Abstract
Guanine nucleotide-exchange factors (GEFs) stimulate guanine nucleotide exchange and the subsequent activation of Rho-family proteins in response to extracellular stimuli acting upon cytokine, tyrosine kinase, adhesion, integrin, and G-protein coupled receptors (GPCRs). Upon Rho activation, several downstream events occur, such as morphological and cytokskeletal changes, motility, growth, survival, and gene transcription. The RhoGEF Leukemia-Associated RhoGEF (LARG) is a member of the Regulators of G-protein Signaling Homology Domain (RH) family of GEFs originally identified as a result of chromosomal translocation in acute myeloid leukemia. Using a novel fluorescence polarization guanine nucleotide binding assay utilizing BODIPY-Texas Red-GTPγS (BODIPY-TR-GTPγS), we performed a ten-thousand compound high-throughput screen for inhibitors of LARG-stimulated RhoA nucleotide binding. Five compounds identified from the high-throughput screen were confirmed in a non-fluorescent radioactive guanine nucleotide binding assay measuring LARG-stimulated [35S] GTPγS binding to RhoA, thus ruling out non-specific fluorescent effects. All five compounds selectively inhibited LARG-stimulated RhoA [35S] GTPγS binding, but had little to no effect upon RhoA or Gαo [35S] GTPγS binding. Therefore, these five compounds should serve as promising starting points for the development of small molecule inhibitors of LARG-mediated nucleotide exchange as both pharmacological tools and therapeutics. In addition, the fluorescence polarization guanine nucleotide binding assay described here should serve as a useful approach for both high-throughput screening and general biological applications.
Keywords: high-throughput screening, fluorescence polarization, RhoGEF, RhoA, LARG, Drug Discovery
Introduction
The Rho-family of small GTPases are a main subset of the Ras superfamily of small GTPases (∼ 20-25 kDa) 1. The Rho-family of GTPases is further subdivided into ten subgroups based upon their identity. In this work, we focus on the small GTPase RhoA, which is a member of the Rho subgroup (RhoA, RhoB, and RhoC) within the Rho-family of proteins. In the cell, Rho GTPases are activated upon ligand activation of cell surface receptors, such as cytokine, tyrosine kinase, adhesion, integrin, and G-protein coupled receptors (GPCRs) 2. As a result, this causes Rho GTPases to switch from their inactive GDP-bound form to their active GTP-bound form leading to various downstream cellular events, such as cytoskeletal and morphological changes, migration, growth, survival, and gene transcription 1.
The activation state of Rho GTPases is controlled by several classes of regulatory proteins. The GTPase-activating proteins (GAPs) catalyze GTP hydrolysis by active Rho proteins leading to their inactivation. The guanine nucleotide-dissociation inhibitors (GDIs) sequester GDP-bound Rho GTPases in the cytosol, thus preventing their subcellular localization to the plasma membrane and activation. Lastly, the guanine nucleotide exchange factors (GEFs) stimulate the exchange of GTP for GDP upon Rho GTPases leading to their activation, and are the focus of this work. There are more than sixty identified human Rho GEFs that specifically activate Rho GTPases 1.
Rho GEFs are characterized by their Dbl homology (DH) (∼200 residues) and C-terminal adjacent pleckstrin homology (PH) (∼100 residues) domains. DH domains are primarily responsible for catalyzing the guanine nucleotide exchange (GTP for GDP) upon Rho GTPases 1. PH domains are responsible for membrane localization of Rho GEFs and aid the DH domain in catalysis of guanine nucleotide exchange 1. The focus of this work is upon the leukemia-associated RhoGEF (LARG), which was originally identified as a chromosomal translocation fusion protein to mixed-lineage leukemia (MLL) in a patient with acute myeloid leukemia 3. LARG is a member of the regulators of G-protein signaling homology domain (RH) containing RhoGEF family of proteins (LARG, p115- RhoGEF, PDZ-RhoGEF) 4. This family of Rho GEFs is linked to Rho-dependent signaling pathways controlled by heterotrimeric G-protein coupled receptors coupled to the Gα12 and Gα13 subunits, such as the lysophosphatidic acid, bombesin, and thrombin receptors 4-7.
LARG is an important regulatory protein in several clinical disorders. Considering its original identification in acute myeloid leukemia, LARG has largely been thought to be a key player in cancer progression. In conjunction with the Raf-1 kinase, LARG has been shown to transform mouse fibroblasts 8. It has also been shown to play a role in cell migration and growth of head and neck squamous cancer cells through adhesion and tyrosine kinase receptor interaction and subsequent RhoA activation 9. LARG has also been shown, through overexpression and siRNA knockdown, to play a role in thrombin and bombesin receptor-mediated prostate cancer cell morphology changes and migration 6, 7. In addition to cancer, LARG has been implicated in lectin signaling in dendritic cells, which is important in sequestration of human immunodeficiency virus (HIV) virions leading to the progression of the HIV disease 10. LARG also has been shown to play a role in vascular biology and calcium signaling. It has been directly linked to a role in salt-induced hypertension in transgenic mice and it has been speculated to play a role in erectile dysfunction 11-13. As a result, considering the implication of LARG in multiple diseases and disorders, it is an attractive molecular target for drug discovery.
Currently, there are limited pharmacological tools targeting RhoGEFs and RhoGTPases. To date, most of the efforts have focused upon inhibiting the lipid modification of RhoGTPases, which is necessary for plasma membrane localization and activation. The inhibitors of this modification include farnesyltransferase and geranylgeranyl transferase inhibitors and the cholesterol-lowering statin drugs 14, 15. Unfortunately, these inhibitors and drugs are not specific to RhoGTPases, which complicates the mechanistic interpretation of results using these inhibitors. To date, there are only two reported specific inhibitors of RhoGTPases. Both inhibitors selectively inhibit Rac1 but they differ in their mechanism of inhibition. The inhibitor NSC23766 (IC50, ∼50 μM) inhibits by binding to the GEF binding pocket on Rac1 16, and EHT 1864 (Kd, 40 nM) inhibits by binding to the nucleotide binding pocket on Rac1 17, 18. Although, there are a few examples of selective RhoGTPase inhibitors, there are no published examples to date of inhibitors that target RhoGEFs.
