation of the phagosomal membrane [29],[30],[31], indicating that cytosolic recognition might be critical for IFNα/β responses to diverse bacterial pathogens including Mtb.
The primary pathways leading to IFNα/β induction upon bacterial infection remain obscure. Since transfection of DNA into the cytosol of macrophages can induce a Tbk-1 and Irf3-dependent IFNα/β response similar to that seen upon L. monocytogenes infection, bacterial DNA has been implicated as the eliciting stimulus [32]. Two different cytosolic DNA sensors have been identified, DAI [33] and AIM2 [34], but their importance during bacterial infections remains to be demonstrated. While Nod2 recognition of MDP is not absolutely required for IFNα/β production [35], it has been shown to synergize with the cytosolic DNA response and enhance IFN production during both L. monocytogenes and Mtb infection [36]. However, Nod2 stimulation alone is thought to be insufficient to induce type I IFN production [36].
In sum, while a large fraction of the macrophage response to Mtb infection depends on type I IFN [24] and therefore is likely to rely on a cytosolic signaling pathway, the bacterial products recognized and the pathways involved remain unknown. We previously found that Mtb infection of macrophages triggers Rip2 polyubiquitination in a TLR and MyD88 independent manner [21]. We now show that this stimulation is due to the ESX1-dependent entry of bacterial products into the cytosol where they are recognized by Nod2, implicating MDP as the relevant PAMP. Unexpectedly, this results in IFNα/β production that is dependent on a novel pathway consisting of Nod2, Rip2, Tbk1, and Irf5. This work is the first to implicate NLRs in IRF activation and to suggest a role for Irf5 in anti-bacterial innate immune responses. Furthermore, we found that the unusual N-glycolyl MDP produced by Mtb was 10–100 fold more potent than the commonly studied N-acetylated MDP produced by most bacteria, and that only N-glycolyl MDP could stimulate Rip2-dependent IFNα/β transcription in the absence of other stimulants. Thus, the mammalian Nod2 pathway appears to be remarkably sensitive to mycobacterial MDP and responds to infection by triggering the production of type I interferon, which is responsible for a significant component of the transcriptional response to Mtb infection.
Results
Mycobacterium tuberculosis infection stimulates the ubiquitin modification of Rip2 via the Nod2 protein
The ability of Mtb to rapidly modify macrophage signaling and vesicular sorting pathways [2] suggests that bacterial products gain access to the cytosol soon after phagocytosis. These products are, in turn, likely to be sensed by the host and trigger the innate immune response. Previously, we demonstrated that Mtb rapidly induces the TLR2/4 independent polyubiquitination of the Rip2 protein [21], an event that could represent the initiation of cytosolic recognition. To characterize these events in more detail, we infected the mouse macrophage cell line RAW 264.7 or primary bone marrow derived macrophages (BMDM) with live or heat killed Mtb. In both cell types, we observed that infection with live, but not heat-killed, Mtb stimulated the rapid polyubiquitination of Rip2. The Mtb-induced ubiquitin modification reached maximal levels within 1 hour post-infection and declined by 4 hours (Figure 1A). Furthermore, pretreatment of cells with cytochalasin D to inhibit phagocytosis reduced Rip2 polyubiquitination in a dose-dependent manner (Figure 1B), indicating that the bacteria must be both live and intracellular to initiate this response.
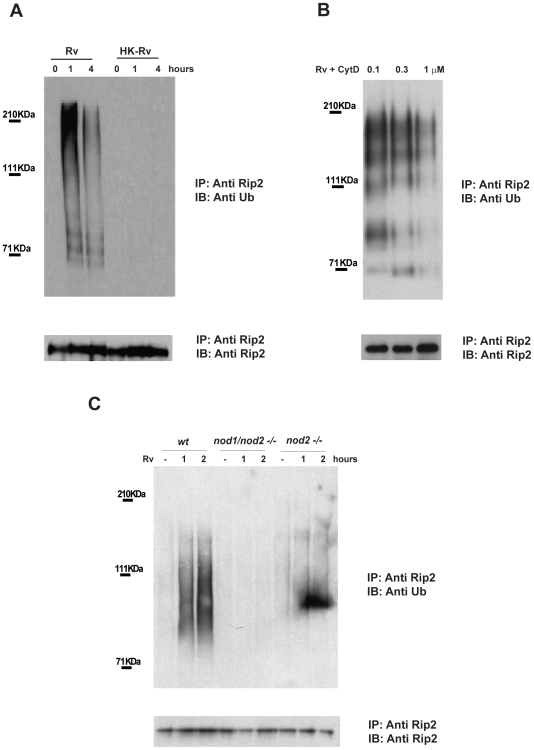
Figure 1. Live, intracellular Mycobacterium tuberculosis stimulates Rip2 polyubiquitination.
A. Live, but not heat killed Mtb (Rv) stimulates Rip2 polyubiquitination. Bone marrow-derived macrophages (BMDM) were infected with live virulent (Rv) or heat killed (HK-Rv) strains of M. tuberculosis (strain H37Rv) for the indicated times. Polyubiquitinated Rip2 protein was detected by immunoprecipitating the cell lysates with an anti-Rip2 antibody followed by immunoblotting with an anti-ubiquitin antibody. Immunoprecipitates were also immunoblotted with a Rip2 antibody (lower panel) to insure that equal amounts of protein were immunoprecipitated. B. The cell permeable mycotoxin Cytochalasin D inhibits Rip2 polyubiquitination upon Mtb infection. The murine RAW 264.7 macrophage cell line was pretreated with cytochalasin D (cytD) for 1 hour at the indicated concentrations before being infected with virulent Mtb H37Rv (Rv) for 1 hour. Polyubiquitinated Rip2 proteins were detected as described above. Treatment with 1 µM cytD caused a 95% decrease in phagocytosis, as described in the Materials and Methods section. C. Mycobacterium tuberculosis-induced Rip2 polyubiquitination is Nod2-dependent. Wildtype, nod1/nod2−/− or nod2−/− bone marrow-derived macrophages cell lines were generated (see Materials and Methods) and either left uninfected (−) or infected with live virulent M. tuberculosis (H37Rv) for the indicated times. Polyubiquitinated Rip2 proteins were detected as described above.
Since Nod1 and Nod2 have been implicated in the cytosolic recognition of mycobacterial components [23], we sought to determine if Rip2 polyubiquitination depended on these proteins. In contrast to cells from wild type mice, inducible Rip2 polyubiquitination was not observed in macrophages derived from mice lacking Nod1 and Nod2, and was greatly reduced in cells lacking only Nod2 (Figure 1C). These data confirmed that intracellular Mtb is recognized by a Nod2-dependent pathway and that this protein is required for the stable ubiquitination of Rip2.
The mycobacterial ESX1 system is required for Nod recognition
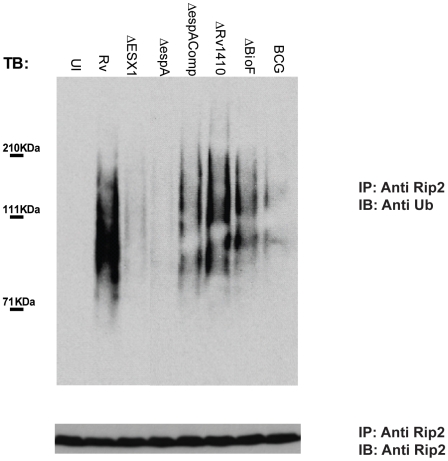
Live intracellular Mycobacteria were required to stimulate the Nod-Rip2 pathway, indicating that the bacterium actively participated in this process, likely via the translocation of bacterial products into the cytosol. A specialized protein secretion system, encoded by the ESX1 locus, has been implicated in the perturbation of the host membranes [29],[30],[37] and for stimulation of the type I IFN response [28] and inflammasome activation [38], suggesting that this system might contribute to cytosolic recognition via Nod proteins. In order to test this hypothesis, we infected the mouse macrophage cell line RAW 264.7 with wild type Mtb or mutants lacking ESX1 function. No induction in Rip2 polyubiquitination was observed upon infection with a strain of Mtb harboring the “RD1” mutation, which deletes a portion of the ESX1 locus [39]. Similarly, a mutant lacking espA, a distally-encoded gene that is required for ESX1-mediated secretion [40], also failed to elicit this response (Figure 2). The phenotype of the latter mutant could be complemented by the expression of espA from a plasmid vector, demonstrating that the inability to stimulate Rip2 polyubiquitination was linked to the espA mutation. Furthermore, M. bovis BCG, an attenuated vaccine strain carrying the RD1 deletion and therefore lacking ESX1 function [39], was unable to stimulate Rip2 polyubiquitination. While all of these ESX1 mutants are less virulent than wild type bacteria, the lack of Nod2-Rip2 stimulation did not appear to be a nonspecific effect of attenuation. Two unrelated bacterial mutants that are unable to grow intracellularly, a biotin auxotroph (ΔbioF [41]) and a small molecule efflux mutant (TN::rv1410c [42]), robustly stimulated this response (Figure 2). Taken together, these observations indicate that a functional ESX1 secretion system is specifically required for Nod2 stimulation.
Figure 2. Rip2 polyubiquitination upon Mtb infection requires ESX-1.
