Abstract
Anionic lipids and cholesterols (CHOL) are critical to the function of nicotinic acetylcholine receptors (nAChR). We investigated their interactions with an open- and closed-channel α4β2 nAChR by over 10-ns molecular dynamics simulations in a ternary lipid mixture of 1-palmitoyl-2-oleoyl phosphatidylcholine (POPC), 1-palmitoyl-2-oleoyl phosphatidic acid (POPA), and CHOL with a ratio of 3:1:1. On average there were 65 and 74 interfacial lipids around the closed- and open-channel α4β2 nAChR, respectively, in the equilibrated simulation systems. 42% of the interfacial POPA in the open-channel system had acyl chains partially inserted into intra- or inter-subunit cavities, as compared to only 7% in the closed-channel α4β2. No CHOL was found in cavities within single subunits, though some CHOL infiltrated into the gaps between subunits. Because of its smaller head group, POPA could access some non-annular sites where POPC could not easily reach due to steric exclusion. Furthermore, POPA not only acted as an acceptor for hydrogen bonding (H-bonding) as POPC did, but also as a donor through its hydroxyl group for H-bonding with the backbone of the protein. The charged head group of POPA allowed the lipid to form stable salt bridges with conserved Arg and Lys residues at the interfaces of the transmembrane (TM) and extracellular (EC) or intracellular (IC) domains of the α4β2. A higher number of salt bridges and hydrogen bonds (H-bonds) between POPA and the α4β2 nAChR were found in the open system than in the closed system, suggesting a potential role of POPA in the equilibrium between different channel states. Most interfacial POPA molecules showed lower order parameters than the bulk POPA due to the mixed effect of gauche defects, hydrophobic mismatch, and the lipid orientations near the magic angle. These unique properties enable the interfacial POPA to achieve what POPC cannot with regard to specific interactions with the protein, thereby making POPA essential for the function of nAChR.
Keywords: Anionic phospholipid, Cholesterol, Lipid binding sites, α4β2 nAChR
Introduction
Anionic lipids and cholesterol (CHOL) are often critical membrane components to ensure ion channel stability and functionality. Their importance to the channel functions is often explained via indirect and direct mechanisms. The indirect mechanism proposes that anionic lipids and CHOL alter lipid packing and fluidity that ultimately affect the internal protein dynamics and functions.1,2 The direct mechanism states that specific interactions between lipid molecules and ion channels modulate channel functions, such as those exhibited in the mechanosensitive channel3 and the crystal structure of KcsA.4 The recent X-ray structure of the bacterial Gloeobacter violaceus pentameric ligand-gated ion channel homologue (GLIC)5 also showed ∼15 lipids surrounding the protein. These lipids appeared to be phosphatidylcholine (PC) molecules, which do not exist naturally in the bacterial membrane. Some of these lipid head groups were closely associated with the charged residues near the intracellular and extracellular domains of GLIC.
Nicotinic acetylcholine receptors (nAChR), pentameric ligand-gated cation-selective channels, require proper lipid compositions in order to function properly. Normally, agonist binding to the extracellular (EC) domains could trigger opening of the ion channels and stimulate the cation permeation. However, without the presence of anionic phosphatidic acid (PA) and/or CHOL in the membrane, reconstituted Torpedo californica nAChR would not be able to undergo an agonist-induced conformation change to retain the channel gating activity.6-8 It has been demonstrated that membranes containing phosphatidylcholine (PC)/PA/CHOL in 3:1:1 are particularly effective at maintaining the stability of functional reconstituted nAChR.9 A lingering question remains; what do PA and CHOL actually do to nAChR that cannot be accomplished by PC alone? The ability of PA and CHOL to reduce lipid fluidity has been proposed to indirectly affect the nAChR function,1,2 but other lines of experimental evidence point to direct effects. For example, photolabeling studies using [3H] Azicholesterol found that CHOL might interact directly with the transmembrane M4, M3, and M1 segments of each subunit of the Torpedo nAChR.10 It has also been suggested, both experimentally and computationally, that the presence of nAChR facilitates lipid lateral phase separation to form PA-rich domains.11-13 It is believed that these PA domains can help stabilize nAChR in a functional resting state.11-13
Despite some successes, experimental investigations on the underlying cause of nAChR sensitivity to PA and CHOL are often challenged by technique difficulties. Unlike in the cases of the mechanosensitive channel3 and KcsA,4 there is to date no high-resolution experimental nAChR structure showing protein-lipid interactions. Although recent X-ray structures of GLIC5,14 and ELIC15 offered valuable information about ion permeation and channel gating that might also be applicable to nAChR, the structures did not reveal anything about PA or CHOL, because these lipids do not naturally co-exist with prokaryotic channels. Computational studies have proven useful to provide insights that are difficult to capture experimentally. A recent molecular dynamics (MD) simulation study16 has suggested that nAChR possesses both superficial and deeply buried sites for CHOL binding. Furthermore, CHOL occupation stabilizes the protein structure and supports contacts between the agonist-binding domain and the pore, which are thought to be essential for activation of the receptor.16 MD simulations on nAChR embedded in PC and PA lipids have found that only PA could form visible domains around the protein, though it is not entirely clear why the PA domain could help stabilize the nAChR in a resting state.13 Other computational studies regarding PA or CHOL effects on nAChR have not included both PA and CHOL in the same system; therefore it is unknown if the presence of PA would affect CHOL or vice versa. A possible competition between PA and CHOL for the same binding sites on nAChR has been suggested based on fluorescence quenching.12