Therefore, in this study, we performed a ten-thousand compound high-throughput screen for small molecule inhibitors of LARG-stimulated RhoA nucleotide exchange. In order to do this screen, we first developed a novel fluorescence polarization (FP) RhoA nucleotide binding assay using BODIPY-TR-GTPγS. We chose fluorescence polarization rather than both the standard fluorescence intensity and radioactive [35S] GTPγS binding approaches because it proved to be more environmentally friendly, robust, reliable, and reproducible. Out of ten-thousand compounds screened, six compounds were confirmed to inhibit LARG-mediated guanine nucleotide exchange. Out of these six compounds, five confirmed in a non-fluorescent, radioactive [35S] GTPγS guanine nucleotide binding assay with IC50 values in the micromolar range. These compounds showed selectivity for LARG-stimulated RhoA [35S] GTPγS binding by having little to no effect upon RhoA and Gαo [35S] GTPγS binding. Therefore, as a result of the high-throughput screen with this novel fluorescence polarization guanine nucleotide binding assay, five promising compounds were identified as inhibitors of LARG-stimulated RhoA nucleotide binding. Therefore, with further synthetic chemistry follow-up, these compounds may lead to useful inhibitors of LARG-stimulated RhoA nucleotide binding for both research and therapeutic purposes.
Materials and Methods
Plasmids, Protein Purification, and Chemical Reagents
Human RhoA (residues 1-189, C189S) was expressed in Escherichia coli as described previously 19. Human LARG encoding the DH/PH domains (residues 765-1138) was expressed in Escherichia coli as described previously 20. Gαo expression and purification in Escherichia coli was described previously 21. BODIPY® Texas-Red (TR) guanosine 5′-O-(3-thiotriphospahte) (GTPγS) was obtained from Molecular Probes – Invitrogen (Eugene, OR). [35S] GTPγS was obtained from Perkin Elmer (Waltham, MA). GTPγS was obtained from EMD Biosciences (San Diego, CA). The non-ionic detergents IGEPAL and Lubrol were from Sigma (St. Louis, MO). The 10,000 structurally diverse chemical compounds were obtained from ChemBridge (San Diego, CA) as part of the collection of the University of Michigan Center for Chemical Genomics (CCG). The chemical similarity was low – at 80% similarity calculated with the ICMPro (Molsoft LLC, La Jolla, CA) clustering algorithm there were 4390 clusters with a median size of 1 compound and mean size of 2.28 compounds.
Guanine Nucleotide Binding Fluorescence Polarization Assays
Exchange buffer (20mM Tris HCl pH 7.5, 150 mM NaCl, 10 mM MgCl2, 10% Glycerol, 0.01% IGEPAL, freshly prepared 1mM DTT) was added to each well of a black 96-well plate. Purified full-length human RhoA(C189S), purified DH/PH domain of human LARG, and BODIPY-FL-GTPγS or BODIPY-TR-GTPγS were added sequentially to each well to a final volume of 100 μL per well. Fluorescein or Texas-Red fluorescence polarization was read in a Victor2 plate reader using excitation at 485 nm and emission at 535 nm for fluorescein, or an excitation at 560 nm and emission at 630 nm for Texas-Red. The measured values of polarization (mP) were calculated by using the formula: mP = (F‖ - F⊥)/(F‖ + F⊥) where F‖ = fluorescence intensity parallel to the excitation plane, F⊥ = fluorescence intensity perpendicular to the excitation plane. The statistical Z′ – factor used to assess assay suitability for high-throughput screening was calculated by using the formula, Z′ = 1 − [(3σc+ + 3σc-)/(|μc+ - μc-|)] where σ = standard deviation, μ = mean, c+ = with LARG, c- = without LARG).
RhoA [35S] GTPγS Guanine Nucleotide Binding Assay
The indicated concentrations of purified DH/PH domain of human LARG (0.5-2 nM, final) are added to a tube in Buffer I (20 mM Tris pH 7.5, 1 mM EDTA, 1 mM DTT, 50 mM NaCl, 0.1% Lubrol, 2 mM MgCl2) in a final volume of 180 μL. To this mixture, 45 μl of purified human RhoA (C189S) in Buffer I is added to yield a final concentration of 500 nM. The reaction was initiated by adding 225 μL of 2X Binding Buffer (100 mM Tris pH 7.5, 1 mM EDTA, 2 mM DTT, 100 mM NaCl, 10 mM MgCl2, 5 μM GTPγS, 0.1% Lubrol, 6.75 μCi [35S] GTPγS) for a final reaction volume of 450 μL. Reaction mixtures were incubated at room temperature for 1, 5, 10, 30, 60, 120, and 180 minutes. 50 μL of reaction mixture was removed and diluted in a tube containing 4 mL of ice-cold Wash Buffer (20 mM Tris pH 8.0, 100 mM NaCl, 25 mM MgCl2) to stop the reaction. An additional 4 mL of Wash Buffer was added to the tube and the sample filtered on a BA85 25mm nitrocellulose filter using a Hoeffer filtration system. Filters were washed two times with 4 mL of Wash Buffer. Filters were dried under a heat lamp for 5 minutes. Filters were counted in 4 mL of scintillation fluid (Scintiverse) for 1 min using a Beckman LS 5801 Scintillation Counter. The identical method was followed for RhoA [35S] GTPγS binding studies without LARG, except reaction mixtures were incubated at room temperature for 1, 5, 10, 30, 60, 130, and 180 minutes.
Gαo [35S] GTPγS Guanine Nucleotide Binding Assay
Purified Gαo was diluted to a final concentration of 10 μM in 180 μL of Gαo dilution buffer (10 mM HEPES pH 7.7, 1mM EDTA, 0.1% Lubrol, 1 mM DTT). [35S] GTPγS binding was initiated by adding 180 μL of Gαo binding cocktail (50 mM HEPES pH 7.7, 1 mM EDTA, 40 mM MgCl2, 200 mM NaCl, 2 μM GTPγS, 6.75 μCi [35S] GTPγS, 1 mM DTT) and incubated at room temperature for 1, 5, 10, 30, 60, 90, and 120 minutes. 40 μL of the reaction mixture was diluted in a tube containing 4 mL of ice-cold Wash Buffer (20 mM Tris pH 8.0, 100 mM NaCl, 25 mM MgCl2). This reaction mixture was then filtered on a BA85 25 mm nitrocellulose filter using a Hoeffer filtration system. Filters were washed twice with 4 mL of Wash Buffer, then dried under a heat lamp for 5 minutes. Filters were counted in 4 mL of scintillation fluid for 1 min using a Beckman LS 5801 Scintillation Counter.