The murine RAW 264.7 macrophage cell line was left uninfected (UI), infected with Mtb H37Rv (Rv), with ESX1 mutant strains of Mtb (ΔESX1, ΔespA), with a complemented ΔespA-C strain, with the attenuated vaccine strain Mycobacterium bovis BCG or with the unrelated attenuated mutants Tn::rv1410 and ΔbioF. Cell lysates were immunoprecipitated with an anti-Rip2 antibody followed by immunoblotting with an anti-ubiquitin antibody. Immunoprecipitates were also immunoblotted with a Rip2 antibody to insure that equal amounts of protein were immunoprecipitated.
Membrane damage allows Nod2-mediated recognition of ESX1 mutants
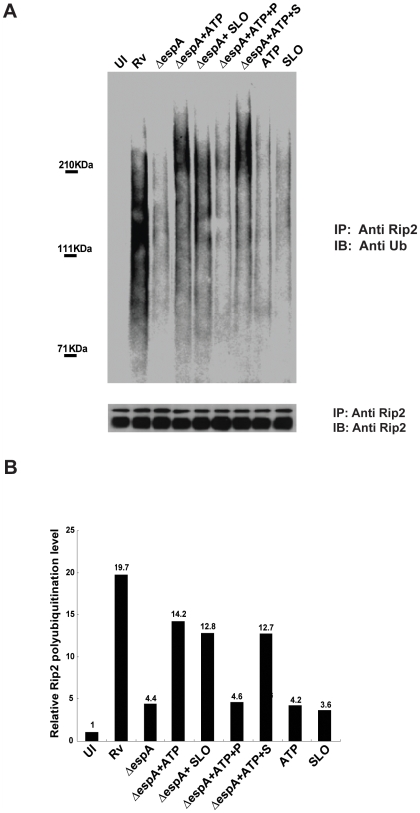
Since the Mtb-induced Rip2 polyubiquitination required ESX1, we hypothesized that this system might be responsible for the release of Nod2 ligands into the cytosol, perhaps via the disruption of vacuolar membrane integrity. However, it also remained possible that ESX1-deficient strains simply lacked a critical PAMP or other Nod2 stimulating activity. To distinguish between these possibilities, we investigated whether ESX1 function could be complemented by two exogenous membrane-disruptive activities. Streptolysin O (SLO) is a cholesterol-dependent toxin that introduces pores directly into mammalian membranes. Pores can also be introduced by adding ATP to macrophages, resulting in stimulation of the P2X7 receptor and the subsequent opening of the hemichannel, pannexin-1 (PANX1) [43]. We observed that membrane perturbation by either of these two methods resulted in robust Rip2 polyubiquitination upon infection with espA-deficient bacteria, which were otherwise unable to induce this response (Figure 3). The involvement of PANX1 in the ATP-facilitated Rip2 ubiquitination was verified by the addition of a competitive inhibitory peptide of the PANX1 pore. This peptide, but not a scrambled control peptide, inhibited Rip2 polyubiquitination to levels observed in cells infected with the ΔespA mutant (Figure 3). While the K+ flux subsequent to membrane damage has been found to stimulate NLRs in some circumstances [5], we found that the addition of ATP or SLO alone resulted in a minimal response. These data indicate that SLO, PANX1 and ESX1 are all likely to promote Nod2 pathway activation via a similar mechanism, by facilitating the release of a stimulatory mycobacterial component into the cytosol. Since this pathway appears to be specific for peptidoglycan fragments, mycobacterial MDP-containing fragments were the most likely candidates.
Figure 3. Membrane damage allows Nod2-mediated recognition of ESX1 mutants.
A. The mouse RAW 264.7 macrophage cell line was was treated with ATP (5 mM) or SLO (5 µg/ml) for 15 minutes before infection with virulent wild type (Rv) or ESX1 mutant (ΔespA) strains of Mtb. After one hour, cell lysates were immunoprecipitated with a Rip2 antibody followed by immunoblotting with an anti-ubiquitin antibody. Immunoprecipitates were immunoblotted with a Rip2 antibody to insure that equal amounts of protein were immunoprecipitated. To verify that ATP was acting via the PANX1 protein, cells were pretreated with 500 µM of either a PANX1 blocking peptide (+P) or control scrambled peptide (+S) for 15 minutes prior to ATP addition. B. Relative abundance of Rip2 polyubiquitination in each sample as determined by densitometry of the results in panel A. All values were calculated relative to the uninfected sample (UI).
The cytosolic Nod2-Rip2 pathway contributes to type I IFN production upon Mtb infection
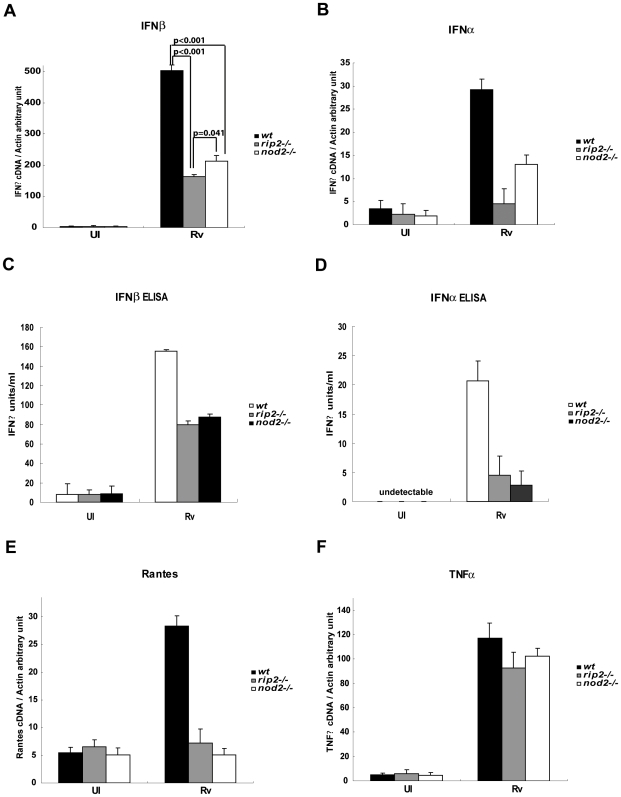
The inability of ESX1 mutants to stimulate either the Nod2-Rip2 pathway or the type I IFN response [28] led us to hypothesize that the Nod2 pathway may mediate type I IFN expression in this system. To investigate a potential link between Nod2 and IFNα/β, we infected Nod2- or Rip2-deficient macrophages with Mtb, and measured the induction of IFNα and IFNβ mRNAs using real time PCR (qRT-PCR). In the absence of Rip2, IFNβ induction was reproducibly reduced approximately 3-fold, whereas IFNα induction was almost completely abrogated (Figure 4A and B). Nod2 deficiency had a similar effect on both IFNα and IFNβ transcription, consistent with its requirement for Rip2 polyubiquitination. Nod1 appears to play no role in this pathway, as nod1−/− macrophages produced wild type levels of IFNβ (Figure S1). The decreases in mRNA abundance observed in rip2−/− and nod2−/− cells were reflected in a similar decrease in protein production, as measured by ELISA (Figure 4C and D).
Figure 4. Type I Interferon production upon Mtb infection is reduced in Rip2- and Nod2-deficient macrophages.
A, B, E, and F. BMDM derived from wt, rip2−/− and nod2−/− mice were infected with Mtb (MOI 10) for 4 h. RNA was harvested and IFNα, IFNβ, RANTES and TNFα mRNA levels were quantified using real time PCR. Gene expression is reported as copy number per 1,000 copies of β-actin. Samples were assayed in triplicate; error bars represent the standard deviation. The experiment shown is representative of at least three. Statistical evaluation was performed using an unpaired Student's t test. p-values>0.05 are reported as “n.s.” (i.e. not significant). C and D. BMDM derived from wt, rip2−/− and nod2−/− mice were infected with Mtb (MOI 10) for 18 h, the amount of IFNα and IFNβ released in the supernatant was quantified by ELISA. Samples were assayed in triplicate; error bars represent the standard deviation. N.D. indicates not detected, that is the actual value is below zero in standard curve.
In order to assess the importance of Nod2 and Rip2 to the downstream IFNα/β-dependent macrophage response, we quantified the induction of RANTES mRNA, which depends on type I IFN secretion and signaling via the IFNαβ receptor (IFNAR1) in this infection model [24]. We found that in the absence of Rip2 or Nod2, Mtb infection failed to induce RANTES expression (Figure 4E). These data suggest that the effect of a Rip2 deficiency on downstream type I IFN responses may be even more pronounced than the IFNβ mRNA levels indicate. In contrast, TNFα mRNA levels were unaffected by Nod2- or Rip2-deficiency (Figure 4F) indicating that other pattern recognition pathways remained responsive to Mtb in these cells.