In the present study, we intended to determine how PA and CHOL interact with nAChR when both PA and CHOL exist in the system, and how the interactions could potentially affect nAChR functions. A ternary lipid mixture comprised of 1-palmitoyl-2-oleoyl phosphatidylcholine (POPC), 1-palmitoyl-2-oleoyl phosphatidic acid (POPA), and CHOL in a 3:1:1 molar ratio17 was used for embedding α4β2 nAChR, which is one of the most abundant nAChR subtypes in the brain and composed of two α4 and three β2 subunits. Structure models of the α4β2 nAChR in the closed- and open-channel states were generated through homology modeling of the known structure of the Torpedo nAChR (PDB: 2BG9)18 and subsequently applying the lowest frequency eigenvector of normal mode analysis19 to the closed-channel structure. The resulting closed and open channels had the minimum pore radius of 2.7 and 3.4 Å, respectively. Both closed- and open-channel models were subjected to over ten nanoseconds of MD simulations in a fully hydrated and pre-equilibrated ternary lipid patch.17 Na+ permeability and agonist binding properties of these models were tested and showed good agreement with previous experimental data. A detailed comparison of the open- and closed-channel structures of the α4β2 nAChR was given in our recent publication,20 in which the simulations of α4β2 nAChR provided valuable insights into the channel gating mechanism that shares considerable similarity to the channel gating inferred from the recent X-ray structures of pentameric ligand-gated ion channels (LGICs) in the open5,14 and closed states.15 Here, the focus is on the protein-lipid interactions. Using the same simulation data,20 we have identified unique interactions between POPA and α4β2 nAChR that might be relevant to the modulation of nAChR functions. We have also identified residues that interact directly with POPA or CHOL. Some of these residues have been suggested previously by experimental data.21
Methods
System preparations and MD simulations
Detailed descriptions of the open- and closed-channel α4β2 nAChR structures embedded in a fully hydrated POPC-POPA-CHOL (in 3:1:1 molar ratio) lipid mixture17 have been reported previously.20 The closed-channel system had 1 protein, 162 POPC, 55 POPA, 55 CHOL lipid molecules, 122 ions (108 Na+ and 14 Cl-) and 33,641 water molecules. The open-channel system had 1 protein, 160 POPC, 50 POPA, 54 CHOL lipid molecules, 115 ions (96 Na+ ions and 9 Cl-), 2 nicotine molecules, and 28,382 water molecules. The upper and lower leaflets of lipids had similar lipid composition in both models.
MD simulations were performed using CHARMM27 force-field parameters22 and NAMD 2.623 on BigBen (a Cray XT3 MPP machine) at the Pittsburgh Supercomputing Center. The systems were simulated under a constant pressure (1 atm) and a constant temperature (303 K), which were regulated via Nosé-Hoover Langevin piston pressure control24,25 and the Langevin damping dynamics.26 Periodic boundary conditions and water wrapping were applied. The bonded interactions and the short-range non-bonded interactions were calculated at every time-step (1 fs) and every two time-steps, respectively. Electrostatic interactions were calculated at every four time-steps using the particle mesh Ewald method. 27 The cutoff distance for non-bonded interactions was 12 Å and 1-4 scaling factor was 1.0. A smoothing function was employed for the van der Waals interactions at a distance of 10 Å. The pair-list of the non-bonded interaction was calculated every 20 time-steps with a pair-list distance of 13.5 Å. A harmonic restraint initially applied to the Cα atoms of the proteins was gradually removed over the course of ∼2 ns of MD simulations. A total of 10 ns NPT simulation was performed on each simulation system.
Data analysis
The majority of data analysis used the scripts developed within the VMD software environment.28 H-bonding between lipids and protein was defined as the distance between donor and acceptor atoms not greater than 3 Å and the donor-hydrogen-acceptor angle not smaller than 160°. Salt bridging between the protein and POPA was counted if the distance between a nitrogen atom of a basic residue and an oxygen atom of POPA was within 3.2 Å. The lipid ordering in the ternary system was measured using both order parameters and the fraction of gauche dihedral angles (defined here as between 50° and 70° or between -50° and -70°) along the hydrocarbon chains. Lipid order parameter was calculated as:
| (1) |
where n denotes the nth carbon counted from the carbonyl carbon atom along the acryl chain, and βn is the angle made by a C-H bond and the lipid bilayer normal. Bracket denotes averaging over time and the ensemble of the same C-H bonds in the same class of lipids.