High-Throughput LARG-stimulated RhoA Guanine Nucleotide Binding Fluorescence Polarization Screen
Using a Multidrop 384 (Thermo), exchange buffer was added to each well of a low volume black 384 well plate (Corning, cat.#: 3676); - 20 μL was added for the positive control wells (i.e. no LARG) and 15 μL was added to all other wells. Using a 384-well pin tool on a Biomek FX liquid handling workstation (Beckman Coulter), approximately 250 nL of compound (stock concentration 0.75-4 mM - ChemBridge) or DMSO, for control wells, was added (producing final compound concentrations of ∼ 6-30 μM and 0.8% DMSO). Plates were then incubated at room temperature for 5 minutes. Using a Multidrop micro (Thermo) 5 μL of purified DH/PH domain of human LARG in exchange buffer was added to the wells of the 384-well plate to produce a final concentration of 100 nM. Plates were then incubated for 5 minutes at room temperature. Using a Multidrop micro 5 μL of purified human RhoA (C189S) in exchange buffer was added to produce a final concentration of 2 μM. Plates were then incubated for 2 minutes at room temperature. The reaction was initiated by using a Multidrop micro to add 5 μL of BODIPY-TR GTPγS in exchange buffer to a final concentration of 500 nM and a final reaction volume of 30 μL. Plates were incubated for 20 minutes at room temperature, then read for fluorescence polarization with a PHERAstar (BMG LabTech) plate reader with a 575 nm band pass excitation filter, a 620 nm band pass emission filter, a dichroic mirror, and a dual emission beam splitter to permit simultaneous recording of parallel and perpendicular fluorescence readings. Plates were individually incubated after each addition so that all plates would be read in the PHERAstar plate reader precisely at 20 minutes after GTPγS addition (i.e. prior to saturation of the reaction time course). For follow-up dose-response studies of the primary hits, the same method was carried out as for the high-throughput screen, except reagents were added by hand with a multichannel pipet. In addition, compounds were added to the wells of the 384 well plate using serial two-fold dilutions over a range of concentrations between 100 μM and 3.125 nM.
Compound Properties
Compound properties were calculated with ChemAxon JChem software. The LogP value method is described in Viswanadhan et al. 22.
Results
Fluorescence Polarization Guanine Nucleotide Binding Assay
In order to study RhoA guanine nucleotide exchange in vitro, we used a novel fluorescence polarization guanine nucleotide binding assay adapted from the standard fluorescence intensity guanine nucleotide binding assay described previously by our laboratory 23, 24. This fluorescence polarization assay utilizes the non-hydrolysable fluorescently labeled guanine nucleotide BODIPY-GTPγS fluorophores. LARG-stimulated BODIPY-GTPγS fluorophore binding to RhoA results in fluorescence polarization that can be read by a plate reader (Fig. 1A). The structures of the BODIPY-GTPγS fluorophores used in this study are shown in figure 1C.
Fig. 1. GTPase Cycle, Mechanisms of Inhibition, and Structure of GTP Analogs.

(A) In its inactive form, the small GTPase RhoA is bound to both GDP and Mg+2. Upon the binding of the DH/PH domain of the RhoGEF LARG (which is the catalyst of the guanine nucleotide exchange reaction), a conformational change occurs in the switch regions of the small GTPase RhoA causing an intermediate to form that is free of nucleotide and Mg+2. This enables the higher concentrated nucleotide GTP and Mg+2 to bind to the small GTPase RhoA. In our in vitro assays described in this report, we utilize the BODIPY-Texas Red fluorescently labeled or [35S] radioactive labeled non-hydrolysable form of the guanine nucleotide, GTPγS. As a result, the GTP-bound RhoA can be released from the RhoGEF LARG in an active state. In our in vitro assays described in this report, this results in a polarized light signal in our fluorescence polarization assay, or a radioactive signal in our [35S] radioactive assay that can be quantitatively measured. (B) There are several potential mechanisms of inhibition of LARG stimulated RhoA nucleotide binding, which include: (I) competitive inhibition at the site of interaction between LARG and RhoA, (II) allosteric inhibition by drug binding to LARG, (III) competitive inhibition at the site of nucleotide binding, or (IV) allosteric inhibition by drug binding to RhoA. (C) The chemical structures of the three non-hydrolysable forms of the guanine nucleotide, GTPγS, used in both the fluorescence and radioactive in vitro assays described in this report are depicted.
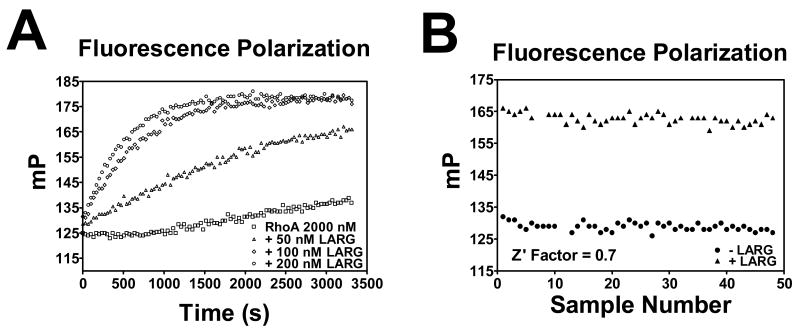
Due to the importance of having a robust and reproducible assay for high-throughput screening, we compared the suitability of nucleotide binding to RhoA measured by fluorescence intensity (data not shown) versus fluorescence polarization (Fig. 2). Although both intensity and polarization assays gave signals for RhoA nucleotide binding, only the fluorescence polarization assay (Z′ – factor = 0.7) gave suitably reproducible data appropriate for doing a high-throughput screen (Fig. 2). The fluorescence polarization assay takes into account fluorescence that is both parallel and perpendicular to the excitation plane and the ratiometric nature of the measurement cancels many contributions to the noise 25. This most likely accounts for the difference between the Z′-factors of the two assays. As a result, fluorescence polarization should be a useful assay for high-throughput screening for small molecule inhibitors of GEF stimulated small GTPase guanine nucleotide binding. In addition, the assay has recently been used for general biological applications studying GEF activity upon small GTPases 26.
Fig. 2. Fluorescence Polarization RhoA Nucleotide Binding Assays.
Purified DH/PH domain of LARG (200 nM) stimulated nucleotide stimulation of purified RhoA (C189S) (2μM) utilizing BODIPY-FL-GTPγS (1μM) and BODIPY-TR-GTPγS (1 μM). Values were measured in a Victor2 plate reader as described in the Materials and Methods. (A) LARG-dependent nucleotide binding to RhoA (C189S) (□) (2 μM) stimulated by LARG [50 nM (Δ), 100 nM (◊), 200 nM (○)] utilizing BODIPY-TR-GTPγS (1 μM) was measured over a time course of 55 minutes for fluorescence polarization by using a Victor2 plate reader as described in the Materials and Methods. (B) Fluorescence polarization measurements were taken from 48 wells not containing LARG [- LARG (●)] and 48 wells containing LARG [+ LARG (▲)] in a 96-well black plate after incubating for 20 minutes at room temperature. Data in panels A and B represent experiments performed in duplicate. Data in panel A and B are represented as mP polarization values. The Z′ – factor was calculated for the fluorescence polarization measurement. The Z′ – factor for the fluorescence polarization assay was 0.7 indicating an assay suitable for high-throughput screening.