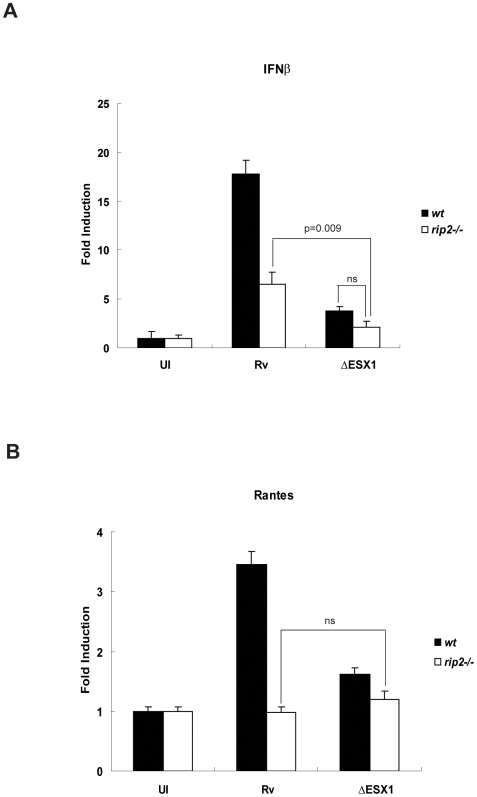
Consistent with previous work [28], we found that infection with ESX1 mutant bacteria induced significantly less IFNβ and RANTES expression than wild type bacteria (Figure 5). To test whether ESX1-mediated type I IFN expression was mediated solely via Rip2, we infected Rip2-deficient macrophages with ESX1 mutant bacteria and quantified IFNβ and RANTES mRNA levels. We found that in the absence of Rip2, the loss of ESX1 function resulted in a further decrease in IFNβ mRNA levels (Figure 5A), suggesting the presence of an additional host pathway(s) that contribute to IFNβ induction. However, Rip2 deletion had no significant effect in the absence of ESX1 (Figure 5B), supporting our biochemical evidence that NOD2 stimulation depends entirely the ESX1-dependent delivery of stimulants into the cytosol.
Figure 5. Multiple cytosolic pathways lead to IFNβ induction.
A. BMDM derived from wt, rip2−/− and nod2−/− mice were infected with virulent Mtb H37Rv (Rv) or with an ESX1 mutant (ΔESX1) at an MOI of 10 for 4 h. RNA was harvested, and IFNβ mRNA levels were quantified using real time PCR. Gene expression of IFNβ is reported as copy number per 1,000 copies of β-actin. Samples were assayed in triplicate; error bars represent the standard deviation. The experiment shown is representative of at least three. B. BMDM derived from wt, rip2−/− and nod2−/− mice were infected with virulent Mtb H37Rv (Rv) or with an ESX1 mutant (ΔESX1) at an MOI of 10 for 4 h. RNA was harvested, and RANTES mRNA levels were quantified using real time PCR. Gene expression of RANTES is reported as copy number per 1,000 copies of β-actin. Samples were assayed in triplicate; error bars represent the standard deviation. The experiment shown is representative of at least three.
The N-glycolylated MDP produced by Mtb is a potent stimulator of the Nod2-mediated type I IFN response
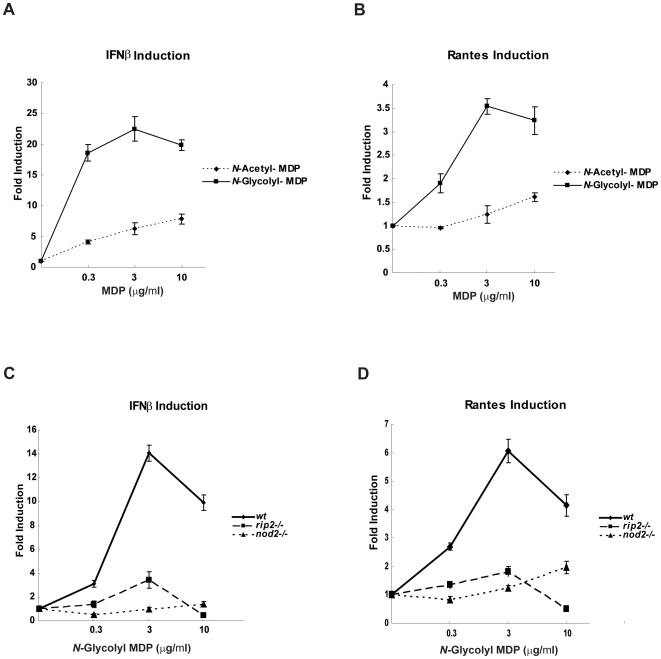
While our data indicated that a significant fraction of the IFNα/β response could be attributed to the Nod2-Rip2 pathway, it has been suggested that MDP stimulation alone is unable to induce type I IFNs and can only augment responses triggered by other pathways [36]. Indeed, we also found that the N-acetylated MDP that is commonly used to stimulate Nod2 was a very poor inducer of IFNβ and RANTES expression (Figure 6A and B). However, our preliminary studies investigating Rip2 polyubiquitination indicated that Mtb was a particularly potent stimulator of this response [21], and therefore we reasoned that this could be due to the N-glycolylated form of MDP produced by Mtb. To determine if this form of MDP was sufficient to induce type I IFN responses, we compared the ability of N-acetyl- and N-glycolyl-MDP to stimulate IFNβ expression. In contrast to N-acetyl MDP, treatment with the N-glycolylated form stimulated a robust IFNβ response, which was entirely dependent on Rip2 and Nod2 (Figure 6). In addition, at least 30-fold less N-glycolyl-MDP was necessary to stimulate the IFNβ transcription. Thus, the Nod2/Rip2 pathway alone is sufficient to induce the production of the IFN response when stimulated with this potent form of MDP.
Figure 6. Nod2 stimulation is sufficient for type I IFN induction.
A and B. N-Glycolyl MDP is more potent than the more common N-Acetylated derivative. The macrophage cell line RAW 264.7 was treated with the indicated concentrations of N-Glycolyl-MDP or N-Acetyl-MDP for 4 h. RNA was harvested, and IFNβ and RANTES mRNA levels were quantified using real time PCR. Gene expression of IFNβ (A) and RANTES (B) was normalized to β-actin then normalized to untreated control to estimate fold induction. Samples were assayed in triplicate; error bars represent the standard deviation. The experiment shown is representative of at least three. C and D. The N-Glycolyl-MDP-induced type I IFN response is Rip2- and Nod2- dependent. Wt, rip2−/− and nod2−/− transformed macrophage cell lines were treated for 4 hours with increasing concentrations of N-Glycolyl-MDP. RNA was harvested and IFNβ and RANTES mRNA levels were quantified using real time PCR. Gene expression of IFNβ (C) and RANTES (D) was normalized to β-actin and compare to untreated control to establish the fold induction. Samples were assayed in triplicate; error bars represent the standard deviation. The experiment shown is representative of at least three.
Induction of the host type I IFN response upon Mtb infection requires the Tbk1 kinase and Irf5
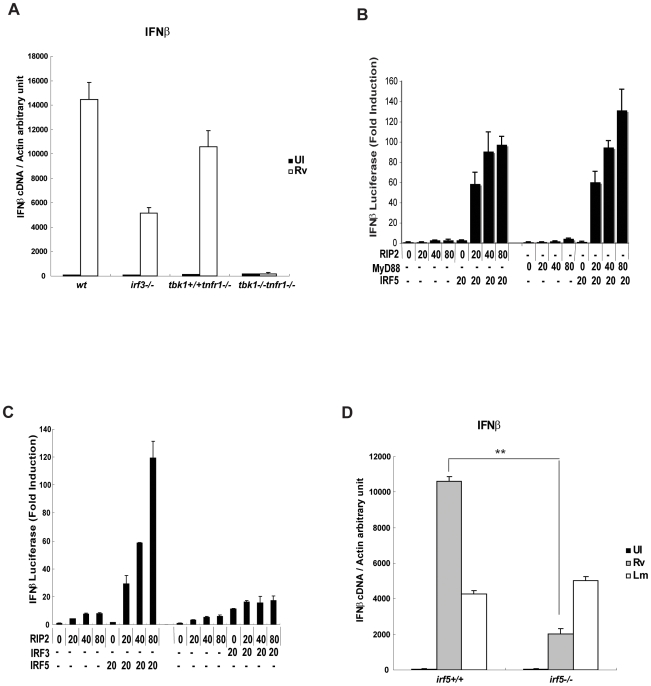
Listeria monocytogenes infection induces a potent host type I IFN response mediated by the Tbk1 kinase and Irf3 [27],[32],[35],[44]. To test whether Mtb infection triggered similar pathways, we infected Irf3-deficient and Tbk1/Tnfr1-deficient macrophages with Mtb and measured IFN induction. The Tnfr1 deficiency was necessary to suppress the embryonic lethality of Tbk1 deletion [45]. Similar to the L. monocytogenes model, we found that IFNβ induction by Mtb infection was completely dependent upon Tbk1, and the loss of Tnfr1 had little effect (Figure 7A). However, in contrast to the complete dependence on Irf3 observed for L. monocytogenes [27],[32],[35], we found IFNβ expression was reduced, but not ablated when Irf3-deficient macrophages were infected with M. tuberculosis (Figure 7A). This partial dependence on Irf3 was not changed by varying the multiplicity of infection (Figure S2). These data prompted us to test whether other IRFs mediate Nod2-dependent type I IFN responses.
Figure 7. Mtb-induced type I IFN response is Tbk1-dependent and mediated through both Irf3 and Irf5.