Results and Discussion
Lipids exposed to the α4β2 nAChR
The lipids became well packed around the α4β2 nAChR in both open- and closed-channel systems soon after ∼2-ns MD simulations. A recent experimental study found that the functionality of nAChR reconstituted into PA/CHOL membranes decreased rapidly below approximately 65 lipids per receptor,8 indicating a minimum number of lipids required for surrounding the nAChR. In this study, the interfacial lipids are defined broadly as those exposed to any protein atoms within a radius of 2.4 Å. For lipids penetrating into cavities within a subunit or between subunits, we follow the traditional definition29 and call them non-annular lipids. In our closed-channel α4β2 system after 10-ns simulation, there was an average of 65 interfacial lipids surrounding the protein. In the open-channel system, 74 interfacial lipids were observed on average. A slightly higher number of interfacial lipids in the open channel system was expected because of a moderate surface expansion in the open-channel nAChR in comparison to the closed one.
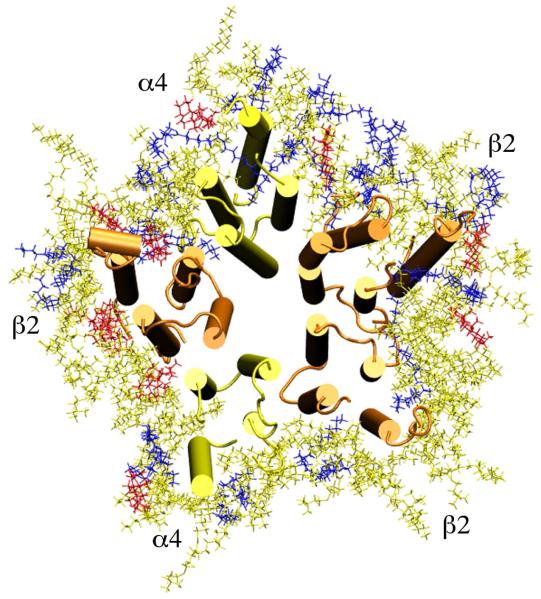
Fig.1 shows lipids exposed to the open-channel α4β2 nAChR after a 10-ns simulation. Although the interfacial lipids could exchange with bulk lipids, we found that less than 20% of interfacial lipids experienced such exchanges over the course of MD simulations. To fully sample the interfacial lipid exchange will require an extension of the simulation time by at least an order of magnitude, considering that interfacial lipids might exchange at a rate slower than the lateral diffusion rate of 10-11-10-12m2/s for the bulk lipids30,31 due to interactions with the protein. The composition of interfacial lipids in Fig. 1 more or less reflected that of the original ternary lipid patches after embedding the α4β2 nAChR for MD simulations. There were no additional modifications prior to simulations to adjust POPA and CHOL populations at the protein-lipid interface. Table 1 summarizes the number of interfacial lipids, averaged over 500 snapshots near the end of the 10-ns MD simulations, in both open- and closed-channel systems. The percentage of protein-facing POPA is comparable to 20% of POPA composition in the bulk lipids, but ∼12% of CHOL at the interface is lower than the bulk value. Nevertheless, the amount of POPA and CHOL surrounding the protein provides enough information about their interactions with the α4β2 nAChR.
Figure 1.
Top view of CHOL (red), POPA (blue), and POPC (yellow) lipids exposed to the open-channel α4β2 nAChR after a 10-ns simulation. The α4 and β2 subunits are in yellow and orange cartoon format, respectively.
Table 1. Average number of lipids exposed to the protein.
| POPC | POPA | CHOL | TOTAL | |
|---|---|---|---|---|
| Open α4β2 | 52 | 14 | 8 | 74 |
| Close α4β2 | 42 | 14 | 9 | 65 |
It is noticeable in Fig. 1 that some lipids intruded into the intra- or inter-subunit cavities of the α4β2 nAChR, acting as the so-called “non-annular” lipids.29 A closer inspection reveals that only the tails of acyl chains of POPA or POPC occupied non-annular binding sites, not an entire lipid molecule. 42% of the interfacial POPA in the open-channel system had their acyl chains (with the majority being the unsaturated sn-2 chains) partially inserted into intra- or inter-subunit cavities, compared to only 7% in the closed-channel α4β2. No CHOL was found in cavities within a single subunit, though some CHOL infiltrated into the gaps between subunits. Lack of CHOL in the deeply buried sites of the α4β2 nAChR seems to conflict with the previous observation by Brannigan et al..16 However, we did not place CHOL molecules into potential binding sites in the protein in advance of simulations as was done in their study.16 One could not rule out the possibility that CHOL molecules would “naturally” migrate into the deeply buried sites of the α4β2 if the simulation time was a few orders of magnitude longer.
POPA
The majority of interfacial POPA lipids interacted with α4β2 nAChR through H-bonding or salt bridging. POPA had nearly three times more H-bonds in the open-channel system than in the closed system. The H-bonds in the open-channel system also lasted two times longer on average. The same tendency held true for salt bridges. All five subunits of α4β2 nAChR in the open-channel system had salt bridges contributed by POPA. On average, the open-channel system had five times more POPA-protein salt bridges than the closed-channel system. Moreover, the salt bridges could last several nanoseconds in the open-channel model whereas they lasted only ∼100 ps in the closed channel.