Comparison between the Fluorescence Polarization and [35S] Guanine Nucleotide Binding Assays
The standard [35S] GTPγS nucleotide binding assay is generally not suitable for high-throughput screening due to time consumption, accumulation of environmental hazardous waste, and the complexity of adapting the assay to a 384-well plate format. Therefore, we chose the fluorescence polarization assay to use for our high-throughput screen. We found that in both assays LARG was able to stimulate RhoA (C189S) nucleotide binding in a concentration-dependent manner (Fig. 3A-B). However, the time courses for saturation in the two methods differed with saturation occurring at 40 minutes for the fluorescence polarization assay, and 180 minutes for the [35S] radioactive assay. The basal rate constant for nucleotide exchange was slower with the [35S] GTPγS method. The faster kinetics of binding in the fluorescence polarization assay may be due, in part, to: higher concentrations of RhoA and nucleotide, the lower amount of nucleotide relative to the amount of RhoA, the higher NaCl or MgCl2 concentrations or the non-native BODIPY nucleotide structure in the fluorescence polarization assay. However, plotting the rate constants on a graph both give a linear plot (Fig. 3C-D) suggesting that both the fluorescence polarization and radioactive nucleotide binding assays are appropriate for measuring GEF-stimulated nucleotide binding to the Rho-family of proteins.
Fig. 3. Dose-Dependent LARG-stimulated RhoA Nucleotide Binding.
(A) Dose-dependent nucleotide binding to RhoA (C189S) (□) (2 μM) stimulated by LARG [50 nM (Δ), 100 nM (◊), 200 nM (○)] utilizing BODIPY-TR-GTPγS (500 nM) was measured over a time course of 40 minutes in a Victor2 plate reader as described in the Materials and Methods. (B) Dose-dependent nucleotide binding to RhoA (C189S) (■) (500 nM) stimulated by LARG [0.5 nM (▲), 1nM (✦), 2nM (●)utilizing [35S] GTPγS was measured over a time course of 180 minutes by a Beckman LS 5801 Scintillation Counter as described in the Materials and Methods. The black dotted vertical line and the solid black circles indicate the time point (incomplete saturation point) used for the high-throughput screen. (C, D) Rate constants for the reactions were plotted versus the concentration of LARG yielding a linear plot for both the FP and radioactive binding assays. A linear regression plot was performed using Graphpad Prism 5, yielding R2 values of 0.9. Rate constants were calculated using the one phase exponential association equation Y = Ymax * (1-exp(-k*X) (k = rate constant for the reaction). Data in panel A were measured in duplicate and are representative data of N = 3. Data in panel A were background subtracted from BODIPY-TR-GTPγS fluorescence alone. Data in panel B represent the mean ± SEM of three separate experiments (N = 3).
High-Throughput Screen for Inhibitors of LARG-stimulated RhoA Nucleotide Exchange
A ten-thousand diverse chemical compound collection from ChemBridge was used to screen for small molecule inhibitors of LARG-stimulated RhoA nucleotide exchange. The fluorescence polarization guanine nucleotide binding assay utilizing the purified DH/PH domain of LARG and RhoA (C189S) was used for the screen. Compounds were added to a black 384-well low volume plate containing exchange buffer with a 384-well pin tool on the Biomek FX workstation. Then, the purified DH/PH domain of LARG, RhoA (C189S), and BODIPY-TR-GTPγS were added to the plate in sequential order. After a twenty minute incubation (non-saturating time point), polarization was read in a PHERastar plate reader as shown in figure 4A. The assay was tested for its suitability for high-throughput screening and reliability by using the Z′-factor statistical measurement. Utilizing purified RhoA (C189S) alone plus DMSO (positive control for screen – 100% inhibition) and the purified DH/PH domain of LARG and RhoA (C189S) plus DMSO (negative control for the screen – 0% inhibition), the Z′-factor in this high-throughput screening format was determined to be 0.52 indicating a suitable and reliable screen (Fig. 4B) 27. While many FP-type screens give higher Z′ factors than this, consideration should be taken of the fact that this is a kinetic assay done at a pre-saturation time point so it would not be expected to have quite as good a Z′ as a standard equilibrium FP binding assay.
Fig. 4. Fluorescence Polarization Screen for Inhibitors of LARG-stimulated RhoA Nucleotide Binding.

(A) Schematic of the protocol used for the 10,000 ChemBridge diverse chemical compound high-throughput screen (HTS) as described in the Materials and Methods. All samples, compounds and controls, contain 0.8% DMSO. (B) Control samples run on each plate include (100 nM) LARG as a negative (0% inhibition) control (●) and no LARG as a positive (100% inhibition) control ( ). The statistical Z′ – Factor measurement for the HTS was calculated to be 0.52 for this entire screen indicating suitable reproducibility. (C) Data from the 10,000 ChemBridge compound HTS (CCG compounds 5029-15028) are shown (●) with a 30 percent inhibition line indicated in red. (D) Dose-response follow-up of the positive hits from the HTS in the LARG-stimulated RhoA nucleotide BODIPY-TR-GTPγS binding assay measured at a time point of 20 minutes. Data in panel D were measured in duplicate and are represented as percent change from the +LARG negative control (0%) comapred to the no LARG positive control (100%).
). The statistical Z′ – Factor measurement for the HTS was calculated to be 0.52 for this entire screen indicating suitable reproducibility. (C) Data from the 10,000 ChemBridge compound HTS (CCG compounds 5029-15028) are shown (●) with a 30 percent inhibition line indicated in red. (D) Dose-response follow-up of the positive hits from the HTS in the LARG-stimulated RhoA nucleotide BODIPY-TR-GTPγS binding assay measured at a time point of 20 minutes. Data in panel D were measured in duplicate and are represented as percent change from the +LARG negative control (0%) comapred to the no LARG positive control (100%).