A. M. tuberculosis-induced type I IFN response is Tbk1-dependent and only partially mediated through Irf3. BMDM derived from wt, irf3−/− and tbk1−/−tnfr1−/− mice and littermate controls were infected with virulent Mtb H37Rv (Rv) at an MOI of 10 for 4 h. RNA was harvested and IFNβ mRNA level was quantified using real time PCR. Gene expression is reported as copy number per 1,000 copies of β-actin. Samples were assayed in triplicate; error bars represent the standard deviation. The experiment shown is representative of at least three. B. Co-expression of RIP2 and IRF5 stimulate IFNβ luciferase reporter activity. HEK293T cells were co-transfected with IFNβ-luciferase reporter plasmid (40 ng) together with the indicated concentrations of MyD88, IRF5 and RIP2 expression plasmids. Luciferase activity was measured 24 h later using Dual Luciferase reporter assay system (Promega). Renilla luciferase gene (40 ng) was co-transfected and used as an internal control. Each experiment was repeated three times. Data are expressed as mean±s.d. of three replicates. C. Co-expression of RIP2 and IRF3 does not stimulate IFNβ luciferase reporter activity. HEK293T cells were co-transfected with IFNβ-luciferase reporter plasmid (40 ng) together with the indicated concentrations of IRF5, IRF3 and RIP2 expression plasmids. Luciferase activity was measured 24 h later using Dual Luciferase reporter assay system (Promega). The Renilla luciferase gene (40 ng) was co-transfected and used as an internal control. Each experiment was repeated three times. Data are expressed as mean±s.d. of three replicates. D. Irf5 is required for an optimal type I IFN response upon Mtb infection. BMDM from irf5−/− or control littermates were infected with virulent Mtb H37Rv (Rv) at an MOI of 10, or with Listeria monocytogenes (Lm) strain 10403S (MOI 10) for 4 hours. RNA was harvested and IFNβ mRNA level was quantified by real time-PCR. IFNβ mRNA levels are reported as copy number per 1,000 copies of β-actin. Samples were assayed in triplicate; error bars represent standard deviation. Data shown is representative of at least three independent experiments.
Induction of IFNβ expression is dependent on the formation of the enhancesome which includes the NF-κB, ATF-2, c-jun, Irf3 and Irf7 transcription factors [46]. Irf5 is a related family member that has also been shown to contribute to induction of type I IFN responses triggered by TLRs, and overexpression of MyD88 has been shown to synergize with Irf5 to induce IFNβ expression [47]. Based on these studies, we tested whether RIP2 collaborates with IRF5 or IRF3 to stimulate IFNβ luciferase reporter activity. HEK293 cells were transfected with an IFNβ-luciferase reporter construct, along with increasing amounts of expression plasmids encoding RIP2, MyD88, IRF3 or IRF5. RIP2 and IRF5 coexpression stimulated IFNβ promoter activity in a dose dependent manner and to a similar extent as MyD88 and IRF5 (Figure 7B). In contrast, RIP2 and IRF3 expression failed to induce this robust response (Figure 7C). RIP2 and IRF5 expression also stimulated IFNα4 promoter activity as well as a reporter construct containing multimerized ISRE elements (data not shown).
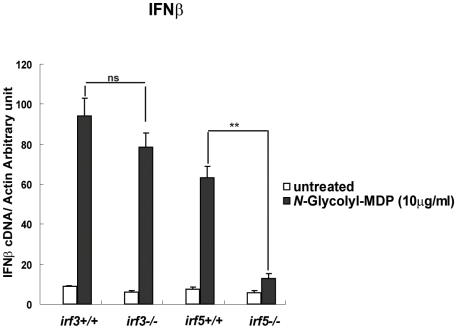
To further investigate the contribution of Irf5 to the anti-bacterial type I IFN response, we infected macrophages from Irf5-deficient mice and control littermates with either Mtb or L. monocytogenes, and measured IFNβ expression. Consistent with the luciferase reporter studies, we found that Mtb-induced IFNβ (Figure 7D) and IFNα (Figure S3) expression was impaired in the absence of Irf5. In contrast, the response to Listeria was unaffected by the loss of Irf5 (Figure 7D). While the related Rip1 adaptor protein regulates Irf7 activity in innate anti-viral signaling [48], we found that IFNβ induction after Mtb infection was unaffected by Irf7 deficiency (data not shown). To rule out the possibility that Irf3 expression levels may also be affected in irf5−/− macrophages, we verified that the Irf3 protein level was unchanged in Irf5-deficient cells (Figure S4). These results indicated that Mtb infection stimulates type I IFN expression via a pathway that depends on Nod2, Rip2, Tbk1, and Irf5. This contrasts with the pathway triggered by L. monocytogenes, which depends entirely on Irf3 and not Irf5. We reasoned that this dependence on different Irf proteins might be explained by the preferential stimulation of a Nod2-Rip2-Irf5 pathway by mycobacterial peptidoglycan. Consistent with this model, we found that the IFNβ induction triggered by N-glycolyl MDP was entirely dependent on Irf5 and independent of Irf3 (Figure 8), functionally linking Irf5 with the Nod2 pathway.
Figure 8. The N-Glycolyl-MDP-induced type I IFN response is Irf5-dependent and Irf3 independent.
BMDM derived from irf3−/− and irf5−/− mice and their littermate controls were left untreated or treated for 6 hours with 10 µg/ml of N-Glycolyl-MDP. RNA was harvested, and IFNβ mRNA levels were quantified using real time PCR. IFNβ mRNA levels are reported as copy number per 1,000 copies of β-actin. Samples were assayed in triplicate; error bars represent the standard deviation.
Discussion
Mammals first detect microbial infections via an array of PRRs that include both cell surface TLRs and cytosolic NLRs. However, not all microbial interactions represent a pathological state, and the immune system must be able to discriminate to some degree between colonization by commensal organisms and dangerous infection. One level of discrimination is provided by the desensitization or anatomical sequestration of TLRs at sites of chronic stimulation, such as the gut, which presumably allows for tolerance to normal flora [49],[50]. Bacterial pathogens can still be recognized at these sites via NLRs, since these systems rely on the specific ability of pathogens to translocate PAMPs into the host cytosol.
The concept that NLRs are specific for pathogenic organisms that disrupt host membranes is supported in a number of bacterial systems in which the loss of specific virulence functions abrogates NLR signaling. For example, in resting macrophages, cytosolic recognition of L. monocytogenes requires the pore-forming toxin, listeriolysin O [26],[27]. Similarly, Helicobacter pylori [51] and Legionella pneumophila [32] mutants lacking a functional type IV secretion system (T4SS), and Shigella flexneri [52] or Salmonella enterica serovar typhimurium [52] mutants lacking a functional type III secretion system (T3SS) fail to stimulate NLR pathways. In each case, the virulence system in question is responsible for host membrane damage and the likely translocation of bacterial products into the cytosol where they can be recognized by NLRs and/or other cytosolic surveillance systems.
Similarly, we found that the ESX1 specialized protein secretion system of Mtb is required for Nod2 recognition. While it has been suggested that type I IFN induction via ESX1 might represent a specific immunomodulatory virulence strategy [28], analogies to these other pathogens suggests that perhaps NLR recognition is simply a byproduct of a membrane damaging function that allows bacterial products to enter the cytosol. This model is supported by our observations that other membrane perturbing agents, such as SLO and PANX1 can substitute for ESX1 function and allow cytosolic recognition. Thus, in a number of cases it appears that NLRs can be considered as sentinels for pathogens that rely on membrane damage as a pathogenic strategy.
Based on their common role in protein secretion and in facilitating cytosolic recognition, it is tempting to speculate that ESX1 and Gram-negative T3SS and T4SS function analogously to deliver effector proteins into the host cytosol. Despite these similarities, the role played by ESX1 during infection remains unclear, since no translocated effectors have been identified to date. In both Mtb and M. marinum, a related pathogen of ectotherms, ESX1 has been implicated in host membrane disruption and one of the major substrates of this system, EsxA, has been proposed to possess a membrane-lytic activity [30],[37]. This single activity could be sufficient to account for the delivery of MDP and other PAMPs to the cytosol. It remains to be determined whether perturbing host membranes is the only role played by ESX1 during infection, or if this system also serves additional functions analogous to the specialized secretion systems of other pathogens.
A major consequence of the cytosolic recognition of Mtb is the induction of type I IFN. While the importance of this response in viral defense is clear and virtually universal, its role in antibacterial immunity appears to vary. Mice deficient in the type I IFN receptor, Ifnar1, are significantly more susceptible to several Gram-positive and -negative bacterial infections [53],[54],[55],[56], indicating that IFNα/β are important for immunity to many bacteria. However, Ifnar1 mutation has the opposite effect on the outcome of L. monocytogenes infection [57], suggesting that IFNα/β can also exacerbate disease. The role played by IFNα/β in Mtb infection remains somewhat uncertain. The induction of several immunologically important genes, including NOS2, depend on IFNα/β, suggesting a protective role. Initial studies of mouse and human infections appeared to support this view [58],[59]. However, like the L. monocytogenes system, mutation of the IFNα/β receptor has in most cases been associated with decreased bacterial burden in mouse models of tuberculosis [28],[59],[60],[61],[62]. IFNα/β may fail to protect against disease because Mtb inhibits the response to these cytokines in infected macrophages [63]. The ultimate influence of IFNα/β on Mtb infection appears to depend on a number of experimental factors, which might include host species, bacterial strain, route of infection and dose. Despite these differences however, some important themes emerge from these studies. Most importantly, the effect of IFNα/β is most apparent after the onset of adaptive immunity and not before, suggesting that the major role-played by type I IFNs during tuberculosis may be to instruct the priming or maintenance of the adaptive immune response and perhaps to control the differentiation of regulatory T cells [59].