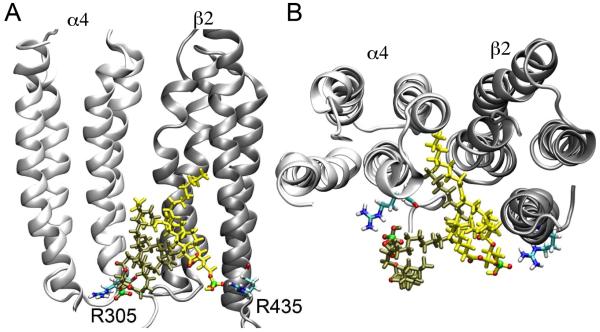
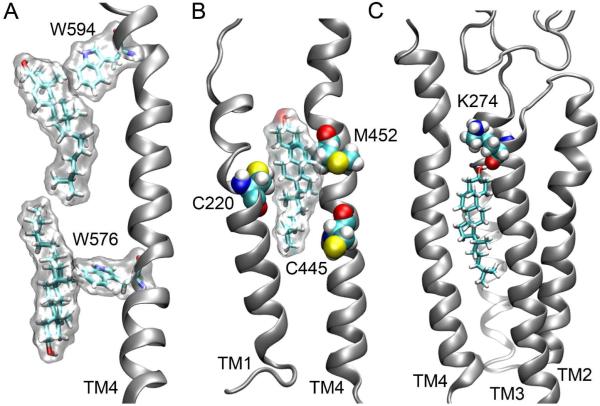
H-bonds and salt bridges were formed almost exclusively at the interfaces of TM and EC or TM and IC domains of the α4β2. In the open-channel model, more than 30% of the interfacial POPA formed salt bridges with highly conserved Arg residues near the IC domains, including α4-R305, β2-R299, α4-R572, and β2-R435. These salt bridges often lasted multiple nanoseconds. Fig. 2 shows that two POPA molecules formed salt bridges with two adjacent Arg residues (α4-R305 and β2-R435) in the open-channel system. One of the POPA partially intruded into a cavity between the α4 and β2 subunits. This situation did not occur in the closed-channel system, presumably due to a smaller cavity size. We also noticed that the aforementioned Arg residues were more exposed to the lipids in the open-channel than in the closed-channel nAChR. This explains why we observed more salt bridges between POPA and Arg residues in the open channel. Interestingly, the X-ray structure of KcsA4 also revealed key Arg residues in each of the four subunits that are particularly attractive to anionic phospholipids.32 Occupancy of anionic lipids at non-annular sites of the KcsA homotetramer has been suggested to be critical for channel opening.33,34
Figure 2.
(A) Side view of two POPA lipids forming salt bridges with two adjacent Arg residues (α4-R305 and β2-R435); and (B) Top-view of these two POPA lipids penetrating into an inter-subunit protein cavity between the α4 (white) and β2 (silver) subunits. Two POPA lipids (colored in yellow and tan) are shown in licorice format with oxygen and phosphor atoms highlighted in red and green, respectively. Arg residues are plotted in licorice format. Nitrogen, oxygen, hydrogen, and carbon atoms are shown in blue, red, white, and cyan, respectively.
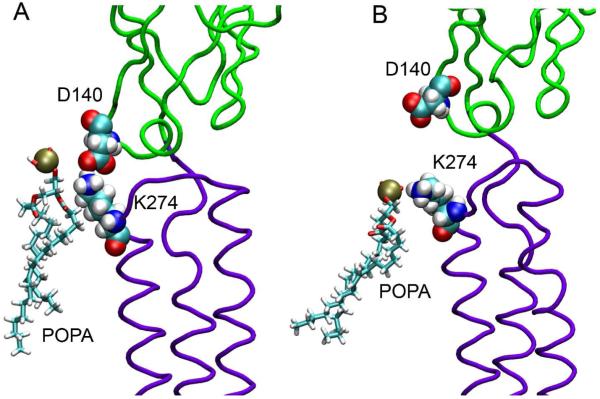
Lys residues (β2-K274) at the TM2-TM3 linker can potentially form salt bridges with D140 in the Cys-loop, as depicted in Fig. 3. Formation and breakage of the salt bridges at the EC-TM interface of Cys-loop receptors, such as nAChR, was believed to alter receptor channel functions.35 Furthermore, the involvement of POPA in this region changed the overall charging pattern of the gating interface, whose effects on the channel gating seems inevitable.36 While the α subunits provide agonist-binding sites, the β subunits also contribute to channel gating, as suggested by both in vitro and in vivo studies.37,38 Fig. 3 shows that the negatively-charged POPA can compete with D140 for salt bridging with β2-K274, leading to a change of the overall charging pattern at the gating interface of the α4β2 nAChR.
Figure 3.