Compounds which showed more than 30% inhibition or values of % inhibition more than 3 standard deviations from the negative control were considered to be actives. Figure 4C shows 19 hits from the 30% inhibition criterion and an additional 12 were found with the 3 standard deviation criterion providing 31 initial actives (or a “hit” rate of 0.3%). There were several plates with clusters of compounds that just met the cutoff and they were retested in duplicate using the primary screening assay methodology. Any that confirmed in either of the two measurements were retained in the actives list and were studied further in a concentration response study using the BODIPY-TR-GTPγS fluorescence polarization assay. Out of those thirteen compounds, seven (0.07% - hit rate) did inhibit in this follow-up assay and six compounds did not (<50% inhibition at 100 μM) (Fig. 4D). Of the 7 confirmed actives, six were available for re-supply from ChemBridge for follow-up studies. The three most potent of these six compounds, CCG-14631 (4-{5-[(4,6-dioxo-2-sulfanylidene-1,3-diazinan-5-ylidene)methyl]furan-2-yl}-N-(1,3-thiazol-2-yl)benzene-1-sulfonamide) (Chembridge, cat.#: 6192873), CCG-12529 (7-(3-nitrophenyl)-8-azatricyclo[7.4.0.0ˆ{2,6}]trideca-1(13),3,9,11-tetraene-10-carboxylic acid) (ChemBridge, cat.#: 5584249) and CCG-5849 (4-amino-2-(5-amino-1,3-benzothiazol-2-yl)phenol) (ChemBridge, cat.#: 5312639)) had IC50 values between 3 μM and 10 μM (Fig. 4D, Table 1). The three remaining compounds, CCG-14113 ((N′Z)-2-[(3-methylphenyl)amino]-N′-[(2,4,6-tribromo-3-hydroxyphenyl)methylidene]acetohydrazide) (ChemBridge, cat#: 6119878), CCG-13528 ((5Z)-3-(2H-1,3-benzodioxol-5-yl)-2-imino-5-{[5-(2-nitrophenyl)furan-2-yl]methylidene}-1,3-thiazolidin-4-one) (ChemBridge, cat.#: 5354792), were less potent with IC50 values between 32 μM and 65 μM (Fig. 4D, Table 1) in the fluorescence polarization assay.
Table 1. Chemical Compound Properties and IC50 Values.
This table describes the chemical compound properties along with the IC50s and Hill Slopes for the 6 compounds identified in the HTS. The IC50 and Hill Slope values were taken from the dose-dependent data in the [35S] GTPγS assay in Fig. 4D and Fig. 5B. NA indicates that the compound had no inhibitory activity.
| Compound ID | Molecular Weight | Loq P | IC50 (μM) [TR-GTPγS] | Hill Slope [TR-GTPγS] | IC50 (μM) [GTPγS35] | Hill Slope [GTPγS35] |
|---|---|---|---|---|---|---|
| CCG-13528 | 435.4 | 4.13 | 39.8 | -3.4 | 4.6 | -1.8 |
| CCG-14631 | 460.5 | 1.89 | 3.1 | -0.7 | 7.0 | -1.1 |
| CCG-7167 | 397.4 | 5.52 | 65.1 | -1.8 | 4.2 | -2.2 |
| CCG-14113 | 520.0 | 5.40 | 32.2 | -1.0 | 32.8 | -7.1 |
| CCG-12529 | 336.3 | 4.33 | 4.9 | -1.0 | 27.5 | -2.3 |
| CCG-5849 | 257.3 | 1.97 | 10.7 | -1.6 | NA1 | NA1 |
Secondary Screen in [35S] GTPγS Radioactive Guanine Nucleotide Binding Assay
As mentioned previously, a potential mechanism for false positives in a fluorescence-based assay is quenching of the fluorescence signal. To address this concern, we utilized the traditional radioactive [35S] GTPγS guanine nucleotide filter binding assay as a secondary follow-up assay for our 6 confirmed active compounds. For these experiments, we utilized a time point of twenty-five minutes, prior to saturation on the kinetic curve in figure 5A. Five of the six candidate compounds showed dose-dependent inhibition in the [35S] GTPγS guanine nucleotide binding assay. The one compound that was not active in the radioligand assay showed a marked effect on the total fluorescence as calculated by Fparallel+2*Fperpendicular (54% reduction). The three most potent compounds in this radioactive-based assay, CCG-13528, CCG-14631, and CCG-7167, had IC50 values between 4 μM and 7 μM (Fig. 5B, Table 1). The other two compounds, CCG-14113 and CCG-12529, were less potent with IC50 values around 30 μM (Table 1). These IC50 values for several of the compounds differ between the [35S] GTPγS and fluorescence polarization methods. These differences can most likely be attributed to experimental differences between the assays such as NaCl and MgCl2 concentrations and the different nature of the detergent or nucleotide used. Despite these differences, the fact that 5 of the 6 tested compounds did show some activity in the [35S] GTPγS method is encouraging.
Fig. 5. Dose Response Inhibition of LARG-stimulated RhoA [35S] GTPγS Binding by HTS Hits.
(A) Time-course of guanine nucleotide binding of RhoA (C189S) (■) (500 nM) stimulated by LARG [2nM (●)] utilizing [35S] GTPγS was measured over a time course of 180 minutes by a Beckman LS 5801 Scintillation Counter as described in the Materials and Methods. (B) Compound dose-dependent inhibition of LARG (2 nM) stimulated RhoA (C189S) (500 nM) guanine nucleotide binding at a 25 minute time point. (C) Chemical structures of the 5 compounds that show dose-dependent inhibition in panel B. Data in panel A and B were measured in duplicate and represent mean ± SEM of three separate experiments (N = 3). Data in panel B is represented as percent of DMSO control for [35S] GTPγS binding.
CCG-13528 and CCG-14631 satisfy all of Lipinski's rules-of-five for chemical compounds that have the most potential in becoming a drug with respect to their physical properties 28. CCG-7167 had only one violation of Lipinski's Rule of Five due to a partition coefficient (log P) of 5.52 (Table 1). Interestingly, our three most potent compounds are all extended aromatic molecules. They have 4 (CCG-14631) or 5 (CCG-13528, CCG-7167) rings and are 16 (CCG-13528) or 18 (CCG-14631, CCG-7167) atoms in length (Fig. 5C). This suggests a potential pharmacophore for inhibiting LARG-stimulated RhoA nucleotide exchange which may be explained by the long shallow pocket of the LARG/RhoA interaction site 29.
Compound Selectivity for LARG-stimulated RhoA nucleotide binding
Inhibition of LARG stimulation of RhoA nucleotide exchange can occur by two primary mechanisms. The compound could directly inhibit GTP binding to RhoA or it could block LARG-mediated nucleotide exchange. Determining the exact mechanism of the inhibition of LARG-stimulated RhoA nucleotide exchange is beyond the scope of the present study, however, we did determine whether the compounds inhibited GTP binding or LARG-mediated exchange in the [35S] GTPγS guanine nucleotide binding assay. [35S] GTPγS binding to the small GTPase RhoA and the G-protein alpha subunit Gαo was determined at sixty minutes and twenty-five minutes, respectively. (Fig. 6A-B). [35S] GTPγS binding to RhoA and Gαo was tested in the absence of GEFs to determine whether the compounds inhibited GTP binding directly. As expected, all five compounds inhibited LARG-stimulated RhoA [35S] GTPγS binding (≥ 50%) yet they had only modest or no effect upon direct RhoA and Gαo [35S] GTPγS binding at a maximal concentration of 100 μM (Fig. 6C). Therefore, the five compounds identified in the high-throughput screen described here do appear to inhibit the LARG-mediated nucleotide exchange in some manner.