A variety of bacterial pathogens trigger the type I IFN response, and a paradigm has begun to emerge regarding the induction of this response by bacteria. One current model suggests that bacterial DNA translocated into the host cytosol is the major eliciting agent. This model is based largely on the observations that infection with L. monocytogenes or L. pneumophilla, or transfection of DNA into the cytosol induces a similar IFNβ response that is Rip2 independent, and Tbk1- and Irf3-dependent [32]. Other PAMPs, such as MDP, can provide a synergistic IFN-inducing stimulus, but have not appeared to be sufficient for induction of IFNβ in the absence of other triggers [36].
In contrast, our data support a model whereby Nod2 stimulation by Mtb infection induces the polyubiquitination of Rip2, which acts via the Tbk1 kinase to stimulate the activity of Irf5 and induce transcription of IFNα/β. This differs from the pathway triggered by other bacteria such as L. monocytogenes, which depends entirely on Irf3 in resting macrophages [32] and does not involve Irf5 (Figure 7). Although Irf5 has previously been shown to be activated by the MyD88-dependent TLR7 and TLR9 pathways, this work reveals a novel role for this protein in Nod2 signaling, and a new link between Nod proteins and the type I IFN response. Furthermore, we found that unlike the N-acetylated MDP found in many bacteria, stimulation with the N-glycolylated MDP derivative found in mycobacteria was sufficient to stimulate the IFN response in the absence of other stimuli.
A significant component of IFNβ induction remains intact upon Mtb infection of Rip2-deficient macrophages (Figures 4 and 5), indicating that additional pathways are also involved. Since virtually all IFNβ expression is ESX1-dependent, it appears that the residual induction observed in rip2−/− macrophages also depends on cytosolic recognition pathways. These pathways could certainly include a DNA sensor that acts via Irf3, as proposed for other infections, since Irf3 deficiency had a moderate effect on IFNβ expression in our experiments (Figure 7A and S2). Thus, our data do not imply that Mtb is stimulating IFNα/β in a fundamentally different manner from other bacteria. Instead, it is likely that bacterial pathogens stimulate the IFN response via multiple, partially redundant pathways, and that the relative importance of each is determined by the unique biology of the infection. In the case of Mtb, we speculate that the N-glycolylation of its peptidoglycan, and perhaps a paucity of other stimulants such as DNA, favor recognition via Nod2. It is also possible that the balance of these pathways might be affected by the activation state of the macrophage. When resting macrophages are infected with L. monocytogenes, the IFN response requires LLO and is completely Irf3 dependent. In contrast, IFNγ-stimulated cells are able to deliver this bacterium to the lysosome, where the cell wall is degraded to produce abundant peptidoglycan fragments. In this situation, a significant component of the IFNβ induction depends on Nod2 and not Irf3 [64]. While Irf5 was not investigated in this study, it is possible that this represents another situation in which robust Nod2 signaling promotes a Nod2- and Irf5- dependent type I IFN response.
While we found that loss of Nod2-Rip2 signaling only partially reduces the induction of IFNβ, Rip2 deletion completely abrogated IFNα and RANTES expression. These results can be explained by the structure of the IFN regulatory circuit. Initially, only IFNβ is expressed, and subsequently IFNα and other interferon regulated genes (IRGs), such as RANTES, are induced via an Ifnar1 and Irf7-dependent autocrine/paracrine signaling pathway [65]. Thus, it appears that the decrease in IFNβ expression that we observe is sufficient to severely impair downstream IRG induction, at least in this cell culture model.
Multiple steps of this pathway are likely to depend on stable ubiquitin modifications. Not only did we observe that Rip2 is polyubiquitinated upon infection, but we also found that a Rip2 point mutant that cannot be stably ubiquitin modified is unable to mediate IFNα/β induction in response to Mtb infection (Figure S5). Collectively, these data suggest that polyubiquitinated Rip2 is required for Mtb-induced type I IFN expression via Irf5. Interestingly, MyD88-dependent activation of Irf5 involves formation of a tertiary complex that includes the E3 ubiquitin ligase, Traf6 [66],[67]. This E3 ubiquitin ligase associates with Rip2 upon MDP stimulation, raising the possibility that a Rip2-Traf6-Irf5 complex might exist and that the activity of Irf5 might also be regulated by ubiquitin.
The specificity of the innate immune system has been shaped by the very powerful natural selection imposed by microbial pathogens. Our work suggests that upon infection with Mtb, a particularly potent form of MDP is translocated into the host cell cytosol where it triggers a novel signaling pathway leading to the robust induction of the type I IFN response. It is unlikely to be coincidental that the active component of our most potent adjuvant, complete Freund's adjuvant (CFA), consists of mycobacterial cell fragments. The specific pathway described in this work might play a major role in this adjuvant's effectiveness, since IFNα/β production is required for CFA to promote antigen-specific immune responses (55). Thus, while PAMPs are often regarded as invariant microbial components, it is clear that functionally important pathogen-specific differences exist in the composition of these molecules, and that the immune system can differentiate these subtly distinct structures.
Given the potent adjuvant activity of mycobacterial components, it is somewhat surprising that the attenuated vaccine strain M. bovis BCG, which produces the same PAMPs present in CFA, provides poor protection against pulmonary TB in adults [68],[69]. The lack of ESX1 function in this strain appears to be at least partially responsible, since the reconstitution of ESX1 improves the efficacy of this vaccine [70],[71]. While this effect has previously been attributed to either the secretion of additional antigens or altered antigen presentation, it is also possible that ESX1 activity improves immunity by delivering crucial PAMPs into the cytosol where they are optimally recognized. Understanding both the details of PAMP trafficking, as well as the precise specificity of PAMP recognition, promises to aid in both the design of improved adjuvants and more effective tuberculosis vaccines.
Materials and Methods
Mice
C57BL/6 mice ages 8–12 weeks were obtained from the Jackson Laboratory. rip2−/− mice were a kind gift from Dr. Vishva M. Dixit (Genentech, Inc. South San Francisco, CA). nod2−/− mice were provided by Dr. Peter J. Murray (Department of Infectious Diseases, St. Jude Children's Research Hospital, Memphis, TN). nod1−/− and nod1−/−nod2−/− mice were provided by Dr. Gabriel Nunez (University of Michigan Medical School, Ann Arbor, MI). irf3−/−, irf5−/−, tbk1+/+tnfr1−/− and tbk1−/−tnfr1−/− mice and their littermate controls were provided by Dr. Kate A. Fitzgerald (University of Massachusetts Medical School, Worcester, MA). Mice were housed under specific pathogen-free conditions, and in accordance with the University of Massachusetts Medical School, IACUC guidelines.
Bacteria
The WT strain of M. tuberculosis used in these studies was the H37Rv strain. All the mutants were derived from the wild type strain. ΔESX-1 was obtained from D. Sherman (SBRI, Seattle.WA) [39]. ΔBioF, ΔRv3616 and ΔRv3616-complemented strains have been described previously [40],[41]. TN::Rv1410 contains a himar-1 transposon inserted at nucleotide #688 of the 1557 bp predicted open reading frame [72]. All strains were cultured in 7H9 medium containing 0.05% Tween 80 and OADC enrichment (Becton Dickinson). Pre-titered stocks of Listeria monocytogenes strain 10403 stored at −80°C (kindly provided by Victor Boyartchuk) were recovered for 1 hr at 37°C in 9 ml of Tryptic Soy Broth (BD Biosciences). Bacteria were then washed and resuspended in PBS prior to infection.
Antibodies and reagents
Anti-Rip2 (Rabbit) and anti-ubiquitin (Mouse) antibodies were obtained from Santa Cruz Biotechnology. Anti-Irf3 antibody was obtained from Zymed. Anti-Irf5 antibody was obtained from Abcam. Anti-β-actin antibody was obtained from Sigma. MDP was obtained from InvivoGen. Mouse TNF-α was obtained from Sigma. LPS derived from Escherichia coli strain 0111.B4 was purchased from Sigma, dissolved, treated with deoxycholate, and re-extracted with phenol/chloroform as described in [73]. The pannexin-1 mimetic blocking peptides panx1 (WRQAAFVDSY) and the scrambled peptide control were synthesized by GeneScript Corporation (Piscataway, NJ) and have been described previously [74]. Streptolysin O (SLO) a pore forming protein derived from Streptococcus and Adenosine 5′- triphosphate (ATP) were purchased from Sigma. N-glycolyl muramyl dipeptide (N-glycolyl MDP) was custom synthesized (Carbohydrate Synthesis, Oxford, UK) and shown to be more than 95% pure by NMR spectrometry. This preparation was found to be free of endotoxin contamination using the Limulus amebocyte lysate assay (Pyrotell, Cape Cod Inc., MA).