POPA competes with D140 to form a salt bridge with K274 in the β2 subunit. (A) A salt bridge formed between D140 and K274; (B) A salt bridge formed between K274 and POPA. EC and TM domains are colored in green and purple, respectively. Nitrogen, oxygen, hydrogen, phosphor, and carbon atoms are shown in blue, red, white, tan, and cyan, respectively.
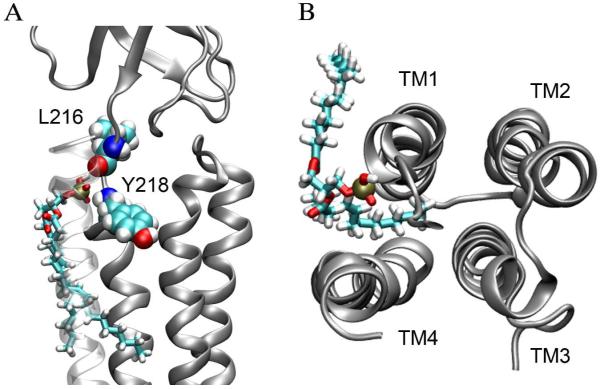
Besides its privilege for salt bridging, POPA molecules also demonstrated their uniqueness in H-bonding. They not only acted as H-bond acceptors using their oxygen atoms (just like POPC), but also acted as H-bond donors using their hydroxyl group. POPC almost exclusively formed H-bonds with side-chains of the protein, but POPA could form H-bonding with backbone amides or carbonyls of residues near the pre-TM1 region of the open-channel nAChR. Fig. 4 demonstrates such an example of POPA forming H-bonds with α4-L216 (backbone carbonyl) and α4-Y218 (backbone amide) simultaneously. Intermittently, this POPA also formed H-bonds with the backbone amide of the nearby α4-T219 and α4-I220 residues. This type of H-bonding with the protein backbone exhibited a much long lifetime (∼10 ns). Strong POPA H-bonds with residues in the pre-TM1 region of the open channel enhance the conformation favorable for channel opening.
Figure 4.
(A) Side view of a POPA lipid bound to a putative binding pocket near the pre-TM1 segment in the α4 subunit; and (B) top view of the POPA lipid penetrating into an intra-subunit protein pocket. The POPA lipid is shown in licorice format and the protein subunit is shown in silver. Nitrogen, oxygen, hydrogen, phosphor, and carbon atoms are shown in blue, red, white, tan, and cyan, respectively.
Overall, the interfacial POPA can achieve what POPC cannot with regard to the interactions with the α4β2 nAChR. The salt bridging through negative charges of POPA and the H-bonding through the hydroxyl group of POPA form specific interactions at the protein sites that are potentially important for channel gating. Furthermore, the smaller head group enables POPA to interact with the protein more closely and access non-annular sites (i.e. near pre-TM1; see Fig. 4) that are not reachable by POPC due to steric exclusion. This observation is consistent with a recent report on the role of anionic lipid head group size in stabilizing a functional nAChR.39 The existence of more long-lasting salt bridges and H-bonds between POPA and the α4β2 nAChR in the open-channel system than the closed-channel system suggested a possibility that POPA could shift the equilibration between open- and closed-channel states toward the open states. All the aforementioned POPA effects could be further amplified when POPA-rich domains11,13 form around the nAChR.
CHOL
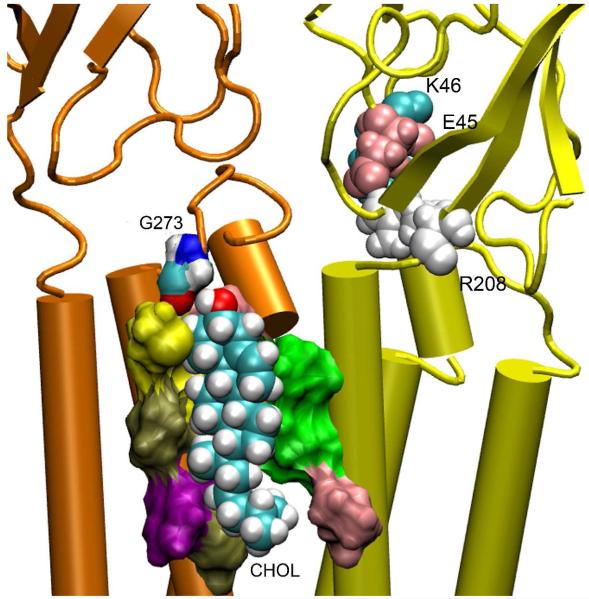
There were on average nine CHOL molecules exposed to the protein in both open- and closed-channel systems over the course of MD simulations. The majority were found close to TM4. This observation is consistent with photoaffinity labeling studies using Azicholesterol.10 CHOL molecules typically interact with the protein via H-bonding and hydrophobic interactions. They also interact with aromatic residues through π-stacking. Fig. 5 demonstrates several snapshots of such interactions. Early fluorescence-quenching and energy-transfer measurements suggested that CHOL might occupy sites in the vicinity of transmembrane cysteine residues of the γ subunit of Torpedo nAChR.40 In our open-channel system, we found a putative CHOL-binding pocket involving transmembrane cysteines (C445, C220) of the β2 subunit, as shown in Fig. 5B. We also found that CHOL could make special efforts for strong H-bonding at non-annular sites. At the beginning of the simulation for the open-channel system, a CHOL molecule was ∼10 Å away from K274 in one of the β2 subunits, but three POPC lipids were 6 Å closer than CHOL to this residue. Within a 5 ns simulation, the CHOL moved 8 Å for H-bonding with the backbone carbonyl of β2-K274, as shown in Fig. 5C. Non-annular CHOL binding sites were also found in the closed-channel system. An example is shown in Fig. 6. A CHOL fits well into the groove between α4 and β2 subunits. This CHOL molecule formed H-bonding with β2-G273 intermittently. The CHOL binding at the interface of α4 and β2 might have stabilized the adjacent salt bridge embracing α4-E50 (homolog to α1-E45 in 2BG9) and α4-R213 (homolog to α1-R209 in 2BG9). This salt bridge remained formed throughout a 20 ns MD simulation, while the same type of salt-bridge in other α4 subunits broke in both the open and closed models. The electrostatic interactions (i.e. α1-E45 and α1-R209 in 2BG9) at the gating interface are particularly important for channel functions.36
Figure 5.