Fig. 6. Compound Selectivity for LARG-stimulated RhoA [35S] GTPγS Binding.
(A) Time-course of RhoA (C189S) (■) (500 nM) [35S] GTPγS binding over 180 minutes. (B) Time-course of Gαo (●) (250 nM) [35S] GTPγS binding over 120 minutes. Both no RhoA and no Gαo or [35S] GTPγS alone are represented by (■). (C) Bar graph representation of compound (100 μM) selectivity for LARG-stimulated [35S] GTPγS binding versus RhoA (C189S) and Gαo [35S] GTPγS binding. Binding was measured at a 60 minute time point (indicated by the dotted vertical line) for RhoA (C189S) and 25 minute time point (indicated by the dotted vertical line and open circles) for both Gαo and LARG-stimulated RhoA [35S] GTPγS binding. Data in panel A, B, and C were measured in duplicate and represent mean ± SEM for three separate experiments (N = 3). Data in panel C is represented as percent of DMSO control for [35S] GTPγS binding.
Discussion
Radioactive and fluorescence assays are the primary approaches utilized to measure guanine nucleotide exchange of small GTPases. These approaches typically use radioactive [3H] or [35S] or fluorescent N-methyl-3′-O-anthranoyl (MANT) and boron dipyrromethene (BODIPY) analogs of guanine nucleotides. They either measure binding of the non-hydrolysable GTP analog, GTPγS, to G-proteins or release of labeled GDP from G-proteins 23, 24, 30. Upon nucleotide exchange, the guanine nucleotide analogs yield either a measurable radioactive or fluorescence signal. Here, we describe a novel assay that utilizes fluorescence polarization rather than the standard fluorescence intensity approach to measure LARG-stimulated RhoA nucleotide binding. We chose the fluorescence polarization assay due to its better, initial baseline kinetics, signal to noise ratio, and Z′-factor (data not shown). The Z′-factor measures the reliability of a positive screening hit based on the dynamic range and intrinsic variability of the assay 27, 31. The Z′-factor for the fluorescence polarization assay was 0.7 in pilot studies and showed a sustained level of 0.52 in our 10,000 compound screen. This should be sufficient for high-throughput screening, and for general biological applications.
One major issue in fluorescence-based high-throughput screening assays is the type of fluorophore one chooses. The most commonly used fluorophores used to label guanine nucleotides, MANT and BODIPY-FL, have the drawback of being detected in the blue and green regions of the electromagnetic spectrum 32. This enhances the chance of false positives in high-throughput screens due to compounds absorbing light in this region of the electromagnetic spectrum. Therefore, in the work described here, we utilize a GTPγS nucleotide labeled with the BODIPY-TR fluorophore (Fig. 1C), which is detected in the red region of the electromagnetic spectrum. This, along with the use of the ratiometric FP method, should decrease the chance of false positives due to non-specific compound absorbance. We compared the fluorescence polarization BODIPY-TR-GTPγS binding assay as a measure of nucleotide exchange with the traditional radioactive [35S] GTPγS binding assay. We found both assays to be able to show concentration-dependent LARG-stimulated RhoA nucleotide binding and linear rate constants with respect to LARG concentration (Fig. 3). However, the two methods were different with respect to experimental conditions (i.e. buffers, protein concentrations, type of detergent and nucleotide) and reaction time. Despite the differences between assays, we chose the fluorescence polarization assay for a high-throughput screen for inhibitors of LARG-mediated nucleotide exchange based upon its reliability, adaptability to a 384-well plate format, lack of production of environmental waste, and time efficiency.
There are several ways that one can molecularly target the function of RhoGEFs and Rho GTPases. One can inhibit the membrane localization of Rho GTPases through blockade of lipid modifications. Interestingly, using a high-throughput screening approach, Peterson et. al. describe the identification of a geranylgeranyl transferase type I small molecule inhibitor (IC50 – 10 μM) that inhibits Rho GTPase function 33. Another way to obstruct Rho function is through small molecule stimulation of GAPs, in order to stimulate intrinsic hydrolysis of Rho GTPases promoting inhibition of function. Small molecule stabilization of the association of GDIs with Rho GTPases can also be effective in disrupting the activation of Rho GTPases by preventing membrane localization 1. Also, small molecule inhibition of GEFs would be effective in disrupting Rho function by preventing activation of Rho GTPases. Function can be abolished by directly targeting the RhoGTPase. Interestingly, Gao et.al. describe the identification of a direct small molecule inhibitor of Rac1, NSC23766 (IC50 – 50 μM), through structure-based virtual screening approach 16. Another direct Rac1 inhibitor, EHT 1864 (Kd – 40 nM), selectively inhibits Rac1 through blocking the binding of the guanine nucleotide to Rac1 17, 18. Also, we recently identified an inhibitor downstream in the rho-stimulated gene transcription pathway using a cell-based luciferase reporter screen 34. As a result, we describe here a fluorescence polarization high-throughput screen of a ChemBridge diverse chemical library for inhibitors of LARG-mediated RhoA nucleotide exchange.
LARG was originally discovered as a chromosomal translocation fusion to MLL in a patient with Acute Myeloid Leukemia 3. Therefore, from its initial identification, LARG has been linked to cancer. LARG gene expression was shown to be upregulated in patients with the preleukaemic disorder Scwachman-Diamond syndrome (SDS) 35. LARG was first shown to have oncogenic activity in a NIH3T3 transformation assay 8. Since then, LARG- mediated Rho activation has been shown to be important in prostate cancer (PC-3) and breast cancer (MDA-MB-435) cell morphology, motility, and invasion upon extracellular stimulation by the GPCR ligands bombesin, thrombin, and lysophosphatidic acid 6, 7, 36. In addition, LARG interaction with the adhesion receptor, CD44, or tyrosine kinase receptor, epidermal growth factor receptor (EGFR) and subsequent Rho activation has been shown to be necessary for cytoskeletal modification, motility, and invasion in head and neck squamous cell carcinoma (HSC-3) cells 9. Therefore, a small molecule inhibitor of LARG-mediated RhoA nucleotide exchange would be a useful tool to elucidate cancer mechanisms and as a potential cancer therapeutic.
In addition to cancer, LARG also has the potential to play an important role in HIV or vascular diseases. LARG has been shown to be upregulated in rat vascular smooth muscle by Angiotensin II and it is basally expressed in rat corpus cavernosum, thus potentially playing a vital role in contraction and calcium sensitization 11, 13, 37. Recently, LARG was determined to be the key RhoGEF involved in the development of salt-induced hypertension 12. Interestingly, Hodges et. al. describe a role for LARG in HIV-1 pathogenesis via forming a complex with the c-type lectin receptor, DC-SIGN 10. Therefore, small molecule inhibition of LARG-mediated RhoA nucleotide exchange may prove to be useful in the development of pharmacological tools and therapeutics for better understanding of disease pathogenesis and treatment of diseases and disorders, including HIV, hypertension, and erectile dysfunction (ED).