Macrophage infections
Bone marrow from 8- to 10-week-old mice was harvested from femurs and differentiated into macrophages for 7 days in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% L929-cell conditioned medium, 10% fetal bovine serum, 2 mM L-glutamine and 1 mM sodium pyruvate. After 7 days in culture, bone marrow derived macrophages (BMDMs) were washed with phosphate-buffered saline (PBS) and seeded into tissue culture plates for infection. RAW 264.7 macrophage cell line was cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum. All Mtb strains were cultivated in 7H9 broth, grown to exponential phase and washed thoroughly in DMEM media prior to infection. Bacterial clumps were removed by passing the washed suspension through a 5 µm syringe filter. For the peptide blocking studies, the cells were pre incubated with the desired peptides for 30 minutes followed by ATP or SLO for additional 15 minutes. Macrophages were infected at an MOI of 10 for 1 or 2 hours after which filtered cell lysates were immunoprecipitated with anti-Rip2 antibody (Santa Cruz). Heat inactivation was achieved by incubating the bacteria at 80°C for 30 minutes. Immortalized macrophage cell lines from wild type, rip2−/−, nod2−/− and nod1−/−nod2−/− mice were established by infecting bone marrow cells with a v-raf/mil and v-myc retrovirus in the presence of GM-CSF and polybrene [75],[76]. These rip2−/−, nod2−/− and nod1−/−nod2−/− macrophage cell lines express CD11b and Gr-1 and are capable of phagocytosing antibody coated beads. To determine the effect of cytochalasin D on the phagocytic function of the macrophages, we used the Vybrant phagocytosis assay kit to quantify the uptake of fluorescent E. coli. This assay was performed according to the protocol provided by the manufacturer.
Immunoprecipitation and Western blot analysis
For the immunoprecipitation and ubiquitination assays, cell lysates were prepared in radioimmune precipitation assay (RIPA) buffer (150 mM NaCl, 50 mM Tris-HCl (pH 7.5), 1% NP40, 0.25% deoxycholate, 0.1% SDS, 1 mM EDTA), supplemented with protease inhibitors (Roche Applied Science) and 5 mM N-Ethylmaleimide (Sigma), immunoprecipitated with anti-Rip2 antibody (Santa Cruz). Polyubiquitinated Rip2 proteins were detected by immunoblotting with an anti-ubiquitin antibody (Santa Cruz). Total immunoprecipitated Rip2 protein was measured by immunoblotting with anti-Rip2 antibodies (Santa Cruz).
Luciferase reporter assay
HEK293 cells (2×104) seeded in 96 well plates were transfected with 40 ng of the IFNβ luciferase reporter plasmid together with a total of 100 ng of various expression plasmids using GeneJuice (Novagen). The total amounts of transfected DNA were kept constant in all experiments by adjustment with empty vector. Luciferase activity was measured 24 h later using Dual Luciferase reporter assay system (Promega). The Renilla luciferase gene (40 ng) was co-transfected and used as an internal control plasmid. IFNβ luciferase reporter activity was normalized to Renilla luciferase reporter activity. Each experiment was repeated three times. Data are expressed as mean±s.d. of three replicates.
Real time quantitative PCR analysis
To measure IFNα/β mRNA levels upon MDP treatment or Mtb infection, total RNA was extracted from the macrophage cultures using Trizol reagent (Invitrogen) according to the manufacturer's directions. cDNA was prepared from 2 µg of total RNA and quantitative real-time PCR performed using SYBR green as a label with the following primers: mIFNα-F, 5′-AAGATGCCCTGCTGGCTG; mIFNα-R, 5′-TTCTGCTCTGACCACCTCCC; mIFNβ-F, 5′-CGTCTCCTGGATGAACTCCAC; mIFNβ-R, TGAGGACATCTCCCACGTCA; β-actin-F, 5′-CGAGGCCCAGAGCAAGAGAG; β-actin-R, 5′-CGGTTGGCCTTAGGGTTCAG; mTNFα-F, CAGTTCTATGGCCCAGACCCT; mTNFα-R, CGGACTCCGCAAAGTCTAAG; mRANTES-F, GCCCACGTCAAGGAGTATTTCTA; mRANTES-R, ACACACTTGGCGGTTCCTTC. Results shown are representative of more than three separate infection experiments, with each PCR performed in triplicate. All values reported were in the linear range of the experiment and were normalized to β-actin values. Standard curves were generated by linear dilution of a cDNA sample generated from poly I∶C-stimulated macrophages.
ELISA
IFNα protein in cell culture supernatants was performed using a custom ELISA as described previously [77]. IFNα concentrations were calculated using a recombinant IFNα (HyCult, Biotechnology, Uden, Netherlands) standard curve performed in quadruplicate using linear regression, and expressed in units per ml. IFNβ protein in cell culture supernatants was measured similarly using a custom ELISA as described in [78].
Supporting Information
Type I Interferon production upon Mtb infection is reduced in Rip2- and Nod2-deficient macrophages but not in Nod1-deficient macrophages. BMDM derived from wt, nod1−/−, rip2−/− and nod2−/− mice were infected with Mtb (MOI 10) for 4 h. RNA was harvested, and IFNβ mRNA levels were quantified using real time PCR. Gene expression is reported as copy number per 1,000 copies of β-actin. Samples were assayed in triplicate; error bars represent the standard deviation.
(0.23 MB PDF)
Mtb-induced type I IFN response is only partially mediated through Irf3. BMDM derived from wt and irf3−/− mice were infected with virulent Mtb H37Rv (Rv) at an MOI of 1, 3 and 10 for 4 h. RNA was harvested, and IFNβ mRNA level was quantified using real time PCR. Gene expression is reported as copy number per 10,000 copies of β-actin. Samples were assayed in triplicate; error bars represent the standard deviation.
(0.23 MB PDF)
Irf5 is required for an optimal type I IFNα response upon Mtb infection. BMDM from irf5−/− or control littermates were infected with virulent Mtb H37Rv (Rv) at an MOI of 10, or with Listeria monocytogenes (Lm) strain 10403S (MOI 10) for 4 hours. RNA was harvested, and IFNα mRNA level was quantified by real time-PCR. Gene expression of IFNα is reported as copy number per 1,000 copies of β-actin. Samples were assayed in triplicate; error bars represent standard deviation.
(0.23 MB PDF)
Irf3 and Irf5 expression levels in irf3−/− and irf5−/− macrophages. BMDM derived from irf3−/− and irf5−/− mice and their littermate controls were lysed in RIPA buffer and the Irf3 and Irf5 expression levels was determined by immunoblotting of anti Irf3 (Zymed) and Irf5 (Abcam) antibodies. Protein loading level was measured by β-actin antibody (Sigma).
(0.40 MB PDF)
Rip2 polyubiquitination is required for the Mtb-induced Type I IFN response. The rip2−/− transformed macrophage cell line was infected with the retroviral vector alone or with retroviruses expressing wild type Rip2 or a form of Rip2 (K209R) that cannot be ubiquitin modified [22]. The rip2−/− reconstituted macrophage cell lines were then infected with Mtb (MOI 10) for 4 h. RNA was harvested, and IFNβ mRNA levels were quantified using real time PCR. Gene expression is reported as copy number per 1,000 copies of β-actin. Samples were assayed in triplicate; error bars represent the standard deviation. Rip2 expression levels in each of the rip2−/− reconstituted macrophage cell lines were examined by immunoblotting to insure that equivalent expression levels of Rip2 were achieved.