(A) π-stacking ineraction (W594) and hydrophobic interaction (W576) formed between Trp residues in the α4 subunit and CHOL molecules; (B) CHOL bound to three residues containing sulphur atoms (shown in yellow) in the β2 subunit; and (C) in the open channel model, CHOL formed H-bonding with K-274 intermittently in the β2 subunit. Nitrogen, oxygen, hydrogen, and carbon atoms are shown in blue, red, white and cyan, respectively.
Figure 6.
A CHOL occupied an inter-subunit groove between the α4 (yellow) and β2 (orange) subuints in the closed model. The CHOL (shown in VDW format) formed H-bonding with β2-G273 (shown in surface format) intermittently. Other residues forming the binding pocket are colored differently. The salt bridge between α4-R213 and α4-E50 in the α4 subunit was retained during the entire MD simulations.
Order parameters of interfacial lipids
Lipid fluidity and elasticity were previously proposed to affect nAChR functions.1,2 However, the correlation between bulk lipid order and nAChR function was questioned by the observation9 that the nAChR in higher ordering 3:2 POPC/CHOL was predominately desensitized (non-functional), whereas the majority of nAChR in 3:2 POPC/POPA was functional, even though 3:2 POPC/POPA membranes exhibited lower ordering. The dynamic property of lipids, especially interfacial lipids, in relation to protein function remains obscure.
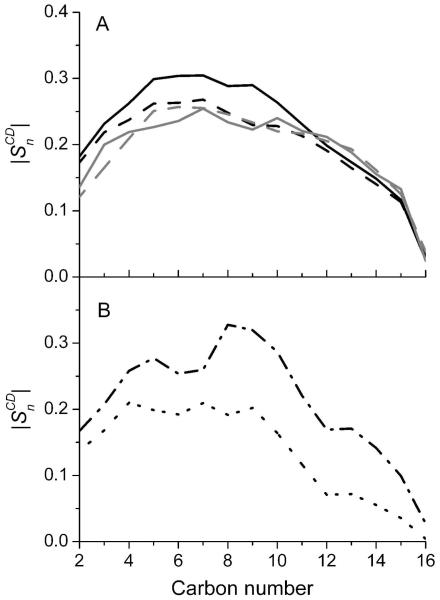
Fig. 7A gives a comparison of the order parameters of all POPA and POPC molecules in the open- and closed-channel systems. POPA shows higher SCD values than POPC in both systems due to its higher interaction probability with CHOL as found in the previous ternary lipid simulations.17 The interfacial POPC had slightly lower SCD values than the bulk POPC, but the averaged SCD values of 14 interfacial POPA molecules, as shown in Fig. 7B, were 20-30% lower than that of overall POPA.
Figure 7.
(A) Comparison of the order parameters for POPA (black) and POPC (gray) in the open channel model (solid line) and closed model (dashed line). (B) Comparison of the order parameters for those POPA molecules forming salt bridges (black dash-dotted line) and the overall interfacial POPA (black dotted line) in the open channel.