The five compounds identified in this study would be of interest for further development based on the multiple diseases and disorders that an inhibitor of LARG-mediated RhoA nucleotide exchange may have a positive impact upon. All five compounds have potency in the micromolar range, which make them reasonable candidates for synthetic chemistry follow-up. In addition, the compounds do not inhibit GTP binding to RhoA or to Gαo. Therefore, they most likely inhibit LARG-mediated nucleotide binding by: (i) binding to the LARG binding surface on RhoA, (ii) binding to the RhoA binding surface on LARG, (iii) allosterically binding to either LARG or RhoA causing a conformational change, (iv) binding to both RhoA and LARG, or (v) covalently modifying RhoA or LARG. Further development, specificity studies, and mechanistic characterization are still needed, but the compounds identified in this study inhibit LARG-mediated RhoA nucleotide exchange with micromolar potency in vitro.
Acknowledgments
We thank Dr. John Tesmer (University of Michigan, Ann Arbor, MI) for kindly providing the LARG DNA expression plasmid for protein expression and for helpful discussion. We thank Dr. David L. Roman (University of Michigan, Ann Arbor, MI) for assistance with the [35S] GTPγS binding studies, and for helpful advice and discussion. We thank Dr. Stuart J. Decker (Life Sciences Institute, University of Michigan, Ann Arbor, MI) for helpful suggestions for our fluorescence polarization high-throughput screening protocol. We thank Renju Jacob for help with the chemoinformatic analysis of the high-throughtput screening data.
Funding: NIH 5R01GM039561 (to RRN), NIH/NCI 5F31CA113268 (to CRE)
Footnotes
NA represents No Activity in the assay
References
- 1.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 2.Kjoller L, Hall A. Signaling to Rho GTPases. Exp Cell Res. 1999;253:166–179. doi: 10.1006/excr.1999.4674. [DOI] [PubMed] [Google Scholar]
- 3.Kourlas PJ, Strout MP, Becknell B, Veronese ML, Croce CM, Theil KS, Krahe R, Ruutu T, Knuutila S, Bloomfield CD, Caligiuri MA. Identification of a gene at 11q23 encoding a guanine nucleotide exchange factor: evidence for its fusion with MLL in acute myeloid leukemia. Proc Natl Acad Sci U S A. 2000;97:2145–2150. doi: 10.1073/pnas.040569197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- 5.Booden MA, Siderovski DP, Der CJ. Leukemia-associated Rho guanine nucleotide exchange factor promotes G alpha q-coupled activation of RhoA. Mol Cell Biol. 2002;22:4053–4061. doi: 10.1128/MCB.22.12.4053-4061.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q, Liu M, Kozasa T, Rothstein JD, Sternweis PC, Neubig RR. Thrombin and lysophosphatidic acid receptors utilize distinct rhoGEFs in prostate cancer cells. J Biol Chem. 2004;279:28831–28834. doi: 10.1074/jbc.C400105200. [DOI] [PubMed] [Google Scholar]
- 7.Zheng R, Iwase A, Shen R, Goodman OB, Jr, Sugimoto N, Takuwa Y, Lerner DJ, Nanus DM. Neuropeptide-stimulated cell migration in prostate cancer cells is mediated by RhoA kinase signaling and inhibited by neutral endopeptidase. Oncogene. 2006;25:5942–5952. doi: 10.1038/sj.onc.1209586. [DOI] [PubMed] [Google Scholar]
- 8.Reuther GW, Lambert QT, Booden MA, Wennerberg K, Becknell B, Marcucci G, Sondek J, Caligiuri MA, Der CJ. Leukemia-associated Rho guanine nucleotide exchange factor, a Dbl family protein found mutated in leukemia, causes transformation by activation of RhoA. J Biol Chem. 2001;276:27145–27151. doi: 10.1074/jbc.M103565200. [DOI] [PubMed] [Google Scholar]
- 9.Bourguignon LY, Gilad E, Brightman A, Diedrich F, Singleton P. Hyaluronan-CD44 interaction with leukemia-associated RhoGEF and epidermal growth factor receptor promotes Rho/Ras co-activation, phospholipase C epsilon-Ca2+ signaling, and cytoskeleton modification in head and neck squamous cell carcinoma cells. J Biol Chem. 2006;281:14026–14040. doi: 10.1074/jbc.M507734200. [DOI] [PubMed] [Google Scholar]
- 10.Hodges A, Sharrocks K, Edelmann M, Baban D, Moris A, Schwartz O, Drakesmith H, Davies K, Kessler B, McMichael A, Simmons A. Activation of the lectin DC-SIGN induces an immature dendritic cell phenotype triggering Rho-GTPase activity required for HIV-1 replication. Nat Immunol. 2007;8:569–577. doi: 10.1038/ni1470. [DOI] [PubMed] [Google Scholar]
- 11.Linder AE, Webb RC, Mills TM, Ying Z, Lewis RW, Teixeira CE. Rho-kinase and RGS-containing RhoGEFs as molecular targets for the treatment of erectile dysfunction. Curr Pharm Des. 2005;11:4029–4040. doi: 10.2174/138161205774913390. [DOI] [PubMed] [Google Scholar]
- 12.Wirth A, Benyo Z, Lukasova M, Leutgeb B, Wettschureck N, Gorbey S, Orsy P, Horvath B, Maser-Gluth C, Greiner E, Lemmer B, Schutz G, Gutkind JS, Offermanns S. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat Med. 2008;14:64–68. doi: 10.1038/nm1666. [DOI] [PubMed] [Google Scholar]
- 13.Teixeira CE, Ying Z, Webb RC. Proerectile effects of the Rho-kinase inhibitor (S)-(+)-2-methyl-1-[(4-methyl-5-isoquinolinyl)sulfonyl]homopiperazine (H-1152) in the rat penis. J Pharmacol Exp Ther. 2005;315:155–162. doi: 10.1124/jpet.105.086041. [DOI] [PubMed] [Google Scholar]
- 14.Collisson EA, Kleer C, Wu M, De A, Gambhir SS, Merajver SD, Kolodney MS. Atorvastatin prevents RhoC isoprenylation, invasion, and metastasis in human melanoma cells. Mol Cancer Ther. 2003;2:941–948. [PMC free article] [PubMed] [Google Scholar]