(0.23 MB PDF)
Acknowledgments
We gratefully acknowledge the contributions of Victor Boyartchuck and Jinhee Lee, who were instrumental in facilitating the Listeria experiments and phagocytosis assays.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by NIH/AI RO1 AI075118 grant to MAK, NIH RO1 AI064282 to CMS and NIH RO1 AI067497 to KAF. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Aziz MA, Wright A. The World Health Organization/International Union Against Tuberculosis and Lung Disease Global Project on Surveillance for Anti-Tuberculosis Drug Resistance: a model for other infectious diseases. Clin Infect Dis. 2005;41(Suppl 4):S258–262. doi: 10.1086/430786. [DOI] [PubMed] [Google Scholar]
- 2.Rohde K, Yates RM, Purdy GE, Russell DG. Mycobacterium tuberculosis and the environment within the phagosome. Immunol Rev. 2007;219:37–54. doi: 10.1111/j.1600-065X.2007.00547.x. [DOI] [PubMed] [Google Scholar]
- 3.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 4.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Franchi L, Park JH, Shaw MH, Marina-Garcia N, Chen G, et al. Intracellular NOD-like receptors in innate immunity, infection and disease. Cell Microbiol. 2008;10:1–8. doi: 10.1111/j.1462-5822.2007.01059.x. [DOI] [PubMed] [Google Scholar]
- 6.Sirard JC, Vignal C, Dessein R, Chamaillard M. Nod-like receptors: cytosolic watchdogs for immunity against pathogens. PLoS Pathog. 2007;3:e152. doi: 10.1371/journal.ppat.0030152. doi:10.1371/journal.ppat.0030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delbridge LM, O'Riordan MX. Innate recognition of intracellular bacteria. Curr Opin Immunol. 2007;19:10–16. doi: 10.1016/j.coi.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 9.Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jehanno M, et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 10.Girardin SE, Travassos LH, Herve M, Blanot D, Boneca IG, et al. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J Biol Chem. 2003;278:41702–41708. doi: 10.1074/jbc.M307198200. [DOI] [PubMed] [Google Scholar]
- 11.Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J Biol Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 12.Boneca IG. The role of peptidoglycan in pathogenesis. Curr Opin Microbiol. 2005;8:46–53. doi: 10.1016/j.mib.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Cloud-Hansen KA, Peterson SB, Stabb EV, Goldman WE, McFall-Ngai MJ, et al. Breaching the great wall: peptidoglycan and microbial interactions. Nat Rev Microbiol. 2006;4:710–716. doi: 10.1038/nrmicro1486. [DOI] [PubMed] [Google Scholar]
- 14.Mengin-Lecreulx D, Lemaitre B. Structure and metabolism of peptidoglycan and molecular requirements allowing its detection by the Drosophila innate immune system. J Endotoxin Res. 2005;11:105–111. doi: 10.1179/096805105X35233. [DOI] [PubMed] [Google Scholar]
- 15.Boneca IG, Dussurget O, Cabanes D, Nahori MA, Sousa S, et al. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc Natl Acad Sci U S A. 2007;104:997–1002. doi: 10.1073/pnas.0609672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azuma I, Thomas DW, Adam A, Ghuysen JM, Bonaly R, et al. Occurrence of N-glycolylmuramic acid in bacterial cell walls. A preliminary survey. Biochim Biophys Acta. 1970;208:444–451. doi: 10.1016/0304-4165(70)90217-5. [DOI] [PubMed] [Google Scholar]
- 17.Raymond JB, Mahapatra S, Crick DC, Pavelka MS., Jr Identification of the namH gene, encoding the hydroxylase responsible for the N-glycolylation of the mycobacterial peptidoglycan. J Biol Chem. 2005;280:326–333. doi: 10.1074/jbc.M411006200. [DOI] [PubMed] [Google Scholar]
- 18.Meylan E, Tschopp J. The RIP kinases: crucial integrators of cellular stress. Trends Biochem Sci. 2005;30:151–159. doi: 10.1016/j.tibs.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, et al. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem. 2001;276:4812–4818. doi: 10.1074/jbc.M008072200. [DOI] [PubMed] [Google Scholar]
- 20.McCarthy JV, Ni J, Dixit VM. RIP2 is a novel NF-kappaB-activating and cell death-inducing kinase. J Biol Chem. 1998;273:16968–16975. doi: 10.1074/jbc.273.27.16968. [DOI] [PubMed] [Google Scholar]
- 21.Yang Y, Yin C, Pandey A, Abbott D, Sassetti C, et al. NOD2 pathway activation by MDP or Mycobacterium tuberculosis infection involves the stable polyubiquitination of Rip2. J Biol Chem. 2007;282:36223–36229. doi: 10.1074/jbc.M703079200. [DOI] [PubMed] [Google Scholar]
- 22.Hasegawa M, Fujimoto Y, Lucas PC, Nakano H, Fukase K, et al. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-kappaB activation. Embo J. 2008;27:373–383. doi: 10.1038/sj.emboj.7601962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferwerda G, Girardin SE, Kullberg BJ, Le Bourhis L, de Jong DJ, et al. NOD2 and toll-like receptors are nonredundant recognition systems of Mycobacterium tuberculosis. PLoS Pathog. 2005;1:e34. doi: 10.1371/journal.ppat.0010034. doi:10.1371/journal.ppat.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi S, Blumenthal A, Hickey CM, Gandotra S, Levy D, et al. Expression of many immunologically important genes in Mycobacterium tuberculosis-infected macrophages is independent of both TLR2 and TLR4 but dependent on IFN-alphabeta receptor and STAT1. J Immunol. 2005;175:3318–3328. doi: 10.4049/jimmunol.175.5.3318. [DOI] [PubMed] [Google Scholar]
- 25.Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J Exp Med. 2007;204:987–994. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Riordan M, Yi CH, Gonzales R, Lee KD, Portnoy DA. Innate recognition of bacteria by a macrophage cytosolic surveillance pathway. Proc Natl Acad Sci U S A. 2002;99:13861–13866. doi: 10.1073/pnas.202476699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stockinger S, Materna T, Stoiber D, Bayr L, Steinborn R, et al. Production of type I IFN sensitizes macrophages to cell death induced by Listeria monocytogenes. J Immunol. 2002;169:6522–6529. doi: 10.4049/jimmunol.169.11.6522. [DOI] [PubMed] [Google Scholar]
- 28.Stanley SA, Johndrow JE, Manzanillo P, Cox JS. The Type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J Immunol. 2007;178:3143–3152. doi: 10.4049/jimmunol.178.5.3143. [DOI] [PubMed] [Google Scholar]
- 29.Gao LY, Guo S, McLaughlin B, Morisaki H, Engel JN, et al. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol Microbiol. 2004;53:1677–1693. doi: 10.1111/j.1365-2958.2004.04261.x. [DOI] [PubMed] [Google Scholar]
- 30.Hsu T, Hingley-Wilson SM, Chen B, Chen M, Dai AZ, et al. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc Natl Acad Sci U S A. 2003;100:12420–12425. doi: 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, et al. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 2007;129:1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- 32.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 33.Muruve DA, Petrilli V, Zaiss AK, White LR, Clark SA, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 34.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009 doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stockinger S, Reutterer B, Schaljo B, Schellack C, Brunner S, et al. IFN regulatory factor 3-dependent induction of type I IFNs by intracellular bacteria is mediated by a TLR- and Nod2-independent mechanism. J Immunol. 2004;173:7416–7425. doi: 10.4049/jimmunol.173.12.7416. [DOI] [PubMed] [Google Scholar]
- 36.Leber JH, Crimmins GT, Raghavan S, Meyer-Morse NP, Cox JS, et al. Distinct TLR- and NLR-mediated transcriptional responses to an intracellular pathogen. PLoS Pathog. 2008;4:e6. doi: 10.1371/journal.ppat.0040006. doi:10.1371/journal.ppat.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Jonge MI, Pehau-Arnaudet G, Fretz MM, Romain F, Bottai D, et al. ESAT-6 from Mycobacterium tuberculosis dissociates from its putative chaperone CFP-10 under acidic conditions and exhibits membrane-lysing activity. J Bacteriol. 2007;189:6028–6034. doi: 10.1128/JB.00469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koo IC, Ohol YM, Wu P, Morisaki JH, Cox JS, et al. Role for lysosomal enzyme beta-hexosaminidase in the control of mycobacteria infection. Proc Natl Acad Sci U S A. 2008;105:710–715. doi: 10.1073/pnas.0708110105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis KN, Liao R, Guinn KM, Hickey MJ, Smith S, et al. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. J Infect Dis. 2003;187:117–123. doi: 10.1086/345862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fortune SM, Jaeger A, Sarracino DA, Chase MR, Sassetti CM, et al. Mutually dependent secretion of proteins required for mycobacterial virulence. Proc Natl Acad Sci U S A. 2005;102:10676–10681. doi: 10.1073/pnas.0504922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farrow MF, Rubin EJ. Function of a mycobacterial major facilitator superfamily pump requires a membrane-associated lipoprotein. J Bacteriol. 2008;190:1783–1791. doi: 10.1128/JB.01046-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanneganti TD, Lamkanfi M, Kim YG, Chen G, Park JH, et al. Pannexin-1-mediated recognition of bacterial molecules activates the cryopyrin inflammasome independent of Toll-like receptor signaling. Immunity. 2007;26:433–443. doi: 10.1016/j.immuni.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 44.O'Connell RM, Vaidya SA, Perry AK, Saha SK, Dempsey PW, et al. Immune activation of type I IFNs by Listeria monocytogenes occurs independently of TLR4, TLR2, and receptor interacting protein 2 but involves TNFR-associated NF kappa B kinase-binding kinase 1. J Immunol. 2005;174:1602–1607. doi: 10.4049/jimmunol.174.3.1602. [DOI] [PubMed] [Google Scholar]
- 45.Perry AK, Chow EK, Goodnough JB, Yeh WC, Cheng G. Differential requirement for TANK-binding kinase-1 in type I interferon responses to toll-like receptor activation and viral infection. J Exp Med. 2004;199:1651–1658. doi: 10.1084/jem.20040528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wathelet MG, Lin CH, Parekh BS, Ronco LV, Howley PM, et al. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 47.Schoenemeyer A, Barnes BJ, Mancl ME, Latz E, Goutagny N, et al. The interferon regulatory factor, IRF5, is a central mediator of toll-like receptor 7 signaling. J Biol Chem. 2005;280:17005–17012. doi: 10.1074/jbc.M412584200. [DOI] [PubMed] [Google Scholar]