Gauche defects might be partially responsible for the lower order parameters of the interfacial lipids. We determined gauche defects by measuring the fraction of gauche dihedrals of the interfacial and bulk lipids using the same method as previously reported for the ternary lipids.17 We found that the total number of gauche defects in the interfacial POPC or POPA was ∼2.2 for the sn1 and sn2 chains, ∼10% to 20% greater than the corresponding number in the bulk lipids. An increase in the gauche content of the lipid tails induced by the presence of a hydrophobic transmembrane helix was also observed previously using sum-frequency vibrational spectroscopy, where an increase in the lipid flip-flop rate was found to associate with the raise in the gauche content.41 Besides gauche defects, we suspected that there might be other factors causing considerably lower SCD values of interfacial POPA molecules. After carefully inspecting our simulation data, we noticed a mixed effect of protein-lipid interactions on SCD. The five interfacial POPA molecules that formed salt bridges with nAChR had higher SCD values than those of bulk POPA at a number of carbon positions (Fig. 7B). However, several interfacial POPA molecules had their sn1 chains protrude to the protein surface or cavities. The orientations of C-H bonds in these lipid chains relative to the bilayer normal were only 10 to 20° from the magic angle (57.4°), resulting in extremely low SCD values. Two of the 14 interfacial POPA molecules were found to have their hydrophobic tails located close to hydrophilic residues. Such a hydrophobic mismatch induced enormous lipid chain motion over the course of simulation and caused low SCD values. We also noticed that the interfacial POPA forming stable H-bonds with the protein could have very low SCD values due to the aforementioned reasons. A recent simulation study by Dickey and Faller13 also showed that a POPA lipid could lay almost orthogonal to the bilayer normal in order to form H-bonds with the TM4 helix of the δ-subunit of Torpedo nAChR.
In summary, the interfacial lipids had more gauche defects than the same types of lipids in bulk. The lipid-protein interaction drove lipids, especially POPA, into situations of hydrophobic mismatch and being oriented close to the magic angle, resulting in the apparent lower SCD values.
Conclusions
Agonist binding to the EC domains of nAChR induces conformational changes that could lead to channel opening in the TM domains. The coupling of the agonist binding to the channel gating is believed to proceed through two pathways: (a) covalent bonding of the β10 strand in the EC domain and the TM1 helix (i.e, pre-TM1 segment), and (b) interactions of the Cys-loop and β1-β2 linker in the EC domain with the linker connecting the TM2 and TM3 domains.35,36,42 Our simulation study has demonstrated that interfacial POPA and CHOL could potentially engage in the channel gating process through direct interactions with critical residues involved in both pathways, as shown in Figs. 3-6. Particularly, POPA and CHOL interact specifically with residue β2-K274 in different subunits, either through salt bridging or H-bonding. Since β2-K274 is a highly conserved residue in the TM2-TM3 linker of various subtypes of nAChR (i.e., α1-K274, β1-R282, γ1K285, and δ1-K290 in Torpedo nAChR), what occurred here in the α4β2 nAChR could also happen in other subtypes of nAChR.
A small head group of negative charges is unique for POPA; the small size makes it more accessible to some non-annular sites that could not be easily reached by POPC due to steric exclusion. POPA can associate with the protein more closely and occupy intra- or inter-subunit protein cavities more effectively than POPC. The charged head group of POPA allows the lipid to form stable salt bridges with conserved Arg and Lys residues that may be critical to channel conformational changes, whereas apparently POPC does not possess such an ability. POPC can act as an H-bond acceptor and form H-bonds with side chains of the protein. POPA can accomplish the same, but it can also act as H-bond donor through its hydroxyl group. POPA form H-bonds not only with the side chains, but also the backbone of the protein. These unique features enable the interfacial POPA to achieve what POPC cannot with regard to specific interactions with the protein, thereby making POPA essential for the function of nAChR.
Acknowledgements
The authors would like to thank Dr Esmael Haddadian for helpful discussions. The computation was supported in part by the National Science Foundation through TeraGrid resources provided by the Pittsburgh Supercomputing Center. TeraGrid systems are hosted by Indiana University, LONI, NCAR, NCSA, NICS, ORNL, PSC, Purdue University, SDSC, TACC and UC/ANL. This research was supported in part by grants from NIH (R01GM066358, R01GM056257, and R37GM049202).