- 15.Sebti SM, Hamilton AD. Farnesyltransferase and geranylgeranyltransferase I inhibitors and cancer therapy: lessons from mechanism and bench-to-bedside translational studies. Oncogene. 2000;19:6584–6593. doi: 10.1038/sj.onc.1204146. [DOI] [PubMed] [Google Scholar]
- 16.Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci U S A. 2004;101:7618–7623. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desire L, Bourdin J, Loiseau N, Peillon H, Picard V, De Oliveira C, Bachelot F, Leblond B, Taverne T, Beausoleil E, Lacombe S, Drouin D, Schweighoffer F. RAC1 inhibition targets amyloid precursor protein processing by gamma-secretase and decreases Abeta production in vitro and in vivo. J Biol Chem. 2005;280:37516–37525. doi: 10.1074/jbc.M507913200. [DOI] [PubMed] [Google Scholar]
- 18.Shutes A, Onesto C, Picard V, Leblond B, Schweighoffer F, Der CJ. Specificity and mechanism of action of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. J Biol Chem. 2007;282:35666–35678. doi: 10.1074/jbc.M703571200. [DOI] [PubMed] [Google Scholar]
- 19.Rossman KL, Cheng L, Mahon GM, Rojas RJ, Snyder JT, Whitehead IP, Sondek J. Multifunctional roles for the PH domain of Dbs in regulating Rho GTPase activation. J Biol Chem. 2003;278:18393–18400. doi: 10.1074/jbc.M300127200. [DOI] [PubMed] [Google Scholar]
- 20.Kristelly R, Earnest BT, Krishnamoorthy L, Tesmer JJ. Preliminary structure analysis of the DH/PH domains of leukemia-associated RhoGEF. Acta Crystallogr D Biol Crystallogr. 2003;59:1859–1862. doi: 10.1107/s0907444903018067. [DOI] [PubMed] [Google Scholar]
- 21.Roman DL, Talbot JN, Roof RA, Sunahara RK, Traynor JR, Neubig RR. Identification of small-molecule inhibitors of RGS4 using a high-throughput flow cytometry protein interaction assay. Mol Pharmacol. 2007;71:169–175. doi: 10.1124/mol.106.028670. [DOI] [PubMed] [Google Scholar]
- 22.Vishwanadan VN, G AK, Revankar GR, Robins RK. Atomic Physicochemical Parameters for Three Dimensional Structure Directed Quantitative Structure-Activity Relationships; Additional Parameters for Hydrophobic and Dispersive Interactions and Their Application for an Automated Superposition of Certain Naturally Occurring Nucleoside Antibiotics. Journal of Chemical Information and Computer Sciences. 1989;29:163–172. [Google Scholar]
- 23.McEwen DP, Gee KR, Kang HC, Neubig RR. Fluorescent BODIPY-GTP analogs: real-time measurement of nucleotide binding to G proteins. Anal Biochem. 2001;291:109–117. doi: 10.1006/abio.2001.5011. [DOI] [PubMed] [Google Scholar]
- 24.McEwen DP, Gee KR, Kang HC, Neubig RR. Fluorescence approaches to study G protein mechanisms. Methods Enzymol. 2002;344:403–420. doi: 10.1016/s0076-6879(02)44730-1. [DOI] [PubMed] [Google Scholar]
- 25.Owicki JC. Fluorescence polarization and anisotropy in high throughput screening: perspectives and primer. J Biomol Screen. 2000;5:297–306. doi: 10.1177/108705710000500501. [DOI] [PubMed] [Google Scholar]
- 26.Lutz S, Shankaranarayanan A, Coco C, Ridilla M, Nance MR, Vettel C, Baltus D, Evelyn CR, Neubig RR, Wieland T, Tesmer JJ. Structure of Galphaq-p63RhoGEF-RhoA complex reveals a pathway for the activation of RhoA by GPCRs. Science. 2007;318:1923–1927. doi: 10.1126/science.1147554. [DOI] [PubMed] [Google Scholar]
- 27.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 28.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 29.Kristelly R, Gao G, Tesmer JJ. Structural determinants of RhoA binding and nucleotide exchange in leukemia-associated Rho guanine-nucleotide exchange factor. J Biol Chem. 2004;279:47352–47362. doi: 10.1074/jbc.M406056200. [DOI] [PubMed] [Google Scholar]
- 30.Remmers AE, Posner R, Neubig RR. Fluorescent guanine nucleotide analogs and G protein activation. J Biol Chem. 1994;269:13771–13778. [PubMed] [Google Scholar]
- 31.Verkman AS. Drug discovery in academia. Am J Physiol Cell Physiol. 2005;286:C465–474. doi: 10.1152/ajpcell.00397.2003. [DOI] [PubMed] [Google Scholar]
- 32.Rojas RJ, Kimple RJ, Rossman KL, Siderovski DP, Sondek J. Established and emerging fluorescence-based assays for G-protein function: Ras-superfamily GTPases. Comb Chem High Throughput Screen. 2003;6:409–418. doi: 10.2174/138620703106298509. [DOI] [PubMed] [Google Scholar]
- 33.Peterson YK, Kelly P, Weinbaum CA, Casey PJ. A novel protein geranylgeranyltransferase-l inhibitor with high potency, selectivity and cellular activity. J Biol Chem. 2006;281:12445–12450. doi: 10.1074/jbc.M600168200. [DOI] [PubMed] [Google Scholar]
- 34.Evelyn CR, Wade SM, Wang Q, Wu M, Iniguez-Lluhi JA, Merajver SD, Neubig RR. CCG-1423: a small-molecule inhibitor of RhoA transcriptional signaling. Mol Cancer Ther. 2007;6:2249–2260. doi: 10.1158/1535-7163.MCT-06-0782. [DOI] [PubMed] [Google Scholar]
- 35.Rujkijyanont P, Beyene J, Wei K, Khan F, Dror Y. Leukaemia-related gene expression in bone marrow cells from patients with the preleukaemic disorder Shwachman-Diamond syndrome. Br J Haematol. 2007;137:537–544. doi: 10.1111/j.1365-2141.2007.06608.x. [DOI] [PubMed] [Google Scholar]
- 36.Kitzing TM, Sahadevan AS, Brandt DT, Knieling H, Hannemann S, Fackler OT, Grosshans J, Grosse R. Positive feedback between Dia1, LARG, and RhoA regulates cell morphology and invasion. Genes Dev. 2007;21:1478–1483. doi: 10.1101/gad.424807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ying Z, Jin L, Palmer T, Webb RC. Angiotensin II Up-Regulates the Leukemia-Associated Rho Guanine Nucleotide Exchange Factor (RhoGEF), a Regulator of G Protein Signaling Domain-Containing RhoGEF, in Vascular Smooth Muscle Cells. Mol Pharmacol. 2006;69:932–940. doi: 10.1124/mol.105.017830. [DOI] [PubMed] [Google Scholar]