- 48.Huye LE, Ning S, Kelliher M, Pagano JS. Interferon regulatory factor 7 is activated by a viral oncoprotein through RIP-dependent ubiquitination. Mol Cell Biol. 2007;27:2910–2918. doi: 10.1128/MCB.02256-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abreu MT, Fukata M, Arditi M. TLR signaling in the gut in health and disease. J Immunol. 2005;174:4453–4460. doi: 10.4049/jimmunol.174.8.4453. [DOI] [PubMed] [Google Scholar]
- 50.Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2001;167:1882–1885. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 51.Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5:1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki T, Franchi L, Toma C, Ashida H, Ogawa M, et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3:e111. doi: 10.1371/journal.ppat.0030111. doi:10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niesel DW, Hess CB, Cho YJ, Klimpel KD, Klimpel GR. Natural and recombinant interferons inhibit epithelial cell invasion by Shigella spp. Infect Immun. 1986;52:828–833. doi: 10.1128/iai.52.3.828-833.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Freudenberg MA, Merlin T, Kalis C, Chvatchko Y, Stubig H, et al. Cutting edge: a murine, IL-12-independent pathway of IFN-gamma induction by gram-negative bacteria based on STAT4 activation by Type I IFN and IL-18 signaling. J Immunol. 2002;169:1665–1668. doi: 10.4049/jimmunol.169.4.1665. [DOI] [PubMed] [Google Scholar]
- 55.Weigent DA, Huff TL, Peterson JW, Stanton GJ, Baron S. Role of interferon in streptococcal infection in the mouse. Microb Pathog. 1986;1:399–407. doi: 10.1016/0882-4010(86)90071-9. [DOI] [PubMed] [Google Scholar]
- 56.Gold JA, Hoshino Y, Hoshino S, Jones MB, Nolan A, et al. Exogenous gamma and alpha/beta interferon rescues human macrophages from cell death induced by Bacillus anthracis. Infect Immun. 2004;72:1291–1297. doi: 10.1128/IAI.72.3.1291-1297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O'Connell RM, Saha SK, Vaidya SA, Bruhn KW, Miranda GA, et al. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J Exp Med. 2004;200:437–445. doi: 10.1084/jem.20040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Giosue S, Casarini M, Alemanno L, Galluccio G, Mattia P, et al. Effects of aerosolized interferon-alpha in patients with pulmonary tuberculosis. Am J Respir Crit Care Med. 1998;158:1156–1162. doi: 10.1164/ajrccm.158.4.9803065. [DOI] [PubMed] [Google Scholar]
- 59.Ordway D, Henao-Tamayo M, Harton M, Palanisamy G, Troudt J, et al. The hypervirulent Mycobacterium tuberculosis strain HN878 induces a potent TH1 response followed by rapid down-regulation. J Immunol. 2007;179:522–531. doi: 10.4049/jimmunol.179.1.522. [DOI] [PubMed] [Google Scholar]
- 60.Bouchonnet F, Boechat N, Bonay M, Hance AJ. Alpha/beta interferon impairs the ability of human macrophages to control growth of Mycobacterium bovis BCG. Infect Immun. 2002;70:3020–3025. doi: 10.1128/IAI.70.6.3020-3025.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manca C, Tsenova L, Bergtold A, Freeman S, Tovey M, et al. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha/beta. Proc Natl Acad Sci U S A. 2001;98:5752–5757. doi: 10.1073/pnas.091096998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reutterer B, Stockinger S, Pilz A, Soulat D, Kastner R, et al. Type I IFN are host modulators of strain-specific Listeria monocytogenes virulence. Cell Microbiol. 2008;10:1116–1129. doi: 10.1111/j.1462-5822.2007.01114.x. [DOI] [PubMed] [Google Scholar]
- 63.Prabhakar S, Qiao Y, Hoshino Y, Weiden M, Canova A, et al. Inhibition of response to alpha interferon by Mycobacterium tuberculosis. Infect Immun. 2003;71:2487–2497. doi: 10.1128/IAI.71.5.2487-2497.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Herskovits AA, Auerbuch V, Portnoy DA. Bacterial ligands generated in a phagosome are targets of the cytosolic innate immune system. PLoS Pathog. 2007;3:e51. doi: 10.1371/journal.ppat.0030051. doi:10.1371/journal.ppat.0030051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Remoli ME, Giacomini E, Lutfalla G, Dondi E, Orefici G, et al. Selective expression of type I IFN genes in human dendritic cells infected with Mycobacterium tuberculosis. J Immunol. 2002;169:366–374. doi: 10.4049/jimmunol.169.1.366. [DOI] [PubMed] [Google Scholar]
- 66.Kawai T, Sato S, Ishii KJ, Coban C, Hemmi H, et al. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat Immunol. 2004;5:1061–1068. doi: 10.1038/ni1118. [DOI] [PubMed] [Google Scholar]
- 67.Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 68.Colditz GA, Berkey CS, Mosteller F, Brewer TF, Wilson ME, et al. The efficacy of bacillus Calmette-Guerin vaccination of newborns and infants in the prevention of tuberculosis: meta-analyses of the published literature. Pediatrics. 1995;96:29–35. [PubMed] [Google Scholar]
- 69.Fine PE. Variation in protection by BCG: implications of and for heterologous immunity. Lancet. 1995;346:1339–1345. doi: 10.1016/s0140-6736(95)92348-9. [DOI] [PubMed] [Google Scholar]
- 70.Brodin P, Majlessi L, Brosch R, Smith D, Bancroft G, et al. Enhanced protection against tuberculosis by vaccination with recombinant Mycobacterium microti vaccine that induces T cell immunity against region of difference 1 antigens. J Infect Dis. 2004;190:115–122. doi: 10.1086/421468. [DOI] [PubMed] [Google Scholar]
- 71.Pym AS, Brodin P, Majlessi L, Brosch R, Demangel C, et al. Recombinant BCG exporting ESAT-6 confers enhanced protection against tuberculosis. Nat Med. 2003;9:533–539. doi: 10.1038/nm859. [DOI] [PubMed] [Google Scholar]
- 72.Sassetti CM, Boyd DH, Rubin EJ. Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci U S A. 2001;98:12712–12717. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hirschfeld M, Ma Y, Weis JH, Vogel SN, Weis JJ. Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine toll-like receptor 2. J Immunol. 2000;165:618–622. doi: 10.4049/jimmunol.165.2.618. [DOI] [PubMed] [Google Scholar]
- 74.Pelegrin P, Surprenant A. Pannexin-1 mediates large pore formation and interleukin-1beta release by the ATP-gated P2X7 receptor. EMBO J. 2006;25:5071–5082. doi: 10.1038/sj.emboj.7601378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roberson SM, Walker WS. Immortalization of cloned mouse splenic macrophages with a retrovirus containing the v-raf/mil and v-myc oncogenes. Cell Immunol. 1988;116:341–351. doi: 10.1016/0008-8749(88)90236-5. [DOI] [PubMed] [Google Scholar]
- 76.Gandino L, Varesio L. Immortalization of macrophages from mouse bone marrow and fetal liver. Exp Cell Res. 1990;188:192–198. doi: 10.1016/0014-4827(90)90159-8. [DOI] [PubMed] [Google Scholar]
- 77.Geeraedts F, Goutagny N, Hornung V, Severa M, de Haan A, et al. Superior immunogenicity of inactivated whole virus H5N1 influenza vaccine is primarily controlled by Toll-like receptor signalling. PLoS Pathog. 2008;4:e1000138. doi: 10.1371/journal.ppat.1000138. doi:10.1371/journal.ppat.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roberts ZJ, Goutagny N, Perera PY, Kato H, Kumar H, et al. The chemotherapeutic agent DMXAA potently and specifically activates the TBK1-IRF-3 signaling axis. J Exp Med. 2007;204:1559–1569. doi: 10.1084/jem.20061845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Type I Interferon production upon Mtb infection is reduced in Rip2- and Nod2-deficient macrophages but not in Nod1-deficient macrophages. BMDM derived from wt, nod1−/−, rip2−/− and nod2−/− mice were infected with Mtb (MOI 10) for 4 h. RNA was harvested, and IFNβ mRNA levels were quantified using real time PCR. Gene expression is reported as copy number per 1,000 copies of β-actin. Samples were assayed in triplicate; error bars represent the standard deviation.
(0.23 MB PDF)
Mtb-induced type I IFN response is only partially mediated through Irf3. BMDM derived from wt and irf3−/− mice were infected with virulent Mtb H37Rv (Rv) at an MOI of 1, 3 and 10 for 4 h. RNA was harvested, and IFNβ mRNA level was quantified using real time PCR. Gene expression is reported as copy number per 10,000 copies of β-actin. Samples were assayed in triplicate; error bars represent the standard deviation.
(0.23 MB PDF)
Irf5 is required for an optimal type I IFNα response upon Mtb infection. BMDM from irf5−/− or control littermates were infected with virulent Mtb H37Rv (Rv) at an MOI of 10, or with Listeria monocytogenes (Lm) strain 10403S (MOI 10) for 4 hours. RNA was harvested, and IFNα mRNA level was quantified by real time-PCR. Gene expression of IFNα is reported as copy number per 1,000 copies of β-actin. Samples were assayed in triplicate; error bars represent standard deviation.
(0.23 MB PDF)
Irf3 and Irf5 expression levels in irf3−/− and irf5−/− macrophages. BMDM derived from irf3−/− and irf5−/− mice and their littermate controls were lysed in RIPA buffer and the Irf3 and Irf5 expression levels was determined by immunoblotting of anti Irf3 (Zymed) and Irf5 (Abcam) antibodies. Protein loading level was measured by β-actin antibody (Sigma).
(0.40 MB PDF)
Rip2 polyubiquitination is required for the Mtb-induced Type I IFN response. The rip2−/− transformed macrophage cell line was infected with the retroviral vector alone or with retroviruses expressing wild type Rip2 or a form of Rip2 (K209R) that cannot be ubiquitin modified [22]. The rip2−/− reconstituted macrophage cell lines were then infected with Mtb (MOI 10) for 4 h. RNA was harvested, and IFNβ mRNA levels were quantified using real time PCR. Gene expression is reported as copy number per 1,000 copies of β-actin. Samples were assayed in triplicate; error bars represent the standard deviation. Rip2 expression levels in each of the rip2−/− reconstituted macrophage cell lines were examined by immunoblotting to insure that equivalent expression levels of Rip2 were achieved.
(0.23 MB PDF)