References
- 1.Baenziger JE, Darsaut TE, Morris ML. Biochemistry. 1999;38:4905–4911. doi: 10.1021/bi990181l. [DOI] [PubMed] [Google Scholar]
- 2.Baenziger JE, Morris ML, Darsaut TE, Ryan SE. J. Biol. Chem. 2000;275:777–784. doi: 10.1074/jbc.275.2.777. [DOI] [PubMed] [Google Scholar]
- 3.Powl AM, East JM, Lee AG. Biochemistry. 2005;44:5873–5883. doi: 10.1021/bi047439e. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 5.Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux JP, Delarue M, Corringer PJ. Nature. 2009;457:111–114. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- 6.Fong TM, McNamee MG. Biochemistry. 1986;25:830–840. doi: 10.1021/bi00352a015. [DOI] [PubMed] [Google Scholar]
- 7.Barrantes FJ. Brain Res. Brain Res. Rev. 2004;47:71–95. doi: 10.1016/j.brainresrev.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Hamouda AK, Sanghvi M, Sauls D, Machu TK, Blanton MP. Biochemistry. 2006;45:4327–4337. doi: 10.1021/bi052281z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.daCosta CJ, Ogrel AA, McCardy EA, Blanton MP, Baenziger JE. J. Biol. Chem. 2002;277:201–208. doi: 10.1074/jbc.M108341200. [DOI] [PubMed] [Google Scholar]
- 10.Hamouda AK, Chiara DC, Sauls D, Cohen JB, Blanton MP. Biochemistry. 2006;45:976–986. doi: 10.1021/bi051978h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poveda J, Encinar J, Fernndez A, Mateo C, Ferragut J, Gonzlez-Ros J. Biochemistry. 2002;41:12253–12262. doi: 10.1021/bi0200099. [DOI] [PubMed] [Google Scholar]
- 12.Wenz JJ, Barrantes FJ. Biochemistry. 2005;44:398–410. doi: 10.1021/bi048026g. [DOI] [PubMed] [Google Scholar]
- 13.Dickey AN, Faller R. Biophys. J. 2008;95:5637–5647. doi: 10.1529/biophysj.108.136895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilf RJ, Dutzler R. Nature. 2009;457:115–118. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- 15.Hilf RJ, Dutzler R. Nature. 2008;452:375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- 16.Brannigan G, Henin J, Law R, Eckenhoff R, Klein ML. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14418–14423. doi: 10.1073/pnas.0803029105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng MH, Liu LT, Saladino AC, Xu Y, Tang P. J. Phys. Chem. B. 2007;111:14186–14192. doi: 10.1021/jp075467b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Unwin N. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 19.Suhre K, Sanejouand YH. Nucleic Acids Res. 2004;32:W610–614. doi: 10.1093/nar/gkh368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haddadian EJ, Cheng MH, Coalson RD, Xu Y, Tang P. J. Phys. Chem. B. 2008;112:13981–13990. doi: 10.1021/jp804868s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamouda AK, Sanghvi M, Chiara DC, Cohen JB, Blanton MP. Biochemistry. 2007;46:13837–13846. doi: 10.1021/bi701705r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 23.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nose S. J. Chem. Phys. 1984;81:511–519. [Google Scholar]
- 25.Hoover WG. Phys. Rev. A. 1985;31:1695. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 26.Brünger A. X-PLOR, Version 3.1: A system for X-ray crystallography and NMR. Yale University; New Haven: 1992. [Google Scholar]
- 27.Darden T, York D, Pedersen L. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 28.Humphrey W, Dalke A, Schulten K. J. Mol. Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 29.Jones OT, McNamee MG. Biochemistry. 1988;27:2364–2374. doi: 10.1021/bi00407a018. [DOI] [PubMed] [Google Scholar]
- 30.Christova Y, James PS, Cooper TG, Jones R. J Androl. 2002;23:384–392. [PubMed] [Google Scholar]
- 31.Gaede HC, Gawrisch K. Biophys J. 2003;85:1734–1740. doi: 10.1016/S0006-3495(03)74603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deol SS, Domene C, Bond PJ, Sansom MS. Biophys. J. 2006;90:822–830. doi: 10.1529/biophysj.105.071407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marius P, Alvis SJ, East JM, Lee AG. Biophys. J. 2005;89:4081–4089. doi: 10.1529/biophysj.105.070755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marius P, Zagnoni M, Sandison ME, East JM, Morgan H, Lee AG. Biophys. J. 2008;94:1689–1698. doi: 10.1529/biophysj.107.117507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kash TL, Jenkins A, Kelley JC, Trudell JR, Harrison NL. Nature. 2003;421:272–275. doi: 10.1038/nature01280. [DOI] [PubMed] [Google Scholar]
- 36.Xiu X, Hanek AP, Wang J, Lester HA, Dougherty DA. J. Biol. Chem. 2005;280:41655–41666. doi: 10.1074/jbc.M508635200. [DOI] [PubMed] [Google Scholar]
- 37.Maskos U, Molles BE, Pons S, Besson M, Guiard BP, Guilloux JP, Evrard A, Cazala P, Cormier A, Mameli-Engvall M, Dufour N, Cloez-Tayarani I, Bemelmans AP, Mallet J, Gardier AM, David V, Faure P, Granon S, Changeux JP. Nature. 2005;436:103–107. doi: 10.1038/nature03694. [DOI] [PubMed] [Google Scholar]
- 38.Dahan DS, Dibas MI, Petersson EJ, Auyeung VC, Chanda B, Bezanilla F, Dougherty DA, Lester HA. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10195–10200. doi: 10.1073/pnas.0301885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baenziger JE, Medaghlia SA, daCosta CJ, Lavigne N. Biophys. J. 2008;94:1408. Pos. [Google Scholar]
- 40.Narayanaswami V, McNamee MG. Biochemistry. 1993;32:12420–12427. doi: 10.1021/bi00097a021. [DOI] [PubMed] [Google Scholar]
- 41.Anglin TC, Liu J, Conboy JC. Biophys J. 2007;92:L01–03. doi: 10.1529/biophysj.106.096057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grosman C, Salamone FN, Sine SM, Auerbach A. J. Gen. Physiol. 2000;116:327–340. doi: 10.1085/jgp.116.3.327. [DOI] [PMC free article] [PubMed] [Google Scholar]